Abstract
The chitinase-like proteins YKL-39 (chitinase 3-like-2) and YKL-40 (chitinase 3-like-1) are highly expressed in a number of human cells independent of their origin (mesenchymal, epithelial or haemapoietic). Elevated serum levels of YKL-40 have been associated with a negative outcome in a number of diseases ranging from cancer to inflammation and asthma. YKL-39 expression has been associated with osteoarthritis. However, despite the reported association with disease, the physiological or pathological role of these proteins is still very poorly understood. Although YKL-39 is homologous to the two family 18 chitinases in the human genome, it has been reported to lack any chitinase activity. In the present study, we show that human YKL-39 possesses a chitinase-like fold, but lacks key active-site residues required for catalysis. A glycan screen identified oligomers of N-acetylglucosamine as preferred binding partners. YKL-39 binds chitooligosaccharides and a newly synthesized derivative of the bisdionin chitinase-inhibitor class with micromolar affinity, through a number of conserved tryptophan residues. Strikingly, the chitinase activity of YKL-39 was recovered by reverting two non-conservative substitutions in the active site to those found in the active enzymes, suggesting that YKL-39 is a pseudo-chitinase with retention of chitinase-like ligand-binding properties.
Keywords: chitinase, chitinase-like proteins, glycan, glycan array, glycobiology, protein structure, lectin, X-ray crystallography
INTRODUCTION
The human genome possesses two genes coding for active chitinases, hCHT (chitotriosidase) and the hAMCase (acidic mammalian chitinase), both from the CAZy GH18 (glycoside hydrolase 18) family [1]. hCHT is a phagocyte-specific chitinase that exists in two isoforms, a major 50 kDa and a C-terminally truncated 39 kDa isoform [2]. The C-terminal domain of the 39 kDa isoform has been shown to be involved in the binding of chitin [3], whereas the 50 kDa form is secreted from macrophages and the truncated form is not. Rather, it is routed to the lysosomes and can be used as a marker for measuring the progression of Gaucher’s disease [4–6]. AMCase has a similar domain structure, but a distinct expression pattern, being expressed in the gastrointestinal tract and lung by tissue macrophages and endothelial cells [7,8]. Expression is increased during Th2-driven pathologies, and consequently inhibition of enzymatic activity has been considered as a therapeutic strategy for allergic inflammation and asthma [8–10].
Along with active chitinases, a number of closely related proteins without detectable chitinase activity have been identified in mammalian genomes. These include YKL-40 (chitinase 3-like-1), YKL-39 (chitinase 3-like-2), Ym1/2 (chitinase 3-like-3/4), oviduct-specific glycoprotein and stabilin-1-interacting chitinase-like protein [11–17]. YKL-40 (also termed HC-gp39 or Chi3l1) was found to be secreted, along with chitotriosidase, from activated human macrophages and it was later detected in the culture supernatant of chondrocytes, synovial cells and osteoblasts [2,11]. In vitro experiments demonstrated YKL-40 induction through the cell-stress pathway when chondrocytes were exposed to LPS (lipopolysaccharide) [18]. This lectin has also been identified as a protein overexpressed in inflamed tissues in vivo [19,20]. Clinical research has shown that high levels of YKL-40 are found in the serum of patients suffering from chronic asthma and also in patients with severe arthritis [21–23]. Immune response studies have linked YKL-40 to a down-regulation of the inflammatory mediators MMP (matrix metalloprotease) 1 and MMP3 and IL-8 (interleukin-8), suggesting a protective influence under innate immune response conditions [24]. YKL-40 has been shown to have the ability to act as a growth factor for skin and fetal lung fibroblasts [25]. YKL-40 is also used as a disease marker in Type 1 Gaucher’s disease and in solid-state tumour progression (reviewed in [26]). Knockout studies of the mouse orthologue of YKL-40 [BRP-39 (breast regression protein 39)] revealed a significant reduction in the Th2 inflammatory response and an increase in cellular apoptosis under challenge with ovalbumin, which was rescued by supplementing the BRP-39 protein [27]. There is a paucity of information about the biological function of YKL-39; nevertheless, the protein has been suggested as a diagnostic marker for the diagnosis and management of osteoarthritis based on increased expression levels in osteoarthritic cartilage [28,29].
Despite a relatively high sequence identity and predicted structural similarity to the family 18 chitinases such as chitotriosidase and AMCase, chitinase-like proteins lack glycosyl hydrolase activity [30]. The loss of enzymatic activity is attributed to the substitution of the catalytic residues of the DxxDxDxE motif, which characterizes the active site of family 18 chitinases [13,31–34]. Although YKL-39 appears to have an active site incompatible with chitin hydrolysis, it may have retained the ability to bind chitin-like molecules, although the identity of the physiological ligand, if any, is currently unknown.
In the present study, we have investigated the ligand preferences of YKL-39 by screening a carbohydrate microarray, identifying chitooligosaccharides as the most likely ligands. Furthermore, YKL-39 showed micromolar binding affinity for chitooligosaccharides and chitinase inhibitors, but no measurable chitinase activity. The crystal structure of YKL-39 reveals the molecular basis for this affinity as well as for the lack of hydrolytic activity. Interestingly, the hydrolytic activity of YKL-39 can be generated by reconstructing the catalytic DxxDxDxE motif. Thus we show that YKL-39 is a pseudo-chitinase, having retained the ability to bind chitin, yet lost the ability to hydrolyse it.
MATERIALS AND METHODS
Molecular cloning
The coding sequence for YKL-39 residues 27–390 (lacking the signal peptide) was inserted into the pPIC9 Pichia pastoris expression vector. The following oligonucleotides were used as primers to amplify the 1145 bp fragment and introduce additional restriction sites (in bold letters and indicated): forward, 5′-CGGCAAGCTTACAAACTGGTTTGCTAC-3′ (HindIII) and reverse 5′-ACATACGCGTCATCTTGCCTGCTTCT-3′ (MluI). Point mutations were introduced by site-directed mutagenesis: N35Q (forward, 5′-GTTTGCTACTTTACCCAATGGTCCCAGGACCGG-3′ and reverse, 5′-CCGGTCCTGGGACCATTGGGTAAAGTAGCAAAC-3′) and S143D/I145E (forward, 5′-GATGATCTGGATGTAAGCTGGGAGTACCC-3′ and reverse 5′-CTACTAGACCTACATTCGACCCTCATGGG-3′).
The plasmid vectors were linearized with SacI before transforming into P. pastoris GS115 cells (Invitrogen) using the LiCl method according to the manufacturer’s instructions. Briefly, a 50-ml culture was grown to an D600 of 0.8–1.0 at 30°C with shaking. The cells were pelleted at 3000 g, and washed with 25 ml of sterile water. The cell pellet was resuspended in 1 ml of 100 mM LiCl and 50 μl of cells were used per transformation. The 100 mM LiCl solution was removed by centrifugation at maximum speed for 12 s and the transformation solution was added in the following order: 240 μl sterile 50% PEG [poly(ethylene glycol)] 3000, 36 μl of 1 M LiCl, 25 μl of single-stranded salmon sperm DNA (2 mg/ml) and 5 μg of linearized plasmid DNA. After gentle resuspension the cultures were incubated at 37°C for 30 min, heat shocked at 42°C for 25 min and then plated on MD (minimal dextrose) selection plates. The plates were incubated for 2–4 days at 30°C.
Protein expression and purification
Expression cell lines were cultured in BMGY (buffered glycerol complex) medium at room temperature (22°C) with shaking until the D600 value reached 40–80. The BMGY growth medium was removed by centrifugation and replaced by BMMY (buffered minimum methanol) medium to induce the overexpression and secretion of YKL-39. Protein expression was carried out over 72 h. Methanol was added on the second and third day to a final concentration of 1%. The supernatant was concentrated by cross-flow membrane filtration (25 kDa molecular-mass cut-off) and dialysed into 50 mM NaAc (sodium acetate; pH 5.5) for subsequent ion-exchange chromatography on CM (carboxymethyl)–Sepharose FF (fast flow). The protein eluted at approximately 30% of a 0–1 M NaCl elution gradient. Fractions containing YKL-39 were pooled and dialysed into 50 mM NaAc and 150 mM NaCl. Size-exclusion chromatography was used as a final purification step to obtain crystallization-grade protein. The protein was passed over a XK26/60 Superdex 75 column in 50 mM NaAc (pH 5.5) and 150 mM NaCl. Pure protein was dialysed into 50 mM NaAc (pH 5.5) and concentrated to 30 mg/ml.
Glycan screen
YKL-39 was analysed for glycan binding by Core H of the Consortium of Functional Glycomics (http://www.functionalglycomics.org) using the Printed Array Version 2. The microarray contained 264 natural and synthetic glycans with amino linkers, printed on to chemically modified glass microscope slides in replicates of six. YKL-39 was labelled with AlexaFluor 488 (Invitrogen protein-labelling kit) and exposed to the glycan screen for 1 h at room temperature. Binding was determined fluorimetrically after washing steps in the following three solutions: (i) 20 mM Tris/HCl (pH 7.4), 150 mM NaCl, 2 mM CaCl2, 2 mM MgCl2 and 0.05% Tween 20; (ii) the same buffer without Tween 20; and (iii) deionized water. Slides were spun dry and then scanned on the ProScanArray Express from PerkinElmer (excitation at 495 nm and emission measured at 520 nm).
Structure solution
Wild-type YKL-39 protein expressed in P. pastoris cells contains a small proportion of N-glycosylated product. In the interest of obtaining a homogenous sample for crystallography, the single glycosylation site (Asn35) was mutated (N35Q). YKL-39 N35Q protein crystals in complex with chitohexaose (GlcNAc)6 were obtained through co-crystallization of 30 mg/ml protein with 1 mM (GlcNAc)6 in conditions containing 23% PEG 3000 and 0.1 M sodium citrate (pH 6). Crystals were cryoprotected in 30% ethylene glycol, 23% PEG 3000 and 0.1 M sodium citrate (pH 6), cryocooled and diffraction data were collected at the ESRF BM14 (European Synchrotron Radiation Facility Bending Magnet beamline 14). They belonged to space group C2 and provided data sets to 1.95 Å (1 Å = 0.1 nm) resolution. The structure was solved by molecular replacement using a monomer of the YKL-40 structure (deposited amino acid sequence Swiss-Prot P36222, PDB code 1HJX) [34] using MolRep [35], where it was determined there were 12 monomers in the asymmetric unit. Following rigid-body refinement and restrained refinement using Refmac5 [36], model building was conducted in Coot [37] with refinements using non-crystallographic symmetry restraints. Water molecules were added and the chitooligosaccharide ligand was fitted into the positive density in the substrate-binding site of each monomer. Final refinement including a description of anisotropy with TLS (Translation–Libration–Screw-rotation) [38] resulted in an R factor of 0.191 (Rfree = 0.239). Data collection and refinement statistics are summarized in Table 1. The co-ordinates and structure factors have been deposited in the PDB under accession code 4AY1.
Table 1. Details of data collection and structure refinement for YKL-39 in complex with (GlcNAc)6.
Values in parentheses are for the highest resolution shell. Rmerge, Rwork and Rfree were calculated according to the standard equations: Rmerge= ∑∑i|Ii – <I|/∑∑i Ii, where Ii and <I> are the observed intensity and the average intensity of multiple observations of symmetry identical reflections respectively; Rwork = ∑∥Fobs| – |Fcalc∥/∑ |∑ |Fobs|, where Fobs and Fcalc are the observed and calculated structure factors respectively; and Rfree was computed as in Rwork, but only for (2%) randomly selected reflections, which were omitted from refinement.
| Parameter | Value |
|---|---|
| Data collection | |
| Space group | C121 |
| Cell dimensions | |
| a, b, c (Å) | 255.7, 152.5, 138.2 |
| α, β, γ (°) | 90.00, 94.62, 90.00 |
| Resolution (Å) | 20.00–1.95 (2.02–1.95) |
| R merge | 0.055 (0.424) |
| I/σI | 17.2 (2.2) |
| Completeness (%) | 99.6 |
| Redundancy | 3.4 (3.2) |
| Refinement | |
| Resolution (Å) | 20.00–1.95 |
| Number of unique reflections | 383206 |
| Rwork/Rfree (%) | 19.1/23.9 |
| Number of atoms | |
| Protein | 37103 |
| Ligand/ion | 2437 |
| Water | 1894 |
| B-factors | |
| Protein | 26.3 |
| Ligand/ion | 25.9 |
| Water | 30.4 |
| RMSD | |
| Bond lengths (Å) | 0.012 |
| Bond angles (°) | 1.36 |
Chitinase activity assay
Reactions were conducted in 0.2 M Na2HPO4/0.1 M citric acid (pH 5.5) with a final reaction volume of 50 μl. The final enzyme concentrations were 2 nM for hCHT and 20 nM for YKL-39. The fluorigenic substrate 4MU-(GlcNAc)3 (4-methylumbelliferyl-β-D-N,N′,N′’-triacetylchitotriose; Sigma) was used. Reactions were quenched with 100 μl of 3 M glycine/NaOH (pH 10) after a 60 min incubation at 37°C. Product formation was quantified on a microtiterplate fluorescence reader, FLX800 (Bio-Tek), at excitation/emission wavelengths of 360 nm and 460 nm respectively. All of the reactions were carried out in triplicate. The readings for samples containing no enzyme were subtracted to control for non-enzymatic substrate hydrolysis. Data were analysed and plotted using GraphPad Prism software.
Tryptophan fluorescence
Tryptophan fluorescence measurements were carried out in triplicate using 1.25 ml of protein solution per scan at a concentration of 1 μM in 50 mM Tris (pH 7.5). The final added volume of ligand stock was less than 1% of the total experimental sample. Control scans were conducted in which the ligand buffer (H2O) was used to determine the effect of dilution. Peak scans were measured between 337 and 357 nm. All of the scans were taken in duplicate with a 5 min rest period in between scans. Controls were subtracted from the experimental measurements to normalize, and data were analysed and plotted using the GraphIt software (Gecces).
Preparation of chitinase inhibitors
General remarks
Chemical reagents were purchased from Sigma–Aldrich, Fluka, Acros and Lancaster. Anhydrous DMF (dimethylformamide) was obtained from Sigma–Aldrich. All of the other solvents were obtained from Sigma–Aldrich and used as received. Analytical TLC was performed using silica gel 60 F254 pre-coated on aluminium sheets (0.25 mm thickness). Flash chromatography was performed on columns of silica gel 60 (35–70 μm) from Fisher Scientific. Melting points were determined with a Reichert–Jung apparatus and are uncorrected. 1H NMR spectra were recorded on a JEOL JMN GX-270 (270 MHz) or on a Bruker Avance III (500 MHz) spectrometer. 13C NMR spectra were recorded on a Varian Mercury VX 400 (100 MHz) or on a Bruker Avance III (125 MHz) spectrometer. Chemical shifts are reported in ppm (δ) and J values are in Hz. Mass spectra were recorded using a Bruker MicroTOF autospec electrospray ionization mass spectrometer. All of the compounds submitted to biological analysis had a purity >95%, as judged by analytical HPLC. Analytical HPLC was performed on a Dionex Ultimate 3000 Instrument (Dionex), equipped with a Phenomenex Gemini 5 μm C-18 (150 mm × 4.6 mm) column.
Bisdionin C [39] and F [10] were prepared as described previously. For the preparation of bisdionin G, 8-ethyl-3-methyl-1H-purine-2,6(3H, 7H)-dione [40] was first regioselectively alkylated at N-7 with 4-methoxybenzyl chloride, according to the method of van Muijlwijk-Koezen et al. [42]. This intermediate mono-xanthine was then alkylated at N-1 with 1-(3-bromopropyl)-3, 7-dimethyl-1H-purine-2,6(3H, 7H)-dione [41], followed by removal of the 4-methoxybenzyl group under acidic conditions [10], to give bisdionin G, which was characterized by 1H and 13C NMR and HRMS (high-resolution MS). The purity of all three bisdionins was >95% as judged by analytical HPLC.
8-Ethyl-7-(4-methoxybenzyl)-3-methyl-1H-purine-2,6(3H, 7H)-dione (1)
A solution of 8-ethyl-3-methyl-1H-purine-2,6(3H, 7H)-dione [40] (0.98 g, 5.03 mmol) in anhydrous DMF (20 ml) was treated with anhydrous potassium carbonate (1.04 g, 7.55 mmol), and the resulting suspension was stirred at room temperature under nitrogen for 2 h. The mixture was treated with 4-methoxybenzyl chloride (0.88 ml, 6.50 mmol) and stirred overnight at room temperature. The reaction mixture was poured into H2O (75 ml), extracted with dichloromethane (2×50 ml), and the organic extracts were dried (Na2SO4), filtered and evaporated to give a yellow solid. Purification by column chromatography (0–70% acetone in dichloromethane) gave 1 (Scheme 1) as a pale yellow solid (0.80 g, 50%). Mp > 230°C; δH (CDCl3, 270 MHz): 1.25 (3H, t, J 7.4, CH3CH2), 2.71 (2H, q, J 7.4, CH3CH2), 3.53 (3H, s, 3H, N-3 CH3), 3.77 (3H, s, OCH3), 5.41 (2H, s, NCH2Ar), 6.84 (2H, d, J 8.8, 2 × ArH), 7.14 (2H, d, J 8.8, 2× ArH), 7.86 (1H, s, H-1); δC (CDCl3, 100 MHz): 11.62, 20.53, 28.88, 47.69, 55.20, 106.96, 113.90, 114.25, 128.06, 128.58, 150.08, 151.18, 154.66, 155.92, 159.37; found (ES+ ) 315.1449 [M + H]+ , C16H19N4O3 requires 315.1452.
Scheme 1.
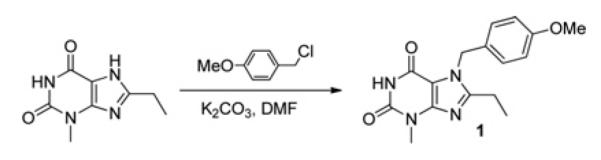
8-Ethyl-7-(4-methoxybenzyl)-3-methyl-1H-purine-2,6(3H, 7H)-dione (1)
1-[3-(3,7-dimethyl-2,6-dioxo-2,3,6,7-tetrahydro-1H-purin-1-yl)propyl]-8-ethyl-7-(4-methoxybenzyl)-3-methyl-1H-purine-2,6(3H, 7H)-dione (2)
A solution of 1 (0.19 g, 0.60 mmol) in anhydrous DMF (10 ml) was treated with anhydrous potassium carbonate (83 mg, 0.60 mmol), and the resulting suspension was stirred at 120°C under nitrogen for 2 h. The mixture was treated with 1-(3-bromopropyl)-3,7-dimethyl-1H-purine-2,6(3H, 7H)-dione [41] (0.88 ml, 6.50 mmol) and stirred overnight at this temperature. The cooled reaction mixture was poured into H2O (15 ml), and the precipitate that formed was filtered off, washed with H2O and dried in vacuo over NaOH pellets at 50°C. This gave 2 (Scheme 2) as a yellow solid (0.28 g, 87%) which was used without further purification. Mp > 230°C; δH (CDCl3, 270 MHz): 1.22 (3H, t, J 7.7, CH3CH2), 2.00–2.05 (2H, m, CH2CH2CH2), 2.67 (2H, q, J 7.7, CH3CH2), 3.49 (6H, s, 2× N-3 CH3), 3.72 (3H, s, N-7 CH3), 3.93 (3H, s, OCH3), 4.06 (4H, t, J 6.9, CH2CH2CH2), 5.43 (2H, s, NCH2Ar), 6.80 (2H, d, J 8.8, 2× ArH), 7.11 (2H, d, J 8.8, 2× ArH), 7.46 (1H, s, H-8).
Scheme 2.
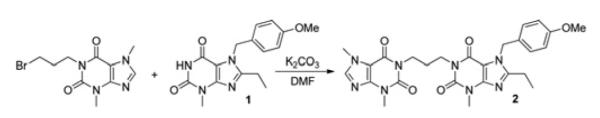
1-(3-(3,7-dimethyl-2,6-dioxo-2,3,6,7-tetrahydro-1H-purin-1-yl)propyl)-8-ethyl-7-(4-methoxybenzyl)-3-methyl-1H-purine-2,6(3H, 7H)-dione (2)
1-{3-[3,7-dimethyl-2,6-dioxo-2,3-dihydro-6H-purin-1(7H)-yl]propyl}-8-ethyl-3-methyl-1H-purine-2,6(3H, 7H)-dione (3)–Bisdionin G
A solution of 3 (Scheme 3; 0.25 g, 0.47 mmol) in trifluoracetic acid (1.5 ml) was treated with anisole (66 μl, 0.61 mmol) and concentrated sulfuric acid (2 drops). The mixture was heated at 75°C overnight, then the solvent was evaporated off. The residue was dissolved in dichloromethane and was washed with saturated aqueous NaHCO3. The organic phase was dried (Na2SO4), filtered and evaporated. The crude product was purified by column chromatography (10–100% acetone in dichloromethane) to give 3 as a white solid (0.12 g, 61%). Mp > 230°C; δH (CDCl3/CD3OD, 1:1, 500 MHz): 1.33 (3H, t, J 7.8, CH3CH2), 2.02 (2H, quintet, J 7.3, CH2CH2CH2), 2.79 (2H, q, J 7.8, CH3CH2), 3.52 (3H, s, N-3 CH3), 3.53 (3H, s, N-3 CH3), 3.96 (3H, s, N-7 CH3), 4.05–4.10 (4H, m, CH2CH2CH2), 7.76 (1H, s, H-8); δC (CDCl3/CD3OD, 1:1, 125 MHz): 12.69, 22.82, 27.60, 30.07, 30.39, 33.91, 40.05, 40.15, 108.50, 143.238, 149.44, 152.46, 156.10; found (ES+ ) 415.1833 [M + H]+ , C18H23N8O4 requires 415.1837.
Scheme 3.
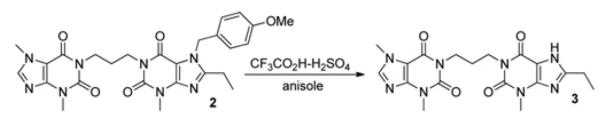
1-(3-(3,7-dimethyl-2,6-dioxo-2,3-dihydro-6H-purin-1(7H)-yl)propyl)-8-ethyl-3-methyl-1H-purine-2,6(3H, 7H)-dione (3)–Bisdionin G
RESULTS AND DISCUSSION
YKL-39 selectively binds chitooligosaccharides with micromolar affinity
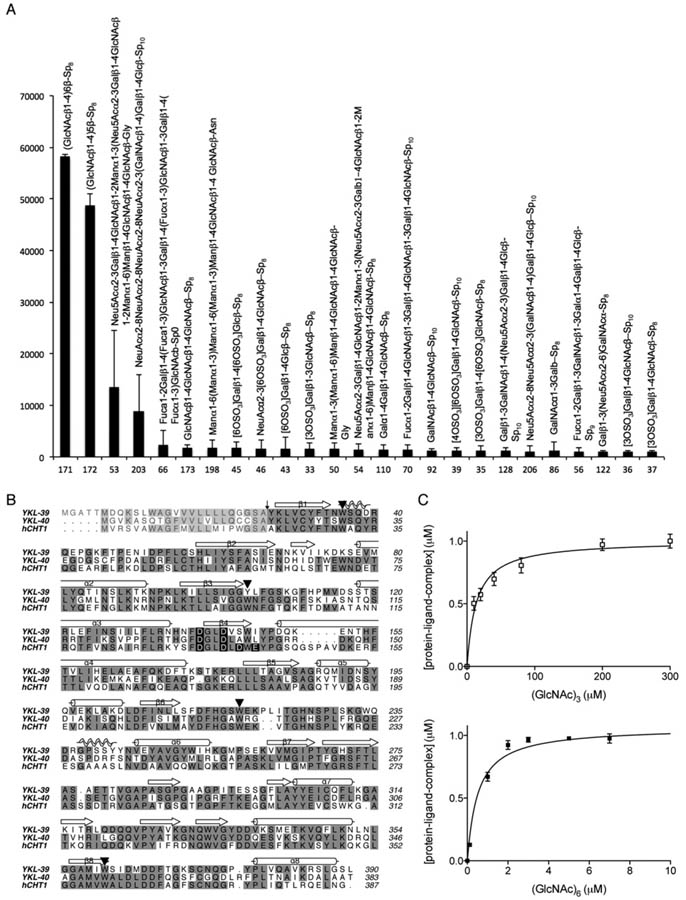
In an attempt to discover potential carbohydrate ligands for the chitinase-like protein YKL-39, a glycan array screen was carried out by Core H of the Consortium of Functional Glycomics. The screen revealed several potential binding partners for YKL-39 (Figure 1A). A common characteristic of these glycan ligands was the presence of β(1,4) glycosidic bonds, and the strongest binders were the chitooligosaccharides (GlcNAc)5 and (GlcNAc)6. To quantify the strength of this interaction, intrinsic tryptophan fluorescence was used, exploiting the presence of several conserved tryptophan residues (Figure 1B), which have been shown to play a role in substrate binding by family 18 chitinase [31,43,44]. Indeed, dose-dependent changes in intrinsic tryptophan fluorescence were observed in the presence of chitooligosaccharides (Figure 1C). This allowed determination of the dissociation constants for these ligands, indicating that YKL-39 has a higher affinity for (GlcNAc)6 (Kd 0.6 ± 0.1 μM) than for the shorter ligand (GlcNAc)3 (32 ± 4 μM).
Figure 1. Ligand binding and sequence conservation of YKL-39.
(A) The carbohydrate specificity of YKL-39 was investigated by screening an array of 264 glycans (Consortium of Functional Glycomics, Printed Array Version 2). YKL-39 was labelled with AlexaFluor 488 and binding was determined fluorimetrically. Relative fluorescence units (means ± S.D. for four observations) are shown for the top 25 synthetic carbohydrate structures. (B) Sequence alignement of the active chitinases hCHT and the chitinase-like proteins YKL-40 and YKL-39. Identical residues are shaded in grey and conserved tryptophan residues are marked with ▾. The active sitemotif DxxDxDxE is highlighted in black shading. (C) Changes in intrinsic tryptophan flourescence for YKL-39 and (GlcNAc)3 and (GlcNAc)6 fitted to a single-site-binding isotherm. Results are means ± S.D. for three independent experiments.
YKL-39 adopts a chitinase-like fold but does not possess a catalytically competent active site
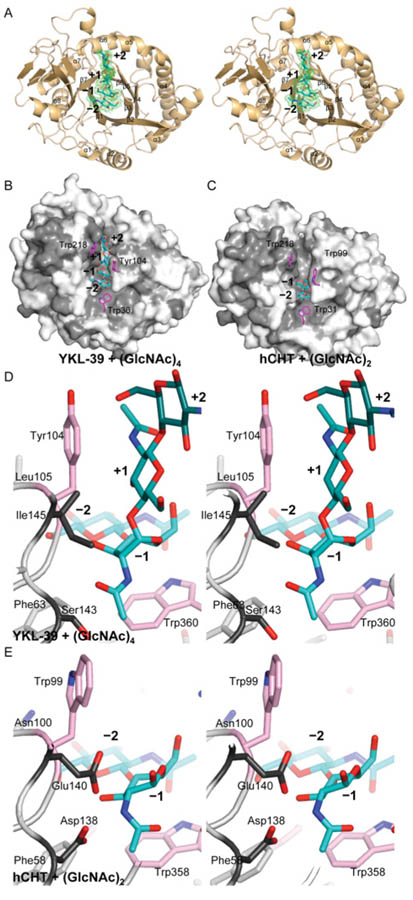
To investigate how YKL-39 binds chitooligosaccharide ligands without catalysing their hydrolysis, we determined the structure of a YKL-39–chitooligosaccharide complex by X-ray crystallography. A recombinant expression system of an N-glycosylation-deficient YKL-39 mutant was developed in P. pastoris, producing 3–5 mg of protein per litre of culture. Crystals of a YKL-39–(GlcNAc)6 complex were obtained through co-crystallisation and diffracted to 1.95 Å (Table 1). The YKL-39 structure was solved by molecular replacement, and refined to an R factor of 0.191 (Rfree = 0.239) with good geometry (Table 1). The asymmetric unit contains 12 monomers with very similar conformations [RMSD (root mean square deviation) 0.2 352 = Å over Cα atoms]. The structure (Figure 2A) reveals a (β/α)8 barrel, similar to YKL-40 (RMSD = 1.1 Å for 352 Cαs) and the catalytic domain of hCHT (RMSD = 1.1 Å for 364 Cαs), with well-defined electron density for sugars in the −2 to +2 subsites (Figure 2A). The short α/β domain inserted after strand β7, which forms the substrate-binding groove in family 18 chitinases, is likewise conserved in YKL-39 (Figure 2A).
Figure 2. Structure of the YKL-39 chitooligosaccharide complex.
(A) Stereo image of a cartoon representation of the structure of YKL-39 in complex with (GlcNAc)6, showing four monosaccharide units of the ligand (cyan carbon atoms) with ordered electron density. Unbiased Fo-Fc, Φcalc electron density for the ligand is shown in green (contoured at 2.25 σ). (B) Surface representation of the YKL-39 chitooligosaccharide complex, coloured by sequence conservation with hCHT (light grey = similarity, grey identity). The side chains of key solvent exposed aromatic residues are shown as sticks with = pink carbon atoms. The ligand is shown as a sticks model with cyan carbon atoms. (C) Surface representation of the hCHT chitobiose complex (PDB code 1LQ0 [31]), coloured by sequence conservation with YKL-39 (light grey = similarity, grey = identity). The side chains of key solvent exposed aromatic residues are shown as sticks with pink carbon atoms. The ligand is shown as a sticks model with cyan carbon atoms. (D) and (E) Stereo images of the ligand binding site of YKL-39 (D) and the active site of hCHT (E). The sugar ligand is shown as sticks with cyan carbon atoms. Catalytic residues in hCHT and their equivalents in YKL-39 are shown with dark grey carbon atoms. The side chains of key solvent exposed aromatic residues are shown as sticks with pink carbon atoms.
Previous studies have reported the structures of chitooligosaccharide complexes of chitinases and chitinase-like proteins (e.g. [31,34,44]). In YKL-39, the −2 to +2 sugars of the chitooligosaccharide ligand adopt a similar conformation to that observed in the YKL-40 complex (RMSD of 0.3 Å over 57 atoms). Binding of the oligosaccharide is achieved through a combination of hydrogen-bonding interactions and hydrophobic stacking between the sugar rings and surface-exposed aromatic amino acids, similar to that reported in previous chitinase–oligosaccharide complexes (Figure 2). These aromatic residues are conserved between hCHT, YKL-40 and YKL-39, except for a single tryptophan-to-tyrosine substitution in YKL-39 (Tyr104 in Figures 1B and 2). Stacking between sugar ligands and tryptophans in their cognate binding proteins/enzymes is known to be an important contribution to binding affinity in protein–glycan complexes [33,45,46]. Previous mutational studies of the conserved tryptophans in active chitinases have shown a reduction or complete loss of activity [45,47,48]. Owing to the conservation of the chitinase solvent-exposed/ligand stacking tryptophans in YKL-39, and their extensive interactions with the co-crystallized chitooligosaccharide ligand, it is likely that these residues are important for chitooligosaccharide binding.
Analysis of the active site region reveals the reason for the lack of chitinase activity: the catalytic machinery is severely disrupted (Figures 2D and 2E). The active site motif DxxDxDxE, essential for the activity of GH18 chitinases [31,33,34], is not conserved in YKL-39 (Figure 1B). Catalysis involves participation of the acetamido group of N-acetylglucosamine and proceeds via a bicyclic oxazoline intermediate [31,33,44]. The last aspartate residue in the DxxDxDxE motif (Asp138 in hCHT) positions the acetamido group for nucleophilic attack, whereas the glutamate residue (Glu140) performs general acid/base catalysis (Figure 2E). Notably, in YKL-39 these catalytic residues are substituted to a serine (Ser143) and an isoleucine (Ile145) residue (Figure 2D). Strikingly, although the hCHT oligosaccharide complex shows the sugar in the −1 subsite to be in the boat conformation with the N-acetyl group aligned for nucleophilic attack, in the YKL-39–(GlcNAc)6 complex the N-acetyl group assumes a catalytically incompetent conformation, instead occupying a pocket created by the hydrophobic substitutions in the DxxDxDxE motif (Figures 2D and 2E).
Two point mutations suffice to generate catalytically competent YKL-39
To confirm that the substitutions within the DxxDxDxE motif are the sole cause for the lack of hydrolytic activity, Ser143 and Ile145 were mutated to the corresponding active site residues (aspartate and glutamate respectively) in active chitinases. Strikingly, the S143D/I145E double mutant showed significant chitinase activity (Figure 3). Both human chitotriosidase and YKL-39 S143D/I145E showed similar affinity for the fluorigenic pseudosubstrate 4MU-(GlcNAc)3 with a Michaelis constant (Km) of 53 ± 3 μM for hCHT and 9 ± 1 μM for the active YKL-39 mutant. However, substrate turnover by the active YKL-39 mutant is significantly slower, with a kcat value of 0.02 ± 0.0002 s−1, as compared with 15.7 ± 0.3 s−1 for hCHT (Figure 3). Slight conformational differences induced by mutations in the vicinity of the active site may account for the difference in turnover rate. Nevertheless, the implications of this experiment are that YKL-39 is a pseudo-chitinase, that is, in the course of evolution, YKL-39 appears to have retained the ability to bind to chitooligosaccharides, but lost the ability to hydrolyse them. Although chitooligosaccharides are the tightest YKL-39-binding ligands identified to date, it is not yet clear whether these represent physiological ligands for this protein.
Figure 3. Steady-state kinetics of hCHT and an active YKL–39 mutant.
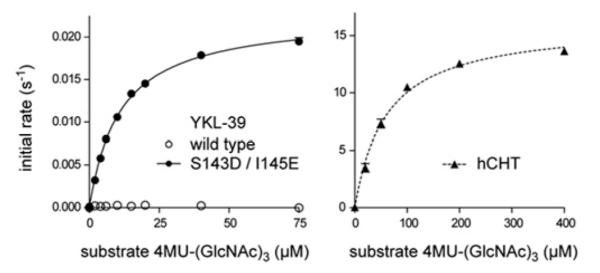
Initial velocity measurements of chitinase activity of YKL-39 active mutant (S143D/I145E) and hCHT on the fluorigenic pseudosubstrate 4MU-(GlcNAc)3, fitted to the standard Michaelis–Menten equation. Results are means ± S.E.M. for three independent experiments. From the fit the following steady-state kinetics parameters were obtained: Km 53 ± 3 μM and kcat = 15.7 ± 0.3 s−1 for hCHT, and Km = 9 ± 1 μM and kcat 0.0200 ± 0.0002 s−1 for the active YKL-39 mutant.
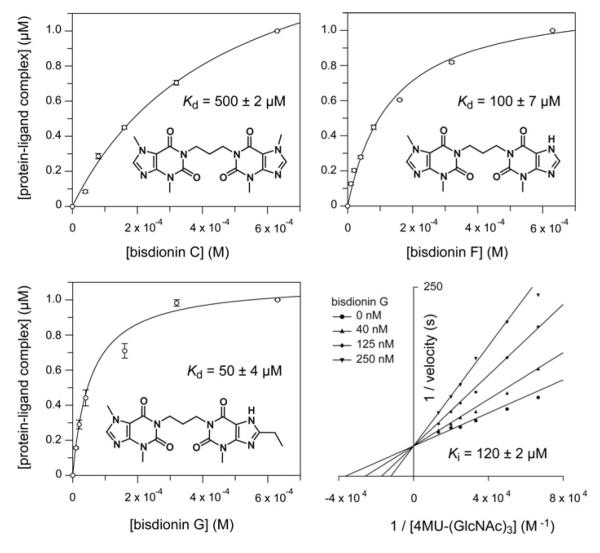
A novel bisdionin-based compound binds YKL-39 with micromolar affinity
To enable cell biological studies towards the role of YKL-39, a potent antagonist that competes with glycan binding would be a useful chemical biological tool. Rationally designed dixanthine derivatives, bisdionins, have been reported as micromolar range inhibitors of family 18 chitinases [49] (Figure 4). Structure-guided modifications to either xanthine ring can be readily incorporated to fine-tune the affinity and selectivity of these ligands, as illustrated by the recently reported bisdionin F derivative (Figure 4), that possesses submicromolar activity against hAMCase with selectivity over hCHT [10]. The bisdionins have been shown to tightly bind the active site of family 18 chitinases by forming extensive stacking interactions with the solvent-exposed tryptophan residues lining the chitooligosaccharide-binding site. To investigate whether the bisdionins would also potently bind YKL-39, we conducted tryptophan fluorescence binding assays (Figure 4). Both bisdionin C and F bind YKL-39 with Kd values in the high micromolar range, Kd = 500 ± 2 and 100 ± 7 μM respectively (Figure 4). By screening a small library of synthetic bisdionin derivatives, we identified bisdionin G, a derivative of bisdionin F carrying an ethyl substituent on the 8-position of the second xanthine moiety, that binds YKL-39 with a Kd value of 500 ± 4 μM (Figure 4). The binding mode for this compound was further confirmed by inhibition studies with the active mutant (S143D/I145E) of YKL-39. Competitive inhibition with a Ki of 120 ± 2 μM was observed (Figure 4). The structural information provided here will aid the development of YKL-39 targeted bisdionin derivatives as cell biological tools to study the function of these proteins.
Figure 4. The bisdionins and their affinity for YKL-39.
The chemical structures of the bisdionin family of chitinase inhibitors are shown, together with bisdionin-induced changes in YKL-39 intrinsic tryptophan fluorescence fitted to a single-site-binding isotherm. Results are means ± S.D. for three independent experiments. The lower right-hand panel shows inhibition of the active mutant of YKL-39, S143D/I145E, by bisdionin G. A double-reciprocal plot is shown to visualize the competitive nature of the inhibition. The Ki value was determined by non-linear regression as described in the Materials and methods section.
Conclusions
YKL-39 shares sequence homology with mammalian chitinases, but differences in expression pattern and the absence of any hydrolytic activity suggest an independent function. A glycan microarray, performed at the Consortium of Functional Glycomics, identified chitooligosaccharides as the best ligands among a selection of 264 synthetic carbohydrates. They are somewhat unlikely physiological ligands: chitin is not synthesized by mammals, and the tissue expression of YKL-39 largely precludes contact with nutrition or pathogens. Nevertheless, chitin and short fractions of the polymer remain the only binding partners of YKL-39 reported to date, and we proceeded to study the binding mode by determining the structure of a complex between YKL-39 and chitohexaose. The conformation of the ligand is strikingly similar to that observed in structures of active chitinases, including the distortion of the sugar in the −1 position, which assumes the boat conformation characteristic of enzymes utilizing a substrate-assisted catalysis mechanism involving neighbouring group participation. The results of the present study demonstrate that the lack of enzymatic activity is attributable to substitutions in the DxxDxDxE motif, which characterizes the active site of mammalian chitinases. Indeed, restoration of this motif by site-directed mutagenesis at just two positions resulted in recovery of chitinase activity.
A distinctive feature of the chitinase active site groove is a series of solvent-exposed aromatic side chains acting as hydrophobic stacking-interfaces for sugar binding. These are conserved without exception in YKL-39. In contrast with hydrogen bonding, such interactions are less specific, suggesting that other glycans possessing β-1,4 linkages could potentially be accommodated in a similar manner. The bisdionin family of chitinase inhibitors was identified as micromolar binders and may be useful tools in functional studies to compete with ligand binding. Such studies will reveal more of the enigmatic function of the chitinase-like proteins. From the conservation of glycan binding and loss of hydrolase activity of YKL-39, we may conclude that its physiological role is that of a pseudo-chitinase, whose physiological ligand is yet to be identified.
ACKNOWLEDGEMENTS
We acknowledge G. Penman’s technical assistance with protein purification.
FUNDING This work was supported by a Wellcome Trust Senior Research Fellowship (to D.v.A.).
Abbreviations used
- AMCase
acidic mammalian chitinase
- BMGY
buffered glycerol complex
- BRP-39
breast regression protein 39
- DMF
dimethylformamide
- GH18
glycoside hydrolase 18
- hAMCase
human AMCase
- hCHT
human chitotriosidase
- MMP
matrix metalloprotease
- NaAc
sodium acetate
- PEG
poly(ethylene glycol)
- RMSD
root mean square deviation
- YKL-39
chitinase 3-like-2
- YKL-40
chitinase 3-like-1
Footnotes
The co-ordinates and structure factors have been deposited in the PDB under accession code 4AY1.
REFERENCES
- 1.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Renkema GH, Boot RG, Au FL, Donker-Koopman WE, Strijland A, Muijsers AO, Hrebicek M, Aerts JM. Chitotriosidase, a chitinase, and the 39-kDa human cartilage glycoprotein, a chitin-binding lectin, are homologues of family 18 glycosyl hydrolases secreted by human macrophages. Eur. J. Biochem. 1998;251:504–509. doi: 10.1046/j.1432-1327.1998.2510504.x. [DOI] [PubMed] [Google Scholar]
- 3.Boot RG, Verhoek M, Donker-Koopman W, Strijland A, van Marle J, Overkleeft HS, Wennekes T, Aerts JM. Identification of the non-lysosomal glucosylceramidase as β-glucosidase 2. J. Biol. Chem. 2007;282:1305–1312. doi: 10.1074/jbc.M610544200. [DOI] [PubMed] [Google Scholar]
- 4.Korolenko TA, Djanayeva SJ, Falameyeva OV, Wevers RA, Filjushina EE, Buzueva II, Kaledin VI, Sandula J, Nowicky J. Chitotriosidase as a new marker of macrophage stimulation in a tumor model treated with cyclophosphamide and Ukrain. Drugs Exp. Clin. Res. 2000;26:279–283. [PubMed] [Google Scholar]
- 5.Malaguarnera L. Chitotriosidase: the yin and yang. Cell. Mol. Life Sci. 2006;63:3018–3029. doi: 10.1007/s00018-006-6269-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boot RG, Hollak CE, Verhoek M, Sloof P, Poorthuis BJ, Kleijer WJ, Wevers RA, van Oers MH, Mannens MM, Aerts JM, van Weely S. Glucocerebrosidase genotype of Gaucher patients in The Netherlands: limitations in prognostic value. Hum. Mutat. 1997;10:348–358. doi: 10.1002/(SICI)1098-1004(1997)10:5<348::AID-HUMU3>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 7.Boot RG, Blommaart EF, Swart E, Ghauharali-van der Vlugt K, Bijl N, Moe C, Place A, Aerts JM. Identification of a novel acidic mammalian chitinase distinct from chitotriosidase. J. Biol. Chem. 2001;276:6770–6778. doi: 10.1074/jbc.M009886200. [DOI] [PubMed] [Google Scholar]
- 8.Zhu Z, Zheng T, Homer RJ, Kim YK, Chen NY, Cohn L, Hamid Q, Elias JA. Acidic mammalian chitinase in asthmatic Th2 inflammation and IL-13 pathway activation. Science. 2004;304:1678–1682. doi: 10.1126/science.1095336. [DOI] [PubMed] [Google Scholar]
- 9.Matsumoto T, Inoue H, Sato Y, Kita Y, Nakano T, Noda N, Eguchi-Tsuda M, Moriwaki A, Kan OK, Matsumoto K, et al. Demethylallosamidin, a chitinase inhibitor, suppresses airway inflammation and hyperresponsiveness. Biochem. Biophys. Res. Commun. 2009;390:103–108. doi: 10.1016/j.bbrc.2009.09.075. [DOI] [PubMed] [Google Scholar]
- 10.Sutherland TE, Andersen OA, Betou M, Eggleston IM, Maizels RM, van Aalten D, Allen JE. Analyzing airway inflammation with chemical biology: dissection of acidic mammalian chitinase function with a selective drug-like inhibitor. Chem. Biol. 2011;18:569–579. doi: 10.1016/j.chembiol.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hakala BE, White C, Recklies AD. Human cartilage gp-39, a major secretory product of articular chondrocytes and synovial cells, is a mammalian member of a chitinase protein family. J. Biol. Chem. 1993;268:25803–25810. [PubMed] [Google Scholar]
- 12.Sendai Y, Komiya H, Suzuki K, Onuma T, Kikuchi M, Hoshi H, Araki Y. Molecular cloning and characterization of a mouse oviduct-specific glycoprotein. Biol. Reprod. 1995;53:285–294. doi: 10.1095/biolreprod53.2.285. [DOI] [PubMed] [Google Scholar]
- 13.Hu B, Trinh K, Figueira WF, Price PA. Isolation and sequence of a novel human chondrocyte protein related to mammalian members of the chitinase protein family. J. Biol. Chem. 1996;271:19415–19420. doi: 10.1074/jbc.271.32.19415. [DOI] [PubMed] [Google Scholar]
- 14.Sun YJ, Chang NC, Hung SI, Chang AC, Chou CC, Hsiao CD. The crystal structure of a novel mammalian lectin, Ym1, suggests a saccharide binding site. J. Biol. Chem. 2001;276:17507–17514. doi: 10.1074/jbc.M010416200. [DOI] [PubMed] [Google Scholar]
- 15.Webb DC, McKenzie AN, Foster PS. Expression of the Ym2 lectin-binding protein is dependent on interleukin (IL)-4 and IL-13 signal transduction: identification of a novel allergy-associated protein. J. Biol. Chem. 2001;276:41969–41976. doi: 10.1074/jbc.M106223200. [DOI] [PubMed] [Google Scholar]
- 16.Kzhyshkowska J, Mamidi S, Gratchev A, Kremmer E, Schmuttermaier C, Krusell L, Haus G, Utikal J, Schledzewski K, Scholtze J, Goerdt S. Novel stabilin-1 interacting chitinase-like protein (SI-CLP) is up-regulated in alternatively activated macrophages and secreted via lysosomal pathway. Blood. 2006;107:3221–3228. doi: 10.1182/blood-2005-07-2843. [DOI] [PubMed] [Google Scholar]
- 17.Guo L, Johnson RS, Schuh JC. Biochemical characterization of endogenously formed eosinophilic crystals in the lungs of mice. J. Biol. Chem. 2000;275:8032–8037. doi: 10.1074/jbc.275.11.8032. [DOI] [PubMed] [Google Scholar]
- 18.Haglund L, Bernier SM, Onnerfjord P, Recklies AD. Proteomic analysis of the LPS-induced stress response in rat chondrocytes reveals induction of innate immune response components in articular cartilage. Matrix Biol. 2008;27:107–118. doi: 10.1016/j.matbio.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 19.Mizoguchi E. Chitinase 3-like-1 exacerbates intestinal inflammation by enhancing bacterial adhesion and invasion in colonic epithelial cells. Gastroenterology. 2006;130:398–411. doi: 10.1053/j.gastro.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 20.Lang M, Schlechtweg M, Kellermeier S, Brenmoehl J, Falk W, Scholmerich J, Herfarth H, Rogler G, Hausmann M. Gene expression profiles of mucosal fibroblasts from strictured and nonstrictured areas of patients with Crohn’s disease. Inflamm. Bowel Dis. 2009;15:212–223. doi: 10.1002/ibd.20735. [DOI] [PubMed] [Google Scholar]
- 21.Chupp GL, Lee CG, Jarjour N, Shim YM, Holm CT, He S, Dziura JD, Reed J, Coyle AJ, Kiener P, et al. A chitinase-like protein in the lung and circulation of patients with severe asthma. N. Engl. J. Med. 2007;357:2016–2027. doi: 10.1056/NEJMoa073600. [DOI] [PubMed] [Google Scholar]
- 22.Trudel G, Recklies A, Laneuville O. Increased expression of chitinase 3-like protein 1 secondary to joint immobility. Clin. Orthop. Relat. Res. 2007;456:92–97. doi: 10.1097/BLO.0b013e3180307c0a. [DOI] [PubMed] [Google Scholar]
- 23.Ober C, Tan Z, Sun Y, Possick JD, Pan L, Nicolae R, Radford S, Parry RR, Heinzmann A, Deichmann KA, et al. Effect of variation in CHI3L1 on serum YKL-40 level, risk of asthma, and lung function. N. Engl. J. Med. 2008;358:1682–1691. doi: 10.1056/NEJMoa0708801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ling H, Recklies AD. The chitinase 3-like protein human cartilage glycoprotein 39 inhibits cellular responses to the inflammatory cytokines interleukin-1 and tumour necrosis factor-α. Biochem. J. 2004;380:651–659. doi: 10.1042/BJ20040099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Recklies AD, White C, Ling H. The chitinase 3-like protein human cartilage glycoprotein 39 (HC-gp39) stimulates proliferation of human connective-tissue cells and activates both extracellular signal-regulated kinase- and protein kinase B-mediated signalling pathways. Biochem. J. 2002;365:119–126. doi: 10.1042/BJ20020075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kzhyshkowska J, Gratchev A, Goerdt S. Human chitinases and chitinase-like proteins as indicators for inflammation and cancer. Biomark Insights. 2007;2:128–146. [PMC free article] [PubMed] [Google Scholar]
- 27.Lee CG, Hartl D, Lee GR, Koller B, Matsuura H, Da Silva CA, Sohn MH, Cohn L, Homer RJ, Kozhich AA, et al. Role of breast regression protein 39 (BRP-39)/chitinase 3-like-1 in Th2 and IL-13-induced tissue responses and apoptosis. J. Exp. Med. 2009;206:1149–1166. doi: 10.1084/jem.20081271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knorr T, Obermayr F, Bartnik E, Zien A, Aigner T. YKL-39 (chitinase 3-like protein 2), but not YKL-40 (chitinase 3-like protein 1), is up regulated in osteoarthritic chondrocytes. Ann. Rheum. Dis. 2003;62:995–998. doi: 10.1136/ard.62.10.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steck E, Breit S, Breusch SJ, Axt M, Richter W. Enhanced expression of the human chitinase 3-like 2 gene (YKL-39) but not chitinase 3-like 1 gene (YKL-40) in osteoarthritic cartilage. Biochem. Biophys. Res. Commun. 2002;299:109–115. doi: 10.1016/s0006-291x(02)02585-8. [DOI] [PubMed] [Google Scholar]
- 30.Bussink AP, Speijer D, Aerts JM, Boot RG. Evolution of mammalian chitinase(-like) members of family 18 glycosyl hydrolases. Genetics. 2007;177:959–970. doi: 10.1534/genetics.107.075846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fusetti F, von Moeller H, Houston D, Rozeboom HJ, Dijkstra BW, Boot RG, Aerts JM, van Aalten DM. Structure of human chitotriosidase. Implications for specific inhibitor design and function of mammalian chitinase-like lectins. J. Biol. Chem. 2002;277:25537–25544. doi: 10.1074/jbc.M201636200. [DOI] [PubMed] [Google Scholar]
- 32.Lu Y, Zen KC, Muthukrishnan S, Kramer KJ. Site-directed mutagenesis and functional analysis of active site acidic amino acid residues D142, D144 and E146 in Manduca sexta (tobacco hornworm) chitinase. Insect Biochem. Mol. Biol. 2002;32:1369–1382. doi: 10.1016/s0965-1748(02)00057-7. [DOI] [PubMed] [Google Scholar]
- 33.Terwisscha van Scheltinga AC, Hennig M, Dijkstra BW. The 1.8Å resolution structure of hevamine, a plant chitinase/lysozyme, and analysis of the conserved sequence and structure motifs of glycosyl hydrolase family 18. J. Mol. Biol. 1996;262:243–257. doi: 10.1006/jmbi.1996.0510. [DOI] [PubMed] [Google Scholar]
- 34.Houston DR, Recklies AD, Krupa JC, van Aalten DM. Structure and ligand-induced conformational change of the 39-kDa glycoprotein from human articular chondrocytes. J. Biol. Chem. 2003;278:30206–30212. doi: 10.1074/jbc.M303371200. [DOI] [PubMed] [Google Scholar]
- 35.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 36.Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 38.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. D Biol. Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 39.Schüttelkopf AW, Andersen OA, Rao FV, Allwood M, Rush CL, Eggleston IM, Van Aalten DMF. Bisdionin C: a rationally designed, submicromolar inhibitor of family 18 chitinases. ACS Med. Chem. Lett. 2011;2:428–432. doi: 10.1021/ml200008b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salas-Peregrin JM, Sanchez–Martinez E, Colacio-Rodriguez E. Metal ion–purine interactions: preparation and study of some metal complexes of 8-ethyl-xanthine and 8-ethyl-3-methyl-xanthine. Inorg. Chim. Acta. 1985;107:23–27. [Google Scholar]
- 41.Fischer B, Yefidoff R, Major DT, Rutman-Halili I, Shneyvays V, Zinman T, Jacobson KA, Shainberg A. Characterization of ‘mini-nucleotides’ as P2X receptor agonists in rat cardiomyocyte cultures. An integrated synthetic, biochemical, and theoretical study. J. Med. Chem. 1999;42:2685–2696. doi: 10.1021/jm990085i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Muijlwijk-Koezen JE, Timmerman H, van der Sluis RP, van de Stolpe AC, Menge WM, Beukers MW, van der Graaf PH, de Groote M, IJzerman AP. Synthesis and use of FSCPX, an irreversible adenosine A1 antagonist, as a ‘receptor knock-down’ tool. Bioorg. Med. Chem. Lett. 2001;11:815–818. doi: 10.1016/s0960-894x(01)00069-5. [DOI] [PubMed] [Google Scholar]
- 43.Perrakis A, Tews I, Dauter Z, Oppenheim AB, Chet I, Wilson KS, Vorgias CE. Crystal structure of a bacterial chitinase at 2.3 Å resolution. Structure. 1994;2:1169–1180. doi: 10.1016/s0969-2126(94)00119-7. [DOI] [PubMed] [Google Scholar]
- 44.van Aalten DM, Komander D, Synstad B, Gaseidnes S, Peter MG, Eijsink VG. Structural insights into the catalytic mechanism of a family 18 exo-chitinase. Proc. Natl. Acad. Sci. U.S.A. 2001;98:8979–8984. doi: 10.1073/pnas.151103798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Watanabe T, Ishibashi A, Ariga Y, Hashimoto M, Nikaidou N, Sugiyama J, Matsumoto T, Nonaka T. Trp122 and Trp134 on the surface of the catalytic domain are essential for crystalline chitin hydrolysis by Bacillus circulans chitinase A1. FEBS Lett. 2001;494:74–78. doi: 10.1016/s0014-5793(01)02317-1. [DOI] [PubMed] [Google Scholar]
- 46.Zhang H, Huang X, Fukamizo T, Muthukrishnan S, Kramer KJ. Site-directed mutagenesis and functional analysis of an active site tryptophan of insect chitinase. Insect Biochem. Mol. Biol. 2002;32:1477–1488. doi: 10.1016/s0965-1748(02)00068-1. [DOI] [PubMed] [Google Scholar]
- 47.Itoh Y, Watanabe J, Fukada H, Mizuno R, Kezuka Y, Nonaka T, Watanabe T. Importance of Trp59 and Trp60 in chitin-binding, hydrolytic, and antifungal activities of Streptomyces griseus chitinase C. Appl. Microbiol. Biotechnol. 2006;72:1176–1184. doi: 10.1007/s00253-006-0405-7. [DOI] [PubMed] [Google Scholar]
- 48.Lienemann M, Boer H, Paananen A, Cottaz S, Koivula A. Toward understanding of carbohydrate binding and substrate specificity of a glycosyl hydrolase 18 family (GH-18) chitinase from Trichoderma harzianum. Glycobiology. 2009;19:1116–1126. doi: 10.1093/glycob/cwp102. [DOI] [PubMed] [Google Scholar]
- 49.Schüttelkopf AW, Andersen OA, Rao FV, Allwood M, Lloyd C, Eggleston IM, van Aalten DM. Screening-based discovery and structural dissection of a novel family 18 chitinase inhibitor. J. Biol. Chem. 2006;281:27278–27285. doi: 10.1074/jbc.M604048200. [DOI] [PubMed] [Google Scholar]