Abstract
A number of practical advantages have made the nematode Caenorhabditis elegans a useful model for genetic and developmental biological research. These same advantages, along with conservation of disease and stress response pathways, availability of mutant and transgenic strains, and wealth of biological information, have led to the increased use of C. elegans in toxicological studies. Although the potential to study the mechanisms of developmental toxicology in C. elegans is promising, embryonic and larval growth tests to identify compounds that affect the nematode have remained the primary use of C. elegans in developmental toxicology. Here, we describe a C. elegans larval growth and development assay for medium and high throughput screening using the COPAS Biosort flow cytometer and provide descriptions of the data and subsequent analysis.
Keywords: C. elegans, high throughput screen, growth and development, COPAS Biosort, developmental toxicity
1. Introduction
Caenorhabditis elegans is a free living soil nematode that has been used extensively as a model organism for developmental biology (Figure 1) (1). C. elegans reproduce quickly and in large numbers. At 20° C, development proceeds from embryo through four distinct larval stages (L1-L4) to gravid adult hermaphrodites in approximately 72 h (2). Because they are transparent, the internal development of all C. elegans cells and organs can be easily monitored using light microscopy. Maintenance and culturing of C. elegans is inexpensive and straight forward. In the laboratory, C. elegans feed on E. coli (3). There are several resources available describing the general characteristics and development of C. elegans (4–7).
Figure 1. Adult C. elegans hermaphrodite.
Image was acquired at 20× magnification.
C. elegans is becoming a popular toxicological test organism, as evidenced by recent reviews (8–10). Toxicity endpoints recently reported include reproduction (11), feeding (12), DNA damage and repair (13, 14), and gene expression (15, 16). Despite the advantages of C. elegans as a model of development and disease, its use in developmental toxicology has largely focused on observational rather than mechanistic studies (17). Observational studies include neuronal development using GFP-labeled neurons (18, 19) as well as whole organism growth and development (20–22). Here we describe a standardized C. elegans growth assay useful for screening large numbers of chemicals for potential developmental effects.
2. Materials
2.1 Nematode culture and maintenance
K-agar plates: 2.36 g KCl, 3 g NaCl, 2.5 g Bacto-peptone, 17 g Bacto-agar, 1 mL cholesterol (5 mg/mL), 1 mL 1 M CaCl2, 1 mL 1 M MgSO4 in 1 L dH2O
K-medium: 2.36 g KCl, 3 g NaCl, in 1 L dH2O
Complete K-medium: 1 L K-medium, 1 mL cholesterol (5 mg/mL), 1 mL 1 M CaCl2, 1 mL 1 M MgSO4
Bleaching solution:1 g NaOH, 20 mL 5.25% NaOCl, 80 mL dH2O
Freezing solution: 20 mL 1 M NaCl, 10 mL 1 M KH2PO4 (pH 6.0), 60 mL glycerol, 0.6 mL 0.1 M MgSO4
M9 Buffer: 3 g KH2PO4, 6 g Na2HPO4, 5 g NaCl, 1 ml 1 M MgSO4, in 1 L dH2O
2.2 C. elegans growth assay
Biomek 2000 Laboratory Automation Workstation (Beckman Coulter, Inc., Brea, CA)
COPAS Biosort with REFLX option (Union Biometrica, Inc., Holliston, MA)
3. Methods
3.1. Nematode culture and maintenance
3.1.1.
Nematode strains: The wild-type N2 strain of C. elegans was obtained from the Caenorhabditis Genetics Center (Minneapolis, MN) and used for all growth assays. Transgenic and mutant C. elegans strains may also be used to address specific experimental questions and are handled as described for N2. C. elegans cultures are fed the OP50 strain of E. coli seeded either onto K-agar plates or added to complete K-medium.
3.1.2.
Storage of C. elegans: L1 nematodes can be frozen and stored in liquid nitrogen (23). K-agar plates that are cleared of bacteria (usually four days after embryo preparation) containing numerous L1s are rinsed with K-medium. Nematodes are transferred with sterile glass Pasteur pipets to 15-mL centrifuge tubes. Tubes are then centrifuged at 2,000×g for 1 min and all but 1.5 mL of the supernatant is aspirated and discarded. An equal volume of freezing solution is added to the tube. This solution is then gently mixed with the Pasteur pipet and transferred to 3×2-mL cryogenic tubes. The cryogenic tubes are labeled and placed in a Mr. Frosty isopropanol bath (Nalgene Labware) and placed in a −80 °C freezer overnight. The next day, two tubes are transferred to liquid nitrogen for long term storage. The third tube is thawed and the nematode pellet in the bottom of the tube is transferred to a 60-mm K-agar plate seeded with bacteria. The following day, nematode survival is verified using a dissecting microscope. One stock tube is thawed every three months to prevent contamination and genetic drift of the C. elegans population.
3.1.3.
Preparation of L1 larva - Embryo preparations are performed daily to obtain age-synchronized cultures. Gravid three-day-old adult nematodes and embryos are rinsed from K-agar plates with K-medium and transferred to 15-mL centrifuge tubes. After centrifugation at 2,000×g for 2 min, the supernatant is carefully aspirated from the nematode pellet and then approximately 10 mL of bleaching solution is added. After 10 min of gentle agitation, the tube is centrifuged at 2000×g for 2 min and the supernatant is removed, leaving a white pellet in the bottom of the tube. The pellet is resuspended in approximately 10 mL K-medium and pelleted by centrifugation at 2,000×g for 2 min. After removing all but approximately 0.5 ml of the supernatant, the embryos are gently resuspended with a sterile glass pipet and transferred to a sterile 25 cm2 culture flask (with vented cap) containing 5 mL complete K-medium. Embryos are allowed to hatch in the absence of food overnight, yielding a synchronized L1 culture.
3.2. C. elegans growth assay
3.2.1.
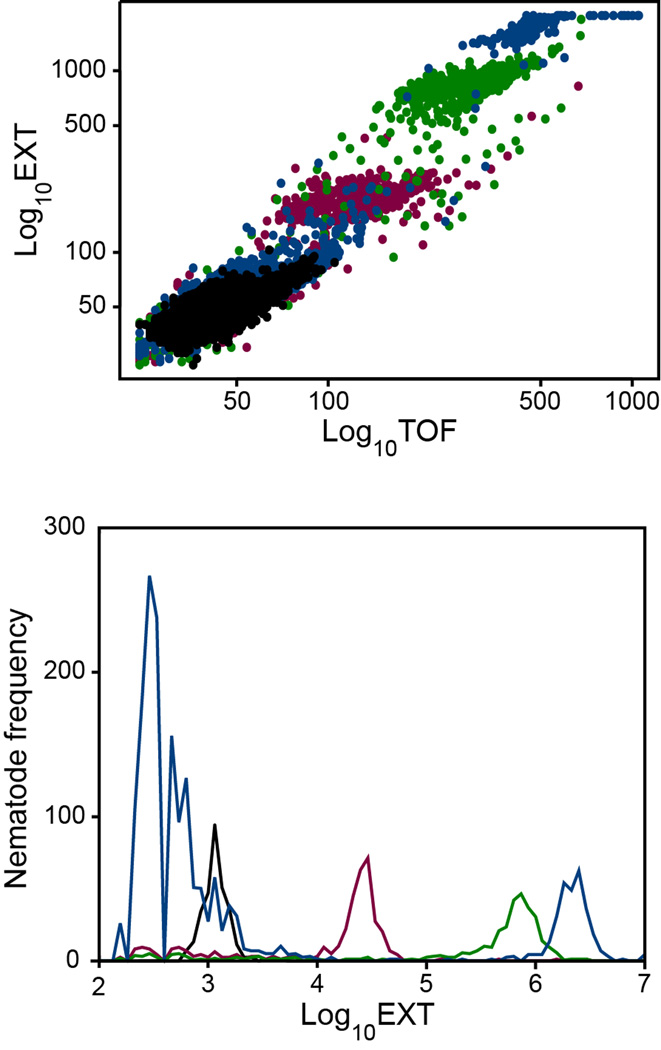
Development of C. elegans from L1 to L4 larvae takes approximately 48 h at 20 °C (20) (Figure 2). This time period was chosen for growth assays to allow for maximum development, while avoiding the production of offspring at later times.
Figure 2. C. elegans growth from L1 to adult.
Untreated L1 nematodes were incubated at 20°C and sampled at 0 h (black), 24 h (red), 48 h (green), and 72 h (blue). Upper panel, scatter-plot of optical density (log(EXT)) versus length (log(TOF)). Each point corresponds to an individual nematode. Lower panel, frequency distributions of log(EXT) versus numbers of nematodes. At 72 h, adult nematodes (high EXT, TOF) and their offspring (low EXT, TOF) were observed.
3.2.2.
At the start of the growth assay, L1s are transferred to the sample cup of the COPAS Biosort and diluted with complete K-medium to approximately 1 nematode/µL. Fifty L1s are dispensed into each well of a 96-well plate containing E. coli and the test chemical, over a range of eleven concentrations, to a final volume of 100 µL. Initial optical densities (ODs) are recorded to monitor bacteria concentrations at the start of the exposure period. After the 48-h exposure, ODs are recorded and C. elegans are aspirated from each well of the exposure plate using the REFLX option of the COPAS Biosort. The COPAS Biosort is a flow cytometer that measures, sorts and dispenses individual nematodes based on nematode length (referred to as time of flight (TOF)), absorbance (referred to as extinction (EXT)), and two channels of fluorescence (24). After aspirating the C. elegans samples, measurement of nematode TOF, EXT, and fluorescence are recorded for individual animals.
3.2.3.
Day 1: prepare embryos for synchronized L1 culture (3.1.3).
3.2.4.
Day 2: prepare the 96-well exposure plates using the Biomek 2000. Pipet complete K- medium to each of 12 wells of a 24-well tissue culture plate (total volume of each well = 500 µl). Because exposure concentrations are diluted by the addition of nematodes (~1 µL per nematode), prepare chemical stocks at 2x the desired final concentration. The initial stock solution is typically 100x the highest concentration to be tested and is dissolved in K-medium or DMSO (see Note 4.1). Measure the pH of the stock solution (see Note 4.2). Pipet appropriate volumes of test chemical stock solution to each of the 12 wells. Pipet 75 µL bacteria suspended in complete K-medium into each well. After thoroughly mixing, pipet 50 µL into each exposure well of the 96-well plate, leaving rinse wells between exposure concentrations to prevent carryover between treatments (see Note4.3).
3.2.5.
Add 50 L1 nematodes into exposure wells using the COPAS Biosort. Read OD570 to monitor bacteria concentration. Seal plate with Breathe-Easy gas permeable sealing membranes (USA Scientific, Ocala, FL) and incubate for 48 h at 20°C.
3.2.6.
Day 4: remove the sealing membrane and measure the OD570. Observe nematodes using a dissecting microscope to verify the health of the negative controls, absence of contamination, and concentration response to the test chemical. Using COPAS Biosort, aspirate and measure the TOF and EXT of individual C. elegans.
3.2.7.
Verify that the COPAS Biosort is aspirating all of the exposure media from the sample wells and the absence of air bubbles or other sampling errors (see Note 4.4).
3.3. Analysis of C. elegans growth data
The COPAS Biosort EXT measurement reflects the amount of light absorbed as an object passes through the laser and has been successfully used as an indicator of size for C. elegans (20). Over the duration of the growth assay, unexposed nematodes grow at a near exponential rate; so the log transform is taken to keep the variance of the measurements stable across concentrations (22). Observations at 48 h include nematodes as well as detritus built up over the exposure period. The detritus may include chemical precipitates, bacteria clumps, or biological detritus from the nematodes (see Note 4.5). The detritus size distribution will be referred to as the noise distribution and may depend on the exposure chemical and concentration. For some exposures, the nematode size distribution may overlap with the noise distribution.
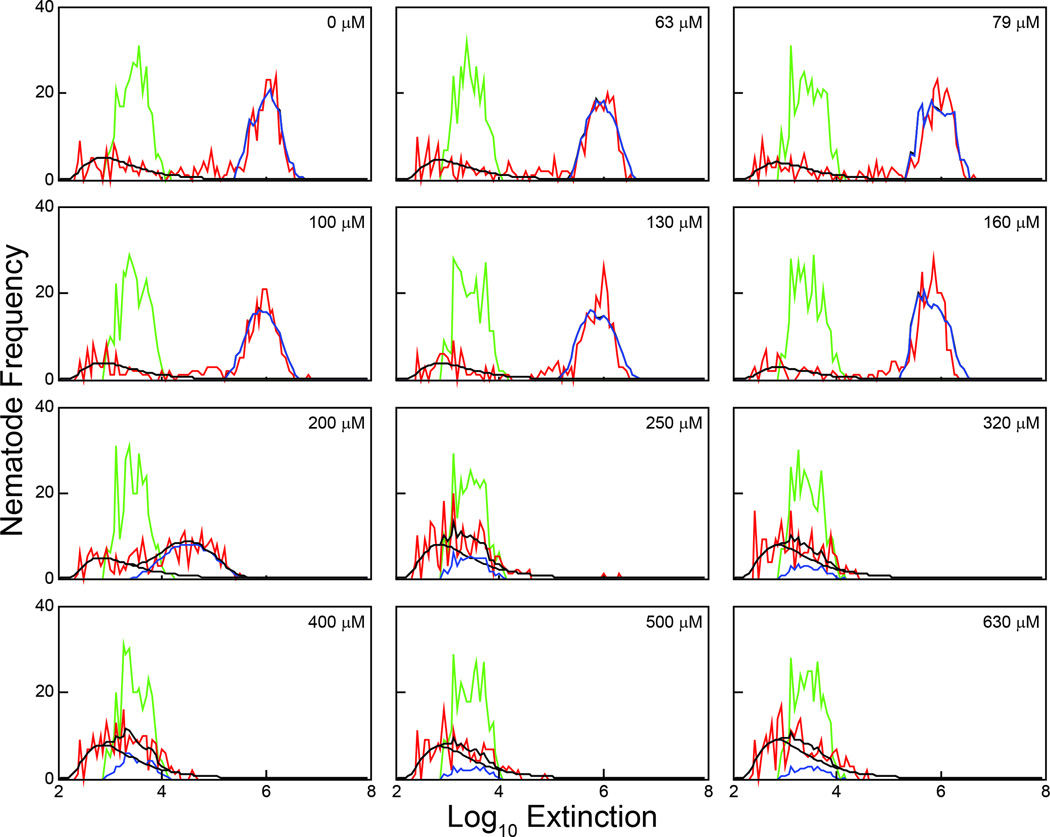
To identify nematode measurements quickly for further analysis, a grid is imposed on the log(EXT) range of all observations, dividing the range into a fixed number of equally sized bins. The grid is chosen so that the resolution of the histograms of observations is high enough to distinguish important features. Figure 3 shows the results of a growth assay with exposures to eleven concentrations of cadmium chloride plus an untreated control. Using the same grid, red lines indicate histograms of all aspirated observations at 48 h after loading, while green lines indicate histograms of the loaded observations. Nematode measurements are identifiable for the first six concentrations, but the higher cadmium concentrations interfere with nematode growth, keeping sizes within the range of detritus.
Figure 3. Observed and predicted frequency distributions of log(EXT) measurements after 48-h exposure to cadmium chloride.
The loading distribution (green line) and the observed measurements (red line) after 48-h for untreated and groups exposed to eleven concentrations of cadmium chloride are presented. The estimated lognormal distribution describing the extraneous noise measurements for each cohort (black line) and the predicted frequency distribution for the nematode measurements based on the growth model (blue line) are also presented.
An earlier growth model (20, 22) is adapted to the 48 h growth assay, predicting the 48 h size distribution by multiplying the loaded size distribution (the numbers corresponding to the green lines in Figure 3) by a transition matrix. The matrix is determined by two integer-valued parameters that indicate the domain of the size distribution of the aspirated nematodes at 48 h. A lognormal distribution is used for the noise observations, and a weighted average of the two distributions is fit to all aspirated measurements (red curves in Figure 3) for each concentration. The parameters estimated determine the predicted joint distribution, shown in Figure 3 as the blue lines, including the two integer-valued parameters determining the domain of the nematode size distribution at 48 h. All observations within that domain are used as nematode observations in later analyses. Once the aspirated nematode measurements have been determined, the mean of the loaded measurement may be subtracted from all aspirated measurements, so that the numbers reflect nematode growth over the duration of the assay. Additional analytical details are described in Smith et al. (22).
4. Notes
4.1.
Compounds are typically diluted in complete K-medium or 1% DMSO, depending on aqueous solubility. A number of other vehicles have also been evaluated and may be useful for solubilizing certain chemicals including ethanol (≤ 0.1%), cyclodextrins, and PEG-60.
4.2.
For chemicals that alter the pH of the exposure solution, an alternative buffer should be used in place of complete K-medium. For chemicals that increase the pH above 8.5, M9 buffer is substituted for complete K-medium. In cases for which the pH decreases below 4.5, complete K-medium should be buffered with 1N KOH up to a pH of 5.5.
4.3.
One limitation of the COPAS Biosort REFLX tool is carryover, the cross-contamination of a sample well with nematodes from previously sampled sample well(s) or treatment group(s). As nematodes are aspirated from the wells, they are trapped against a filter within the Biosort and then washed into the flow cell. Carryover nematodes may be stuck to the aspirating tool, bubbling filter, or tubing, especially if compounds affect the viscosity of the exposure solution. To minimize carryover, rinse wells containing 1% DMSO are placed between samples of different treatment groups.
4.4.
A number of conditions may affect COPAS Biosort sampling efficiency or data quality including clogging of the flow cell, sample tubing, or aspirating tool; leaking of the waste tubing; disrupted flow due to pressure changes; and excessive sampling due to air bubbles, waste, or chemical precipitates (see Note 4.5). If the Biosort is observed to be clogging or leaking, the run should be aborted and the machine cleaned and primed. If clogging or noise is not observed until after sampling is complete, in some cases, the data may be ‘cleaned’ using mathematical modeling.
4.5.
A certain level of noise is normal in COPAS Biosort data. This routine noise may include dead bacteria, shed cuticles from molting (especially if development is delayed due to chemical exposure), dead nematodes, and air bubbles in the system. Routine noise may be modeled and removed as described in Section 3.3.
5. Current and future applications
5.1.
The C. elegans growth assay is currently being used by the National Toxicology Program’s (NTP’s) WormTox Screening Facility (25) in several capacities. Through collaborations with investigators studying molecular mechanisms of environmental, chemical toxicity or the effects of genetic mutation on growth and development, the growth assay provides one phenotypic descriptor (26). In support of the NTP’s mission to evaluate chemicals of public health concern, a suite of standardized medium throughput C. elegans assays (i.e., growth, reproduction, and feeding) are being used to test chemicals at eleven concentrations. This approach allows full characterization of the concentration-response and has been used to test more than 80 compounds including metals, pesticides, and other organic compounds.
5.2.
The standardized growth assay was modified to allow for high throughput screening of chemical libraries containing hundreds to thousands of chemicals. For high throughput screening, all chemicals must be tested at a pre-determined concentration range rather than across empirically-derived ranges. The C. elegans growth assay is currently being used to screen the U. S. EPA’s ToxCast Phase I and II libraries, which combined contain 1011 chemicals (27, 28). Comparisons of the C. elegans results to these assays and mammalian toxicology data will help model chemical effects on human neurodevelopment. As part of the Tox21 intergovernmental collaboration (29), the C. elegans growth assay will ultimately be included in screens of ~10,000 chemicals.
Acknowledgments
The authors would like to thank Julie R. Rice, Daniel W. Snyder, and Paul E. Dunlap for technical assistance during the development and performance of C. elegans growth assays. We also thank Sandra J. McBride and Grace E. Kissling for assistance in the development of statistical analyses and mathematical modeling tools. This work was supported in part by the National Toxicology Program and by the Intramural Research Program of the National Institute of Environmental Health Sciences, National Institutes of Health (Z01ES102046).
References
- 1.Sulston JE, Horvitz HR. Post-embryonic cell lineages of nematode, Caenorhabditis elegans. Dev. Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- 2.Hope IA. C. elegans: A Practical Approach. Oxford: Oxford University Press; 1999. [Google Scholar]
- 3.Brenner S. The genetics of, Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riddle DL, Blumenthal T, Meyer BJ, Priess JR. C. elegans II. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 5.Wood WB. The Nematode Caenorhabditis elegans. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 6.Strange K. C. elegans: Methods and Applications. Totowa, NJ: Humana Press Inc. ; 2006. [Google Scholar]
- 7. www.wormbase.org.
- 8.Leung MCK, Williams PL, Benedetto A, Au C, Helmcke KJ, Aschner M, Meyer JN. Caenorhabditis elegans: An emerging model in biomedical and environmental toxicology. Toxicol. Sci. 2008;106:5–28. doi: 10.1093/toxsci/kfn121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Helmcke KJ, Avila DS, Aschner M. Utility of Caenorhabditis elegans in high throughput neurotoxicological research. Neurotoxicol. Teratol. 2010;32:62–67. doi: 10.1016/j.ntt.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 10.Boyd WA, Smith MV, Kissling GE, Freedman JH. Medium- and high-throughput screening of neurotoxicants using C. elegans. Neurotoxicol. Teratol. 2010;32:68–73. doi: 10.1016/j.ntt.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boyd WA, McBride SJ, Rice JR, Snyder DW, Freedman JH. A high-throughput method for assessing chemical toxicity using a Caenorhabditis elegans reproduction assay. Toxicol. Appl. Pharmacol. 2010;245:153–159. doi: 10.1016/j.taap.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boyd WA, McBride SJ, Freedman JH. Effects of genetic mutations and chemical exposures on Caenorhabditis elegans feeding: evaluation of a novel, high-throughput screening assay. PLoS One. 2007;2:e1259. doi: 10.1371/journal.pone.0001259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allard P, Colaiacovo MP. Bisphenol A impairs the double-strand break repair machinery in the germline and causes chromosome abnormalities. Proc. Natl. Acad. SciUSA. 2010;107:20405–20410. doi: 10.1073/pnas.1010386107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leung MCK, Goldstone JV, Boyd WA, Freedman JH, Meyer JN. Caenorhabditis elegans Generates Biologically Relevant Levels of Genotoxic Metabolites from Aflatoxin B-1 but Not Benzo[a]pyrene In Vivo. Toxicol. Sci. 2010;118:444–453. doi: 10.1093/toxsci/kfq295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swain S, Wren JF, Stuerzenbaum SR, Kille P, Morgan AJ, Jager T, Jonker MJ, Hankard PK, Svendsen C, Owen J, Hedley BA, Blaxter M, Spurgeon DJ. Linking toxicant physiological mode of action with induced gene expression changes in, Caenorhabditis elegans. BMC Systems Biology. 2010;4 doi: 10.1186/1752-0509-4-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wren JF, Kille P, Spurgeon DJ, Swain S, Sturzenbaum SR, Jager T. Application of physiologically based modelling and transcriptomics to probe the systems toxicology of aldicarb for Caenorhabditis elegans (Maupas 1900) Ecotoxicology. 2011;20:397–408. doi: 10.1007/s10646-010-0591-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hood RD. Developmental and Reproductive Toxicology: A Practical Approach. Boca Raton, FL: CRC Press; 2006. [Google Scholar]
- 18.Brandt R, Gergou A, Wacker I, Fath T, Hutter H. A Caenorhabditis elegans model of tau hyperphosphorylation: Induction of developmental defects by transgenic overexpression of Alzheimer's disease-like modified tau. Neurobiol. Aging. 2009;30:22–33. doi: 10.1016/j.neurobiolaging.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 19.VanDuyn N, Settivari R, Wong G, Nass R. SKN-1/NRF2 inhibits dopamine neuron degeneration in a Caenorhabditis elegans model of methylmercury toxicity. Toxicol. Sci. 2010;118:613–624. doi: 10.1093/toxsci/kfq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boyd WA, Smith MV, Kissling GE, Rice JR, Snyder DW, Portier CJ, Freedman JH. Application of a mathematical model to describe the effects of chlorpyrifos on Caenorhabditis elegans development. PLoS One. 2009;4:e7024. doi: 10.1371/journal.pone.0007024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meyer JN, Lord CA, Yang XYY, Turner EA, Badireddy AR, Marinakos SM, Chilkoti A, Wiesner MR, Auffan M. Intracellular uptake and associated toxicity of silver nanoparticles in, Caenorhabditis elegans. Aquatic Toxicology. 2010;100:140–150. doi: 10.1016/j.aquatox.2010.07.016. [DOI] [PubMed] [Google Scholar]
- 22.Smith MV, Boyd WA, Kissling GE, Rice JR, Snyder DW, Portier CJ, Freedman JH. A discrete time model for the analysis of medium-throughput C. elegans growth data. PLoS One. 2009;4:e7018. doi: 10.1371/journal.pone.0007018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lewis JA, Fleming JT. Basic Culture Methods. In: Epstein HF, Shakes DC, editors. Caenorhabditis elegans: Modern Biological Analysis of an Organism. San Diego, CA: Academic Press; 1995. pp. 3–29. [Google Scholar]
- 24.Pulak R. Techniques for analysis, sorting, and dispensing of C. elegans on the COPAS flow-sorting system. Methods Mol. Biol. 2006;351:275–286. doi: 10.1385/1-59745-151-7:275. [DOI] [PubMed] [Google Scholar]
- 25. http://www.niehs.nih.gov/research/atniehs/labs/bmsb/workgroups.cfm.
- 26.Boyd WA, Crocker TL, Rodriguez AM, Leung MCK, Lehmann DW, Freedman JH, Van Houten B, Meyer JN. Nucleotide excision repair genes are expressed at low levels and are not detectably inducible in Caenorhabditis elegans somatic tissues, but their function is required for normal adult life after UVC exposure. Mutation Research-Fundamental and Molecular Mechanisms of Mutagenesis. 2010;683:57–67. doi: 10.1016/j.mrfmmm.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Judson RS, Houck KA, Kavlock RJ, Knudsen TB, Martin MT, Mortensen HM, Reif DM, Rotroff DM, Shah I, Richard AM, Dix DJ. In Vitro Screening of Environmental Chemicals for Targeted Testing Prioritization: The ToxCast Project. Environ. Health Perspect. 2010;118:485–492. doi: 10.1289/ehp.0901392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knudsen TB, Houck KA, Sipes N, Singh AV, Judson R, Martin MT, Weissman A, Kleinstreuer N, Mortensen HM, Reif D, Rabinowitz JR, Setzer RW, Richard AM, Dix DJ, Kavlock RJ. Activity profiles of 309 ToxCast chemicals evaluated across 292 biochemical targets. Toxicology. 2011;282:1–15. doi: 10.1016/j.tox.2010.12.010. [DOI] [PubMed] [Google Scholar]
- 29.Collins FS, Gray GM, Bucher JR. Toxicology - Transforming environmental health protection. Science. 2008;319:906–907. doi: 10.1126/science.1154619. [DOI] [PMC free article] [PubMed] [Google Scholar]