Abstract
This study utilizes sensitive, modern isothermal titration calorimetric (ITC) methods to characterize the microscopic thermodynamic parameters that drive the binding of basic drugs to α-1-acid glycoprotein (AGP) and thereby rationalize the thermodynamic data in relation to docking models and crystallographic structures of the drug-AGP complexes. The binding of basic compounds from the tricyclic antidepressant series, together with miaserine, chlorpromazine, disopyramide and cimetidine all displayed an exothermically driven binding interaction with AGP. The impact of protonation/deprotonation events, ionic strength, temperature and the individual selectivity of the A and F1*S AGP variants on drug-binding thermodynamics were characterized. A correlation plot of the thermodynamic parameters for all of the test compounds revealed enthalpy-entropy compensation is in effect. The exothermic binding energetics of the test compounds were driven by a combination of favorable (negative) enthalpic (ΔH°) and favorable (positive) entropic (ΔS°) contributions to the Gibbs free energy (ΔG°). Collectively, the data imply that the free energies that drive drug binding to AGP and its relationship to drug-serum residency evolve from the complex interplay of enthalpic and entropic forces from interactions with explicit combinations of hydrophobic and polar side-chain sub-domains within the multi-lobed AGP ligand binding cavity.
Keywords: human α-1-acid glycoprotein, thermodynamics, drug binding
1. Introduction
The lipocalin superfamily is a ubiquitous class of extra-cellular transporters of small hydrophobic molecules around the body [1–3]. α1-Acid glycoprotein (AGP; syn. Orosomucoid), an acute phase protein, is the principle extracellular lipocalin with high concentrations in the blood plasma [4, 5]. One of the major physiological roles of AGP involves the binding and transportation of a range of endogenous (i.e lysophospholipids and biliverdin) and exogenous (i.e drugs) compounds [6–8]. AGP-drug interactions are a focus of great importance in the pharmaceutical sciences as this interaction is a major factor in drug transport to tissue receptors, storage and prevention of metabolism [8]. In healthy individuals, the basal plasma concentration of AGP is approximately 20 μmol/L; whereas in diseased states, such as sepsis, it can increase up to 5-fold[8–12]. Therefore, the effect of AGP binding on the pharmacological activity of highly bound drugs can be significant during acute phase reactions [13–20].
AGP itself is composed of a single polypeptide chain composed of 183 amino acids [4, 5, 8, 21]. The polypeptide component only contributes about a half of its total molecular mass of approximately 41 kDa, the rest of its mass derives from the five N-linked sialyl-glycans which confer AGP with a net negative charge at physiological pH [22–24]. These features also render AGP very soluble and acidic (pI~2.8–3.8) [4, 5, 8, 22].
Human plasma AGP exists as three genetic variants, which include the A, F1 and S forms [8, 12, 25–28]. The expression of human AGP is under the control of two adjacent genes ORM1 (syn. AAG-A) and ORM2 (syn. AAG-B/B′), situated on chromosome 9 [26, 27]. The more active of the two, ORM1, that is induced during acute phase reactions, encodes the F1 and S variants, and ORM2 encodes the A variant [8, 12, 25–28]. The precursor product of the ORM1 gene is a 201 amino acid polypeptide with an 18 residue N-terminal secretory peptide that is cleaved [8, 12, 25–28]. The F1 and S variants, encoded by two alleles of the ORM1 gene differ only in a single amino acid codon (Gln20→Arg), and hereon in shall be referred to collectively as the F1*S variant. The ORM2 gene displays 22 base substitutions, which translates into at least 21 amino acid substitutions between the F1*S and A protein variants (Figure S1) [25–27]. In human plasma, the variants generally occur at a mean molar ratio of 3:1 [F1*S:A], however, the F1*S:A ratio can vary among individuals and during inflammation or illness [8, 28–32].
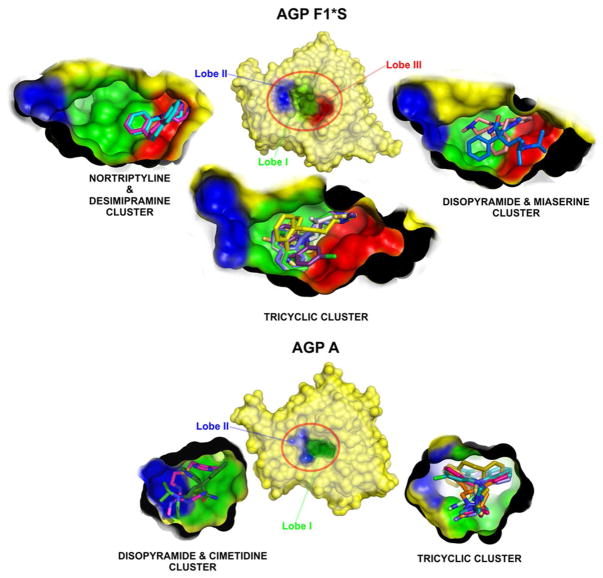
The elucidation of the three dimensional crystal structures of the F1*S and A variants of human AGP revealed that AGP, not unlike other lipocalins, possesses a structural fold consisting of eight anti-parallel β-strands connected by four loops arranged into a β-barrel with three flanking α-helices (Figure S1) [33, 34]. Within the β-barrel motif lies the overtly promiscuous ligand binding pocket [33, 34]. Although both variants display the same tertiary structural configuration, a closer comparison of the crystal structures revealed significant topographical differences between their cavity structures [33, 34]. Compared to the A variant, the ligand binding pocket of the F1*S variant is larger and exhibits three lobes (cf. Figure 5) [34]. Lobe I represents the major sub-compartment and is largely non-polar in character and as such provides the appropriate hydrophobic conditions for accommodating the hydrophobic components of ligands [34]. The flanking lobes, II and III, are smaller and display an acidic character which is due to specific acidic residues, including the Glu64 in lobe II and Asp115 in lobe III [34]. Comparably, the narrower ligand binding pocket of the A variant only displays two lobes, the main central hydrophobic lobe I is present, with a smaller flanking lobe II structure [33]. Investigations into the drug binding selectivity of the individual variants revealed that the A variant displays a stricter drug binding selectivity and an overall higher drug binding affinity [30, 31, 35–43]. In contrast, the F1*S variant generally displayed a broader drug binding selectivity, with an overall lower drug binding affinity [30, 31, 35–43]. The significance of the binding selectivity difference between the A and F1*S variants in relation to their unique binding pocket topology and drug binding energetics is explored in detail in this study.
Figure 5.
Topographic map for the drug docking clusters for the A (PDB ID:3APU) and F1*S (PDB ID: 3KQ0) variants of AGP. The docked ligands are shown in stick representation with AGP shown in surface representation, with each lobe of the binding pocket color coded.
With the exception of ITC, drug binding to human AGP has been extensively characterized by a variety of biophysical methods, including equilibrium dialysis [44], ultra-filtration [43], radio-ligand displacement [30], circular dichroism [45–47], and fluorescence spectroscopy [48–50]. These studies have shown that AGP is largely selective for basic and neutral drugs [8, 51]; however, certain acidic drugs also bind to AGP, albeit, with lower affinities [8, 51]. Despite the wealth of literature on drug-AGP binding interactions, there is a paucity of information in regard to the energetics coupled to these diverse molecular interactions. This study is the first to utilize ITC to characterize the microscopic thermodynamic parameters that drive drug binding to AGP and make correlations with structural information inferred from the docking models and the available crystallographic structures of disopyramide, chlorpromazine and amitriptyline complexes with the A variant of human AGP [33]. Taken together, these principle findings provide a molecular level understanding of the energetics that drives drug-AGP binding interactions in vitro. This detailed appreciation of the binding energetics may in turn facilitate the future development of drugs with targeted modifications aimed at modulating the binding affinity for AGP in order to increase or decrease the unbound plasma fraction [52, 53].
2. Materials and Methods
2.1. Materials
Human AGP (lot # 018K7535) and drug compounds were obtained from Sigma-Aldrich (Sydney, NSW, Australia). All other reagents were of analytical grade or better.
2.2. Re-purification of AGP
All protein chromatographic procedures were performed under fast protein liquid chromatography conditions on an AKTA FPLC system (GE Healthcare, Sydney, N.S.W, Australia). Commercial human AGP preparations from Sigma-Aldrich were re-purified by collecting the void volume from a weak cation (MonoS) exchange chromatography that was pre-equilibrated with Buffer A (20 mM HEPES, pH 7.4), then applying this fraction to a weak anion exchange column (monoQ) and eluting with a gradient from 0→100% Buffer B (consisting of Buffer A +1M NaCl) over 20 min. Approximately 1 ml fractions were collected and run in SDS-PAGE to determine the purity of the preparation (~98% by silver staining)
2.3. Separation of the A and F1*S variants of AGP
The A and F1*S variants of AGP were separated from native (sialylated) mixtures of commercial AGP using the copper(II)-based immobilized metal ion affinity chromatography procedure as previously reported [54]. Briefly, approximately 50 mg of re-purified human AGP in 20 mM HEPES pH 7.4 were applied to a copper(II) Sepharose 5 mL HisTrapHP chromatography column (GE Health Care, Sydney, N.S.W, Australia). The variants were resolved using a gradient of 0→100% 300 mM imidazole at a flow rate of 1 mL/min (4 CV wash-out unbound sample; 0→100% imidazole 5 CV; hold 100% imidazole for 3 CV). The imidazole free fractions were found to contain the F1*S variant(s), whereas the A variant eluted later during the imidazole gradient. The fractions were desalted by ultrafiltration and purity was accessed by isoelectric focusing as previously described [55, 56].
2.4. Isothermal Titration Calorimetry (ITC) assay of drug binding to human AGP
Microcalorimetric measurements of drug binding to AGP were performed on an itc200 or a VP-ITC isothermal titration calorimeter (GE Health Care/Microcal, Northampton, MA, U.S.A). The samples were thoroughly degassed beforehand. AGP samples (140 μM) in 20 mM HEPES pH 7.4 were filled into the microcalorimetric cell and titrated with a 5→16 mM drug solution in protein buffer dialysate at 240 sec intervals. The cell contents were stirred constantly at 360 rpm. Unless otherwise stated the ITC titrations were performed at 30°C. The system was allowed to equilibrate and a stable baseline was recorded before initiating an automated titration. The heat of interaction after each injection measured by the ITC instrument were plotted versus time. As a control for all ITC experiments, drug solution were titrated into buffer under the same injection conditions and the heat of dilution subtracted from the drug-AGP titration data. The total heat signal from each injection were determined as the area under the individual peaks and plotted versus the [drug]/[AGP] molar ratio. The corrected data were analyzed to determine number of binding sites (n), and molar change in enthalpy of binding (ΔH) in terms of a single site model derived as follows:
The quantity r is defined as the moles of drug [D] bound per mole of protein [P] with an association constant (Ka):
| (Eq 1) |
Solving Eq 1 for Ka leads to:
| (Eq 2) |
Since the total concentration of drug [D]T in the cell is known, it can be represented by Eq 3 wherein [P]T equals the total protein concentration in the cell and n the number of binding sites:
| (Eq 3) |
Since Eq 3 shows nr[P]T=[PD], the fraction of sites occupied by the drug, combining Eqs 2 and 3 leads to:
| (Eq 4) |
Solving the quadratic for the fractional occupancy (r) gives:
| (Eq 5) |
The total heat content (Q) of the solution in the volume of the sample cell (Vo; determined relative to zero for the apo-species) at fractional saturation r is given by Eq6, where ΔH represents the molar heat of drug binding:
| (Eq 6) |
Substituting Eq 5 into Eq 6 gives:
| (Eq 7) |
The total heat content, Q can be calculated as function of n, Ka, ΔH because [P]T, [D]T and Vo are known experimental parameters. The parameter Q defined in Eq 7 only applies to the known starting volume of protein solution in the sample cell (Vo). In order to correct for the displaced volume (Vi), the change in heat content Q(i) at the end of the ith injection is defined by Eq 8 to obtain the best fit for n, Ka, and ΔH by standard Marquardt methods until no further significant improvement in fit occurs with continued iteration.
| (Eq 8) |
All data fitting operations were performed with Origin V7.0 (OriginLab, Northampton, MA).
The Gibbs free energy (ΔG0) were calculated from the fundamental equation of thermodynamics Eq 9:
| (Eq 9) |
For determination of the heat capacity (ΔCp) changes upon binding, the amitriptyline and chlorpromazine titrations were performed at 15°C, 20°C, 25°C 30°C and 37°C under the same buffer conditions. The slope of the plot of ΔH versus T(K) was used to calculate the change in heat capacity according to Eq 10:
| (Eq 10) |
2.5. Investigating the thermodynamic parameters associated with protonation/deprotonation events upon the formation of chlorpromazine- and amitriptyline-AGP complexes
This experiment allows for an understanding of the exact number of protons in exchange upon drug-AGP complex formation [57–62]. Fundamentally, the experiment examines the relationship between the apparent enthalpy of binding for drug-AGP complex formation and the change in enthalpies for the ITC titration performed in a series of buffers with differing heats of ionization (ΔHion) (MES ΔHion=3.5 kcal/mol; BisTris ΔHion=6.8 kcal/mol; Imidazole ΔHion=8.8 kcal/mol) [63]. The apparent enthalpy of binding ΔHb is the sum of the intrinsic enthalpy of the reaction ΔHr, which is independent of the choice of buffer used in the experiment, and a term proportional to the enthalpy of ionization of the buffer (ΔHion):
| (Eq 11) |
Where nH+ is the number of protons that are release by the buffer and become associated with the protein (if nH+>0). Conversely, if the protein releases protons to the buffer, then nH+<0.
2.6. Drug docking with the AGP F1*S and A variants
Docking simulations were performed using Glide V4.0 [64] as implemented in the Schrödinger Suite V7.5 following the recommended protocol. The crystallographic coordinates of the apo-human AGP F1*S variant (PDB code: 3KQ0) [34] and the apo-human AGP A variant (PDB code: 3APU) [33] were used at the docking receptors. Drug molecules were prepared using LigPrep [65]. Bound crystalline dihydroxylpropyl acetate and structured water molecules were ignored during docking. The centre of the docking grid was defined by the central lobe of the binding cavity of each AGP variant and the volume of the grid was set to 10 Å3. No further modifications were applied to the default settings (no scaling factor for the vdW radii of nonpolar protein atoms, 0.8 scaling for nonpolar ligand atoms). The GlideScore [64] scoring function was used to select up to 30 poses for each ligand. Poses obtained from Glide were grouped into clusters of poses within 2 Å using the cluster subroutine. A representative structure from each cluster was output in PDB format. Molecular images were generated using the molecular visualization program PYMOL V0.99 (http://www.pymol.org).
3. Results and Discussion
3.1. Characterization of the energetics of drug-AGP interactions by isothermal titration calorimetry
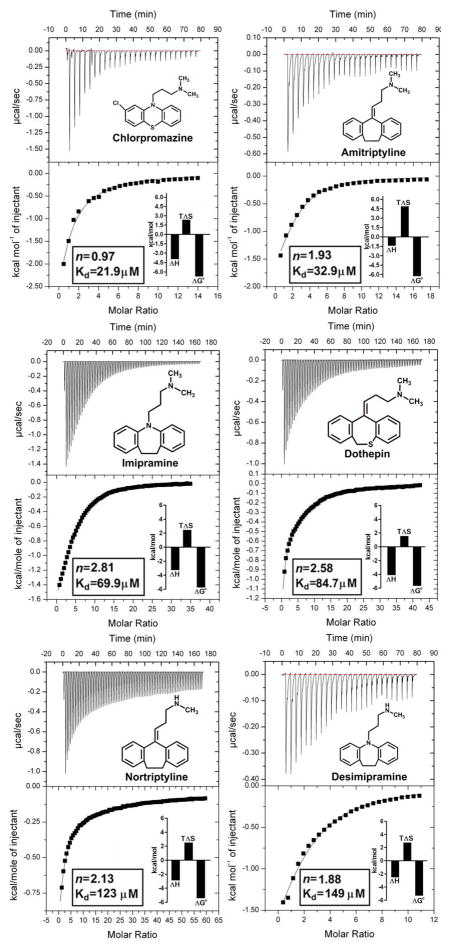
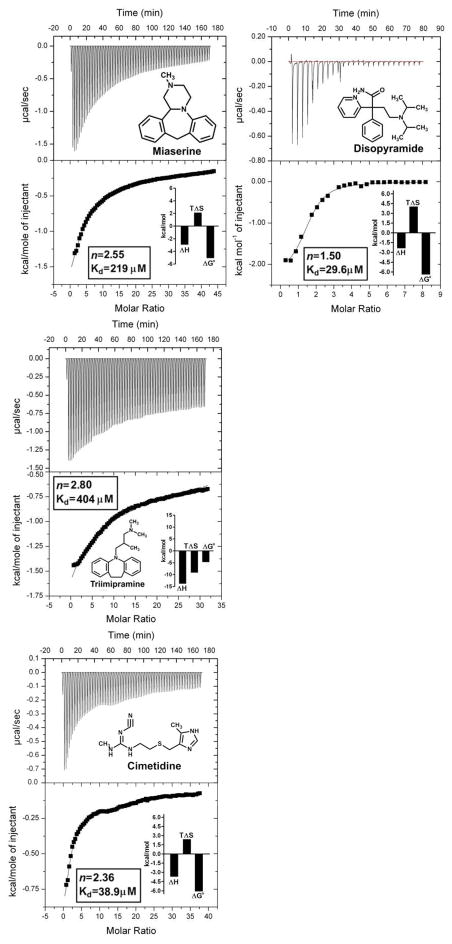
The microscopic thermodynamic forces that drive the binding of a series of basic compounds from the tricyclic antidepressant series, together with miaserine, chlorpromazine, disopyramide and cimetidine to human AGP, were characterized using sensitive modern ITC instrumentation. Examples of the typical drug-AGP microcalorimetric titrations obtained are shown in Figure 1. Titration curves corresponding to an exothermic binding reaction were observed for all of the drug-AGP interactions examined. The calculated thermodynamic parameters from the ITC binding curves are documented in graphical form and tabulated in the supplementary information (Figure 1 insets and Table S1). The ITC curve shape is dependent on the c value (c=nKa[P]T), which is a product of the number of ligand binding site(s) on the protein (n), the binding affinity (Ka) and the concentration of protein ([P]T) used in the experiment [66–69]. It should be noted that due to the moderate-to-low affinity of the drug-AGP interactions the binding curves obtained were hyperbolic in nature. Ideally, for the determination of precise Kd values, the shape of the binding isotherm should be sigmoid in nature [68, 69]. In an attempt to generate a sigmoid binding curve a titration was repeated with amitriptyline (a tight binder with a high aqueous solubility) at a much higher protein concentration (300 μM). Despite the 2-fold greater protein concentration a hyperbolic binding curve was obtained. Situations where the ligand displays a poor aqueous solubility and/or low affinity for the protein, often imposes practical limits on performing titrations at higher ligand and protein concentrations in order to increase the c values to fall within the ideal range (1–1000) [66–69]. The c values for the test compounds ranged from c=12.8 for chlorpromazine (tightest binder, Kd=21.9 μM), c=1.9 for desimipramine (moderate binder, Kd=149 μM) and c=0.7 triimipramine (weakest binder, Kd=404 μM) (Table S1). On account of the limited solubility of triimipramine it is not possible to achieve c values in the preferred range. Notwithstanding, previous studies have confirmed the validity of ITC for studying low affinity systems (Kd > 100 μM) under low c conditions (0.01 < c < 10) [66, 67]. To avoid systematic errors in Ka that can result from freezing the stoichiometric parameter (n), we allowed the n parameter to float during the curve fitting [66]. The neuroleptic, chlorpromazine displayed an approximately 1:1 [drug:AGP] binding stoichiometry (Figure 1. insets and Table S1). Disopyramide displayed an approximately 1.5:1 [drug:AGP] binding stoichiometry. Amitriptyline, nortriptyline and desimipramine displayed an approximately 2:1 [drug:AGP] binding stoichiometry. Whereas, imipramine, trimipramine, dothepin, the tertracyclic class antidepressant miaserine and the antiulcerative, diuretic cimetidine all revealed a binding stoichiometry ranging from 2.5:1 to 3:1 [drug:AGP]. High quality crystallographic evidence has shown that AGP binds tricyclic compounds such as amitriptyline and chlorpromazine in a 1:1 ratio [33]. The basis for the deviation of the binding stoichiometry from unity is likely the differential binding affinity of the drugs for the two major AGP variants present in the preparations. The rank order of affinity from highest to lowest affinity is chlorpromazine (21.9 μM) > disopyramide (29.6 μM) > amitriptyline (32.9 μM) > imipramine (69.9 μM) > dothepin (84.7 μM) > nortriptyline (123μM) > desimipramine (149 μM) > miaserine (219 μM) > trimipramine (404 μM) (Table S1). This is consistent with the rank order previously reported for this series based on binding affinity data generated from a radioligand displacement assay [30].
Figure 1.
Isothermal Titration Calorimetry measurement of drug-AGP binding interactions. Top panels show the heat in μcal/sec per injectant. Bottom panels show the enthalpy (kcal/mol) as a function of the drug-AGP molar ratio. All titrations were performed at 30°C. The insets show the stoichiometry (n), binding affinity (Kd) and thermodynamic parameters (ΔH°, enthalpy; TΔS°, entropy; ΔG°, free energy) for each interaction derived from a one site model fit of the binding isotherms. The chemical structure of each test compound is shown.
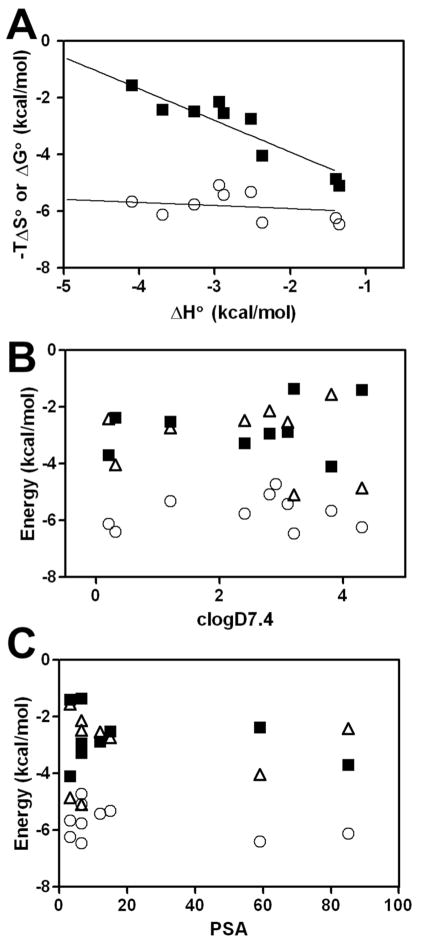
The correlation plot of ΔG° and −TΔS° as a function of ΔH° for all of the test compounds shows that there is marked enthalpy-entropy compensation effect evident (Figure 2A). The slope of the linear regression line of best fit for ΔH° versus −TΔS° was −1.1±0.04, with ΔH° varying between −1.35 and −4.1 kcal/mol (Figure 2A). In comparison, ΔG° remained relatively constant, varying between −5.07 and −6.45 kcal/mol, and showed unremarkable correlation with ΔH° with the slope of the best fit line −0.1±0.04. Moreover, the thermodynamic parameters did not show any correlation with the drug octanol/water partition coefficients or polar surface area (PSA) of the test compounds (Figure 2B,C and Table S1). This observation contrasts the direct correlation between ΔH°, ΔS° and ΔG° and hydrophobicity (logP) reported for the binding of a series of local anesthetics and phenothiazine neuroleptics and AGP [40, 44, 70]. However, it should be noted that these studies employed equilibrium dialysis or fluorometric binding assays to generate affinity constants from which the thermodynamic parameters were indirectly derived. To the best of our knowledge, the only reported attempt to comprehensively characterize the thermodynamics of drug-AGP interactions using microcalorimetric instrumentation involved an early study that examined AGP binding to a series of phenothiazines using a rudimentary differential flow microcalorimeter [71]. Flow microcalorimeters have been associated with systematic errors in microcalorimetry due to substantial superfluous heat generated from the friction produced by the rapid solution flow and viscosity [72]. The reported affinity constants were in the same order of magnitude (104 M−1) as our results [71]. One noticeable difference was the negative (unfavorable) entropy associated with the binding of all of the phenothiazines examined [71]. It should be noted that in addition to the differences in the instrumentation, the aforementioned study did not re-purify the plasma source of AGP used for the microcalorimetric measurements.
Figure 2.
(A) Thermodynamics of drug binding to human AGP. The solid lines represent the linear regression best fit of the data points. ΔG° (○), −TΔS° (■). (B) LogD7.4 and (C) PSA dependence of the drug thermodynamic parameters for ΔG° (○), ΔH° (■) and −TΔS° (△).
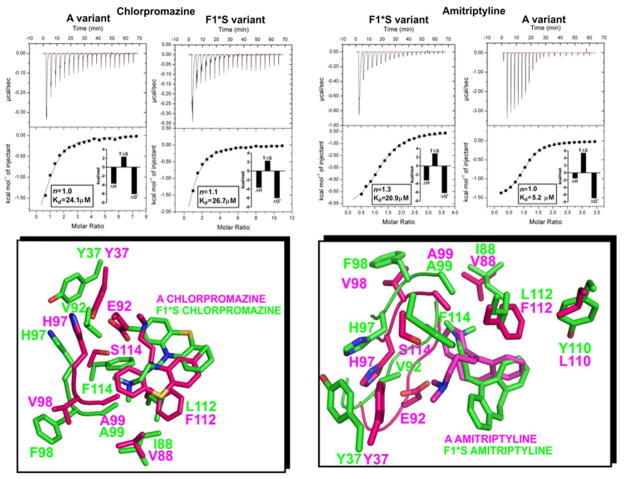
3.2. Characterization of the binding energetics of chlorpromazine and amitriptyline for the separated A and F1*S variants
Commercial preparations of AGP are heterogeneous mixtures of the A and F1*S variants [30, 54], therefore drug binding studies (such as ours) that employ theses heterogeneous preparations provide data representative of the combined binding properties of both variants. Albeit, the data garnered does provide a more faithful picture of the drug binding energetics setting in vivo, where both variants co-exist. In order to examine the variant specific energetics, we separated the two variants and performed titrations with two compounds, amitriptyline, which has been reported to show a >20-fold difference in affinity between A and F1*S and chlorpromazine, that has been reported to display a comparable affinity for both variants [30]. The chlorpromazine binding energetics were comparable for both variants; not surprising given the 1:1 [chlorpromazine:AGP] binding stoichiometry also observed with the heterogenous preparation (Figures 1 and 3). In comparison, amitriptyline displayed a 4-fold greater affinity for the A variant compared to the F1*S variant, owing to the larger (favorable) entropy associated with the amitriptyline-A variant interaction (Figures 3). The comparison of the drug docking results for the A and F1*S variants suggests that ligand binding is dependent on the ability of each drug to adopt favorable stabilizing contacts with the variant specific residues, while maintaining occupancy in the most accommodating region of the pocket based on the drugs molecular size and conformational restraints (Figure 3, bottom panels). In the F1*S docking solution the chlorpromazine molecule is positioned such that it forms a mirror image of the crystallographic chlorpromazine in the A variant structure (Figures 1 and 3). This “mirror” image orientation may allow the chlorpromazine molecule to form contacts that compensate for the residue differences in the pocket of each variant and thereby give rise to the observed binding equivalency. In the case of amitriptyline, the molecule adopts two very different poses within the respective binding pockets of each variant. The tighter interactions seen between the bound amitriptyline and the side chains in the pocket of the A variant may account for the more favorable binding energetics of this variant. The molecular details of these interactions are discussed further in the following sections.
Figure 3.
Isothermal Titration Calorimetry measurement of chlorpromazine and amitriptyline binding interactions with the A and F1*S AGP variants. Top panels show the heat in μcal/sec per injectant. Bottom panels show the enthalpy (kcal/mol) as a function of the drug-AGP molar ratio. All titrations were performed at 30°C. The insets show the stoichiometry (n), binding affinity (Kd) and thermodynamic parameters (ΔH°, enthalpy; TΔS°, entropy; ΔG°, free energy) for each interaction derived from a one site model fit of the binding isotherms. Bottom panels. An overlay of the binding pocket side-chain contacts for each drug-AGP variant complex shown in stick representation.
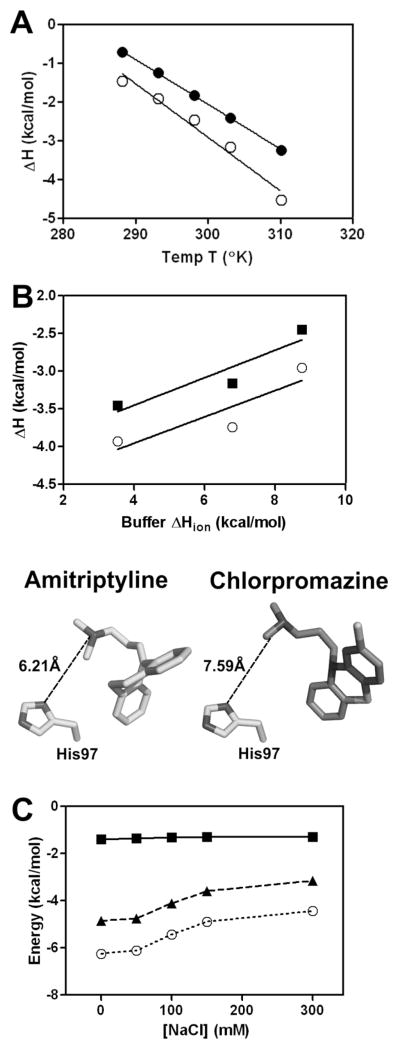
3.3. Dependence on temperature
In order to understand the heat capacity changes associated with drug binding which correlates with the buried solvent accessible surface area upon complex formation, titrations with amitriptyline and chlorpromazine were performed over a series of temperatures (Figure 4A). The heat capacity derived from the measured enthalpies with varying temperature were chlorpromazine ΔCp= −115 cal/K.mol and amitriptyline ΔCp= −138 cal/K.mol, which fall within the typical range for the binding of drug-like molecules to proteins [73]. These negative ΔCp values would suggest some hydrophobic contacts are formed upon association.
Figure 4.
(A) Temperature dependence of the AGP binding enthalpy (ΔH°) for amitriptyline (○) and chlorpromazine (●). The solid line is the linear regression analysis of the data with slopes that yield the heat capacity (ΔCp) change for the binding interaction with chlorpromazine (−115 cal/K.mol) and amitriptyline (−138 cal/K.mol). (B) Dependence of the binding enthalpy for amitriptyline (■) and chlorpromazine (○) binding to AGP on the ionization enthalpy of buffer used in the titration. Titrations were performed at pH 6.0 and 20 mM buffer concentration with buffers of different ionization enthalpies (MES 3.5 kcal/mol; bisTris 6.8 kcal/mol; imidazole 8.8 kcal/mol) to determine the number of protons consumed or released upon drug binding. Bottom panel. Distance in angstroms from the N,N-dimethylpropan-1-amine side chains of amitriptyline (PDB:3APV) and chlorpromazine (PDB:3APX) from the imidazole side chain of His97 seen in the crystal structures with the AGP A variant. (C) The effect of ionic strength on the thermodynamic parameters for amitriptyline binding to AGP at 30°C: ΔG° (○, dotted line), ΔH° (■, solid line) and −TΔS° (▲, dashed line).
3.4. Investigation of the protonation/deprotonation events coupled to the formation of drug-AGP complexes
The number of protonation/deprotonation events linked to drug binding were determined from the slope of a plot of the AGP-drug binding enthalpy measured in a set of buffers as a function of the buffer ionization enthalpy (Figure 4B). The data suggest that for the binding of amitriptyline and chlorpromazine to AGP at pH 6.0 is coupled to a partial proton transfer event from the buffered bulk solution to the drug-AGP complex. The linear regression of the data indicates there is a transfer of 0.17 and 0.18 protons to the chlorpromazine- and amitriptyline-AGP complex, respectively. His97 is situated at the entrance of the ligand binding cavity of AGP [33, 34]. In the crystallographic complexes, the imidazole side chain of His97 is situated 6.2Å and 7.6Å from the N,N-dimethylpropan-1-amine side chains of amitriptyline and chlorpromazine, respectively (Figure 4B) [33, 34]. The imidazole side chain of His has a pKa of approximately 6.0, whereas the N,N-dimethylpropan-1-amine side chains of chlorpromazine and amitriptyline have pKa values of 9.3 and 9.2, respectively [74, 75]. This would mean their N,N-dimethylpropan-1-amine side chains are fully protonated at pH 6.0. According to the pKa values and available structural information it is possible that partial proton transfer to His97 is coupled to the binding of the test drug compounds.
3.5. Dependence on ionic strength
In order to examine the ionic strength dependence of the thermodynamic parameters, the binding of amitriptyline was performed under buffer conditions with increasing sodium chloride (NaCl) concentrations (Figure 4C). The plot of the thermodynamic parameters as a function of the NaCl concentration shows that high salt concentrations have a detrimental effect on the binding interaction of amitriptyline (Figure 4C). The binding affinity decreases with higher salt concentrations seen as a decrease in ΔG°, which plateaus at NaCl concentrations above 100 mM.
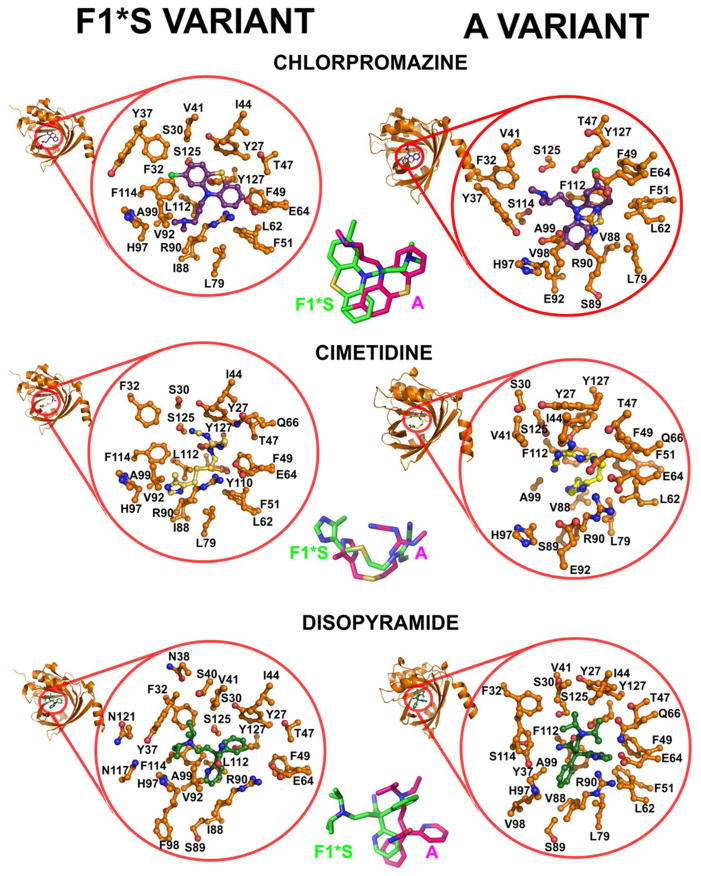
3.6. Molecular topography of drug interactions within AGP binding cavity-analysis of docking models and crystallographic drug-AGP complexes
The highest scoring docking results clustered into the distinctive lobe regions within the pocket of each AGP variant. The drug docking solutions for the F1*S variant grouped into three distinct clusters (Figure 5). The tricyclic antidepressants, chlorpromazine and cimetidine all docked into an area of the pocket that overlaps with lobes I and III (tricyclic cluster). Disopyramide and miaserine docked into an area of the pocket that overlapped with lobes I and III (disopyramide and miaserine cluster). Nortriptyline and desimipramine docked into an area predominantly overlapping with lobe III, with a marginal overlap with lobe I (nortriptyline and desimipramine cluster). The drug docking solutions for the A variant grouped into two distinct clusters (Figure 5). The tricyclic antidepressants and miaserine docked into the same position as the crystallographic chlorpromazine and amitriptyline, which corresponds to a region of the pocket which predominantly overlaps with lobe I (tricyclic cluster). In comparison, disopyramide, and cimetidine docked into a region of the pocket that overlapped with both lobes I and II (disopyramide and cimetidine cluster).
3.7. Thermodynamic mapping of the drug binding sites
In order to gain the most from the thermodynamic data and understand the molecular interactions underlying the drug binding energetics, the available crystallographic complexes of the A variant bound to amitriptyline, chlorpromazine and disopyramide [33] together with drug docking solutions for both variants where analyzed in detail and correlated with the thermodynamic parameters.
3.7.1. Thermodynamic mapping of the chlorpromazine binding site
The thermodynamic binding signature of chlorpromazine was characterized by a dominant favorable enthalpy gain and a smaller (favorable) entropic gain (Figures 1 and 3). The molecular contacts detailed below suggest the favorable negative enthalpy is largely contributed by hydrogen bond formation through the ideal positioning of hydrogen bond donor and acceptor atoms in the protein and ligand; and van der Waals interactions upon complex formation. The lower entropy may be associated with the desolvation of polar groups that originates because hydrogen bond donor and acceptor atoms are hydrogen bonded to water prior to complex formation. In both variants the bound position of the 2-chlorophenothiazin tricyclic ring is stabilized by partial or full π-stacking contacts and tight non-polar interactions with aromatic side chains (Figures 1 and 3). More specifically in the F1*S variant, tight non-polar interactions are seen with the aromatic side chains of Phe32, Tyr37, Phe49, Phe51, Tyr127 and π-stacking interactions with Phe114. In the F1*S structure, the side chain of Tyr37 is pushed out much further than in the A variant pocket, this allows for the accommodation of the large chlorine atom in its docked position within the F1*S pocket. In the chlorpromazine-A variant crystallographic complex tight non-polar interactions interactions are seen between the 2-chlorophenothiazin tricycle and the aromatic side chains of Tyr27, Phe49, Phe51, and Tyr127. In addition, the side chain of Phe112 (Leu112 F1*S variant) at the base of the pocket makes π-stacking contacts with the 2-chlorophenothiazin tricycle. In the A variant pocket Val88 forms part of a hydrophobic sub-domain that cradles the des-chloro phenyl of the phenothiazin tricycle. In the corresponding F1*S position, the side chain of Ile88 makes favorable hydrophobic contacts with the terminus of the N,N-dimethylpropan-1-amine side chain. The β-sheet segment formed by His97, Phe98, and Ala99 is significantly pushed back in the F1*S structure widening this region of the pocket, which allows the N,N-dimethylpropan-1-amine side chain to enter this region of the F1*S pocket. In the A variant the more compact structure of this region only allows for the accommodation of the des-chloro phenyl of the phenothiazin in a “card-and-slot” fashion with the tricycle orientated flat in the vertical (Y) plane. In the A variant crystallographic structure the side chain of Glu92 forms an ionic bridge with His97, coordinating their side chains closer, and consequently narrowing this part of the pocket. Whereas, in the pocket of the F1*S variant the Val92 substitution abolishes this interaction, thereby making this region of the pocket more spacious. The tertiary amide of the N,N-dimethylpropan-1-amine side chain makes polar contacts with Tyr87, Glu92, Ser114, and Ser125 in the pocket of the A variant. In the pocket of the F1*S variant the equivalent Phe114 makes π-stacking contacts with the 2-chlorophenyl of the phenothiazin tricyclic system. In the F1*S variant the alkyl segment of the N,N-dimethylpropan-1-amine side chain is stabilized by hydrophobic contacts with the side chains of Leu79, Ile88, Val92 Ala99, Leu112 and the 3-carbon aliphatic segment of the Arg90 side chain.
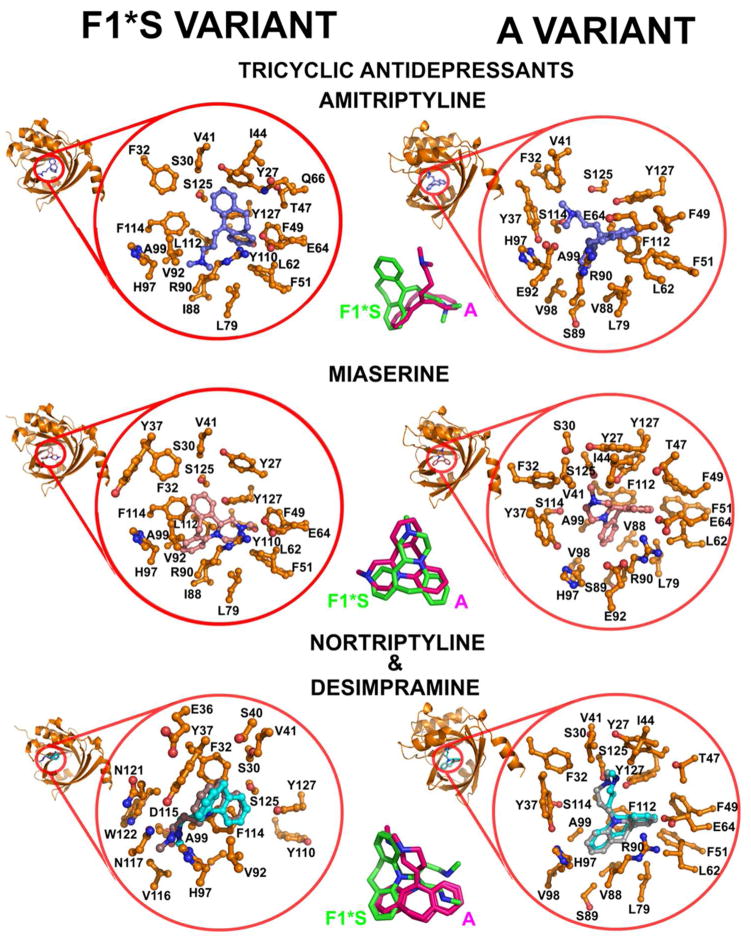
3.7.2. Thermodynamic mapping of the amitriptyline binding site
The thermodynamic binding signature of amitriptyline to the A variant was characterized by a dominant favorable entropy gain and a smaller, albeit favorable enthalpy gain (Figure 3). In comparison the 4-fold lower binding to F1*S was characterized by comparable enthalpy and entropy gains. The thermodynamic signature for binding to the heterogeneous preparation was intelligibly reflective of the combined signatures of each variant (Figure 1). Although both amitriptyline molecules sit in the same horizontal (X) plane, the F1*S docked position is rotated 90° relative to the position of the bound crystallographic amitriptyline in the A variant pocket (Figures 3 and 6). The dibenzocycloheptene tricycle of the crystallographic amitriptyline is stabilized by tight non-polar contacts with the aromatic side chains of Phe49, Phe51 and Phe112, and π-stacking contacts with the side chain of Tyr127. Additionally, stabilizing non-polar contacts are made with the side chains of Leu62, Leu79, Val88 and Ala99. The dibenzocycloheptene tricycle of the docked amitriptyline in the F1*S pocket is stabilized by π-stacking contacts with the aromatic side chains of Tyr27, Phe49 and Tyr127 and tight non-polar contacts with the aromatic side chains of Phe32, Phe51 and Tyr110. The dibenzocycloheptene tricycle also makes stabilizing contacts with the side chains of Val41, Ile44, Thr47, Leu62, Glu64, Gln66 and Leu79. The N,N-dimethylpropan-1-amine side chain of the crystallographic amitriptyline in the A variant pocket is stabilized by contacts with the side chains of Phe32, Tyr37 and Val41. Notably, the substitution of Phe114 in the F1*s variant with Ser114 in the A variant, prevents any steric hindrance and allows the N,N-dimethylpropan-1-amine side chain to be accommodated in this area of the A variant binding pocket. The 90° reorientation places the N,N-dimethylpropan-1-amine side chain of the F1*S docked amitriptyline, precisely over the same region as the dibenzocycloheptene of the crystallographic amitriptyline in the A variant pocket (Figure 6). Similar to chlorpromazine, the N,N-dimethylpropan-1-amine side chain of the docked F1*S amitriptyline is stabilized non-polar contacts with the side chains of Ile88 (Val88 A variant), Leu112 (Phe112 A variant), Phe114 (Ser114 A variant), and the 3-carbon aliphatic segment of the Arg90 side chain. In the pocket of the F1*S the variation of the side chains at positions Phe114 (Ser114 A variant), Leu112 (Phe112 A variant), Ile88 (Val88 A variant) and Tyr110 (Leu110 A variant) would make the orientation seen on the A variant pocket untenable. In particular, the proximal distance (1.77 Å) of the Phe114 side chain would clash sterically with the N,N-dimethylpropan-1-amine side chain of the crystallographic amitriptyline. The more open position of the Tyr37 and His 97 side chains in the F1*S pocket results in the loss of the stabilizing contacts between these residues and the dimethyl terminus of the amitriptyline side chain seen in the A variant pocket. Van der Waals contacts between non-polar groups are coupled to favorable enthalpic gains, the magnitude of which is dependent upon the degree of shape complementarily between the protein pocket and the ligand [76]. Accordingly, the differences in the pocket topology between the two variants may account for the impaired binding energetics and 4-fold lower amitriptyline binding affinity of the F1*S variant. The more compact pocket of the A variant allows the amitriptyline molecule to adopt a tighter fit in which case the desolvation surface is maximized; which would help account for the dominant entropic gain seen with the A variant interaction.
Figure 6.
Molecular docking models of the test compounds bound to the F1*S and A variants of human AGP. The mini-cartoon structure of AGP highlights the cavity region that is expanded in each docking model. The magnified expansion of the cavity shows the AGP side chain contacts within 5 Å of the bound drug. Centre panel. Stick overlays of the drug molecules in their bound orientation within the pocket of each variant.
3.7.3. Thermodynamic mapping of the imipramine, dothepin and trimipramine binding sites
The binding free energies of imipramine, dothepin and trimipramine were driven by a dominant enthalpic component. All three compounds dock into the same position of the F1*S pocket as amitriptyline and make a similar set of contacts. The differences in the thermodynamic signature across the structural series can be construed in terms of the small structural differences between each molecule (Figure 6). Compared to the amtriptyline structure, imipramine and trimipramine display a dibenzazepine tricycle and no unsaturated bond at the base of the N,N-dimethylpropan-1-amine side chain. The F1*S docking solutions suggest the N,N-dimethylpropan-1-amine side chain of imipramine and trimipramine adopt different conformations compared to the side chain of amitriptyline. Trimipramine displays a 2-methyl moiety on the N,N-dimethylpropan-1-amine side chain that sterically clashes with the 3-carbon aliphatic segment of the Arg90 side chain. This is reflected in the loss of favorable binding entropy. The entropy penalty may be associated with the forced solvent exposure of hydrophobic groups which results in a loss of favorable desolvation entropy [76]. The chemical structure of the dothepin molecule is identical to amitriptyline apart from the sulfur atom in the cycloheptene ring. In the F1*S docking solution the sulfur substitution causes the dibenzicycloheptene ring to adopt a more puckered conformation compared to amitriptyline and the other tricyclics. This altered ring conformation results in the loss of many of the favorable π-stacking interactions that are seen between the dibenzicycloheptene ring of amitriptyline and the aromatic side chains in the F1*S pocket. This may account for the ~2.5-fold lower affinity and the unfavorable entropy change associated with the dothepin binding interaction (Figure 1, insets and Table S1).
3.7.4. Thermodynamic mapping of the desimipramine and nortriptyline binding sites
Desimipramine and nortriptyline share an almost identical structure and coincidently docked into almost the same position in the respective pocket of each variant (Figure 6). This may account for the similarities between their thermodynamic signatures that consisted of comparable enthalpic and entropic contributions (Figure 1, insets and Table S1). The In the F1*S docking solutions stabilizing π-π stacking interactions are seen between the tricyclic ring systems of desimipramine and nortriptyline and the aromatic side-chains of Phe32, Tyr37, and Phe114. The N-methylpropan-1-amine side chain of both molecules is stabilized by contacts with the side chains of His97, Val116, Asn117 and Asn121. Together, this combination of non-polar and polar contacts would help account for the comparable entropic and enthalpic gains associated with the desimipramine and nortriptyline binding energetics.
3.7.5. Thermodynamic mapping of the miaserine binding site
The replacement of the alkyl chain with a heterocycle as per miaserine has a detrimental affect on the binding energetics. The loss of binding affinity results from a large entropy loss that over compensates for the enthalpy gain. In the F1*S docking solution the miaserine molecule sits in the same horizontal (X) plane as amitriptyline (Figure 6). The dibenzazepine tricycle of miaserine overlays with the alkyl side-chain of amitriptyline. Many of the π-π stacking interactions seen between the aromatic side chains and the dibenzocycloheptene ring of amitriptyline are lost in the docked position of miaserine, which may account for the ~6-fold lower affinity of miaserine for AGP compared to amitriptyline. Complex formation between the drug molecule and a protein often results in a loss of conformational entropy due to stabilization of protein residues and conformational restraints to the drug molecule itself [76]. The rigidity of the tetracyclic structure would result in a loss of conformational degrees of freedom in the system which may account for the unfavorable entropy change compared to the amitriptyline binding energetics (Figure 1, insets and Table S1). In the F1*S docking solution the N-methyl-pyrazino ring enters a hydrophobic sub-domain of the binding pocket formed by Phe51, Phe49, Leu62, Leu79. Whereas in the A variant docking solution the N-methyl-pyrazino ring is stabilized by contacts with the side chains of Tyr37, Ser30, Val41, Ile44, Ser114 and Ser125. The dibezazepine tricycle in the F1*S variant docking solution is stabilized by π-stacking contacts with the aromatics side chain of Phe114 and tight non-polar contacts with the side chain of Phe32, Tyr27 and Tyr127. Additional contacts are made between the dibezazepine tricycle and the side chains of Val41, Ile88, Val92, Ala99, and the 3-carbon aliphatic segment of the Arg90 side chain. The docking solution for the A variant shows the miaserine molecule occupies the same position of the pocket as the F1*S docking solution, however, the molecules are rotated 90° relative to each other (Figure 6). The repositioning appears to be due to the more open F1*S pocket which is more accommodating to the dibezazepine tricycle within the lobe III space which exists due to the more pushed back position of the Tyr37 and His97 side chains. In comparison the lobe III region is non-existent in the A variant pocket due to the more forward positioning of the Tyr37 and His97 side chains. Accordingly, the 90° reorientation of the miaserine molecule allows for accommodation of the N-methyl-pyrazino ring component into this reduced area of the A variant pocket. In the A variant docking solution the dibezazepine tricycle makes almost the same set of contacts as per the F1*S pocket. Briefly, contacts are seen with the aromatic side chains of Phe49, Phe51, His97 and Phe112. Additional contacts are made with the side chains of Leu62, Glu64, Leu79, Val88, Ala99 and the 3-carbon aliphatic segment of the Arg90 side chain.
3.7.6. Thermodynamic mapping of the disopyramide binding site
Disopyramide has been previously reported to selectively bind to the A variant and displays a very low affinity for the F1*S [30, 31]. This suggests the thermodynamic signature is largely representative of the A variant binding interaction and would also account for the observed binding stoichiometry (n=1.5) (Figure 1, insets and Table S1). The thermodynamic signature of disopyramide was characterized by a dominant entropy that is compensated by a smaller enthalpy. A favorable entropy gain predominantly originates from desolvation of both polar and non-polar groups [76]. However, the desolvation of polar groups is often also coupled to a enthalpic penalty [76]. The structural analysis suggests these changes may originate from sub-optimal positioning of polar groups so they are not forming strong hydrogen bonds within the pocket, such that the penalty due to the desolvation and burial of polar groups predominates. In the F1*S docking solution the disopyramide molecule sits in a very different position compared to the bound disopyramide in the A variant co-crystallographic complex (Figure 6) [33]. In the F1*S docking solution the diisopropylamino moiety is accommodated in lobe III of the pocket, this setting would not be tenable in the A variant pocket. The pyridine ring sits deep within lobe I and is stabilized by tight non-polar contacts with Phe114 (Ser114 A variant) and contacts with the side chains of Leu79 and Leu112. The α-phenyl is stabilized by π-stacking with the aromatic side chain of Tyr27 and non-polar contacts with Ile44. The acetamide moiety is largely solvent exposed. In the A variant co-crystallographic complex, the diisopropylamino moiety is stabilized by contacts with the side chains of Tyr27, Phe32, Val41, Ile44, Glu64 and Gln66. The acetamide moiety is within hydrogen bonding distance to hydroxyl groups on the side chains of Tyr127 and Ser125. The pyridine ring makes non-polar contacts with the aromatic side chains of Phe49 and Phe51. The α-phenyl is stabilized by hydrophobic contacts with Val88, Leu79, Ala99 and non-polar contacts with Phe112 and the 3-carbon aliphatic segment of the Arg90 side chain.
3.7.7. Thermodynamic mapping of the cimetidine binding site
The thermodynamic binding signature of cimetidine was characterized by a dominant favorable enthalpy gain and a smaller, albeit favorable entropy gain (Figure 1, insets and Table S1). In the docking solutions the cimetidine molecules are approximately in the same position in the binding pockets of both variants, being shifted only slightly in the horizontal (X) plane relative to each other (Figure 6). The structural analysis suggests hydrogen binding and van der Waals contacts largely contribute to the favorable enthalpy gain. In the F1*S variant docking solution the 5-methyl-imidazole ring is positioned within lobe III and is stabilized by non-polar contacts with the aromatic side chain of Phe114 (Ser114 A variant) and also makes contacts with the side chains of Ile88, Val92 and Leu112. In the F1*S docking model, the 5-methyl-imidazole ring is flipped in the vertical (Y) plane relative to that of the position docked into the A variant, this re-positioning avoids the steric clash that would be caused between the longer Ile88 (Val88 A variant) side chain and the 5-methyl group. The terminus of the molecule is stabilized by contacts with the side chains of Tyr27, Ser30, Ile44, Phe49, Glu64, Gln66 Ser125 and Tyr127. The N-cyano group is within hydrogen bonding distance to the hydroxyl groups on the side chains of Tyr27, Ser30 and Ser125. In the A variant pocket the entire cimetidine molecule is shifted slightly in the horizontal (X) plane, relative to the F1*S docked position. This is again due to the more compact topology of the A variant pocket which lacks the lobe III, where the 5-methyl-imidazole ring is positioned in the F1*S pocket. Instead, in the A variant pocket, the 5-methyl-imidazole ring is positioned deep within lobe I, with the 5-methyl-group making tight non-polar contacts with the aromatic side chains of Phe112 (Leu112 F1*S), and the side chains of Val88, Leu79, Ala99. The N-cyano terminus of the molecule sits in approximately the same position as in both variants and as such makes a similar set of polar contacts with the side chains of the conserved residues Tyr27, Ser30 and Ser125.
4. Conclusions
Isothermal titration calorimetry has been used to dissect the energetics of drug-AGP interactions and establish correlations with structural data. For a drug to bind to AGP with high affinity and selectivity requires that both the binding enthalpy and entropy contribute favorably to the binding free energy. The thermodynamic signatures of the test compounds were closely coupled to their ligand-specific binding mode within the complex multi-lobed AGP pocket. This would suggest that the AGP ligand binding cavity has evolved into a complex multi-lobed structure so that it is capable of accommodating a chemically diverse range of ligand partners in an energetically favorable manner.
Supplementary Material
Acknowledgments
R.L.N. and J.L. are supported by research grants from the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (R01A1070896 and R01AI079330). T.V, R.L.N, J.L, P.E.T, M.A.B and K.R are also supported by the Australian National Health and Medical Research Council (NHMRC). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health. J.L. is an Australian NHMRC Senior Research Fellow. T.V. and MBare Australian NHMRC Industry Career Development Research Fellows. MC is a NHMRC Australia Fellow supported by AF511105.
Abbreviations
- AGP
human α-1-acid glycoprotein
- ΔG°
Gibbs free energy change
- ΔH°
standard enthalpy change
- ΔS°
standard entropy change
- Ka
association constant
- Kd
dissociation constant
- ΔCp°
standard heat capacity change
- ITC
isothermal titration calorimetry
References
- 1.Flower DR. The lipocalin protein family: structure and function. Biochem J. 1996;318 (Pt 1):1–14. doi: 10.1042/bj3180001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flower DR, North AC, Sansom CE. The lipocalin protein family: structural and sequence overview. Biochim Biophys Acta. 2000;1482:9–24. doi: 10.1016/s0167-4838(00)00148-5. [DOI] [PubMed] [Google Scholar]
- 3.Skerra A. Lipocalins as a scaffold. Biochim Biophys Acta. 2000;1482:337–50. doi: 10.1016/s0167-4838(00)00145-x. [DOI] [PubMed] [Google Scholar]
- 4.Fournier T, Medjoubi NN, Porquet D. Alpha-1-acid glycoprotein. Biochim Biophys Acta. 2000;1482:157–71. doi: 10.1016/s0167-4838(00)00153-9. [DOI] [PubMed] [Google Scholar]
- 5.Hochepied T, Berger FG, Baumann H, Libert C. Alpha(1)-acid glycoprotein: an acute phase protein with inflammatory and immunomodulating properties. Cytokine Growth Factor Rev. 2003;14:25–34. doi: 10.1016/s1359-6101(02)00054-0. [DOI] [PubMed] [Google Scholar]
- 6.Ojala PJ, Hermansson M, Tolvanen M, Polvinen K, Hirvonen T, Impola U, et al. Identification of alpha-1 acid glycoprotein as a lysophospholipid binding protein: a complementary role to albumin in the scavenging of lysophosphatidylcholine. Biochemistry. 2006;45:14021–31. doi: 10.1021/bi061657l. [DOI] [PubMed] [Google Scholar]
- 7.Zsila F, Mady G. Biliverdin is the endogenous ligand of human serum alpha1-acid glycoprotein. Biochem Biophys Res Commun. 2008;372:503–7. doi: 10.1016/j.bbrc.2008.05.090. [DOI] [PubMed] [Google Scholar]
- 8.Kremer JM, Wilting J, Janssen LH. Drug binding to human alpha-1-acid glycoprotein in health and disease. Pharmacol Rev. 1988;40:1–47. [PubMed] [Google Scholar]
- 9.Kristensen CB. Imipramine serum protein binding in healthy subjects. Clin Pharmacol Ther. 1983;34:689–94. doi: 10.1038/clpt.1983.233. [DOI] [PubMed] [Google Scholar]
- 10.Ojala PJ, Hermansson M, Tolvanen M, Polvinen K, Hirvonen T, Impola U, et al. Identification of alpha-1 acid glycoprotein as a lysophospholipid binding protein: A complementary role to albumin in the scavenging of lysophosphatidylcholine. Biochemistry. 2006;45:14021–31. doi: 10.1021/bi061657l. [DOI] [PubMed] [Google Scholar]
- 11.Voulgari F, Cummins P, Gardecki TI, Beeching NJ, Stone PC, Stuart J. Serum levels of acute phase and cardiac proteins after myocardial infarction, surgery, and infection. Br Heart J. 1982;48:352–6. doi: 10.1136/hrt.48.4.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eap CB, Fischer JF, Baumann P. Variations in relative concentrations of variants of human alpha 1-acid glycoprotein after acute-phase conditions. Clin Chim Acta. 1991;203:379–85. doi: 10.1016/0009-8981(91)90312-z. [DOI] [PubMed] [Google Scholar]
- 13.Dudley MN, Blaser J, Gilbert D, Zinner SH. Significance of “extravascular” protein binding for antimicrobial pharmacodynamics in an in vitro capillary model of infection. Antimicrob Agents Chemother. 1990;34:98–101. doi: 10.1128/aac.34.1.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greenblatt DJ, Sellers EM, Koch-Weser J. Importance of protein binding for the interpretation of serum or plasma drug concentrations. J Clin Pharmacol. 1982;22:259–63. doi: 10.1002/j.1552-4604.1982.tb02671.x. [DOI] [PubMed] [Google Scholar]
- 15.Lee BL, Sachdeva M, Chambers HF. Effect of protein binding of daptomycin on MIC and antibacterial activity. Antimicrob Agents Chemother. 1991;35:2505–8. doi: 10.1128/aac.35.12.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Merrikin DJ, Briant J, Rolinson GN. Effect of protein binding on antibiotic activity in vivo. J Antimicrob Chemother. 1983;11:233–8. doi: 10.1093/jac/11.3.233. [DOI] [PubMed] [Google Scholar]
- 17.Nix DE, Matthias KR, Ferguson EC. Effect of ertapenem protein binding on killing of bacteria. Antimicrob Agents Chemother. 2004;48:3419–24. doi: 10.1128/AAC.48.9.3419-3424.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmidt S, Rock K, Sahre M, Burkhardt O, Brunner M, Lobmeyer MT, et al. Effect of protein binding on the pharmacological activity of highly bound antibiotics. Antimicrob Agents Chemother. 2008;52:3994–4000. doi: 10.1128/AAC.00427-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wise R. The relevance of pharmacokinetics to in-vitro models: protein binding--does it matter? J Antimicrob Chemother. 1985;15 (Suppl A):77–83. doi: 10.1093/jac/15.suppl_a.77. [DOI] [PubMed] [Google Scholar]
- 20.Wise R. The clinical relevance of protein binding and tissue concentrations in antimicrobial therapy. Clin Pharmacokinet. 1986;11:470–82. doi: 10.2165/00003088-198611060-00004. [DOI] [PubMed] [Google Scholar]
- 21.Schmid K, Kaufmann H, Isemura S, Bauer F, Emura J, Motoyama T, et al. Structure of α1-acid glycoprotein. Complete amino acid sequence, multiple amino acid substitutions, and homology with the immunoglobulins. Biochemistry. 1973;12:2711–24. doi: 10.1021/bi00738a026. [DOI] [PubMed] [Google Scholar]
- 22.Schmid K, Nimerg RB, Kimura A, Yamaguchi H, Binette JP. The carbohydrate units of human plasma alpha1-acid glycoprotein. Biochim Biophys Acta. 1977;492:291–302. doi: 10.1016/0005-2795(77)90080-0. [DOI] [PubMed] [Google Scholar]
- 23.Nakano M, Kakehi K, Tsai MH, Lee YC. Detailed structural features of glycan chains derived from alpha1-acid glycoproteins of several different animals: the presence of hypersialylated, O-acetylated sialic acids but not disialyl residues. Glycobiology. 2004;14:431–41. doi: 10.1093/glycob/cwh034. [DOI] [PubMed] [Google Scholar]
- 24.Treuheit MJ, Costello CE, Halsall HB. Analysis of the five glycosylation sites of human alpha 1-acid glycoprotein. Biochem J. 1992;283 (Pt 1):105–12. doi: 10.1042/bj2830105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dente L, Ciliberto G, Cortese R. Structure of the human alpha 1-acid glycoprotein gene: sequence homology with other human acute phase protein genes. Nucleic Acids Res. 1985;13:3941–52. doi: 10.1093/nar/13.11.3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dente L, Pizza MG, Metspalu A, Cortese R. Structure and expression of the genes coding for human alpha 1-acid glycoprotein. EMBO J. 1987;6:2289–96. doi: 10.1002/j.1460-2075.1987.tb02503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dente L, Ruther U, Tripodi M, Wagner EF, Cortese R. Expression of human alpha 1-acid glycoprotein genes in cultured cells and in transgenic mice. Genes Dev. 1988;2:259–66. doi: 10.1101/gad.2.2.259. [DOI] [PubMed] [Google Scholar]
- 28.Herve F, Gomas E, Duche JC, Tillement JP. Fractionation of the Genetic-Variants of Human Alpha(1)-Acid Glycoprotein in the Native Form by Chromatography on an Immobilized Copper(Ii) Affinity Adsorbent - Heterogeneity of the Separate Variants by Isoelectrofocusing and by Concanavalin-a Affinity-Chromatography. Journal of Chromatography-Biomedical Applications. 1993;615:47–57. [PubMed] [Google Scholar]
- 29.Hervé F. Fractionation of the genetic variants of human alpha 1-acid glycoprotein in the native form by chromatography on an immobilized copper(II) affinity adsorbent. Heterogeneity of the separate variants by isoelectrofocusing and by concanavalin A affinity chromatography. Journal of chromatography. 1993;615:47–57. [PubMed] [Google Scholar]
- 30.Hervé F, Caron G, Duché J-C, Gaillard P, Abd, Rahman N, Tsantili-Kakoulidou A, et al. Ligand Specificity of the Genetic Variants of Human α1-Acid Glycoprotein: Generation of a Three-Dimensional Quantitative Structure-Activity Relationship Model for Drug Binding to the A Variant Molecular Pharmacology. 1998;54:129–38. doi: 10.1124/mol.54.1.129. [DOI] [PubMed] [Google Scholar]
- 31.Herve F, Duche JC, d’Athis P, Marche C, Barre J, Tillement JP. Binding of disopyramide, methadone, dipyridamole, chlorpromazine, lignocaine and progesterone to the two main genetic variants of human alpha 1-acid glycoprotein: evidence for drug-binding differences between the variants and for the presence of two separate drug-binding sites on alpha 1-acid glycoprotein. Pharmacogenetics. 1996;6:403–15. doi: 10.1097/00008571-199610000-00004. [DOI] [PubMed] [Google Scholar]
- 32.Kushner I, Mackiewicz A. Acute phase proteins as disease markers. Dis Markers. 1987;5:1–11. [PubMed] [Google Scholar]
- 33.Nishi K, Ono T, Nakamura T, Fukunaga N, Izumi M, Watanabe H, et al. Structural Insights into Differences in Drug-binding Selectivity between Two Forms of Human α1-Acid Glycoprotein Genetic Variants, the A and F1*S Forms. Journal of Biological Chemistry. 2011;286:14427–34. doi: 10.1074/jbc.M110.208926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schonfeld DL, Ravelli RB, Mueller U, Skerra A. The 1. 8-A crystal structure of alpha1-acid glycoprotein (Orosomucoid) solved by UV RIP reveals the broad drug-binding activity of this human plasma lipocalin. J Mol Biol. 2008;384:393–405. doi: 10.1016/j.jmb.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 35.Eap CB, Cuendet C, Baumann P. Binding of amitriptyline to alpha 1-acid glycoprotein and its variants. J Pharm Pharmacol. 1988;40:767–70. doi: 10.1111/j.2042-7158.1988.tb05169.x. [DOI] [PubMed] [Google Scholar]
- 36.Eap CB, Cuendet C, Baumann P. Selectivity in the binding of psychotropic drugs to the variants of alpha-1 acid glycoprotein. Naunyn Schmiedebergs Arch Pharmacol. 1988;337:220–4. doi: 10.1007/BF00169251. [DOI] [PubMed] [Google Scholar]
- 37.Eap CB, Cuendet C, Baumann P. Binding of d-methadone, 1-methadone, and dl-methadone to proteins in plasma of healthy volunteers: role of the variants of α1-acid glycoprotein. Clin Pharmacol Ther. 1990;47:338–46. doi: 10.1038/clpt.1990.37. [DOI] [PubMed] [Google Scholar]
- 38.Herve F, Gomas E, Duche JC, Tillement JP. Evidence for differences in the binding of drugs to the two main genetic variants of human alpha 1-acid glycoprotein. Br J Clin Pharmacol. 1993;36:241–9. doi: 10.1111/j.1365-2125.1993.tb04224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakagawa T, Kishino S, Itoh S, Sugawara M, Miyazaki K. Differential binding of disopyramide and warfarin enantiomers to human alpha(1)-acid glycoprotein variants. Br J Clin Pharmacol. 2003;56:664–9. doi: 10.1046/j.1365-2125.2003.01909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taheri S, Cogswell LP, 3rd, Gent A, Strichartz GR. Hydrophobic and ionic factors in the binding of local anesthetics to the major variant of human alpha1-acid glycoprotein. J Pharmacol Exp Ther. 2003;304:71–80. doi: 10.1124/jpet.102.042028. [DOI] [PubMed] [Google Scholar]
- 41.Nishi K, Ueno M, Murakami Y, Fukunaga N, Akuta T, Kadowaki D, et al. A site-directed mutagenesis study of drug-binding selectivity in genetic variants of human alpha(1)-acid glycoprotein. J Pharm Sci. 2009;98:4316–26. doi: 10.1002/jps.21697. [DOI] [PubMed] [Google Scholar]
- 42.Ishizaki J, Fukaishi A, Fukuwa C, Yamazaki S, Tabata M, Ishida T, et al. Evaluation of selective competitive binding of basic drugs to alpha1-acid glycoprotein variants. Biol Pharm Bull. 33:95–9. doi: 10.1248/bpb.33.95. [DOI] [PubMed] [Google Scholar]
- 43.Matsumoto K, Sukimoto K, Nishi K, Maruyama T, Suenaga A, Otagiri M. Characterization of ligand binding sites on the alpha1-acid glycoprotein in humans, bovines and dogs. Drug Metab Pharmacokinet. 2002;17:300–6. doi: 10.2133/dmpk.17.300. [DOI] [PubMed] [Google Scholar]
- 44.Miyoshi T, Sukimoto K, Otagiri M. Investigation of the interaction mode of phenothiazine neuroleptics with alpha 1-acid glycoprotein. J Pharm Pharmacol. 1992;44:28–33. doi: 10.1111/j.2042-7158.1992.tb14358.x. [DOI] [PubMed] [Google Scholar]
- 45.Otagiri M, Yamamichi R, Maruyama T, Imai T, Suenaga A, Imamura Y, et al. Drug binding to alpha 1-acid glycoprotein studied by circular dichroism. Pharm Res. 1989;6:156–9. doi: 10.1023/a:1015936710919. [DOI] [PubMed] [Google Scholar]
- 46.Otagiri M, Miyoshi T, Yamamichi R, Maruyama T, Perrin JH. Effects of tricyclic drug on induced circular dichroism spectra of dicumarol bound to alpha 1-acid glycoprotein. Biochem Pharmacol. 1991;42:729–33. doi: 10.1016/0006-2952(91)90029-5. [DOI] [PubMed] [Google Scholar]
- 47.Otagiri M, Yamamichi R, Imai T, Imamura Y, Takadate A. Study on the binding of dicumarol to alpha 1-acid glycoprotein using circular dichroism spectroscopy. Chem Pharm Bull (Tokyo) 1988;36:4958–62. doi: 10.1248/cpb.36.4958. [DOI] [PubMed] [Google Scholar]
- 48.Zini R, Copigneaux C, Tillement JP. 1-anilino-naphthalene 8-sulfonic acid (ANS) as a probe for the binding of antidepressant drugs to human alpha 1-acid glycoprotein (AAG) Prog Clin Biol Res. 1989;300:417–21. [PubMed] [Google Scholar]
- 49.Maruyama T, Furuie MA, Hibino S, Otagiri M. Comparative study of interaction mode of diazepines with human serum albumin and alpha 1-acid glycoprotein. J Pharm Sci. 1992;81:16–20. doi: 10.1002/jps.2600810104. [DOI] [PubMed] [Google Scholar]
- 50.Maruyama T, Otagiri M, Takadate A. Characterization of drug binding sites on alpha 1-acid glycoprotein. Chem Pharm Bull (Tokyo) 1990;38:1688–91. doi: 10.1248/cpb.38.1688. [DOI] [PubMed] [Google Scholar]
- 51.Israili ZH, Dayton PG. Human alpha-1-glycoprotein and its interactions with drugs. Drug Metab Rev. 2001;33:161–235. doi: 10.1081/dmr-100104402. [DOI] [PubMed] [Google Scholar]
- 52.Kawasaki Y, Freire E. Finding a better path to drug selectivity. Drug Discov Today. 2011;16:985–90. doi: 10.1016/j.drudis.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Freire E. A thermodynamic approach to the affinity optimization of drug candidates. Chem Biol Drug Des. 2009;74:468–72. doi: 10.1111/j.1747-0285.2009.00880.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Herve F, Gomas E, Duche JC, Tillement JP. Fractionation of the genetic variants of human alpha 1-acid glycoprotein in the native form by chromatography on an immobilized copper(II) affinity adsorbent. Heterogeneity of the separate variants by isoelectrofocusing and by concanavalin A affinity chromatography. J Chromatogr. 1993;615:47–57. [PubMed] [Google Scholar]
- 55.Eap CB, Baumann P. Isoelectric focusing of alpha-1 acid glycoprotein (orosomucoid) in immobilized pH-gradients with 8M urea: detection of its desialylated variants using an alkaline phosphatase-linked secondary antibody system. Electrophoresis. 1988;9:650–4. doi: 10.1002/elps.1150091005. [DOI] [PubMed] [Google Scholar]
- 56.Ishizaki J, Fukaishi A, Fukuwa C, Yamazaki S, Tabata M, Ishida T, et al. Evaluation of selective competitive binding of basic drugs to alpha1-acid glycoprotein variants. Biol Pharm Bull. 2010;33:95–9. doi: 10.1248/bpb.33.95. [DOI] [PubMed] [Google Scholar]
- 57.Baker BM, Murphy KP. Evaluation of linked protonation effects in protein binding reactions using isothermal titration calorimetry. Biophys J. 1996;71:2049–55. doi: 10.1016/S0006-3495(96)79403-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baker BM, Murphy KP. Dissecting the energetics of a protein-protein interaction: the binding of ovomucoid third domain to elastase. J Mol Biol. 1997;268:557–69. doi: 10.1006/jmbi.1997.0977. [DOI] [PubMed] [Google Scholar]
- 59.Gomez J, Freire E. Thermodynamic mapping of the inhibitor site of the aspartic protease endothiapepsin. J Mol Biol. 1995;252:337–50. doi: 10.1006/jmbi.1995.0501. [DOI] [PubMed] [Google Scholar]
- 60.Ohtaka H, Velazquez-Campoy A, Xie D, Freire E. Overcoming drug resistance in HIV-1 chemotherapy: the binding thermodynamics of Amprenavir and TMC-126 to wild-type and drug-resistant mutants of the HIV-1 protease. Protein Sci. 2002;11:1908–16. doi: 10.1110/ps.0206402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Czodrowski P, Sotriffer CA, Klebe G. Protonation changes upon ligand binding to trypsin and thrombin: structural interpretation based on pK(a) calculations and ITC experiments. J Mol Biol. 2007;367:1347–56. doi: 10.1016/j.jmb.2007.01.022. [DOI] [PubMed] [Google Scholar]
- 62.Dullweber F, Stubbs MT, Musil D, Sturzebecher J, Klebe G. Factorising ligand affinity: a combined thermodynamic and crystallographic study of trypsin and thrombin inhibition. J Mol Biol. 2001;313:593–614. doi: 10.1006/jmbi.2001.5062. [DOI] [PubMed] [Google Scholar]
- 63.Goldberg RN, Kishore N, Lennen RM. Thermodynamic quantities for the ionization reactions of buffers. Journal of Physical and Chemical Reference Data. 2002;31:231–370. [Google Scholar]
- 64.Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, et al. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem. 2004;47:1739–49. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 65.Chen IJ, Foloppe N. Drug-like bioactive structures and conformational coverage with the LigPrep/ConfGen suite: comparison to programs MOE and catalyst. J Chem Inf Model. 2010;50:822–39. doi: 10.1021/ci100026x. [DOI] [PubMed] [Google Scholar]
- 66.Tellinghuisen J. Isothermal titration calorimetry at very low c. Anal Biochem. 2008;373:395–7. doi: 10.1016/j.ab.2007.08.039. [DOI] [PubMed] [Google Scholar]
- 67.Turnbull WB, Daranas AH. On the Value of c: Can Low Affinity Systems Be Studied by Isothermal Titration Calorimetry? Journal of the American Chemical Society. 2003;125:14859–66. doi: 10.1021/ja036166s. [DOI] [PubMed] [Google Scholar]
- 68.Wiseman T. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Analytical Biochemistry. 1989;179:131–7. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- 69.Broecker J, Vargas C, Keller S. Revisiting the optimal c value for isothermal titration calorimetry. Analytical Biochemistry. 2011;418:307–9. doi: 10.1016/j.ab.2011.07.027. [DOI] [PubMed] [Google Scholar]
- 70.Urien S, Giroud Y, Tsai RS, Carrupt PA, Bree F, Testa B, et al. Mechanism of ligand binding to alpha 1-acid glycoprotein (orosomucoid): correlated thermodynamic factors and molecular parameters of polarity. Biochem J. 1995;306 (Pt 2):545–9. doi: 10.1042/bj3060545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Aki H, Yamamoto M. Thermodynamics of the binding of phenothiazines to human plasma, human serum albumin and alpha 1-acid glycoprotein: a calorimetric study. J Pharm Pharmacol. 1989;41:674–9. doi: 10.1111/j.2042-7158.1989.tb06339.x. [DOI] [PubMed] [Google Scholar]
- 72.Yamamoto M, Aki H. Application of differential flow microcalorimetry for study of drug interactions in the blood system. J Biochem Biophys Methods. 1988;16:271–82. doi: 10.1016/0165-022x(88)90061-9. [DOI] [PubMed] [Google Scholar]
- 73.Ward WH, Holdgate GA. Isothermal titration calorimetry in drug discovery. Prog Med Chem. 2001;38:309–76. doi: 10.1016/s0079-6468(08)70097-3. [DOI] [PubMed] [Google Scholar]
- 74.Whelpton R. Ionization constants, octanol partition coefficients and cholinesterase inhibitor constants for chlorpromazine and its metabolites. J Pharm Pharmacol. 1989;41:856–8. doi: 10.1111/j.2042-7158.1989.tb06387.x. [DOI] [PubMed] [Google Scholar]
- 75.Frisk-Holmberg M, van der Kleijn E. The relationship between the lipophilic nature of tricyclic neuroleptics and antidepressants, and histamine release. Eur J Pharmacol. 1972;18:139–47. doi: 10.1016/0014-2999(72)90235-x. [DOI] [PubMed] [Google Scholar]
- 76.Ladbury JE, Klebe G, Freire E. Adding calorimetric data to decision making in lead discovery: a hot tip. Nat Rev Drug Discov. 2009;9:23–7. doi: 10.1038/nrd3054. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.