Abstract
It has been a decade since the introduction of SH2 profiling, a modular domain-based molecular diagnostics tool. This review covers the original concept of SH2 profiling, different analytical platforms, and their applications, from the detailed analysis of single proteins to broad screening in translational research. Illustrated by practical examples, we discuss the uniqueness and advantages of the approach as well as its limitations and challenges. We provide guidance for basic researchers and oncologists who may consider SH2 profiling in their respective cancer research, especially for those focusing on tyrosine phosphoproteomics. SH2 profiling can serve as an alternative phosphoproteomics tool to dissect aberrant tyrosine kinase pathways responsible for individual malignancies, with the goal of facilitating personalized diagnostics for the treatment of cancer.
Keywords: cancer, tyrosine phosphorylation, phosphoproteomics, SH2 domain, SH2 profiling
Tyrosine Phosphoproteomics in Cancer
The availability of $1,000 whole-genome sequencing will soon fuel personalized medicine for life-threatening human diseases such as cancer.1,2 It is widely accepted that somatic gene alteration is the primary determinant of cancer and can serve as a predictive marker for drug response and patient prognosis.3,4 Proteomics, the large-scale analysis of gene products, has also caught the attention of cancer researchers, with sensitive mass spectrometry as the driving force for large-scale protein profiling of tumors.5,6 To design an effective therapy tailored for each cancer patient based on gene and protein profiles, it is necessary to define the critical proteins responsible for the hallmarks of cancer, such as oncogenic cell growth or resistance to apoptosis.7 An important goal of current research is to exploit these proteins as targets for the rational design of highly specific anticancer agents. However, an obvious problem is that most tumors contain many oncogenic mutations involved in a number of essential signaling pathways,8 and antagonizing all of them may provide no advantage over traditional chemotherapy in terms of adverse effects and toxicity.9 This is why imatinib, a highly successful, rationally designed drug solely targeting the BCR-ABL tyrosine kinase in chronic myelogenous leukemia (CML), has been received with great enthusiasm.10 Tyrosine kinase inhibitors (TKIs) have also been successfully used in the treatment of solid tumors, validating the concept of targeting therapy and the importance of tyrosine kinase pathways in human cancer.11
Tyrosine kinase (TK) signaling pathways are effective targets due to their role as membrane-localized upstream regulators relevant to cancer progression.12 For instance, receptor tyrosine kinases (RTKs) receive extracellular stimuli and transduce the signal to multiple downstream pathways controlling many important biological functions such as cell proliferation, differentiation, and cell motility.13 Consequently, tyrosine phosphorylation controlled by a delicate balance between tyrosine kinases and phosphatases plays an important role in the regulation of many hallmarks of cancer, including cell growth, angiogenesis, invasion, and metastasis.7 Aberrant tyrosine phosphorylation has been reported in many different solid tumors and hematological malignancies.14,15 Because of the prominent role of TK signaling pathways in cancer, a single TKI can have far-reaching effects, providing impetus for the pursuit of effective targets within tyrosine kinase pathways.16 Currently, a major problem is that tumors often acquire resistance to initial TKI therapy through various mechanisms, including the gatekeeper mutation and activation of other TKs, which has led to the development of more potent second-generation multitarget TKIs.17,18 Design of sustainable therapeutic strategies and molecular diagnosis of TK pathways demands comprehensive and versatile analytical platforms to decipher phosphotyrosine-dependent signaling networks in cancer cells.
Mass spectrometry (MS) and site-specific anti-phosphotyrosine antibodies are frequently used for the analysis of tyrosine phosphorylation in cancer.6,19 Due to the low abundance of tyrosine phosphorylated proteins, enrichment is essential prior to the analysis by MS.20 A common strategy in which MS is preceded by the immunoaffinity enrichment of phosphopeptides introduced by Rush et al.21 in 2005 has yielded a rich source of information on dysregulated kinases and other signaling proteins in many types of cancer. One disadvantage of this approach is that relatively large amounts of cells or tissue are required for comprehensive phosphoproteome analysis, limiting this approach especially for clinical applications where only a small amount of cancer tissue is available. Although the sensitivity of MS is greatly improving, typically 100 to 500 mg of cancer tissues has been used in recent phosphoproteomics studies.5,22,23 The requirement of highly specialized instrumentation not available in every laboratory is another disadvantage of MS. Furthermore, it is not always obvious whether tyrosine phosphorylated peptides identified by MS are in fact directly involved in signal transduction or how a pTyr site may affect downstream signaling. As an alternative to comprehensive MS analysis, anti-phosphotyrosine antibodies directed against previously defined sites of phosphorylation should offer unambiguous profiling of phosphotyrosine-dependent signaling. At present, a substantial number of these site-specific anti-phosphotyrosine antibodies are commercially available. However, considering the large number of phosphorylation sites identified in the proteomes of cancer cells, the current panel of antibodies would appear to be insufficient. Moreover, a significant number of commercially available antibodies do not offer sufficient specificity.24
Concept of SH2 Profiling
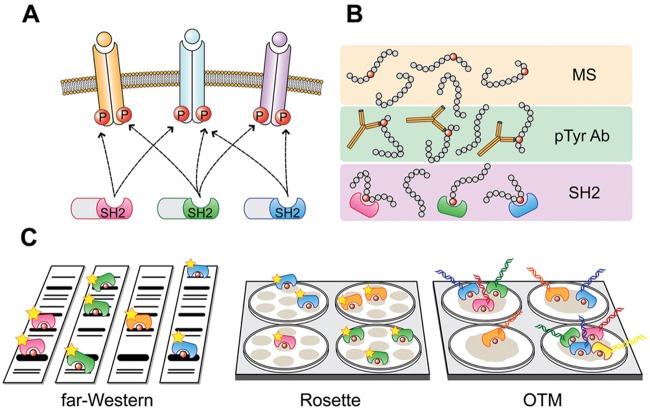
Given the limitations of common phosphoproteomics strategies, we developed SH2 profiling as a complementary approach for the characterization of the global phosphorylation state.25-27 The key components of our analytical platform are Src Homology 2 (SH2) domains, naturally occurring protein interaction domains that recognize specific phosphorylated tyrosine motifs.28 As an integral part of many important signaling proteins, SH2 domains are by far the most prevalent phosphotyrosine-binding module. There are ~120 different SH2 domains encoded in the human genome.29 With a size of approximately 100 amino acids, SH2 domains adopt a characteristic structure composed of a central antiparallel beta sheet flanked by 2 alpha helices. Recognition of phosphotyrosine residues is generally mediated by a conserved, positively charged arginine residue buried in the binding pocket of the SH2 domain. The selectivity of binding to tyrosine phosphorylated ligands is determined by a short stretch of amino acids usually spanning 3 to 5 residues C-terminal to the phosphotyrosine. Additional specificity can be conferred by N-terminal residues of ligands and a secondary binding site.30,31 In addition, it has recently been shown that SH2 binding can be negatively affected by local sequence context such that neighboring positions influence one another.32 Functionally, binding of the SH2 domains to their ligands in the vicinity of the plasma membrane couples the molecular switch of tyrosine phosphorylation (on) or dephosphorylation (off) to downstream effectors. Designated as intracellular “readers” of the state of tyrosine phosphorylation, SH2 domains play a key role in the interpretation, processing, and transduction of cellular signals.33 This unique binding characteristic of SH2 domains thus confers specificity to signal transduction and forms the conceptual basis of SH2 profiling (Fig. 1A).
Figure 1.
Concept and analytical platforms of SH2 profiling. (A) Diagrammatic view of tyrosine kinase signaling state defined by SH2 binding sites. Many extracellular stimuli activate tyrosine kinases, resulting in quantitative changes in tyrosine phosphorylated sites on cellular proteins. These changes are “read” by specific pTyr-recognizing modules such as SH2 domains, to propagate the signal to downstream effectors. These characteristics of SH2 domains form the conceptual basis of SH2 profiling. Red circles represent tyrosine phosphorylated sites (P), and different SH2 domains are distinguished by color. (B) Different analytical platforms of tyrosine phosphoproteomics. Tyrosine phosphorylated peptides are detected by mass spectrometry (MS), anti-phosphotyrosine antibodies (pTyr Ab), or SH2 domains (SH2). MS provides information on the sequence and type of modification, pTyr Ab provides site-specific phosphorylation status, and SH2 detects activated, tyrosine phosphorylated SH2 domain binding sites. (C) SH2 profiling platforms. In far-Western analysis, protein samples are separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and replicate blots are separately probed with horseradish peroxidase (HRP)–labeled SH2 domains. In rosette assay, samples are spotted on membrane, and the binding assay is carried out in a 96-well plate. HRP-labeled SH2 domain probes are incubated with multiple sample spots in a noncompetitive manner (single SH2 per well). In an oligonucleotide-tagged multiplex (OTM) assay, a mixture of SH2 domains with domain-specific DNA tags are incubated with a sample spot allowing for competitive binding (single sample per well). Signal is detected either by chemiluminescence (far-Western and rosette) or real-time polymerase chain reaction (OTM). Quantified values are used to classify samples, such as different cancer tissues, based on SH2 binding preferences (SH2 profiles).
Using a battery of purified SH2 domains for in vitro binding assays, it is possible to quantitatively assess the presence of SH2 domain binding sites (phosphotyrosine motifs) in analytes (e.g., proteins, cell lysates, etc.). In this system (SH2 profiling), a positive SH2 hit suggests that the corresponding SH2 domain containing protein, if expressed in the cell, is involved in tyrosine phosphorylation-dependent protein-protein interaction as part of a tyrosine kinase pathway. Thus, SH2 profiling can provide functional TK pathway information missed in phosphosite cataloguing, making SH2 profiling a unique and complementary approach to conventional tyrosine phosphoproteomics (Fig. 1B). For example, antibody microarray or MS analysis can provide the phosphorylation state of tyrosines on a HER2/ErbB2 receptor in a breast cancer sample, whereas SH2 profiling can show the presence of binding sites for specific SH2 domain–containing proteins on the same receptor. Noteworthy, SH2 profiling assays are not restricted by previous knowledge of phosphorylation sites (i.e., unbiased coverage of tyrosine phosphoproteome), and the sample requirement is equivalent to that for phosphoantibody-based immunoblotting (i.e., lower sample requirement than conventional MS). On the other hand, in SH2 profiling, the actual identification of SH2 ligands requires subsequent antibody or MS analysis. Taken together, SH2 profiling provides qualitatively different information from conventional proteomics approaches. In view of the importance of TK pathways in cancer, we hypothesized that SH2 profiling could serve as an important diagnostic tool that can discern subclasses of tumors based on global tyrosine phosphorylation state.25
Assay Platforms for SH2 Profiling
Modular protein interaction domains have been widely used as tool in proteomics for various purposes such as ligand screening, specificity determination, and domain-ligand interactome analysis.34,35 Similar to antibodies, modular domains can be used in different binding assay platforms defined by immobilization and detection methods. For example, purified SH2 domain proteins can be arrayed on a solid support and incubated with fluorescently labeled samples in solution (e.g., forward-phase array36), or samples can be arrayed on solid phase and incubated with labeled SH2 domains in solution (e.g., reverse-phase array26). Alternatively, interaction can be assessed with both samples and SH2 domains in solution (e.g., fluorescence polarization32). We previously observed that a reverse-phase format has superior sensitivity over a forward-phase format for analyzing complex whole-cell lysates.37 One possible explanation is that SH2 domains may be partially inactivated during the immobilization procedures in forward-phase formats. We also assume that in reverse phase, binding is driven by the relatively high levels of SH2 domain protein in solution. Whereas in forward phase, the concentration of pTyr proteins in lysate solution is extremely low, so signal to noise is poor. Over the past years, we have developed several different analytical platforms for SH2 profiling in the reverse-phase format (Fig. 1C). All platforms are relatively simple, low cost, and amenable to standard laboratories, although experimental setup is a largely manual process in contrast with automated high-throughput phosphoproteomics systems. Technical details have been described in previous publications.26,27,38-40
Quantitative Far-Western Analysis
Far-Western blotting is the original platform of SH2 profiling. Whole cellular protein extracts are separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), transferred to membranes, and subsequently probed with a panel of labeled SH2 domain proteins.25,39 Binding of a SH2 domain to the membrane-immobilized proteins is detected using chemiluminescence. The quantitative SH2 binding profiles of different samples are assessed by apparent molecular size and intensity of bands. An advantage of far-Western over conventional pull-down assays is that interactions between a purified SH2 domain and proteins on the membrane are direct, whereas proteins detected in a SH2 pull-down assay may contain indirect binding partners. Using a cancer cell line expressing various tyrosine mutants of the PDGF receptor, we demonstrated the specificity of SH2 domains, which is improved when a labeled SH2 probe is mixed with several unlabeled domains (competitive binding25). However, we do not usually perform competitive assays, as various degrees of competition in binding may quench relevant signals especially when the concentration of phosphoproteins is low.
Although SH2 profiling aims to quantitatively analyze many cancer samples and SH2 domains in parallel, accurate comparison of far-Western blotting results from multiple experiments is challenging due to gel-to-gel and experimental variations. To overcome the issue, we make multiple efforts, including the use of a multigel apparatus, normalization by internal controls, assay replication, and gel band matching with reference blots.40 For example, pervanadate-treated cell lysates and tyrosine phosphatase-treated lysates can be used as positive and negative controls, respectively, to ensure SH2 probe activity and normalize signal intensity. Reprobing with an anti-pTyr antibody also allows for band matching and quantification of replicate blots using image analysis software. Consequently, far-Western blots of multiple samples with many SH2 probes can be compared and used as the basis for bioinformatics analysis.
Rosette Assay
Although far-Western blotting provides a rich molecular signature based on SH2 binding, it is inherently low throughput and requires relatively large amounts of sample, therefore precluding comprehensive SH2 profiling (>100 SH2 probes). To address this problem, we developed a reverse-phase dot-blotting assay platform (rosette assay). In the rosette assay, nanogram amounts of whole cellular protein extract are serially spotted on a membrane in register with the wells of a 96-well chamber apparatus, allowing parallel probing of ~100 SH2 domains in a single assay. The number of samples analyzed is customizable up to about 100. Since the essential reagents for the far-Western and rosette assays, including labeled SH2 probes and detection system, are identical, they can be coupled; the rosette assay is used for SH2 library screening using minimal amounts of lysate, and subsequently positive hits are confirmed and further analyzed by far-Western. Based on this strategy, a full survey of SH2 interactions using only a few hundred micrograms of lysate, without any phospho-enrichment process, is feasible.26 The library screening result, including positive and negative controls, is digitally captured, quantified using densitometry, and then further processed by bioinformatic analysis.
Oligonucleotide-Tagged Multiplex (OTM) Assay
The oligonucleotide-tagged multiplex (OTM) assay is a distinctive reverse-phase assay from rosette. In the rosette assay, a single SH2 domain is incubated with multiple samples in each well (noncompetitive binding), whereas in OTM, a mixture of SH2 domains differentially tagged with DNA-oligonucleotides is incubated with a single sample. This enables multiplex competitive binding (Fig. 1C). Biotinylated SH2 domains, generated in bacteria using the BirA system, are complexed with biotinylated oligonucleotides via a streptavidin bridge. Following incubation of the DNA-tagged SH2 probes with a sample in a multiwell plate, the bound probes are eluted and quantified by real-time polymerase chain reaction (PCR).27,38 Because it incorporates multiplex SH2 binding and PCR-based detection, OTM offers excellent sensitivity (low femtomole amounts of a phosphopeptide) and low sample requirement (10 µg of cell extract to assess multiple domains). Currently, efforts are under way to reduce the complexity of the quantification process through absolute quantification of the DNA tags by parallel high-throughput sequencing.
Technical Considerations
Since SH2 profiling assays rely on in vitro purified proteins, activity of probes can affect the quality and coverage of the system. We and others have observed that 30% to 40% of SH2 domains purified from bacteria are somewhat or completely insoluble, requiring refolding or alternative expression systems.26,36 These domains tend to be inactive when assessed using a phosphatase inhibitor–treated positive control, presumably due to misfolding or degradation in the purification step. Various strategies to address this issue, including low temperature culture, eukaryotic expression systems, and chaperone coexpression, have resulted in success for a limited number of domains. We also observed some instances of substantial improvement using different domain boundaries, suggesting a contribution to structural stability.
To reduce assay complexity and sample consumption, it may be possible to reduce the number of domains in SH2 profiling based on their overlapping specificity. The known 120 SH2 domains can be classified into about 30 subfamilies according to domain amino acid sequence (e.g., Grb2 family: Grb2, Grap, and Gads).29 Comprehensive SH2 domain studies indicated proteins belonging to a family generally have similar binding preference,26,30,36 although the possibility remains that subtle differences in specificity observed in vitro can affect ligand selection in vivo, such as in the case of SHP-1 and SHP-2.41,42
Applications of SH2 Profiling in Cancer Research
SH2 profiling has become a versatile tool for various levels of cancer studies, from the detailed analysis of single proteins to broad screening in translational research.25-27,40,43-46 First, a tyrosine site on a protein of interest can be readily characterized by rosette assay using synthesized peptides. For example, it is well established that phosphorylation of Y221 of the SH2-SH3 adaptor Crk by the Abl kinase results in an inhibitory conformation via an intermolecular interaction between the Crk SH2 domain and this phosphorylated residue. Recently, another phosphorylation site in Crk (pY251) was found in several human cancer cell lines.46 To explore the biological function of pY251, a rosette screen was performed against a SH2 library. Several SH2 domains, including Abl SH2, were found to bind this site, and this was confirmed by pull-down and surface plasmon resonance (SPR) experiments. The Crk pY251 was then shown to regulate activation of Abl in vitro and in vivo. Interestingly, the binding of Abl SH2 and pY251 was relatively weak, consistent with the fact that the sequence surrounding Y251 does not match the reported Abl SH2 domain consensus, suggesting the interaction between Crk and Abl is dynamic. Similarly, previously uncharacterized carboxy-terminal tyrosine phosphorylation sites in Vav1 were screened by the rosette assay, leading to the hypothesis that Vav1Y826 and Csk are involved in the autoinhibition of NFAT.45 Along with identifying candidate binding proteins, rosette can provide apparent dissociation constants for tyrosine phosphorylated peptides by SH2 binding curve fitting.43
Second, SH2 profiling has been applied to cultured cell samples, providing the tyrosine phosphorylation signature of samples based on the presence of SH2 binding sites. We showed distinctive SH2 binding patterns in response to cell adhesion and growth factor stimulation, as well as in cells expressing different oncogenic PTKs.25-27 Notably, SH2 profiles sometimes can discern a subtle change of tyrosine phosphoproteomes. For example, ectopic expression of oncogenic Src into a mouse fibroblast cell line lacking c-Src, Yes, and Fyn kinases induces pre-invadopodia formation.44 Interestingly, the more invasive phenotype capable of matrix degradation at invadopodia sites requires coexpression of wild-type c-Src. In this case, the global tyrosine phosphorylation levels of both cell lines are equivalent due to the dominant effect of v-Src. However, a rosette assay detected enhanced binding of SH2 domains, including Abl, Crk, and Nck, which are known to interact with cortactin, suggesting they play a role in the cortactin-mediated invadopodia maturation.44
In addition to focused basic research studies, application of SH2 profiling to more complex samples such as a set of human cancer cell lines is important to evaluate assay performance and validate it as a diagnostic tool. To this end, we examined various lung cancer cell lines with different genetic backgrounds such as EGFR and KRAS mutations.40 The SH2 profiles of cell lines were compared using a hierarchical clustering analysis for rosette and far-Western data. The resulting clusters from both assays were overall similar and correlated not only with RTK status (EGFR and MET activity) but also RAS mutation status. For example, one cluster was enriched in cells with high EGFR and MET activity, whereas another cluster contained cells with mutant RAS and low EGFR and MET activity. Interestingly, far-Western–based clustering clearly separated 2 RAS mutant groups, one of which contained an EGF-TKI–sensitive cell line. Since KRAS mutation is known a predictor of EGFR-TKI resistance with rare exception (i.e., KRAS mutant and TKI-sensitive),47,48 it will be intriguing to determine if SH2 profiling can predict these rare cases. Furthermore, assessment of individual probes revealed that a group of SH2 domains bound more strongly to cells with higher sensitivity to EGFR inhibitor (erlotinib), suggesting they may serve as predictive biomarkers for TKI treatment. Taken together, this study illustrates the potential of SH2 profiling to classify cancer cells from the same tumor type independent of oncogene mutation status, protein expression, or marker status information.
Because tumor cell lines do not necessarily recapitulate the signaling state of cancer cells in vivo, we also applied SH2 profiling to patient samples. Using the quantitative 1-tube OTM assay, we analyzed leukemia patient samples (10 acute myeloid leukemia [AML] and 5 CML) in comparison with healthy donor controls.27 A clustering analysis demonstrated correlation between the type of disease or treatment and SH2 binding: One cluster consisted of half of the AML samples and presented strong overall SH2 binding, the rest of AML and untreated CML samples were co-clustered, and a cluster of treated CML and normal controls with weak signal was also clearly separated. By ranking the relative levels of SH2 binding for each leukemia sample, distinct SH2 preferences for different diagnostic classes became apparent, such as the specificity of the GAP SH2 domain toward untreated CML. These data suggested that OTM profiling could be used to distinguish different types of leukemia. Currently, a rosette screen using a larger number of CML and CLL samples is under way.
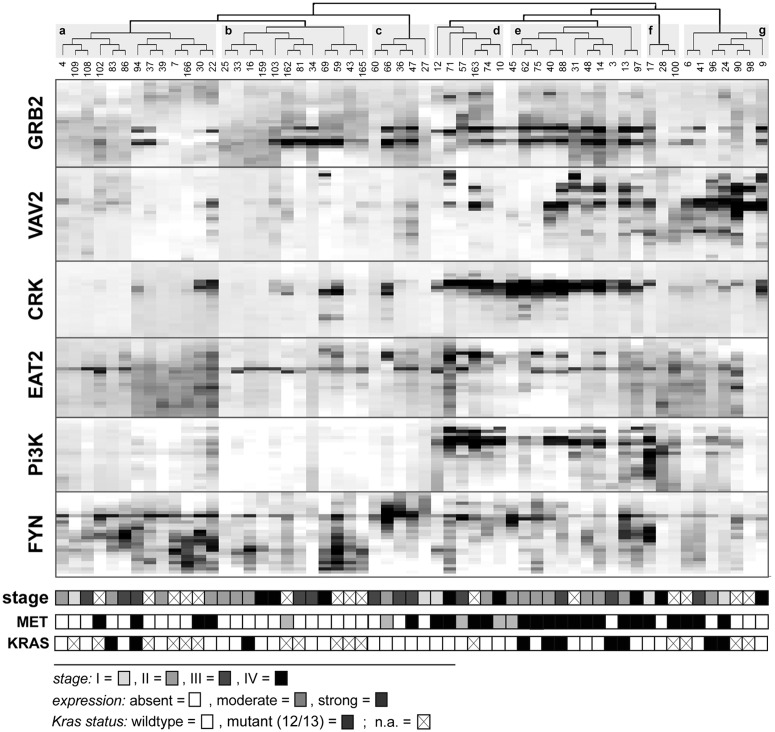
For many of the solid tumors investigated so far, we observed relatively weak levels of phosphotyrosine signal in all SH2 analytical platforms. As a result, some of the subtle but biologically important information may be missed if these differences are too small to be distinguished. Since OTM and rosette assays provide only a single data point for a SH2 domain and a sample, they are particularly affected by low signal and higher noise levels. To discern more subtle differences in the SH2 binding pattern, we performed SH2 profiling of tumor tissues using quantitative far-Western blotting (Figs. 2 and 3). In accordance to our findings in different cancer cell lines, we observed substantial differences in the phosphotyrosine profiles between colon cancer and breast cancer samples, as well as within each tumor type. Based on hierarchical cluster analysis, colon and breast cancer samples were grouped in 7 and 4 major clusters, respectively. To gain deeper insights into the biological relevance of the different clusters, we related the clusters to the pathological stage of disease. For colon cancer, no significant correlation was observed between the 7 major clusters and the different stages of disease. In addition, as far as data were available, no overall correlation was found between SH2 profiles and the mutational status of the KRAS gene (Fig. 2). In contrast, breast cancer samples from advanced stages of disease were significantly overrepresented in 2 of the 4 major clusters (Fig. 3, clusters c and d, respectively; P < 0.001).
Figure 2.
SH2 profiling of colon cancer. Tumors samples (n = 57) were snap frozen immediately after surgical removal and whole cellular extracts were prepared as previously described.25 Far-Western blot analysis was performed with 20 µg of lysate per sample. Lysates were separated on 4% to 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gradient gels, transferred to PDVF membranes, and probed with biotinylated SH2 domains. Streptavidin–horseradish peroxidase (HRP) conjugate was used for detection. Images were scanned and analyzed by the ImageJ software (National Institutes of Health, Bethesda, MD), and digitalized profiles were segmented into bins as previously described.40 Subsequently, correlative, unsupervised hierarchical cluster analysis was performed using the MeV software (version 4.6; MeV, Boston, MA). The 7 major clusters (a-f) identified by cluster analysis are shaded in gray. Levels of MET expression, determined by Western blot analysis, were categorized as moderate or strong; lack of MET expression was categorized as absent. The genomic status of codons 12 and 13, respectively, of the human Kras gene was determined by restriction fragment length polymorphism (RFLP) analysis as previously described.40 Pathological staging is given in accordance with the 2010 American Joint Committee on Cancer classification.
Figure 3.
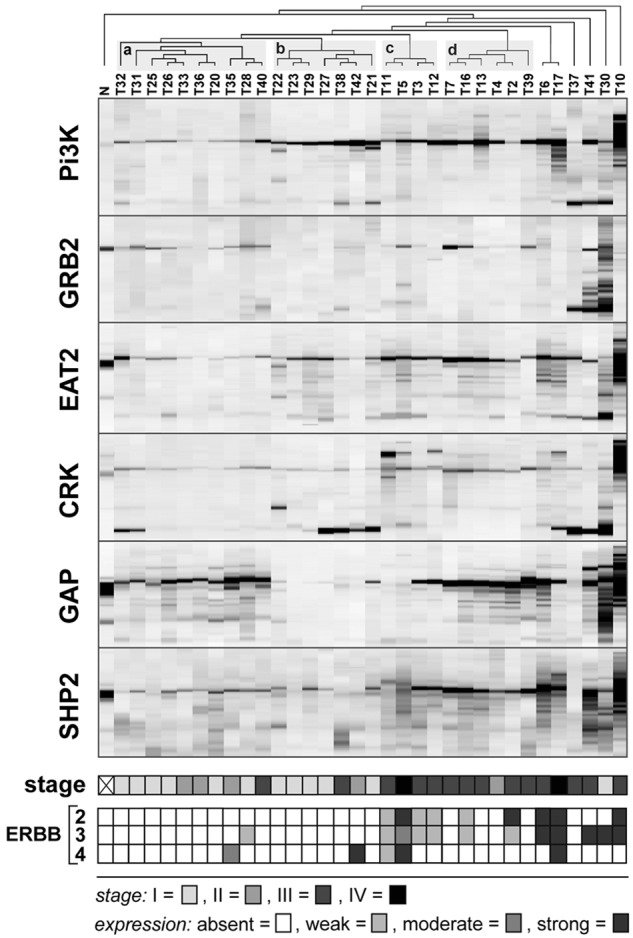
SH2 profiling of native breast cancer samples. Profiling of tumor samples (n = 34) and processing of profiles were performed as described in Fig. 2. The 4 major clusters (a-d) determined by correlative, unsupervised clustering are shaded in gray. Normal breast tissue (N) served as a control. Protein expression levels of ErbB2, ErbB3, and ErbB4 were determined by Western blot analysis.
In an attempt to assign the differences in phosphoactivity depicted by SH2 profiling to tyrosine kinase activity, we determined the protein expression levels of MET and members of the ErbB-family frequently altered or overexpressed in colon and breast cancer, respectively. Strikingly, a high level of MET expression was significantly correlated with 3 closely related colon cancer clusters (Fig. 2, clusters d, e, and f, respectively; P < 0.001). In breast cancer, increased levels of ErbB2 and ErbB3 expression were significantly overrepresented in the 2 clusters (c and d, respectively; P < 0.001), concurrently correlating with advanced stages of disease. Taken together, our data demonstrate that SH2 profiling of clinical samples may provide a rich source of important and potentially predictive information about cancer cell signaling and patient outcomes.
Challenges
Our collective efforts in the past decade have validated the original concept of SH2 profiling: A set of SH2 domains can serve as a means to profile the global tyrosine phosphorylation state. We showed SH2 profiling is a powerful tool to predict biological partners for a specific tyrosine motif on a protein and to classify cancer cells. The 3 analytical platforms are versatile in that rosette assay serves as a rapid SH2 library screening tool, quantitative far-Western analysis provides a functional signature of the tyrosine phosphoproteome, and OTM offers a sensitive 1-tube competitive binding assay. We recognize, however, technical hurdles and challenges exist for the application of SH2 profiling to translational cancer research. Clinical specimens are usually limited in quantity and availability. Considering the future diagnostic application, it is ideal that all analyses be carried out with the amount of tissue provided by a needle biopsy (~10 mg tissue).49 For cultured cell samples, SH2 profiling is sensitive enough without phosphoprotein enrichment, thereby requiring a smaller amount of sample than for MS-based phosphoproteomics. In the case of solid tumor specimens, however, we sometimes observe a significantly lower signal level than with cultured cells, increasing sample requirement. Low signal-to-noise levels affect data quality, particularly for the rosette and OTM screening platforms. In these systems, one SH2 probe gives only one data value as the binding assay is performed within a sample dot. To detect subtle differences in SH2 binding between tumor samples, sufficient signal intensity is necessary. As a result, quantitative phosphotyrosine profiles may not fully be captured by the OTM and rosette assay. On the other hand, quantitative far-Western, which requires 15 to 50 µg per SH2 domain probe, can reveal distinct SH2 binding patterns, and thus tumor-specific clusters are visible (Figs. 2 and 3). This dilemma between sample requirement and data quality is currently a hurdle for implementation of SH2 profiling to large-scale tumor analysis. Nevertheless, the observation that 5 selected SH2 domains were sufficient to provide distinct clusters relevant to breast cancer stages (Fig. 3) suggests that the use of rosette/OTM to exclude domains with low signal prior to far-Western could minimize sample consumption.
Low phosphotyrosine signals can in part be attributed to heterogeneity and impurity of cancer tissue specimens, which contain various amounts of normal cells and connective tissues. This is consistent with the fact that purified hematopoietic cells usually show better quality data, and disease-relevant SH2 profiles have been obtained using the OTM assay.27 In addition, in a preliminary rosette screening, we observed that HER2/ErbB2-positive tumors were co-clustered by their strong binding to known SH2 domains downstream of HER2/ErbB2. Thus, we assume OTM/rosette SH2 platforms should be capable of differentiating tumors containing a higher percentage of cancer cells and/or driven by TK oncogenes. We note, however, that some normal cells such as immune cells associated with inflammatory tumors can contribute to the malignant phenotype of cancer.7
Perspectives
Toward personalized molecular diagnosis using SH2 profiling, we are attempting to assign different SH2-based clusters to clinically relevant parameters associated with cancer development, progression, and effective therapeutic intervention.50As suggested by our data on lung cancer cell lines and native breast cancer samples, SH2 profiling may be of particular value for improved classification of tumors and stratification of patients in combination with established clinical and pathological parameters. Establishing this would require well-controlled retrospective or prospective studies of cancer patients treated with different drug combinations, including tyrosine kinase inhibitors, to demonstrate that therapeutic benefits and improvements in the clinical course of disease can be predicted by SH2 profiling. However, it may be naive to assume phosphotyrosine profiles detected by a single modular domain family can be an independent prognostic marker for human cancer, a complex genetic disease. To fully decipher aberrant signaling in cancer in detail with the aim of identifying specific therapeutic targets, additional information beyond the mere SH2 profile may be needed. We assume TK signaling status as a whole may predict patient outcome, particularly in a subset of tumors where TK signaling drives oncogenesis. How can this comprehensive view of TK signaling status be captured? As tyrosine kinases and phosphatase are the key players controlling the cellular state of phosphorylation, comprehensive profiling of kinase/phosphatase expression may be one promising approach to guide the interpretation and classification of SH2 profiles, as exemplified above for the kinases MET and ErbB in colon and breast cancer. For instance, expression data of particular kinases, phosphatases, and SH2/PTB proteins will be useful in specifying the pTyr-dependent protein interactions that actually occur in cells. SH2 profiling (using purified SH2 domains) detects the presence of binding sites for SH2 domain–containing proteins regardless of their expression in cell, whereas gene or protein expression indicates the protein is actually available to mediate signaling. Expression data can also help in the efficient design of SH2 profiling assays through the exclusion of unexpressed SH2 proteins.
Knowledge of the genomic status and activity of kinases and phosphatases will also without doubt provide important information for the classification of SH2 profiles. Correlations between SH2 profiles and somatic mutation in kinase/phosphatase genes are informative to specify activated TK signaling networks. Furthermore, combining SH2 profiling with other phosphoproteomics tools may also turn out to be a powerful combinatorial strategy. Detailed characterization of tyrosine phosphorylated proteins by MS after pull-down of phosphoproteins with informative SH2 domains should have great potential, enabling the identification of executors and substrates of tyrosine phosphorylation, which may serve as potential targets for the development of rational therapies.
Conclusion
As demonstrated for many different tumor cell lines and native tumor samples, SH2 profiling is a powerful technique for deciphering and classifying aberrant phosphotyrosine-dependent signaling in cancer. Given its sensitivity and relatively small sample requirement, SH2 profiling may be of extraordinary value for the analysis of tumor tissues and monitoring of tumors during therapy where only limited amounts of cancer cells are present. It may also provide “geographical” profiles of the tyrosine phosphorylation state in different areas of the tumor, addressing the important issue of tumor heterogeneity and diversity of signaling. SH2 profiling, in combination with other phosphoproteomics tools and informatics, is evolving as a simple, robust, and highly sensitive technique with both research and clinical applications.
Acknowledgments
The authors thank Bruce Mayer for continuous support for the SH2 profiling project and critical reading of the manuscript, Birgit Klampe for technical assistance, and Joshua Jadwin for help with editing of the manuscript.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was partially supported by the Neag Comprehensive Cancer Center, an American Cancer Society Institutional Research Grant, and NCI challenge grant RC1 CA146843.
References
- 1. Bentley DR. Whole-genome re-sequencing. Curr Opin Genet Dev. 2006;16:545-52 [DOI] [PubMed] [Google Scholar]
- 2. Roychowdhury S, Iyer MK, Robinson DR, et al. Personalized oncology through integrative high-throughput sequencing: a pilot study. Sci Transl Med. 2011;3:111ra121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2008;458:719-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Burbelo PD, Ching KH, Bren KE, Iadarola MJ. Emerging tactical strategies for fighting the war on cancer based on the genetic landscape. Am J Transl Res. 2011;3:251-8 [PMC free article] [PubMed] [Google Scholar]
- 5. Harsha HC, Pandey A. Phosphoproteomics in cancer. Mol Oncol. 2010;4:482-95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kolch W, Pitt A. Functional proteomics to dissect tyrosine kinase signalling pathways in cancer. Nat Rev Cancer. 2010;10:618-29 [DOI] [PubMed] [Google Scholar]
- 7. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-74 [DOI] [PubMed] [Google Scholar]
- 8. Nik-Zainal S, Alexandrov LB, Wedge DC, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979-93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lobell RB, Omer CA, Abrams MT, et al. Evaluation of farnesyl:protein transferase and geranylgeranyl:protein transferase inhibitor combinations in preclinical models. Cancer Res. 2001;61:8758-68 [PubMed] [Google Scholar]
- 10. Kantarjian H, Sawyers C, Hochhaus A, et al. Hematologic and cytogenetic responses to imatinib mesylate in chronic myelogenous leukemia. N Engl J Med. 2002;346:645-52 [DOI] [PubMed] [Google Scholar]
- 11. Giamas G, Man YL, Hirner H, et al. Kinases as targets in the treatment of solid tumors. Cell Signal. 2010;22:984-1002 [DOI] [PubMed] [Google Scholar]
- 12. Pawson T. Regulation and targets of receptor tyrosine kinases. Eur J Cancer. 2002;38(Suppl 5):S3-10 [DOI] [PubMed] [Google Scholar]
- 13. Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Krause DS, Van Etten RA. Tyrosine kinases as targets for cancer therapy. N Engl J Med. 2005;353:172-87 [DOI] [PubMed] [Google Scholar]
- 15. Hunter T. Tyrosine phosphorylation: thirty years and counting. Curr Opin Cell Biol. 2009;21:140-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gschwind A, Fischer OM, Ullrich A. The discovery of receptor tyrosine kinases: targets for cancer therapy. Nat Rev Cancer. 2004;4:361-70 [DOI] [PubMed] [Google Scholar]
- 17. Baselga J. Targeting tyrosine kinases in cancer: the second wave. Science. 2006;312:1175-8 [DOI] [PubMed] [Google Scholar]
- 18. Ou SH. Second-generation irreversible epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs): a better mousetrap? A review of the clinical evidence. Crit Rev Oncol Hematol. 2012;83:407-21 [DOI] [PubMed] [Google Scholar]
- 19. Brennan DJ, O’Connor DP, Rexhepaj E, Ponten F, Gallagher WM. Antibody-based proteomics: fast-tracking molecular diagnostics in oncology. Nat Rev Cancer. 2010;10:605-17 [DOI] [PubMed] [Google Scholar]
- 20. Stasyk T, Huber LA. Mapping in vivo signal transduction defects by phosphoproteomics. Trends Mol Med. 2012;18:43-51 [DOI] [PubMed] [Google Scholar]
- 21. Rush J, Moritz A, Lee KA, et al. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat Biotechnol. 2005;23:94-101 [DOI] [PubMed] [Google Scholar]
- 22. Rikova K, Guo A, Zeng Q, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190-203 [DOI] [PubMed] [Google Scholar]
- 23. Geiger T, Cox J, Ostasiewicz P, Wisniewski JR, Mann M. Super-SILAC mix for quantitative proteomics of human tumor tissue. Nat Methods. 2010;7:383-5 [DOI] [PubMed] [Google Scholar]
- 24. Sevecka M, Wolf-Yadlin A, MacBeath G. Lysate microarrays enable high-throughput, quantitative investigations of cellular signaling. Mol Cell Proteomics. 2011;10:M110.005363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nollau P, Mayer BJ. Profiling the global tyrosine phosphorylation state by Src homology 2 domain binding. Proc Natl Acad Sci U S A. 2001;98:13531-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Machida K, Thompson CM, Dierck K, et al. High-throughput phosphotyrosine profiling using SH2 domains. Mol Cell. 2007;26:899-915 [DOI] [PubMed] [Google Scholar]
- 27. Dierck K, Machida K, Voigt A, et al. Quantitative multiplexed profiling of cellular signaling networks using phosphotyrosine-specific DNA-tagged SH2 domains. Nat Methods. 2006;3: 737-44 [DOI] [PubMed] [Google Scholar]
- 28. Machida K, Mayer BJ. The SH2 domain: versatile signaling module and pharmaceutical target. Biochim Biophys Acta. 2005;1747:1-25 [DOI] [PubMed] [Google Scholar]
- 29. Liu BA, Jablonowski K, Raina M, Arce M, Pawson T, Nash PD. The human and mouse complement of SH2 domain proteins: establishing the boundaries of phosphotyrosine signaling. Mol Cell. 2006;22:851-68 [DOI] [PubMed] [Google Scholar]
- 30. Huang H, Li L, Wu C, et al. Defining the specificity space of the human SRC homology 2 domain. Mol Cell Proteomics. 2008;7:768-84 [DOI] [PubMed] [Google Scholar]
- 31. Bae JH, Lew ED, Yuzawa S, Tome F, Lax I, Schlessinger J. The selectivity of receptor tyrosine kinase signaling is controlled by a secondary SH2 domain binding site. Cell. 2009;138:514-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu BA, Jablonowski K, Shah EE, Engelmann BW, Jones RB, Nash PD. SH2 domains recognize contextual peptide sequence information to determine selectivity. Mol Cell Proteomics. 2010;9:2391-404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lim WA, Pawson T. Phosphotyrosine signaling: evolving a new cellular communication system. Cell. 2010;142:661-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jadwin JA, Ogiue-Ikeda M, Machida K. The application of modular protein domains in proteomics. FEBS Lett. 2012;586:2586-96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu BA, Engelmann B, Nash PD. High-throughput analysis of peptide-binding modules. Proteomics. 2012;10:1527-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jones RB, Gordus A, Krall JA, MacBeath G. A quantitative protein interaction network for the ErbB receptors using protein microarrays. Nature. 2006;439:168-74 [DOI] [PubMed] [Google Scholar]
- 37. Machida K, Mayer BJ, Nollau P. Profiling the global tyrosine phosphorylation state. Mol Cell Proteomics. 2003;2:215-33 [DOI] [PubMed] [Google Scholar]
- 38. Dierck K, Machida K, Mayer BJ, Nollau P. Profiling the tyrosine phosphorylation state using SH2 domains. Methods Mol Biol. 2009;527:131-55 [DOI] [PubMed] [Google Scholar]
- 39. Machida K, Mayer BJ. Detection of protein-protein interactions by far-Western blotting. Methods Mol Biol. 2009;536:313-29 [DOI] [PubMed] [Google Scholar]
- 40. Machida K, Eschrich S, Li J, et al. Characterizing tyrosine phosphorylation signaling in lung cancer using SH2 profiling. PLoS One. 2010;5:e13470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sweeney MC, Wavreille AS, Park J, Butchar JP, Tridandapani S, Pei D. Decoding protein-protein interactions through combinatorial chemistry: sequence specificity of SHP-1, SHP-2, and SHIP SH2 domains. Biochemistry. 2005;44:14932-47 [DOI] [PubMed] [Google Scholar]
- 42. Kuo E, Park DK, Tzvetanova ID, Leiton CV, Cho BS, Colognato H. Tyrosine phosphatases Shp1 and Shp2 have unique and opposing roles in oligodendrocyte development. J Neurochem. 2010;113:200-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dubielecka PM, Machida K, Xiong X, et al. Abi1/Hssh3bp1 pY213 links Abl kinase signaling to p85 regulatory subunit of PI-3 kinase in regulation of macropinocytosis in LNCaP cells. FEBS Lett. 2010;584(15):3279-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kelley LC, Ammer AG, Hayes KE, et al. Oncogenic Src requires a wild-type counterpart to regulate invadopodia maturation. J Cell Sci. 2010;123(Pt 22):3923-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lazer G, Pe’er L, Farago M, Machida K, Mayer BJ, Katzav S. Tyrosine residues at the carboxy-terminus of Vav1 play an important role in regulation of its biological activity. J Biol Chem. 2010;285(30):23075-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sriram G, Reichman C, Tunceroglu A, et al. Phosphorylation of Crk on tyrosine 251 in the RT loop of the SH3C domain promotes Abl kinase transactivation. Oncogene. 2011;30(46):4645-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Linardou H, Dahabreh IJ, Kanaloupiti D, et al. Assessment of somatic k-RAS mutations as a mechanism associated with resistance to EGFR-targeted agents: a systematic review and meta-analysis of studies in advanced non-small-cell lung cancer and metastatic colorectal cancer. Lancet Oncol. 2008;9:962-72 [DOI] [PubMed] [Google Scholar]
- 48. Benesova L, Minarik M, Jancarikova D, Belsanova B, Pesek M. Multiplicity of EGFR and KRAS mutations in non-small cell lung cancer (NSCLC) patients treated with tyrosine kinase inhibitors. Anticancer Res. 2010;30:1667-71 [PubMed] [Google Scholar]
- 49. Cheung YC, Chang JW, Hsieh JJ, Lin G, Tsai YH. Adequacy and complications of computed tomography-guided core needle biopsy on non-small cell lung cancers for epidermal growth factor receptor mutations demonstration: 18-gauge or 20-gauge biopsy needle. Lung Cancer. 2010;67: 166-9 [DOI] [PubMed] [Google Scholar]
- 50. Haura EB. From modules to medicine: How modular domains and their associated networks can enable personalized medicine. FEBS letters. 2012;586:2580-5 [DOI] [PMC free article] [PubMed] [Google Scholar]