Abstract
The CAS (CRK-associated substrate) family of adaptor proteins comprises 4 members, which share a conserved modular domain structure that enables multiple protein-protein interactions, leading to the assembly of intracellular signaling platforms. Besides their physiological role in signal transduction downstream of a variety of cell surface receptors, CAS proteins are also critical for oncogenic transformation and cancer cell malignancy through associations with a variety of regulatory proteins and downstream effectors. Among the regulatory partners, the 3 recently identified adaptor proteins constituting the NSP (novel SH2-containing protein) family avidly bind to the conserved carboxy-terminal focal adhesion–targeting (FAT) domain of CAS proteins. NSP proteins use an anomalous nucleotide exchange factor domain that lacks catalytic activity to form NSP-CAS signaling modules. Additionally, the NSP SH2 domain can link NSP-CAS signaling assemblies to tyrosine-phosphorylated cell surface receptors. NSP proteins can potentiate CAS function by affecting key CAS attributes such as expression levels, phosphorylation state, and subcellular localization, leading to effects on cell adhesion, migration, and invasion as well as cell growth. The consequences of these activities are well exemplified by the role that members of both families play in promoting breast cancer cell invasiveness and resistance to antiestrogens. In this review, we discuss the intriguing interplay between the NSP and CAS families, with a particular focus on cancer signaling networks.
Keywords: migration, invasion, antiestrogen resistance, tyrosine phosphorylation, serine phosphorylation, SRC kinase
Introduction
Transmembrane receptors localized on the cell surface, such as integrin receptors for extracellular matrix proteins, growth factor receptor tyrosine kinases, and cytokine receptors, convey extracellular stimuli into a cascade of intracellular signaling events that lead to cell proliferation and survival, cytoskeletal reorganization, and cell migration. Signaling pathways emanating from cell surface receptors rely on networks of adaptor proteins (which contain multiple protein interaction domains) and scaffold proteins (which bring together multiple components of a signaling pathway).1,2 Although adaptor and scaffold proteins lack enzymatic activity, they represent both crucial links to downstream effector molecules and nodes that interconnect different signaling pathways. By regulating the flow of information in signaling networks, adaptor and scaffold proteins are critical for normal cellular physiology and homeostasis. Consequently, their dysregulation can lead to a spectrum of diseases and particularly cancer development and progression. The members of the CAS (CRK-associated substrate) and NSP (novel SH2-containing protein) families (Table 1) are multidomain proteins that combine both adaptor and scaffold functions to form unique signaling modules in complex signaling networks (Figs. 1 and 2). They achieve this by interacting with each other through their conserved carboxy-terminal domains. In this review, we provide an overview of both families and their shared involvement in cell signaling and cancer malignancy.
Table 1.
CAS and NSP Family Members
| Official gene name | Other names |
|---|---|
| CAS family | |
| BCAR1, breast cancer antiestrogen resistance protein 1 | p130CAS/CAS, CRK-associated substrate |
| HEF1, human enhancer of filamentation 1 | |
| NEDD9, neural precursor cell expressed developmentally downregulated protein 9 | CAS-L/CASL, CAS-related protein lymphocyte type |
| EFS, embryonal FYN-associated substrate | SIN, SRC-interacting or signal-integrating protein |
| CASS4, CAS scaffolding protein family member 4 | HEPL, HEF1-EFS-p130CAS–like protein |
| NSP family | |
| SH2D3A, SH2 domain–containing 3A | NSP1, novel SH2-containing protein 1 |
| BCAR3, breast cancer antiestrogen resistance protein 3 | NSP2, novel SH2-containing protein 2 |
| AND-34, identified from AND TCR transgenic mice clone 34.1 | |
| SH2D3C, SH2 domain–containing 3C | NSP3, novel SH2-containing protein 3 |
| SHEP1, SH2 domain–containing EPH receptor–binding protein 1 | |
| CHAT, CAS/HEF1-associated signal transducer | |
| CHAT-H, hematopoietic cell–specific CHAT | |
Note: Included are the most common names according to UniProtKB/Swiss-Prot (uniprot.org).
Figure 1.
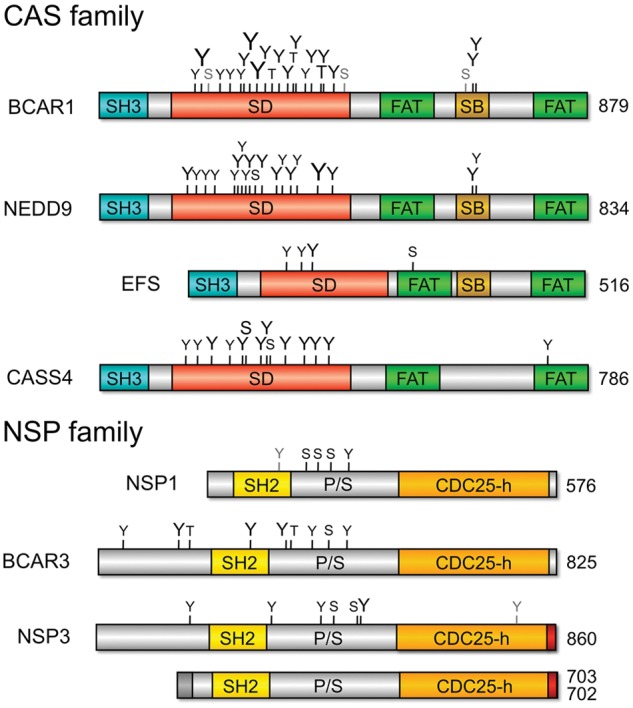
Domain structure and phosphorylation sites of CAS and NSP family proteins. Shown are the main isoforms for the various family members and their amino acid lengths. Isoforms of different lengths are shown only for NSP3; the 2 NSP3 shorter isoforms of 702 and 703 amino acids differ from each other in their most amino-terminal segment (indicated in dark gray). CDC25-h = CDC25 homology domain; FAT = focal adhesion–targeting domain (the central serine-rich region of the CAS proteins has the structure of a FAT domain, although it is not commonly designated as such); P/S = proline/serine-rich region; SB = SRC-binding region (containing binding sites for both SRC SH2 and SH3 domains in BCAR1 and EFS and only for the SRC SH2 domain in NEDD9); SH2 = SH2 domain; SH3 = SH3 domain. Phosphorylation sites (Y = tyrosine; S = serine; T = threonine) were obtained from phosphosite.org; the smallest font indicates sites identified by mass spectrometry in >5 samples, the intermediate-sized font indicates sites identified in >50 samples, and the largest font indicates sites identified in >500 samples. It should be noted that identification of phosphorylation sites by mass spectrometry is not necessarily comprehensive. Indicated in gray are the 3 serine phosphorylation sites in the substrate domain of BCAR1 that depend on BCAR3 expression in MDA-MB-231 breast cancer cells94 (the first of which was also detected in >5 other samples), the tyrosine phosphorylated downstream of the EGF receptor in the SH2 domain of NSP1,81 and the tyrosine phosphorylated downstream of the overexpressed EPHB2 receptor in the CDC25 homology domain of NSP3.90
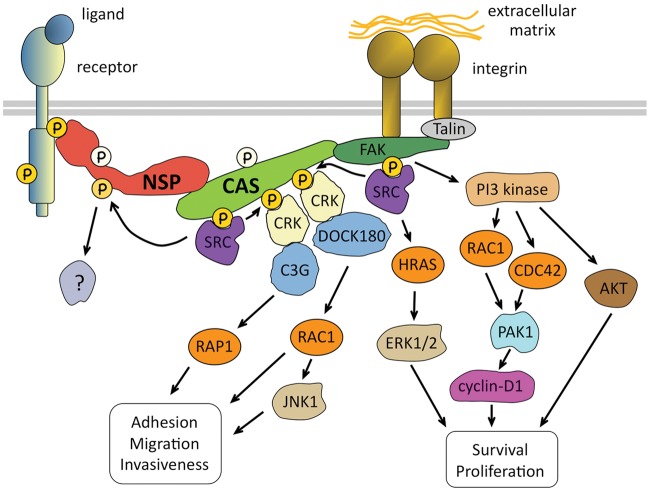
Figure 2.
NSP-CAS signaling networks. The schematic diagram illustrates pathways discussed in the text, depicting how NSP proteins integrate CAS signaling downstream of integrins with signaling pathways downstream of other cell surface receptors. Signaling pathways downstream of cell surface receptors as well as many other known regulators and effectors of CAS family proteins are not shown. Overexpression of several NSP and CAS proteins has been implicated in cancer malignancy and resistance to chemotherapy, which may be due at least in part to perturbation of the signaling interactions shown in the figure.
The CAS Family
Domain organization and general signaling mechanisms
The CAS protein family consists of 4 members, BCAR1, NEDD9, EFS, and CASS4 (see Table 1 for alternative names), which share a conserved multidomain organization (Fig. 1). The amino-terminal SH3 domain of CAS proteins binds polyproline motifs of interacting partners, the most studied of which is the focal adhesion kinase FAK3 (Fig. 2). This is followed by the substrate domain (Fig. 1), which can be “stretched” to expose its many YxxP motifs for phosphorylation, thus functioning as a sensor of cell mechanical stress.4,5 Once phosphorylated by kinases such as SRC, these motifs provide binding sites for the SH2 domains of adaptor proteins, with CRK and CRKL being the most important.6-8 The subsequent serine-rich region can recruit regulatory 14-3-3 proteins through serine-phosphorylated motifs.9 Structurally, this region folds into a 4–helix bundle domain that is reminiscent of the focal adhesion–targeting (FAT) domains found in focal adhesion proteins.10 This is followed by the SRC-binding region, which includes a polyproline motif that can bind the SRC SH3 domain and/or a tyrosine-containing motif that, when phosphorylated by FAK, can bind the SRC SH2 domain.11,12 Notably, features of the SRC-binding region vary among the family members, with CASS4 lacking both SRC-binding motifs. A stretch of approximately 70 amino acids with an unknown structure and function precedes the carboxy-terminal domain, which was only recently revealed to adopt a FAT-type 4–helix bundle fold.13 This domain is responsible for the interaction with NSP proteins as well as several ubiquitin ligases.14,15 Furthermore, in at least some family members, the CAS carboxy-terminal domain can be released by caspases and function independently to promote apoptosis.11,12,16
The detailed signaling mechanisms of CAS proteins have been extensively discussed in several excellent recent reviews.8,11,12,16,17 Therefore, here, we highlight only the main general features of their signaling pathways (Fig. 2). Cell adhesion mediated by integrin receptors is the best characterized event linked to CAS signaling since it leads to the activation of FAK.11,12 FAK can directly recruit SRC family kinases and at the same time phosphorylate the SRC-binding region in CAS proteins, thus generating the binding site for the SRC SH2 domain.11,12 Binding of the SRC SH2/SH3 domains to the FAK-CAS complex relieves their intramolecular inhibition on the kinase domain, leading to SRC activation.18-21 In addition, it positions the activated SRC for phosphorylation of the multiple tyrosines in the CAS substrate domain.22,23 This hyperphosphorylation is a hallmark of the active signaling form of CAS proteins and generates binding sites for CRK adaptors, which then recruit the nucleotide exchange factors DOCK180 and C3G.24-27 DOCK180 is a key activator of the RHO family GTPase RAC1, which in turn controls activation of the JNK MAP kinase and actin cytoskeleton remodeling, while C3G activates the RAS family GTPase RAP1, leading to increased integrin activity.25,28-33 Together, these 2 pathways promote cell substrate adhesion, migration, and invasiveness as well as proliferation.6,8,27,28,31,34-37 Moreover, RAC1 activation by CAS proteins can also occur through PI3 kinase–dependent activation of guanine nucleotide exchange factors, while the PI3 kinase–AKT axis additionally supports cell survival downstream of CAS27,37-39 (Fig. 2).
In addition to FAK, SRC, and the CRK-DOCK180-RAC1, CRK-C3G-RAP1, and PI3 kinase axes, many other signaling molecules have been described as CAS regulators and effectors, accounting for the diverse biological functions of the family.11,12 Among these are the NSP family proteins, which are featured in this review. Although less investigated, the 3 proteins forming the NSP family seem to be among the most avid CAS-binding partners. They are also capable of interacting with receptor tyrosine kinases, thus directly linking CAS proteins to receptor tyrosine kinase–initiated signaling. Before delving into the details of NSP-CAS signaling, we provide an overview of each of the 4 CAS proteins and the distinctive properties that account for their different roles in various malignancies.
BCAR1 (p130CAS)
BCAR1 is the founding member of the CAS family and is ubiquitously expressed in adult human tissues and cancer cell lines11,17 (broadinstitute.org/ccle). It was originally discovered in v-CRK– and v-SRC–transformed cells as p130CAS (Table 1), an abundant and highly phosphorylated 130-kDa protein that interacts with the transforming oncogenes.40-42 Later studies revealed that BCAR1 plays an essential role in cell transformation by SRC and several other oncogenes.37,43-48 The human gene was identified in a genome-wide screen for proteins whose overexpression in estrogen-dependent breast cancer cells confers resistance to antiestrogens (hence, the name breast cancer antiestrogen resistance protein 1, abbreviated to BCAR1) (Table 1).49-51 BCAR1 has also been implicated in resistance to the chemotherapeutic drug doxorubicin.52 Furthermore, high levels of the BCAR1 protein in breast cancer specimens have been correlated with high ERBB2 receptor expression, enhanced proliferation, increased risk for resistance to tamoxifen therapy, and poor clinical outcome.37,53,54 The ability of BCAR1 to support long-term proliferation in the presence of antiestrogens, which appears to be unique among the CAS family proteins, has been linked to tyrosine phosphorylation of its substrate domain.55,56 BCAR1 signaling has also been associated with cell proliferation, survival, and migration/invasion in many tumor types besides breast cancer, including ovarian and prostate cancer, glioblastoma, melanoma, and hematopoietic malignancies.11,12,17,27,57-63 Accordingly, a great abundance of BCAR1 tyrosine- and serine/threonine-phosphorylated peptides have been detected in a wide variety of cancers (phosphosite.org) (Fig. 1).
NEDD9
This member of the CAS family is widely expressed in tissues, and interestingly, its levels are low in the G1 phase of the cell cycle but increase dramatically during mitosis.12,16 NEDD9 is also expressed in most cancer cell lines, with highest levels detected in melanoma and medulloblastoma cell lines12,64 (broadinstitute.org/ccle). Furthermore, many tyrosine-phosphorylated peptides from the NEDD9 substrate domain have been detected by mass spectrometry in a large number and a wide variety of cancer cell lines and tumor samples (phosphosite.org) (Fig. 1), consistent with the reported importance of NEDD9 signaling in most cancers including hematopoietic tumors.12,16,17,65 Interestingly, a distinctive role of NEDD9 in driving invasiveness and metastasis has been extensively documented in melanoma, breast cancer, and other cancers.16,17,64,66-73 In addition, analysis of NEDD9 knockout mice has implicated this CAS family member in tumor initiation in the MMTV–Polyoma middle T mouse mammary tumor model through activation of FAK, SRC, AKT, and ERK.74 However, besides reducing tumor development, over time, the lack of NEDD9 expression can also promote cancer cell aggressiveness by causing genetic instability.75 Consistent with a complex role in tumorigenesis, NEDD9 overexpression may also negatively affect cancer development by causing defects in cytokinesis through its multiple effects on the mitotic machinery as well as by promoting apoptosis through caspase-dependent release of its carboxy-terminal domain.12,16
EFS and CASS4
EFS and CASS4 are the least characterized members of the CAS family, and their roles in cancer have not yet been extensively explored. EFS mRNA is expressed in the adult brain, lung, and thymus as well as in sarcoma cell lines and a subset of cell lines from other cancers, including lung cancer76,77 (broadinstitute.org/ccle). Only a few tyrosine-phosphorylated peptides from the EFS substrate domain have been identified in a variety of tumors and cancer cell lines, including breast cancers and pheochromocytomas (phosphosite.org) (Fig. 1). Therefore, the available evidence suggests that EFS has a more limited role in cancer than the other members of the CAS family.
The most recently identified family member, CASS4, has a restricted tissue distribution and is predominantly found in the lung and spleen.76 In cancer cell lines, CASS4 mRNA expression is highest in leukemia cell lines (broadinstitute.org/ccle). Despite lacking the SRC-binding region, CASS4 is phosphorylated on tyrosines in the substrate domain, likely through FAK-associated SRC.23,76 Multiple tyrosine-phosphorylated peptides derived from the substrate domain of CASS4 have been detected by mass spectrometry in cancers, with the great majority from chronic myelogenous leukemias and a smaller number from lung cancer samples (phosphosite.org) (Fig. 1). Even though CASS4 seems to have similar activities as the other CAS proteins, its carboxy-terminal domain has the most divergent sequence of the 4 family members, and it is not yet known whether it can support a tight interaction with NSP family members.13
The NSP Family
Domain organization and general signaling mechanisms
The NSP family consists of 3 members, NSP1, BCAR3, and NSP3 (see Table 1 for alternative names), which share a similar domain organization (Fig. 1). The amino-terminal segment of NSP proteins is of variable length and subject to alternative splicing in BCAR3 and NSP3.78 Only the longer version of this segment in NSP3 has been shown to promote association with the plasma membrane,79 suggesting that the amino-terminal segment confers distinctive signaling properties to the different isoforms. This region is followed by a SH2 domain that can bind to activated receptor tyrosine kinases, including members of the ERBB and EPH families,80-83 thus recruiting the associated CAS proteins and their signaling partners to the activated receptors (Fig. 2). The proline/serine-rich region that follows the SH2 domain contains putative binding sites for SH3 domain–containing proteins. This region is poorly conserved and may therefore recruit different binding partners to each NSP family member. Although no definite binding partners have yet been characterized, predictions suggest binding of various SH3 domain–containing proteins, including the CDC42 exchange factor intersectin to NSP1 and the ABL nonreceptor tyrosine kinase to NSP3 (scansite.mit.edu). Additionally, cortactin, a SRC substrate and actin cytoskeleton regulator involved in cell invasion,84 is predicted to interact with all 3 NSP proteins (scansite.mit.org).
Perhaps the most intriguing region of the NSP proteins is their large carboxy-terminal domain, which is crucial for interaction with members of the CAS family.81,85,86 This domain has sequence similarity with the CDC25 homology domain of guanine nucleotide exchange factors, which promote the exchange of the GDP nucleotide bound to inactive RAS GTPases with GTP, leading to RAS activation.80,81,85 Hence, nucleotide exchange functions have been proposed for the NSP family.87 However, enzymatic activity could not be confirmed using purified NSP CDC25 homology domains,29,80 which is in line with recent structural studies revealing an architecture incompatible with enzymatic activity but exquisitely suited for CAS binding13 (see below).
In essence, NSP proteins act as sophisticated adaptor molecules that use their SH2 and CDC25 homology domains to connect receptor tyrosine kinases and other cell surface receptors with CAS family members and integrin signaling cascades (Fig. 2). Yet, based on the number and diversity of potential signaling motifs present in the family members, it is likely that other binding partners, mechanisms, and pathways involving the 3 NSP proteins remain to be discovered. For example, little is known about the role of NSP phosphorylation, which represents an additional putative regulatory aspect for the NSP family. Multiple phosphorylation sites have been identified, mostly in the amino-terminal half of all 3 NSP proteins, in normal and cancer cells80,81,86,88-91 (phosposite.org) (Fig. 1). The tyrosine-phosphorylated motifs likely result from the activity of SRC, ABL, and receptor tyrosine kinases, and presumably some of them serve as binding sites for proteins containing SH2 or PTB domains. For example, sequence-based predictions suggest that the ABL kinase SH2 domain may bind to NSP3 (scansite.mit.edu). Furthermore, the NSP amino-terminal segments and proline/serine-rich regions contain multiple serine/threonine-proline (S/TP) motifs that are likely phosphorylated by proline-directed kinases such as ERK86,92 (scansite.mit.edu). Additionally, AMP kinase is predicted to phosphorylate several serines in NSP1 (scansite.mit .edu). A number of serine/threonine-phosphorylated motifs in all 3 NSP proteins are also predicted to bind 14-3-3 proteins.
Consistent with promoting CAS signaling and cancer malignancy, many studies have shown that NSP proteins can enhance SRC activity and CAS tyrosine and serine phosphorylation in a variety of cell culture and in vivo systems.21,79,89,93-99 The amino-terminal half of the NSP proteins, including the SH2 and proline/serine-rich domains, seems to be required to enhance CAS phosphorylation.21,94,96,98,99 CAS-dependent signaling involving CRK and PI3 kinase likely accounts for the ability of all 3 NSP proteins to activate RAC1 and the other RHO family GTPase, CDC42, as well as AKT93,100-103 (Fig. 2). Furthermore, overexpression of NSP1 and NSP3 has been shown to activate the RAC1 downstream effector JNK1.81,86,104
All 3 NSP family members can also increase CAS protein levels, which likely further potentiates CAS signaling.96,103 Although the underlying mechanism for NSP-dependent CAS stabilization is not known, an interesting possibility is that NSP binding might prevent the association with ubiquitin ligases that also interact with the CAS carboxy-terminal FAT domain,14,15 thereby reducing CAS proteolytic degradation. Overall, we are only beginning to understand the signaling pathways and functions of NSP proteins and their CAS complexes in cancer development and progression. Current information on each family member is outlined in the next sections.
NSP1 (SH2D3A)
NSP1 was first identified from a database of human-expressed sequence tags as a new SH2 domain–containing protein.81 NSP1 is the shortest member of the family due to its shorter amino-terminal and proline/serine-rich regions (Fig. 1) and appears to be a pseudogene in the mouse.78 Of the 3 family members, NSP1 has the most restricted expression, and in adult tissues, its transcripts are mainly found in the lung, pancreas, kidney, and liver.78,81 However, NSP1 is present at substantial levels in a variety of cancer cell lines, including pancreatic and lung cancer cells (broadinstitute.org/ccle), where it likely functions downstream of integrins and activated receptor tyrosine kinases including members of the EGF and insulin receptor families.81,82 For example, upon EGF stimulation of COS cells, NSP1 binds to the EGF receptor and becomes phosphorylated on 2 of its 3 tyrosines.81 Both tyrosine and serine/threonine phosphorylation sites have also been detected for NSP1 by mass spectrometry in human tumors and cancer cell lines, most frequently in lung cancers (phosphosite.org) (Fig. 1). This suggests a role for NSP1 in lung cancer, although the functional consequences of NSP1 phosphorylation are not yet known. Interestingly, upregulation of NSP1 expression has been reported as part of the gene expression signature of “KRAS-addicted” lung and pancreatic cancers, suggesting that NSP1 could contribute to KRAS transformation and serve as a biomarker for KRAS-dependent tumors.105 Furthermore, NSP1 overexpression in MCF7 breast cancer cells moderately increases BCAR1 tyrosine phosphorylation and the appearance of a serine-phosphorylated form of BCAR1 with reduced electrophoretic mobility.94 Although NSP1 overexpression in breast cancer cells also activates RAC1, CDC42, and AKT, these signaling activities do not seem sufficient to confer resistance to antiestrogens.103
BCAR3
The BCAR3 gene encodes at least 2 protein isoforms that differ in the amino-terminal segment preceding the SH2 domain,78 but only the longer isoform of 825 amino acids has been studied so far. BCAR3 mRNA is widely expressed in most human tissues78,106 and in cell lines from most types of cancers, with lowest levels in leukemias and lymphomas (broadinstitute.org/ccle) despite the expression of BCAR3 in normal B cells.100 In cell lines from breast, ovarian, and endometrial cancers, BCAR3 mRNA expression seems to be inversely correlated with estrogen receptor expression.103,106 In addition, BCAR3 tyrosine-phosphorylated peptides have been detected by mass spectrometry in many cancers, with a preponderance in lung cancer (phosphosite.org). The widespread expression and phosphorylation of BCAR3 in cancer cells suggest a role in many cancer types. Although the involvement of BCAR3 in lung cancer has not yet been investigated, a number of studies have characterized its role in breast cancer.
BCAR3 exerts a profound influence on various aspects of breast cancer malignancy. It was first identified in the same screen for proteins conferring antiestrogen resistance that also identified the CAS protein BCAR1.106 Later studies revealed that BCAR3-BCAR1 complexes are more evident in more aggressive and estrogen receptor– negative breast cancer cell lines, where BCAR1 signaling is known to promote survival, migration, and invasiveness.39,94,99,103,107 Multiple pathways involved in cell proliferation and survival could contribute to BCAR3-induced estrogen independence in breast cancer cells.108 The ability of BCAR3 to promote proliferation of estrogen-dependent breast cancer cells in the presence of antiestrogens requires both the SH2 and CDC25 homology domains, suggesting that this depends on its ability to link cell surface receptors with CAS signaling networks102 (Fig. 2). A critical event for the antiestrogen resistance phenotype is RAC1 activation, which occurs downstream of FAK–CAS–PI3 kinase as well as CAS-CRK-DOCK180 pathways, leading to increased cyclin D1 levels through the downstream kinase PAK1.101,102 AKT activation downstream of PI3 kinase may also contribute to BCAR3-dependent antiestrogen resistance.102,109
Another key role of BCAR3 likely contributing to estrogen-independent growth involves increasing the activity of SRC recruited to BCAR1, thus leading to not only phosphorylation of the BCAR1 substrate domain and subsequent CRK-RAC1 signaling21,37,89,99,101,102 but likely also other SRC-dependent activities.108,110-112 Interestingly, endogenous BCAR3 has also been found to mediate some of the proliferative effects of the EGF receptor in the nontransformed MCF12A human breast cell line, suggesting that a similar cross-talk with the EGF receptor might also play a role in breast cancer cell proliferation and antiestrogen resistance.83,107,110 Remarkably, although all 3 NSP family members can promote RAC1, CDC42, and AKT activation when overexpressed in breast cancer cells, only BCAR3 can increase cyclin D1 levels and promote robust antiestrogen resistance. BCAR3 is also the most effective of the NSP proteins in inducing BCAR1 and NEDD9 tyrosine and serine phosphorylation.94,98,99,107 Hence, BCAR3 appears to have stronger signaling abilities and/or additional distinctive signaling functions that promote breast cancer cell malignancy.99,103
Estrogen independence is typically accompanied by a pronounced mesenchymal and invasive breast cancer cell phenotype.111,113,114 Interestingly, besides enabling growth in the presence of antiestrogens, BCAR3 promotes mesenchymal attributes as well as cell spreading, migration, and invasiveness. These are also critical aspects of cancer malignancy that encompass the core features of NSP-CAS-CRK signaling (Fig. 2). For example, BCAR3 overexpression in the less aggressive, more epithelial-like MCF7 and T47D breast cancer cells decreases E-cadherin localization at cell-cell junctions with concomitant disruption of junctional integrity and increased fibronectin production.98,103,107 BCAR3 also promotes the formation of membrane ruffles and cell protrusion, which are hallmarks of increased RAC1 signaling and a migratory phenotype.98,103,107 Indeed, BCAR3 can cause BCAR1 relocalization from focal contacts to membrane ruffles and cell protrusions (where BCAR3 is also enriched), thus positioning BCAR1 for the promotion of cell migration and invasiveness.89,99,107 Conversely, siRNA-mediated downregulation of BCAR3 in the more aggressive MDA-MB-231 and BT549 breast cancer cells leads to a less malignant phenotype, including a decrease in SRC activation, BCAR1 tyrosine/serine phosphorylation, and BCAR1 association with CRK accompanied by a transition from a mesenchymal to a more epithelial-like morphology as well as defects in cell spreading, migration, and invasiveness.21,94,107
NSP3 (SH2D3C)
NSP3 was independently identified as an NSP1 family member,81 as a protein that interacts with the EPHB2 receptor tyrosine kinase,80 and as an antigen that localizes at cell-cell junctions.86 Its 2 shorter isoforms of 702 to 703 amino acids are widely expressed,78,81,86 whereas its longer isoform of 860 amino acids is prevalent in hematopoietic cells (Fig. 1). NSP3 is the only family member with a carboxy-terminal PDZ domain–binding motif, which mediates association with the adaptor protein CARD11/CARMA.115 CARD11 is a positive regulator of antigen receptor signaling in B and T cells and forms a signalosome with the NFκB activators BCL10 and MALT1, whose deregulation has been linked to lymphoma development.116
NSP3 mRNA is most highly expressed in the brain, lung, blood vessels, and immune cells.78,81,86 Its transcripts are also preferentially expressed in lymphoma and leukemia cell lines, and thus, NSP3 seems to have complementary expression with BCAR3 in cancer cell lines (broadinstitute.org/ccle). Increased NSP3 protein expression has also been reported in gastric cancers,117 and NSP3-phosphorylated peptides have been detected by mass spectrometry in leukemias, lymphomas, and lung cancers (phosphosite.org), suggesting an involvement in these malignancies. Indeed, NSP3 has been shown to be tyrosine phosphorylated downstream of the BCR-ABL oncogene and may function in concert with NEDD9 in BCR-ABL–transformed cells.16,88
Although the role of NSP3 in chronic myelogenous leukemia and other hematological cancers has not yet been investigated, its activities in B and T cells suggest potential functions in tumorigenesis. The long NSP3 isoform has been implicated in B-cell development and function.95,118 In fact, NSP3 knockout mice almost completely lack a population of B cells in the spleen known as marginal-zone B cells. Furthermore, the knockout B cells have defects in NEDD9 serine phosphorylation and B-cell migration in response to cytokines, although no major defects have been observed in NSP3 knockout T cells. Nevertheless, several observations point to a potential role of NSP3 in T-cell malignancies. One is the impairment of T-cell migration in culture and homing to peripheral tissues in vivo upon siRNA-mediated downregulation of NSP3.77,79 Interestingly, NSP3 membrane localization driven by the unique amino-terminal segment of the long isoform induces the NEDD9 serine phosphorylation, RAP1 activation, and integrin-mediated adhesion underlying T-cell migration. An additional, albeit controversial, factor is the increased production of interleukin-2 (a hallmark of T-cell activation) upon overexpression of the long NSP3 isoform in the Jurkat T-cell leukemia cell line.77,104,115 This effect of NSP3 on interleukin-2 was connected to JNK1 activation through NEDD9 and the FAK-related PYK2 kinase as well as association with the PDZ domain of CARD11.
Similar to the long isoform, a direct involvement of the shorter NSP3 isoforms in cancer malignancy has yet to be proven. However, NSP3 activities downstream of cell surface receptors may play a role in tumor angiogenesis as well as cell migration and invasiveness, which are key factors in cancer development and progression. A potential role in tumor angiogenesis is suggested by the expression and phosphorylation of NSP3 in cultured endothelial cells and in the vasculature of mouse mammary tumors and MDA-MB-231 breast cancer xenografts grown in nude mice78 (phosphosite.org). However, NSP3 knockout mice do not exhibit obvious vascular defects, raising the possibility of a redundancy with BCAR3 or of residual activities of a truncated NSP3 form present in at least 1 of the 2 reported NSP3 knockout mouse lines.78,96,97 NSP3 also interacts with the EPHA4 and EPHB2 receptor tyrosine kinases through its SH2 domain and can be phosphorylated on tyrosine residues downstream of EPHB2 and the EGF receptor.80,90 Additionally, EGF and NGF stimulation in PC12 pheochromocytoma cells and T-cell receptor stimulation in primary mouse T cells cause an upward electrophoretic mobility shift in NSP3, whose appearance is prevented by the ERK pathway inhibitor PD98059, consistent with the presence of multiple potential ERK consensus phosphorylation sites.86,92
Furthermore, NSP3 localizes to membrane ruffles of COS cells stimulated with EGF and promotes cell migration towards EGF.13,86,90 This ability is dependent on the interaction with BCAR1, further exemplifying the importance of NSP-CAS modules in cell migration. NSP3 can also increase RAP1 activity, integrin-mediated cell adhesion and spreading, and membrane ruffling in NIH3T3 cells through CAS-CRK-C3G.92 Interestingly, more prominent effects were observed using engineered myristoylated forms of the protein, emphasizing the importance of membrane targeting, which also leads to SRC-dependent NSP3 phosphorylation.90,92 Finally, NSP3 knockout mice show defects in the olfactory axons, which in vivo are unable to grow through the basal lamina surrounding the brain (as needed to form synapses in the olfactory bulb) and in explant culture have an impaired ability to penetrate the extracellular matrix.97 These findings highlight a proinvasive role of NSP3 in vivo, suggesting a similar role in cancer cell invasiveness.
Structural Features of NSP-CAS Complexes
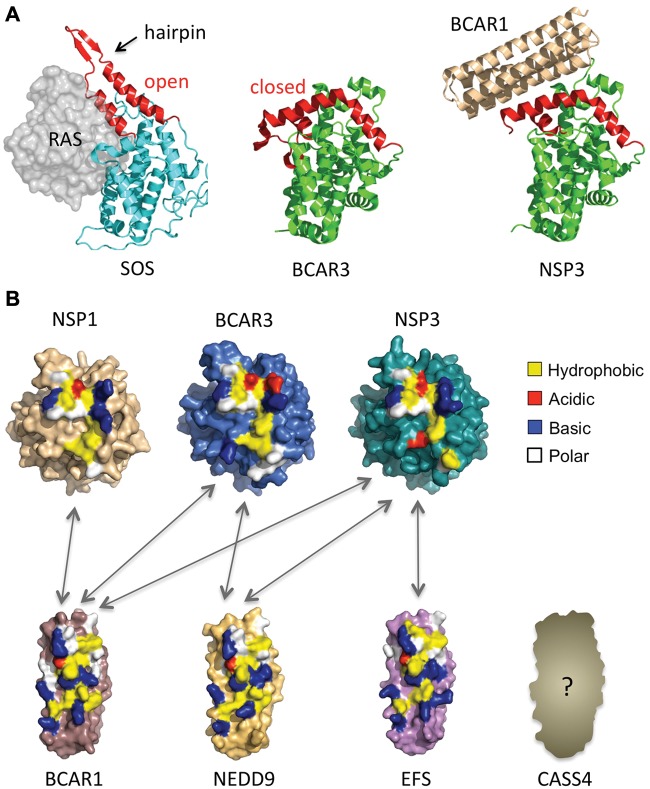
A recent study has provided surprising insight into the structure and function of the NSP carboxy-terminal domain. As mentioned above, in all NSP family members, this domain has a sequence similarity with the CDC25 homology fold found in nucleotide exchange factors for RAS family GTPases.13 The crystal structure of the BCAR3 carboxy-terminal domain has indeed confirmed that a large portion of this domain closely resembles a prototypical CDC25 homology domain13 (Fig. 3A). However, a segment termed the “helical hairpin,” which in enzymatically active exchange factors inserts into the RAS GTPase nucleotide-binding cleft to elicit GDP displacement,119 is grossly distorted in BCAR3 (Fig. 3A). This region folds towards the body of the BCAR3 domain, thus blocking the canonical RAS GTPase-binding site. This results in a novel “closed” form of the CDC25 homology domain. A second structure of the carboxy-terminal domain of NSP3 in complex with BCAR1 shows that NSP3 also shares the closed conformation of the helical hairpin (Fig. 3A). Importantly, the remodeled hairpin forms the majority of the binding site for BCAR1, with further contributions from a sequence insertion that is only found in the CDC25 homology domains of the NSP family. Overall, these findings and the conservation of the NSP- and CAS-interacting domains (Fig. 3B) suggest that NSP proteins have repurposed an enzymatic nucleotide exchange factor domain to serve as an adaptor domain that is highly specific for the recruitment of CAS family members. In addition, the conservation of residues within the binding interfaces of different NSP and CAS proteins suggests that multiple combinations of NSP-CAS pairs should be able to form tight signaling associations. Indeed, multiple NSP-CAS combinations have already been found to function in different signaling contexts77,81,85,86 (Fig. 3B). Therefore, the carboxy-termini of all members of the 2 families (with the possible exception of the less conserved CASS4) enable a highly promiscuous interaction system, creating a spectrum of NSP-CAS complexes that combine the distinctive signaling features of each binding partner to differentially modulate cellular processes and cancer malignancy.
Figure 3.
Conserved interaction domains of NSP and CAS family members enable promiscuous interactions. (A) The NSP CDC25 homology domain is in a “closed” conformation that cannot bind RAS proteins but can tightly interact with CAS proteins. (Left) Structure of the CDC25 homology domain of the active exchange factor SOS bound to its target GTPase, RAS.119 (Middle) The structure of the carboxy-terminal domain of BCAR3 reveals that it also adopts a CDC25 homology domain fold, but in a new closed conformation incapable of RAS binding and exchange factor activity. (Right) The structure of the NSP3-BCAR1 complex reveals that the carboxy-terminal domain of NSP3 also adopts the closed conformation observed for BCAR3. The altered conformation enables strong interaction with the carboxy-terminal FAT domain of the CAS family protein BCAR1 to form an NSP-CAS complex, thus converting the exchange factor domain into an adaptor domain. (B) Surface representations of BCAR1, BCAR3, and NSP3 structures solved by X-ray crystallography,13 and models of NSP1, NEDD9, and EFS. The carboxy-terminus of CASS4 was not modeled due to its greater degree of sequence divergency from the structurally characterized BCAR1. Colors indicate interface residue types identified in the NSP3-BCAR1 complex and the corresponding residues for the other family members. The color coding of interface residues highlights the similarities in the binding interfaces of NSP or CAS family members and thus the potential for promiscuous interactions between the 2 families (with the possible exception of the less conserved CASS4). Arrows indicate experimentally confirmed associations between NSP and CAS family members.
Concluding Remarks
The strong interaction between NSP and CAS family proteins suggests that the 2 families are able to form stable complexes in which NSP proteins are critical modulators of CAS function. Studies with BCAR1 and NEDD9 have demonstrated the paramount importance of NSP-CAS downstream signaling pathways in normal physiology and cancer malignancy, whereas the mechanisms of upstream NSP-CAS regulation are less well understood.
CAS and NSP family proteins can both be phosphorylated on tyrosine and serine/threonine residues in response to cell substrate adhesion as well as cell stimulation with growth factors and cytokines. NSP proteins can potentiate CAS phosphorylation in normal and cancer cells through mechanisms that likely involve both anchoring CAS proteins to receptor tyrosine kinases and recruiting them near plasma membrane–localized SRC. This highlights their dual role as both adaptor and scaffolding proteins that can act in a positive feedback loop to further increase cell adhesion (Fig. 2). The fact that BCAR3, the long NSP3 isoform, and myristoylated NSP3 more readily induce CAS tyrosine and serine phosphorylation as compared to NSP1 or shorter NSP3 isoforms suggests that membrane targeting through the longer amino-terminal segment could act in concert with NSP SH2 domain interactions with cell surface receptors to more effectively promote CAS signaling. It will also be interesting to investigate if NSP proteins bound to the CAS carboxy-terminal FAT domain function together with proteins bound to the CAS SH3 domain as part of the mechanosensory machinery that stretches the central CAS substrate domain, leading to its phosphorylation by SRC.4,5 The mechanisms and kinases responsible for NSP-induced CAS family serine phosphorylation, and whether this phosphorylation may be a consequence of increased substrate adhesion, also remain to be elucidated.
The functional effects of NSP phosphorylation are also unknown, except for phosphorylation of a tyrosine in the NSP3 CDC25 homology domain, which has been reported to prevent interaction with BCAR1.90 A special connection between NSP3 and the ABL nonreceptor tyrosine kinase is suggested by the observed NSP3 phosphorylation downstream of the BCR-ABL oncogene88 (phosphosite.org) and by the predicted binding of the ABL SH2 and SH3 domains to NSP3 but also remains to be explored.
Further characterizing the NSP-CAS association will also help to understand the physiological and pathological activities of the various family members. For example, it will be important to know whether the binding affinities (which are below 30 nM for the BCAR3-BCAR1 and NSP3-BCAR1 complexes13) differ depending on the NSP-CAS pair and if they can be physiologically regulated, for example, by posttranslational modifications.90 It will also be useful to determine the proportion of each protein found in NSP-CAS complexes in the cellular environment and clarify the subcellular localization of the complexes. Structure-based mutations designed to affect NSP-CAS interaction without causing overall conformational changes will be instrumental for these investigations.13 These types of studies have confirmed the importance of NSP-CAS complexes in processes such as the induction of CAS tyrosine phosphorylation and regulation of cell morphology and migration.13,90,98,99 Additionally, the effects of BCAR3 carboxy-terminal deletions support the importance of the CDC25 homology domain and the NSP-CAS association in antiestrogen resistance, SRC activation, CAS relocalization to membrane ruffles, and cell migration.89,101
According to some recent reports, however, substantial impairment of NSP-CAS association by single amino acid changes does not seem to impact NSP protein–dependent SRC and RAC1 activation and antiestrogen resistance induced by BCAR3 overexpression.89,98 Further work will be needed to resolve these discrepancies. Furthermore, deletion of the CDC25 homology domain has demonstrated that at least some BCAR1 serine phosphorylation events do not seem to require interaction with NSP proteins.94 On the other hand, gene transcription analyses and pathway activation mapping have revealed that overexpression of BCAR3 or BCAR1 activates signaling networks that have not only common elements but also distinctive features.109,120 Taken together, these findings highlight the complexities in the activities of NSP and CAS proteins and their modules, which remain to be further unraveled in future studies.
In summary, the role of NSP and CAS proteins in cancer appears to depend on their protein abundance and phosphorylation state and possibly on the particular family member or isoform expressed. Although BCAR1, BCAR3, and SRC mRNA levels may not show a positive correlation with aggressiveness or poor responsiveness to tamoxifen therapy,121 BCAR1 protein levels in breast cancer samples correlate with prognosis and could be useful to decide on treatment options.53,54 Screening for BCAR1 phosphorylation levels could be even more powerful.55,56 On the other hand, only a few mutations have been identified so far in each of the 3 NSP genes as well as in BCAR1 and NEDD9 in different types of cancers (sanger.ac.uk/cosmic), and no information is yet available on the functional effects of the mutations or their relevance to cancer development and malignancy. With regard to therapy, siRNA-based strategies will allow the reduction of NSP-CAS complexes and their malignant activities. Furthermore, better understanding of NSP-CAS signaling mechanisms will enable effective targeting of enzymatic activities associated with the complex or disruption of interactions with upstream regulators and downstream effectors. Developing agents that can disrupt NSP-CAS complexes will be more challenging due to the large interface and strong binding between the 2 classes of molecules.
An article published after submission of this review122 reports that BCAR3 recruits BCAR1 to integrin-mediated focal adhesions by binding through its SH2 domain to a tyrosine phosphorylated sequence of the receptor-like protein tyrosine phosphatase a (PTPa). This leads to BCAR1 phosphorylation by SRC and downstream signals that promote cell migration
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received the following financial support for the research, authorship, and/or publication of this article: National Institutes of Health grants R01CA160457 and P01CA138390 to S.J.R. and E.B.P.; R01CA116099, R21NS067502, and P01HD025938 to E.B.P., and Department of Defense Breast Cancer Research Program postdoctoral fellowship BC100466 to P.D.M.
References
- 1. Pawson T. Dynamic control of signaling by modular adaptor proteins. Curr Opin Cell Biol. 2007;19:112-6 [DOI] [PubMed] [Google Scholar]
- 2. Scott JD, Pawson T. Cell signaling in space and time: where proteins come together and when they’re apart. Science. 2009;326:1220-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Harte MT, Hildebrand JD, Burnham MR, Bouton AH, Parsons JT. p130Cas, a substrate associated with v-Src and v-Crk, localizes to focal adhesions and binds to focal adhesion kinase. J Biol Chem. 1996;271:13649-55 [DOI] [PubMed] [Google Scholar]
- 4. Sawada Y, Tamada M, Dubin-Thaler BJ, et al. Force sensing by mechanical extension of the Src family kinase substrate p130Cas. Cell. 2006;127:1015-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tamada M, Sheetz MP, Sawada Y. Activation of a signaling cascade by cytoskeleton stretch. Dev Cell. 2004;7:709-18 [DOI] [PubMed] [Google Scholar]
- 6. Klemke RL, Leng J, Molander R, Brooks PC, Vuori K, Cheresh DA. CAS/Crk coupling serves as a “molecular switch” for induction of cell migration. J Cell Biol. 1998;140:961-72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Feller SM. Crk family adaptors-signalling complex formation and biological roles. Oncogene. 2001;20:6348-71 [DOI] [PubMed] [Google Scholar]
- 8. Chodniewicz D, Klemke RL. Regulation of integrin-mediated cellular responses through assembly of a CAS/Crk scaffold. Biochim Biophys Acta. 2004;1692:63-76 [DOI] [PubMed] [Google Scholar]
- 9. Garcia-Guzman M, Dolfi F, Russello M, Vuori K. Cell adhesion regulates the interaction between the docking protein p130(Cas) and the 14-3-3 proteins. J Biol Chem. 1999;274: 5762-8 [DOI] [PubMed] [Google Scholar]
- 10. Briknarova K, Nasertorabi F, Havert ML, et al. The serine-rich domain from Crk-associated substrate (p130cas) is a four-helix bundle. J Biol Chem. 2005;280:21908-14 [DOI] [PubMed] [Google Scholar]
- 11. Cabodi S, del Pilar Camacho-Leal M, Di Stefano P, Defilippi P. Integrin signalling adaptors: not only figurants in the cancer story. Nat Rev Cancer. 2010;10:858-70 [DOI] [PubMed] [Google Scholar]
- 12. Tikhmyanova N, Little JL, Golemis EA. CAS proteins in normal and pathological cell growth control. Cell Mol Life Sci. 2010;67:1025-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mace PD, Wallez Y, Dobaczewska MK, et al. NSP-Cas protein structures reveal a promiscuous interaction module in cell signaling. Nat Struct Mol Biol. 2011;18:1381-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Feng L, Guedes S, Wang T. Atrophin-1-interacting protein 4/human Itch is a ubiquitin E3 ligase for human enhancer of filamentation 1 in transforming growth factor-beta signaling pathways. J Biol Chem. 2004;279:29681-90 [DOI] [PubMed] [Google Scholar]
- 15. Nourry C, Maksumova L, Pang M, Liu X, Wang T. Direct interaction between Smad3, APC10, CDH1 and HEF1 in proteasomal degradation of HEF1. BMC Cell Biol. 2004;5:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Singh M, Cowell L, Seo S, O’Neill G, Golemis E. Molecular basis for HEF1/NEDD9/Cas-L action as a multifunctional co-ordinator of invasion, apoptosis and cell cycle. Cell Biochem Biophys. 2007;48:54-72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. O’Neill GM, Seo S, Serebriiskii IG, Lessin SR, Golemis EA. A new central scaffold for metastasis: parsing HEF1/Cas-L/NEDD9. Cancer Res. 2007;67:8975-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Alexandropoulos K, Baltimore D. Coordinate activation of c-Src by SH3- and SH2-binding sites on a novel p130Cas-related protein, Sin. Genes Dev. 1996;10:1341-55 [DOI] [PubMed] [Google Scholar]
- 19. Burnham MR, Bruce-Staskal PJ, Harte MT, et al. Regulation of c-SRC activity and function by the adapter protein CAS. Mol Cell Biol. 2000;20:5865-78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nasertorabi F, Tars K, Becherer K, et al. Molecular basis for regulation of Src by the docking protein p130Cas. J Mol Recognit. 2006;19: 30-8 [DOI] [PubMed] [Google Scholar]
- 21. Schuh NR, Guerrero MS, Schrecengost RS, Bouton AH. BCAR3 regulates Src/p130 Cas association, Src kinase activity, and breast cancer adhesion signaling. J Biol Chem. 2010;285: 2309-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hamasaki K, Mimura T, Morino N, et al. Src kinase plays an essential role in integrin-mediated tyrosine phosphorylation of Crk-associated substrate p130Cas. Biochem Biophys Res Commun. 1996;222:338-43 [DOI] [PubMed] [Google Scholar]
- 23. Ruest PJ, Shin NY, Polte TR, Zhang X, Hanks SK. Mechanisms of CAS substrate domain tyrosine phosphorylation by FAK and Src. Mol Cell Biol. 2001;21:7641-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vuori K, Hirai H, Aizawa S, Ruoslahti E. Introduction of p130cas signaling complex formation upon integrin-mediated cell adhesion: a role for Src family kinases. Mol Cell Biol. 1996;16:2606-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kiyokawa E, Hashimoto Y, Kurata T, Sugimura H, Matsuda M. Evidence that DOCK180 up-regulates signals from the CrkII-p130(Cas) complex. J Biol Chem. 1998;273:24479-84 [DOI] [PubMed] [Google Scholar]
- 26. Ohashi Y, Tachibana K, Kamiguchi K, Fujita H, Morimoto C. T cell receptor-mediated tyrosine phosphorylation of Cas-L, a 105-kDa Crk-associated substrate-related protein, and its association of Crk and C3G. J Biol Chem. 1998;273: 6446-51 [DOI] [PubMed] [Google Scholar]
- 27. Guo W, Giancotti FG. Integrin signalling during tumour progression. Nat Rev Mol Cell Biol. 2004;5:816-26 [DOI] [PubMed] [Google Scholar]
- 28. Kim C, Ye F, Ginsberg MH. Regulation of integrin activation. Annu Rev Cell Dev Biol. 2011;27:321-45 [DOI] [PubMed] [Google Scholar]
- 29. Bos JL, de Rooij J, Reedquist KA. Rap1 signalling: adhering to new models. Nat Rev Mol Cell Biol. 2001;2:369-77 [DOI] [PubMed] [Google Scholar]
- 30. Dolfi F, Garcia-Guzman M, Ojaniemi M, Nakamura H, Matsuda M, Vuori K. The adaptor protein Crk connects multiple cellular stimuli to the JNK signaling pathway. Proc Natl Acad Sci U S A. 1998;95:15394-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Radha V, Mitra A, Dayma K, Sasikumar K. Signalling to actin: role of C3G, a multitasking guanine-nucleotide-exchange factor. Biosci Rep. 2011;31:231-44 [DOI] [PubMed] [Google Scholar]
- 32. Kiyokawa E, Hashimoto Y, Kobayashi S, Sugimura H, Kurata T, Matsuda M. Activation of Rac1 by a Crk SH3-binding protein, DOCK180. Genes Dev. 1998;12:3331-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cote JF, Vuori K. Identification of an evolutionarily conserved superfamily of DOCK180-related proteins with guanine nucleotide exchange activity. J Cell Sci. 2002;115:4901-13 [DOI] [PubMed] [Google Scholar]
- 34. Huang C, Rajfur Z, Borchers C, Schaller MD, Jacobson K. JNK phosphorylates paxillin and regulates cell migration. Nature. 2003;424: 219-23 [DOI] [PubMed] [Google Scholar]
- 35. Shin NY, Dise RS, Schneider-Mergener J, Ritchie MD, Kilkenny DM, Hanks SK. Subsets of the major tyrosine phosphorylation sites in Crk-associated substrate (CAS) are sufficient to promote cell migration. J Biol Chem. 2004;279:38331-7 [DOI] [PubMed] [Google Scholar]
- 36. Bos JL. Linking Rap to cell adhesion. Curr Opin Cell Biol. 2005;17:123-8 [DOI] [PubMed] [Google Scholar]
- 37. Cabodi S, Tinnirello A, Di Stefano P, et al. p130Cas as a new regulator of mammary epithelial cell proliferation, survival, and HER2-neu oncogene-dependent breast tumorigenesis. Cancer Res. 2006;66:4672-80 [DOI] [PubMed] [Google Scholar]
- 38. Almeida EA, Ilic D, Han Q, et al. Matrix survival signaling: from fibronectin via focal adhesion kinase to c-Jun NH(2)-terminal kinase. J Cell Biol. 2000;149:741-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cunningham-Edmondson AC, Hanks SK. p130Cas substrate domain signaling promotes migration, invasion, and survival of estrogen receptor-negative breast cancer cells. Breast Cancer. 2009;2009:39-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Reynolds AB, Kanner SB, Wang HC, Parsons JT. Stable association of activated pp60src with two tyrosine-phosphorylated cellular proteins. Mol Cell Biol. 1989;9:3951-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kanner SB, Reynolds AB, Wang HC, Vines RR, Parsons JT. The SH2 and SH3 domains of pp60src direct stable association with tyrosine phosphorylated proteins p130 and p110. EMBO J. 1991;10:1689-98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Matsuda M, Mayer BJ, Fukui Y, Hanafusa H. Binding of transforming protein, P47gag-crk, to a broad range of phosphotyrosine-containing proteins. Science. 1990;248:1537-9 [DOI] [PubMed] [Google Scholar]
- 43. Honda H, Oda H, Nakamoto T, et al. Cardiovascular anomaly, impaired actin bundling and resistance to Src-induced transformation in mice lacking p130Cas [see comments]. Nat Genet. 1998;19:361-5 [DOI] [PubMed] [Google Scholar]
- 44. Brabek J, Constancio SS, Siesser PF, Shin NY, Pozzi A, Hanks SK. Crk-associated substrate tyrosine phosphorylation sites are critical for invasion and metastasis of SRC-transformed cells. Mol Cancer Res. 2005;3:307-15 [DOI] [PubMed] [Google Scholar]
- 45. Cabodi S, Tinnirello A, Bisaro B, et al. p130Cas is an essential transducer element in ErbB2 transformation. FASEB J. 2010;24:3796-808 [DOI] [PubMed] [Google Scholar]
- 46. Ambrogio C, Voena C, Manazza AD, et al. p130Cas mediates the transforming properties of the anaplastic lymphoma kinase. Blood. 2005;106:3907-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pylayeva Y, Gillen KM, Gerald W, Beggs HE, Reichardt LF, Giancotti FG. Ras- and PI3K-dependent breast tumorigenesis in mice and humans requires focal adhesion kinase signaling. J Clin Invest. 2009;119:252-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wendt MK, Smith JA, Schiemann WP. p130Cas is required for mammary tumor growth and transforming growth factor-beta-mediated metastasis through regulation of Smad2/3 activity. J Biol Chem. 2009;284:34145-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dorssers LC, van Agthoven T, Dekker A, van Agthoven TL, Kok EM. Induction of antiestrogen resistance in human breast cancer cells by random insertional mutagenesis using defective retroviruses: identification of bcar-1, a common integration site. Mol Endocrinol. 1993;7: 870-8 [DOI] [PubMed] [Google Scholar]
- 50. Brinkman A, van der Flier S, Kok EM, Dorssers LC. BCAR1, a human homologue of the adapter protein p130Cas, and antiestrogen resistance in breast cancer cells. J Natl Cancer Inst. 2000;92:112-20 [DOI] [PubMed] [Google Scholar]
- 51. van Agthoven T, Veldscholte J, Smid M, et al. , Functional identification of genes causing estrogen independence of human breast cancer cells. Breast Cancer Res Treat. 2009;114:23-30 [DOI] [PubMed] [Google Scholar]
- 52. Ta HQ, Thomas KS, Schrecengost RS, Bouton AH. A novel association between p130Cas and resistance to the chemotherapeutic drug adriamycin in human breast cancer cells. Cancer Res. 2008;68:8796-804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. van der Flier S, Brinkman A, Look MP, et al. , Bcar1/p130Cas protein and primary breast cancer: prognosis and response to tamoxifen treatment. J Natl Cancer Inst. 2000;92:120-7 [DOI] [PubMed] [Google Scholar]
- 54. Dorssers LC, Grebenchtchikov N, Brinkman A, et al. The prognostic value of BCAR1 in patients with primary breast cancer. Clin Cancer Res. 2004;10:6194-202 [DOI] [PubMed] [Google Scholar]
- 55. Brinkman A, de Jong D, Tuinman S, Azaouagh N, van Agthoven T, Dorssers LC. The substrate domain of BCAR1 is essential for anti-estrogen-resistant proliferation of human breast cancer cells. Breast Cancer Res Treat. 2010;120:401-8 [DOI] [PubMed] [Google Scholar]
- 56. Soni S, Lin BT, August A, Nicholson RI, Kirsch KH. Expression of a phosphorylated p130(Cas) substrate domain attenuates the phosphatidylinositol 3-kinase/Akt survival pathway in tamoxifen resistant breast cancer cells. J Cell Biochem. 2009;107:364-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Salgia R, Pisick E, Sattler M, et al. p130CAS forms a signaling complex with the adapter protein CRKL in hematopoietic cells transformed by the BCR/ABL oncogene. J Biol Chem. 1996;271:25198-203 [DOI] [PubMed] [Google Scholar]
- 58. Eisenmann KM, McCarthy JB, Simpson MA, et al. Melanoma chondroitin sulphate proteoglycan regulates cell spreading through Cdc42, Ack-1 and p130cas. Nat Cell Biol. 1999;1:507-13 [DOI] [PubMed] [Google Scholar]
- 59. Frankel P, Pellet-Many C, Lehtolainen P, et al. Chondroitin sulphate-modified neuropilin 1 is expressed in human tumour cells and modulates 3D invasion in the U87MG human glioblastoma cell line through a p130Cas-mediated pathway. EMBO Rep. 2008;9:983-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hamamura K, Furukawa K, Hayashi T, et al. Ganglioside GD3 promotes cell growth and invasion through p130Cas and paxillin in malignant melanoma cells. Proc Natl Acad Sci U S A. 2005;102:11041-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Feng H, Hu B, Liu KW, et al. Activation of Rac1 by Src-dependent phosphorylation of Dock180(Y1811) mediates PDGFRalpha-stimulated glioma tumorigenesis in mice and humans. J Clin Invest. 2011;121:4670-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Nick AM, Stone RL, Armaiz-Pena G, et al. Silencing of p130cas in ovarian carcinoma: a novel mechanism for tumor cell death. J Natl Cancer Inst. 2011;103:1596-612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Fromont G, Cussenot O. The integrin signalling adaptor p130CAS is also a key player in prostate cancer. Nat Rev Cancer. 2011;11:227. [DOI] [PubMed] [Google Scholar]
- 64. Kim M, Gans JD, Nogueira C, et al. Comparative oncogenomics identifies NEDD9 as a melanoma metastasis gene. Cell. 2006;125:1269-81 [DOI] [PubMed] [Google Scholar]
- 65. Seo S, Ichikawa M, Kurokawa M. Structure and function of cas-L and integrin-mediated signaling. Crit Rev Immunol. 2006;26:391-406 [DOI] [PubMed] [Google Scholar]
- 66. Ahn J, Sanz-Moreno V, Marshall CJ. The metastasis gene NEDD9 product acts through integrin beta3 and Src to promote mesenchymal motility and inhibit amoeboid motility. J Cell Sci. 2012;125:1814-26 [DOI] [PubMed] [Google Scholar]
- 67. Natarajan M, Stewart JE, Golemis EA, et al. HEF1 is a necessary and specific downstream effector of FAK that promotes the migration of glioblastoma cells. Oncogene. 2006;25:1721-32 [DOI] [PubMed] [Google Scholar]
- 68. Ji H, Ramsey MR, Hayes DN, et al. LKB1 modulates lung cancer differentiation and metastasis. Nature. 2007;448:807-10 [DOI] [PubMed] [Google Scholar]
- 69. Giampieri S, Manning C, Hooper S, Jones L, Hill CS, Sahai E. Localized and reversible TGFbeta signalling switches breast cancer cells from cohesive to single cell motility. Nat Cell Biol. 2009;11:1287-96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Sanz-Moreno V, Gadea G, Ahn J, et al. Rac activation and inactivation control plasticity of tumor cell movement. Cell. 2008;135:510-23 [DOI] [PubMed] [Google Scholar]
- 71. Lucas JT, Jr., Salimath BP, Slomiany MG, Rosenzweig SA. Regulation of invasive behavior by vascular endothelial growth factor is HEF1-dependent. Oncogene. 2010;29:4449-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kim SH, Xia D, Kim SW, Holla V, Menter DG, Dubois RN. Human enhancer of filamentation 1 Is a mediator of hypoxia-inducible factor-1alpha-mediated migration in colorectal carcinoma cells. Cancer Res. 2010;70:4054-63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Yang WH, Lan HY, Huang CH, et al. RAC1 activation mediates Twist1-induced cancer cell migration. Nat Cell Biol. 2012;14:366-74 [DOI] [PubMed] [Google Scholar]
- 74. Izumchenko E, Singh MK, Plotnikova OV, et al. NEDD9 promotes oncogenic signaling in mammary tumor development. Cancer Res. 2009;69:7198-206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Singh MK, Izumchenko E, Klein-Szanto AJ, Egleston BL, Wolfson M, Golemis EA. Enhanced genetic instability and dasatinib sensitivity in mammary tumor cells lacking NEDD9. Cancer Res. 2010;70:8907-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Singh MK, Dadke D, Nicolas E, et al. A novel Cas family member, HEPL, regulates FAK and cell spreading. Mol Biol Cell. 2008;19:1627-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Alexandropoulos K, Regelmann AG. Regulation of T-lymphocyte physiology by the Chat-H/CasL adapter complex. Immunol Rev. 2009;232:160-74 [DOI] [PubMed] [Google Scholar]
- 78. Vervoort VS, Roselli S, Oshima RG, Pasquale EB. Splice variants and expression patterns of SHEP1, BCAR3 and NSP1, a gene family involved in integrin and receptor tyrosine kinase signaling. Gene. 2007;391:161-70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Regelmann AG, Danzl NM, Wanjalla C, Alexandropoulos K. The hematopoietic isoform of Cas-Hef1-associated signal transducer regulates chemokine-induced inside-out signaling and T cell trafficking. Immunity. 2006;25:907-18 [DOI] [PubMed] [Google Scholar]
- 80. Dodelet VC, Pazzagli C, Zisch AH, Hauser CA, Pasquale EB. A novel signaling intermediate, SHEP1, directly couples Eph receptors to R-Ras and Rap1A. J Biol Chem. 1999;274:31941-6 [DOI] [PubMed] [Google Scholar]
- 81. Lu Y, Brush J, Stewart TA. NSP1 defines a novel family of adaptor proteins linking integrin and tyrosine kinase receptors to the c-Jun N-terminal kinase/stress-activated protein kinase signaling pathway. J Biol Chem. 1999;274:10047-52 [DOI] [PubMed] [Google Scholar]
- 82. Jones RB, Gordus A, Krall JA, MacBeath G. A quantitative protein interaction network for the ErbB receptors using protein microarrays. Nature. 2006;439:168-74 [DOI] [PubMed] [Google Scholar]
- 83. Oh MJ, van Agthoven T, Choi JE, et al. BCAR3 regulates EGF-induced DNA synthesis in normal human breast MCF-12A cells. Biochem Biophys Res Commun. 2008;375:430-4 [DOI] [PubMed] [Google Scholar]
- 84. Murphy DA, Courtneidge SA. The ‘ins’ and ‘outs’ of podosomes and invadopodia: characteristics, formation and function. Nat Rev Mol Cell Biol. 2011;12:413-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Cai D, Clayton LK, Smolyar A, Lerner A. AND-34, a novel p130Cas-binding thymic stromal cell protein regulated by adhesion and inflammatory cytokines. J Immunol. 1999;163:2104-12 [PubMed] [Google Scholar]
- 86. Sakakibara A, Hattori S. Chat, a Cas/HEF1-associated adaptor protein that integrates multiple signaling pathways. J Biol Chem. 2000;275:6404-10 [DOI] [PubMed] [Google Scholar]
- 87. Gotoh T, Cai D, Tian X, Feig LA, Lerner A. p130Cas regulates the activity of AND-34, a novel Ral, Rap1, and R-Ras guanine nucleotide exchange factor. J Biol Chem. 2000;275:30118-23 [DOI] [PubMed] [Google Scholar]
- 88. Salomon AR, Ficarro SB, Brill LM, et al. Profiling of tyrosine phosphorylation pathways in human cells using mass spectrometry. Proc Natl Acad Sci U S A. 2003;100:443-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Riggins RB, Quilliam LA, Bouton AH. Synergistic promotion of c-Src activation and cell migration by Cas and AND-34/BCAR3. J Biol Chem. 2003;278:28264-73 [DOI] [PubMed] [Google Scholar]
- 90. Dail M, Kalo MS, Seddon JA, Cote JF, Vuori K, Pasquale EB. SHEP1 function in cell migration is impaired by a single amino acid mutation that disrupts association with the scaffolding protein cas but not with Ras GTPases. J Biol Chem. 2004;279:41892-902 [DOI] [PubMed] [Google Scholar]
- 91. Heckel T, Czupalla C, Expirto Santo AI, et al. Src-dependent repression of ARF6 is required to maintain podosome-rich sealing zones in bone-digesting osteoclasts. Proc Natl Acad Sci U S A. 2009;106:1451-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Sakakibara A, Ohba Y, Kurokawa K, Matsuda M, Hattori S. Novel function of Chat in controlling cell adhesion via Cas-Crk-C3G-pathway- mediated Rap1 activation. J Cell Sci. 2002;115: 4915-24 [DOI] [PubMed] [Google Scholar]
- 93. Near RI, Smith RS, Toselli PA, et al. Loss of AND-34/BCAR3 expression in mice results in rupture of the adult lens. Mol Vis. 2009;15: 685-99 [PMC free article] [PubMed] [Google Scholar]
- 94. Makkinje A, Near RI, Infusini G, et al. AND-34/BCAR3 regulates adhesion-dependent p130Cas serine phosphorylation and breast cancer cell growth pattern. Cell Signal. 2009;21:1423-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Browne CD, Hoefer MM, Chintalapati SK, et al. SHEP1 partners with CasL to promote marginal zone B-cell maturation. Proc Natl Acad Sci U S A. 2010;107:18944-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Roselli S, Wallez Y, Wang L, Vervoort V, Pasquale EB. The SH2 domain protein Shep1 regulates the in vivo signaling function of the scaffolding protein Cas. Cell Signal. 2010;22:1745-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Wang L, Vervoort V, Wallez Y, Core N, Cremer H, Pasquale EB. The SRC homology 2 domain protein shep1 plays an important role in the penetration of olfactory sensory axons into the forebrain. J Neurosci. 2010;30:13201-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Vanden Borre P, Near RI, Makkinje A, Mostoslavsky G, Lerner A. BCAR3/AND-34 can signal independent of complex formation with CAS family members or the presence of p130Cas. Cell Signal. 2011;23:1030-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Makkinje A, Vanden Borre P, Near RI, Patel PS, Lerner A. Breast cancer anti-estrogen resistance 3 (BCAR3) augments binding of the c-Src kinase SH3 domain to Crk-associated substrate (p130Cas). J Biol Chem. 2012;287:27703-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Cai D, Felekkis KN, Near RI, et al. The GDP exchange factor AND-34 is expressed in B cells, associates with HEF1, and activates Cdc42. J Immunol. 2003;170:969-78 [DOI] [PubMed] [Google Scholar]
- 101. Cai D, Iyer A, Felekkis KN, et al. AND-34/BCAR3, a GDP exchange factor whose overexpression confers antiestrogen resistance, activates Rac, PAK1, and the cyclin D1 promoter. Cancer Res. 2003;63:6802-8 [PubMed] [Google Scholar]
- 102. Felekkis KN, Narsimhan RP, Near R, et al. AND-34 activates phosphatidylinositol 3-kinase and induces anti-estrogen resistance in a SH2 and GDP exchange factor-like domain-dependent manner. Mol Cancer Res. 2005;3:32-41 [PubMed] [Google Scholar]
- 103. Near RI, Zhang Y, Makkinje A, Vanden Borre P, Lerner A. AND-34/BCAR3 differs from other NSP homologs in induction of anti-estrogen resistance, cyclin D1 promoter activation and altered breast cancer cell morphology. J Cell Physiol. 2007;212:655-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Sakakibara A, Hattori S, Nakamura S, Katagiri T. A novel hematopoietic adaptor protein, Chat-H, positively regulates T cell receptor-mediated interleukin-2 production by Jurkat cells. J Biol Chem. 2003;278:6012-7 [DOI] [PubMed] [Google Scholar]
- 105. Singh A, Greninger P, Rhodes D, et al. A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell. 2009;15:489-500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. van Agthoven T, van Agthoven TL, Dekker A, van der Spek PJ, Vreede L, Dorssers LC. Identification of BCAR3 by a random search for genes involved in antiestrogen resistance of human breast cancer cells. EMBO J. 1998;17:2799-808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Schrecengost RS, Riggins RB, Thomas KS, Guerrero MS, Bouton AH. Breast cancer antiestrogen resistance-3 expression regulates breast cancer cell migration through promotion of p130Cas membrane localization and membrane ruffling. Cancer Res. 2007;67:6174-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Riggins RB, Schrecengost RS, Guerrero MS, Bouton AH. Pathways to tamoxifen resistance. Cancer Lett. 2007;256:1-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. van Agthoven T, Godinho MF, Wulfkuhle JD, Petricoin EF, 3rd, Dorssers LC. Protein pathway activation mapping reveals molecular networks associated with antiestrogen resistance in breast cancer cell lines. Int J Cancer. 2012;131:1998-2007 [DOI] [PubMed] [Google Scholar]
- 110. Riggins RB, Thomas KS, Ta HQ, et al. Physical and functional interactions between Cas and c-Src induce tamoxifen resistance of breast cancer cells through pathways involving epidermal growth factor receptor and signal transducer and activator of transcription 5b. Cancer Res. 2006;66:7007-15 [DOI] [PubMed] [Google Scholar]
- 111. Shah AN, Gallick GE. Src, chemoresistance and epithelial to mesenchymal transition: are they related? Anticancer Drugs. 2007;18: 371-5 [DOI] [PubMed] [Google Scholar]
- 112. Tikhmyanova N, Golemis EA. NEDD9 and BCAR1 negatively regulate E-cadherin membrane localization, and promote E-cadherin degradation. PLoS One. 2011;6:e22102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Hiscox S, Morgan L, Barrow D, Dutkowskil C, Wakeling A, Nicholson RI. Tamoxifen resistance in breast cancer cells is accompanied by an enhanced motile and invasive phenotype: inhibition by gefitinib (‘Iressa’, ZD1839). Clin Exp Metastasis. 2004;21:201-12 [DOI] [PubMed] [Google Scholar]
- 114. Lacroix M, Leclercq G. Relevance of breast cancer cell lines as models for breast tumours: an update. Breast Cancer Res Treat. 2004;83:249-89 [DOI] [PubMed] [Google Scholar]
- 115. Giallourakis C, Cao Z, Green T, et al. A molecular-properties-based approach to understanding PDZ domain proteins and PDZ ligands. Genome Res. 2006;16:1056-72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Rosebeck S, Rehman AO, Lucas PC, McAllister-Lucas LM. From MALT lymphoma to the CBM signalosome: three decades of discovery. Cell Cycle. 2011;10:2485-96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Ryu JW, Kim HJ, Lee YS, et al. The proteomics approach to find biomarkers in gastric cancer. J Korean Med Sci. 2003;18:505-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Al-Shami A, Wilkins C, Crisostomo J, et al. The adaptor protein Sh2d3c is critical for marginal zone B cell development and function. J Immunol. 2010;185:327-34 [DOI] [PubMed] [Google Scholar]
- 119. Boriack-Sjodin PA, Margarit SM, Bar-Sagi D, Kuriyan J. The structural basis of the activation of Ras by Sos. Nature. 1998;394:337-43 [DOI] [PubMed] [Google Scholar]
- 120. Dorssers LC, van Agthoven T, Brinkman A, Veldscholte J, Smid M, Dechering KJ. Breast cancer oestrogen independence mediated by BCAR1 or BCAR3 genes is transmitted through mechanisms distinct from the oestrogen receptor signalling pathway or the epidermal growth factor receptor signalling pathway. Breast Cancer Res. 2005;7:R82-92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. van Agthoven T, Sieuwerts AM, Meijer-van Gelder ME, et al. Relevance of breast cancer antiestrogen resistance genes in human breast cancer progression and tamoxifen resistance. J Clin Oncol. 2009;27:542-9 [DOI] [PubMed] [Google Scholar]
- 122. Sun G, Cheng SY, Chen M, Lim CJ, Pallen CJ. Protein tyrosine phosphatase alpha phosphotyrosyl-789 binds BCAR3 to position Cas for activation at integrin-mediated focal adhesions. Mol Cell Biol 2012;32:3776-89 [DOI] [PMC free article] [PubMed] [Google Scholar]