Abstract
Background
The PER1 circadian-clock gene plays an important role in the regulation of many normal physiological rhythms in vivo. It has been revealed recently that abnormal expression of PER1 correlates closely with the occurrence and development of many cancers. However, the expression and significance of PER1 in oral squamous cell carcinoma (OSCC) remains unknown. The purpose of the present study was to investigate the direct links between aberrant PER1 expression and clinicopathological features of OSCC.
Methods
PER1 expression in cancerous and adjacent noncancerous tissues from 41 patients with OSCC was detected by immunohistochemical staining and real-time reverse transcriptase polymerase chain reaction, and correlations were sought with clinicopathological features in patients.
Results
Expression of PER1 mRNA and protein in OSCC was significantly reduced compared with that in adjacent noncancerous tissue (P < 0.05). Expression of PER1 protein in oral phase III–IV SCC specimens was significantly lower than that in phase I–II specimens (P < 0.05), and stage T1–T2 patients expressed significantly higher levels of PER1 protein than T3–T4 patients (P < 0.05). Expression of PER1 in patients without lymph node metastasis was significantly higher than that in those with lymph node metastasis (P < 0.05). PER1 protein expression showed no significant correlation with patient gender and age, or with degree of tumor cell differentiation (P > 0.05).
Conclusion
Changes in PER1 expression may play an important role in the development, invasion, and metastasis of OSCC, and may also provide novel ideas and methods for investigation of the occurrence, development, and targeted treatment OSCC.
Keywords: oral cancer, squamous cell, circadian gene, PER1, immunohistochemistry, real-time reverse transcriptase polymerase chain reaction
Introduction
Many physiological activities in living subjects, such as body temperature, heart rate, hormone secretion, cell proliferation, and metabolism, change rhythmically with the alternation between day and night. Such circadian rhythm is one of the basic characteristics of an organism’s life activities and is controlled by the circadian system in vivo, which is composed of a series of circadian-clock genes.1 To date, there are at least nine demonstrated major circadian-clock genes, ie, period1 (Per1), period2 (Per2), period3 (Per3), circadian locomotor output cycles kaput (Clock), cryptochrome1 (Cry1), cryptochrome2 (Cry2), brain and muscle Arnt-like protein 1 (Bmal 1), casein kinase 1ɛ (CK1ɛ), and timeless (Tim).2 It has been shown recently that the occurrence, development, prognosis, and treatment of cancer are closely related to the abnormal expression of certain circadian-clock genes.3–9 Therefore, further understanding of the behavior of circadian-clock genes may provide novel ideas and methods for investigation of the occurrence, development, and treatment of tumors.
PER1, a core circadian-clock gene, is located on the short arm of human chromosome 17 (17p12–p13.1), spanning about 16 kb.1,3 The PER1 gene not only plays an important role in the regulation of many normal circadian rhythms in vivo, but also correlates closely with development of cancers.4–9 Studies have shown that the PER1 protein inhibits growth of prostate and colon cancers,4,5 and that abnormal expression of the PER1 gene leads to the occurrence and promotes the development of endometrial cancer, leukemia, and breast cancer.6–9 Data from published research have also indicated that expression and function of the PER1 gene varies significantly between different tumors.4,5,7,10,11 Gery et al have shown that PER1 plays a proapoptotic role and is downregulated in prostate and colon cancers,10 while Sato et al have demonstrated that the PER1 gene is highly expressed in pancreatic and liver cancers and exerts an antiapoptotic effect.11 These findings imply that the PER1 gene may play important but different roles in the occurrence and development of different tumors. However, there is still a lack of studies focusing on the relationship between expression of the PER1 gene and oral cancers.
The overall incidence of oral cancer is approximately 4.7–32.2/10,000 globally, among which more than 90% are SCC.12,13 The current overall 5-year survival rate of oral squamous cell carcinoma (OSCC) post treatment is only 50%–60%,13 so exploring new methods for the treatment of OSCC is of great significance.
The present study detected expression of the PER1 gene in both cancerous and adjacent noncancerous tissues from 41 patients with OSCC and investigated their clinical significance by analyzing the correlation between PER1 expression in OSCC and clinicopathological parameters, trying to, from the perspective of changes in PER1 gene expression, provide new ideas for further investigation of the occurrence, development, and treatment of OSCC.
Materials and methods
Reagents and instruments
A streptavidin-peroxidase immunohistochemistry kit and horseradish peroxidase-labeled goat anti-rabbit IgG antibody were purchased from Beijing Zhongshan Golden Bridge Biotechnology Co, Ltd (Beijing, China). The rabbit anti-human PER1 polyclonal antibody was sourced from Santa Cruz Biotechnology Inc (Santa Cruz, CA). The polymerase chain reaction (PCR) kit was purchased from Takara Bio (Shiga, Japan). The light microscope was a Leica TS-26 (Solms, Germany); the frozen low-speed centrifuge was a Z233MK-2 (Hermle, Wehingen, Germany); conventional PCR reactions were done using a S1000TM thermal cycler (Bio-Rad, Hercules, CA); and quantitative real-time PCR was performed using a Bio-Rad C-1000TM thermal cycler. The UV-GeneQuant spectrophotometer was sourced from Amersham (Uppsala, Sweden).
Clinical data
Cancerous and adjacent noncancerous tissues were obtained from 41 patients with OSCC treated in the Department of Oral and Maxillofacial Surgery, First Affiliated Hospital of Chongqing Medical University from January 2011 to March 2012. The specimens were harvested between 10:50 am and 14:30 pm. The inclusion criteria were as follows: pathological diagnosis of OSCC; no preoperative radiotherapy, chemotherapy, or other treatments; concomitant neck dissection during surgery; and pathological confirmation of cervical lymph node metastasis. In total, 41 specimens were obtained from 24 males and 17 females of mean age 51.6 (range 22–81) years. According to the 2010 Union for International Cancer Control staging criteria,14 six, 12, 16, and seven of the tumor cases were stage T1, T2, T3, and T4, respectively; 14 cases had cervical lymph node metastasis and 27 were without cervical lymph node metastasis. Clinical phase I, II, III, and IV accounted for six, 16, 13, and six cases, respectively. Pathological classification showed 13, 20, and eight cases to be well, moderately, and poorly differentiated, respectively. Each sample was divided into two parts; one part was fixed with 4% formalin and embedded in paraffin, and the other part was placed rapidly in liquid nitrogen. The study was approved by the human research ethics committee of the First Affiliated Hospital of Chongqing Medical University and written informed consent was obtained from all patients who participated.
Immunohistochemistry
The paraffin-embedded tissues were cut into sections 4 μm thick. Streptavidin-peroxidase immunohistochemical staining was performed following the kit instructions. Briefly, the sections were routinely dewaxed, rehydrated, and permeabilized; endogenous peroxidase was blocked with 3% H2O2; antigen retrieval was done under high temperature and pressure to expose the epitopes; and blocking with 5% normal goat serum, incubation with primary and secondary antibodies, and color development with DAB were carried out sequentially in accordance with the kit instructions. The sections were then stained with hematoxylin, dehydrated, cleared, and observed under a light microscope after mounting with neutral resin. Slides with phosphate-buffered saline added instead of primary antibody were used as the negative control. The positive slide provided in the kit was used as the PER1 positive control. Positive PER1staining appeared as a yellow color in the nucleus or cytoplasm. The results were assessed by semiquantitative scoring.15 The intensity of positive staining (no staining, 0; light yellow, 1; brown, 2; and dark brown, 3) and the percentage of positive cells (number of positive cells ≤ 5%, 0; 6%–25%, 1; 26%–50%, 2; 51%–75%, 3; ≥76%, 4) were scored, and the multiplication of both scores was used finally to evaluate PER1 expression as follows: 0, negative; 1–4, positive; >4, strongly positive. All positive and strongly positive sections were expressed as positive.
Real-time reverse transcriptase PCR
Total RNA was extracted from the frozen cancerous tissue and adjacent noncancerous tissues and used for real-time reverse transcriptase PCR detection. The experiment was repeated three times as follows: total RNA was extracted from the cancerous tissue and adjacent noncancerous tissues using an RNAiso Plus kit, and RNA concentration and purity was determined by measuring absorbance at 260 nm and 280 nm. cDNAs were synthesized using a PrimeScript reverse transcriptase reagent kit according to the manufacturer’s manual. Briefly, the reaction (20 μL in total volume) was incubated at 37°C for 15 minutes and then at 85°C for 5 seconds. Real-time reverse transcriptase PCR primers for the target PER1 gene and the internal control gene (β-actin) were designed using Oligo7.0 software (Table 1); the reaction system contained 2 × SYBR Premix Ex Taq™ II 12.5 μL, 0.4 μM forward and reverse primers (1 μL each), a 2 μL DNA template (100 ng), molecular grade H2O 8.5 μL, in a total volume of 25 μL. The reaction was denatured at 95°C for 1.5 minutes, and cycling was carried out at 95°C for 10 seconds and 60°C for 30 seconds, for 40 cycles, the fluorescent signal was collected during a 60°C extension step, and the 2-ΔΔCt method was used to quantify expression of the PER1 gene.
Table 1.
Primer sequence and product size for PER1 and β-actin genes
| Gene | Primer sequence | Product (bp) |
|---|---|---|
| PER1 | Forward: 5′-ACCCTGATGACCCACTCTTCTC-3″ | 170 |
| Reverse: 5′-CTCCTCCATAGCCAAGTCCTGA-3′ | ||
| β-actin | Forward: 5′-CTTCTACAATGAGCTGCGTGTG-3′ | 169 |
| Reverse: 5′-AGAGGCGTACAGGGATAGCACAG-3′ |
Statistical analysis
All data were analyzed by χ2 tests and t-tests using the Statistical Package for the Social Sciences version 17.0 statistical software (SPSS Inc, Chicago, IL), and the results are presented as the mean ± standard deviation. A P value < 0.05 was considered to be statistically significant.
Results
PER1 mRNA and protein expression in OSCC
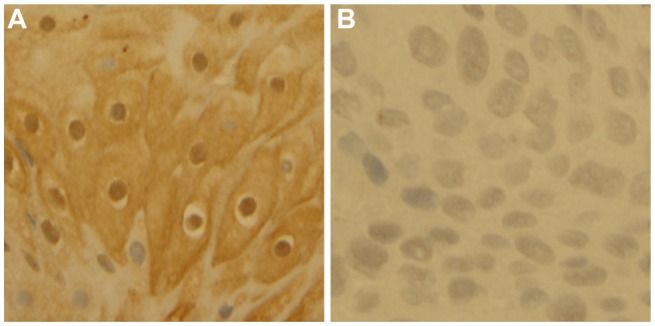
The results of immunohistochemistry show that the rate of positive PER1 protein expression in OSCC and adjacent noncancerous tissues was 82.93% (34/41) and 100% (41/41) respectively, and the former was significantly lower than the latter (χ2 = 7.653, P < 0.05). Expression of PER1 protein in adjacent noncancerous tissues was mainly in the nucleus and cytoplasm and was strongly positive, whereas expression of PER1 was weakly or moderately positive in tumor cells (Figure 1). The results of quantitative real-time PCR also confirm that the relative expression level of PER1 mRNA in OSCC (3.71 ± 0.42) was significantly lower than that in adjacent noncancerous tissues (11.95 ± 0.77, t = −16.171, P < 0.001).
Figure 1.
Immunohistochemical staining of PER1 protein (SP 400×). Expression of PER1 protein in adjacent noncancerous tissues is strongly positive (A) and expression of PER1 protein in oral squamous cell carcinoma is moderately or weakly positive (B).
PER1 protein expression and clinicopathological features of OSCC
Expression of PER1 protein in patients with phase I and II OSCC was significantly higher than that in patients with phase III and IV disease (P < 0.05), while stage T1 and T2 patients expressed significantly higher levels of PER1 protein compared with T3 and T4 patients (P < 0.05). Expression of PER1 protein was significantly higher in patients without lymph node metastasis compared with patients having lymph node metastasis (P < 0.05), but there was no significant correlation with patient age, gender, or degree of differentiation (P > 0.05, Table 2).
Table 2.
The expression of PER1 protein and its relationship with clinicopathological features of patients with OSCC
| Parameters | cases | PER1 expression | χ2 | P | |
|---|---|---|---|---|---|
|
|
|||||
| Negative | Positive | ||||
| Tissue type | |||||
| Tumor | 41 | 7 | 34 | 7.653 | 0.012 |
| Peritumor | 41 | 0 | 41 | ||
| Age | |||||
| ≥60 | 10 | 2 | 8 | 0.131 | 0.937 |
| 45–60 | 20 | 3 | 17 | ||
| <45 | 11 | 2 | 9 | ||
| Gender | |||||
| Female | 17 | 3 | 14 | 0.007 | 1.000 |
| Male | 24 | 4 | 20 | ||
| Tumor differentiation | |||||
| Well | 13 | 3 | 10 | 0.510 | 0.775 |
| Moderate | 20 | 3 | 17 | ||
| Poor | 8 | 1 | 7 | ||
| T staging | |||||
| T1 + T2 | 18 | 0 | 18 | 6.606 | 0.012 |
| T3 + T4 | 23 | 7 | 16 | ||
| Lymph node metastasis | |||||
| No | 27 | 1 | 26 | 9.982 | 0.004 |
| Yes | 14 | 6 | 8 | ||
| Clinical stage | |||||
| I + II | 22 | 1 | 21 | 5.262 | 0.036 |
| III + IV | 19 | 6 | 13 | ||
Abbreviation: OSCC, oral squamous cell carcinoma.
Discussion
The results of this study show that expression of the PER1 gene in OSCC is significantly decreased compared with expression in adjacent noncancerous tissues at both the protein and mRNA levels. Further analysis indicates that expression of the PER1 protein in stage T3 and T4 patients, those with lymph node metastasis, and those with clinical phase III and IV disease, is significantly lower than that in stage T1 and T2 patients, those without lymph node metastasis, and those with clinical phase I and II disease. These results suggest that expression of PER1 protein in OSCC is gradually decreased with tumor development, implying that PER1 may play a tumor suppression role, and its expression may primarily correlate with invasion and metastasis of OSCC. Yeh et al have reported the mechanism of decreased PER1 expression in breast cancer and endometrial carcinoma to be methylation of cytosine-phosphate guanosine sites in PER1 promoter sequences, leading to downregulation and inhibition of PER1 expression.6,16 However, the mechanism needs further investigation in more detail.
A basic characteristic of malignant tumors is uncontrolled proliferation.17PER1 is one of the important circadian-clock genes controlling the normal physiological rhythm of cell proliferation and metabolism in vivo.1–3,18,19 Changes in PER1 expression alter the proliferation rhythm of normal cells, and may easily lead to tumorigenesis. Because a circadian-clock system was not recognized in vivo until recently, the exact mechanism needs to be clarified. In recent reports from Chang et al20 and Klein et al,21PER1 was demonstrated to regulate the proliferation, invasion, and migration of cancer cells by regulating expression of matrix metalloproteinase-2 and intracellular distribution of laminin receptor 1. Decreased PERl protein expression upregulates the expression of matrix metalloproteinase-2 and increases the distribution of laminin receptor 1 onto the cell membrane, thereby enhancing tumor invasion, migration, and metastasis. However, due to the lack of sufficient studies, more detailed mechanisms remain unclear.
In summary, circadian-clock genes form a unique system in living subjects. Further research focusing on these genes may, from the perspective of biological rhythms, provide novel ideas and methods for a better understanding of the occurrence and development of tumors, and for individualized treatment of cancer. The results of this study suggest that the PER1 gene may be used as a marker to determine clinical staging, to evaluate the metastatic risk of OSCC, and as a novel target for the prevention and treatment of OSCC.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Hara Y, Onishi Y, Oishi K, Miyazaki K, Fukamizu A, Ishida N. Molecular characterization of Mybbp1a as a co-repressor on the Period2 promoter. Nucleic Acids Res. 2009;37:1115–1126. doi: 10.1093/nar/gkn1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Badiu C. Genetic clock of biologic rhythms. J Cell Mol Med. 2003;7:408–416. doi: 10.1111/j.1582-4934.2003.tb00243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sahar S, Sassone-Corsi P. Metabolism and cancer: the circadian clock connection. Nat Rev Cancer. 2009;9:886–896. doi: 10.1038/nrc2747. [DOI] [PubMed] [Google Scholar]
- 4.Cao Q, Gery S, Dashti A, et al. A role for the clock gene per1 in prostate cancer. Cancer Res. 2009;69:619–7625. doi: 10.1158/0008-5472.CAN-08-4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mostafaie N, Kállay E, Sauerzapf E, et al. Correlated downregulation of estrogen receptor beta and the circadian clock gene Per1 in human colorectal cancer. Mol Carcinog. 2009;48:642–647. doi: 10.1002/mc.20510. [DOI] [PubMed] [Google Scholar]
- 6.Yeh KT, Yang MY, Liu TC, et al. Abnormal expression of period 1 (PER1) in endometrial carcinoma. J Pathol. 2005;206:111–120. doi: 10.1002/path.1756. [DOI] [PubMed] [Google Scholar]
- 7.Hsu CM, Lin SF, Lu CT, Lin PM, Yang MY. Altered expression of circadian clock genes in head and neck squamous cell carcinoma. Tumour Biol. 2012;33:149–155. doi: 10.1007/s13277-011-0258-2. [DOI] [PubMed] [Google Scholar]
- 8.Yang MY, Yang WC, Lin PM, et al. Altered expression of circadian clock genes in human chronic myeloid leukemia. J Biol Rhythms. 2011;26:136–148. doi: 10.1177/0748730410395527. [DOI] [PubMed] [Google Scholar]
- 9.Yang X, Wood PA, Ansell CM, et al. The circadian clock gene Per1 suppresses cancer cell proliferation and tumor growth at specific times of day. Chronobiol Int. 2009;26:1323–1339. doi: 10.3109/07420520903431301. [DOI] [PubMed] [Google Scholar]
- 10.Gery S, Komatsu N, Baldjyan L, Yu A, Koo D, Koeffler HP. The circadian gene per1 plays an important role in cell growth and DNA damage control in human cancer cells. Mol Cell. 2006;22:375–382. doi: 10.1016/j.molcel.2006.03.038. [DOI] [PubMed] [Google Scholar]
- 11.Sato F, Nagata C, Liu Y, et al. Period1 is an anti-apoptotic factor in human pancreatic and hepatic cancer cells. J Biochem. 2009;146:833–838. doi: 10.1093/jb/mvp126. [DOI] [PubMed] [Google Scholar]
- 12.Warnakulasuriya S. Global epidemiology of oral and oropharyngeal cancer. Oral Oncol. 2009;45:309–316. doi: 10.1016/j.oraloncology.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 13.Johnson NW, Warnakulasuriya S, Gupta PC, et al. Global oral health inequalities in incidence and outcomes for oral cancer causes and solutions. Adv Dent Res. 2011;23:237–246. doi: 10.1177/0022034511402082. [DOI] [PubMed] [Google Scholar]
- 14.Sobin LH, Gospodarowicz MK, Wittekind C. TNM Classification of Malignant Tumors. 7th ed. London, UK: Wiley; 2010. [Google Scholar]
- 15.Xu W, Wang LW, Shi JZ, Gong ZJ. Effects of RNA interference targeting transforming growth factor-beta 1 on immune hepatic fibrosis induced by concanavalin A in mice. Hepatobiliary Pancreat Dis Int. 2009;8:300–308. [PubMed] [Google Scholar]
- 16.Chen ST, Choo KB, Hou MF, et al. Deregulated expression of the PER1, PER2 and PER3 genes in breast cancers. Carcinogenesis. 2005;26:1241–1246. doi: 10.1093/carcin/bgi075. [DOI] [PubMed] [Google Scholar]
- 17.Murphy PJ, Campbell SS. Physiology of the circadian system in animals and humans. J Clin Neurophysiol. 1996;13:2–16. doi: 10.1097/00004691-199601000-00002. [DOI] [PubMed] [Google Scholar]
- 18.Eismann EA, Lush E, Sephton SE. Circadian effects in cancer-relevant psychoneuroendocrine and immune pathways. Psychoneuroendocrinology. 2010;35:963–976. doi: 10.1016/j.psyneuen.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 19.Smolensky MH, Peppas NA. Chronobiology, drug delivery, and chronotherapeutics. Adv Drug Deliv Rev. 2007;59:828–851. doi: 10.1016/j.addr.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 20.Chang L, Wang YH, Liu YY, et al. Key region of laminin receptor 1 for interaction with human period1. Afr J Biotechnol. 2010;9:6401–6407. [Google Scholar]
- 21.Klein G, Vellenga E, Fraaije MW, Kamps WA, de Bont ES. The possible role of matrix metalloproteinase (MMP)-2 and MMP-9 in cancer, eg, acute leukemia. Crit Rev Oncol Hematol. 2004;50:87–100. doi: 10.1016/j.critrevonc.2003.09.001. [DOI] [PubMed] [Google Scholar]