Abstract
Purpose
The goal of this retrospective analysis was to assess how well predictive models could determine which patients would develop liver chemistry signals during clinical trials based on their pretreatment (baseline) information.
Patients and methods
Based on data from 24 late-stage clinical trials, classification models were developed to predict liver chemistry outcomes using baseline information, which included demographics, medical history, concomitant medications, and baseline laboratory results.
Results
Predictive models using baseline data predicted which patients would develop liver signals during the trials with average validation accuracy around 80%. Baseline levels of individual liver chemistry tests were most important for predicting their own elevations during the trials. High bilirubin levels at baseline were not uncommon and were associated with a high risk of developing biochemical Hy’s law cases. Baseline γ-glutamyltransferase (GGT) level appeared to have some predictive value, but did not increase predictability beyond using established liver chemistry tests.
Conclusion
It is possible to predict which patients are at a higher risk of developing liver chemistry signals using pretreatment (baseline) data. Derived knowledge from such predictions may allow proactive and targeted risk management, and the type of analysis described here could help determine whether new biomarkers offer improved performance over established ones.
Keywords: bilirubin, Hy’s Law, ALT, GGT, baseline, prediction
Introduction
According to the US Food and Drug Administration (FDA) Guidance for Industry on Drug Induced Liver Injury (DILI),1 the liver chemistry tests that are primarily evaluated in clinical trials are alanine aminotransferase (ALT), aspartate aminotransferase (AST), alkaline phosphatase (ALP), and total bilirubin (TBL). ALT and AST are sometimes grouped together as aminotransferase (AT). These established liver chemistry tests possess unbalanced sensitivity and specificity as indicators for severe DILI. The most specific signal is obtained when liver chemistry tests are combined into Hy’s law,1,2 with elevations of both AT and bilirubin, but the sensitivity of Hy’s law is poor. While AT elevation > 3 × upper limits of normal (ULN) might be the most sensitive signal, its specificity is low.1,3 Nevertheless, regular measurements of these liver chemistry tests are current practice in clinical trials for detecting DILI, and their elevations above certain thresholds, such as multiples of ULN, are liver chemistry signals of DILI as defined by the FDA guideline.1
We have shown baseline values of liver chemistry tests were predictive for their postbaseline elevations during clinical trials in healthy volunteers treated with placebo.4 The current analysis addresses a follow-up question, which is whether the same pattern exists for patients treated with investigational drugs in clinical trials in late-stage drug development, especially in Phase III clinical trials. The specific goal of the current analysis is to explore available clinical data in an attempt to answer the following questions: (1) can any baseline biomarkers or factors differentiate between patients who developed and those who did not develop liver chemistry signals? (2) Can any baseline biomarkers or factors differentiate between patients who had biochemical Hy’s law versus those who did not? (3) Are there different temporal profiles of liver chemistry signals in biochemical Hy’s law cases (ie, patients with ALT/AST > 3 × ULN, TBL > 2 × ULN, and ALP < 2 × ULN)?
Considering the difficulty of inferring causality on adverse events from historical clinical trials, it was not attempted in this analysis to either assess whether any of the liver chemistry signals, including biochemical Hy’s law cases, were actually related to the investigational drugs, or to conclude if any of the liver chemistry signals were actually due to real liver injury. Both questions are beyond the purpose of this analysis, which is only to predict and examine liver chemistry test results.
Materials and methods
Data
In total, 22 historical Phase III studies (projects A, B, D, and E) and two Phase II studies (project C) were included from five drug projects (Table 1), all selected based on the availability of standard data elements and known occurrences of liver chemistry signals. These projects targeted different therapy areas: central nervous system (project A), cardiovascular (projects B and E), gastrointestinal (project C), and respiratory and inflammation (project D). Data across the 24 trials were standardized and integrated in an Oracle database based on the Study Data Tabulation Model (SDTM) from Clinical Data Interchange Standards Consortium.5 Common data types included laboratory results, demographics, concomitant medications, adverse events, and medical history. Demographics are shown in Table 1.
Table 1.
Patient demographics of included clinical trials
| Project | Study | Age | Sex, n (%) | Race, n (%) | |||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|||||||
| Mean (SD) | Median | Min | Max | Male | Female | White | Other | ||
| A | A.3 | 68.4 (12.1) | 71 | 20 | 97 | 941 (55.4) | 758 (44.6) | 1555 (91.5) | 144 (8.5) |
| A.4 | 68.9 (12.9) | 71 | 21 | 98 | 1750 (54.8) | 1442 (45.2) | 2664 (83.5) | 528 (16.5) | |
| Total | 68.7 (12.6) | 71 | 20 | 98 | 2691 (55) | 2200 (45) | 4219 (86.3) | 672 (13.7) | |
| B | B.1 | 57.5 (11.4) | 58 | 18 | 90 | 766 (51.5) | 721 (48.5) | 1298 (87.3) | 189 (12.7) |
| B.2 | 56.7 (11.2) | 57 | 19 | 86 | 1399 (50.6) | 1368 (49.4) | 2394 (86.5) | 373 (13.5) | |
| B.3 | 57.8 (10.4) | 58 | 20 | 87 | 864 (54.9) | 709 (45.1) | 1247 (79.3) | 326 (20.7) | |
| B.4 | 60.6 (11.1) | 61 | 26 | 92 | 312 (54.9) | 256 (45.1) | 358 (63) | 210 (37) | |
| B.5 | 56.1 (10.5) | 56 | 20 | 83 | 301 (54.4) | 252 (45.6) | 417 (75.4) | 136 (24.6) | |
| B.6 | 57.8 (11.4) | 58 | 20 | 86 | 416 (50.7) | 405 (49.3) | 0 (0) | 821 (100) | |
| B.7 | 56.8 (9.4) | 57 | 29 | 82 | 130 (53.9) | 111 (46.1) | 0 (0) | 241 (100) | |
| B.8 | 62.9 (10.8) | 63 | 29 | 93 | 183 (58.3) | 131 (41.7) | 0 (0) | 314 (100) | |
| Total | 57.6 (11.1) | 58 | 18 | 93 | 4371 (52.5) | 3953 (47.5) | 5714 (68.6) | 2610 (31.4) | |
| C | C.1 | 46.8 (12.7) | 47 | 18 | 72 | 963 (63.2) | 561 (36.8) | 1346 (88.3) | 178 (11.7) |
| C.2 | 46.2 (12.9) | 47 | 18 | 73 | 590 (40.1) | 880 (59.9) | 1195 (81.3) | 275 (18.7) | |
| Total | 46.5 (12.8) | 47 | 18 | 73 | 1553 (51.9) | 1441 (48.1) | 2541 (84.9) | 453 (15.1) | |
| D | D.1 | 33 (12.4) | 32 | 12 | 74 | 232 (40.4) | 279 (48.6) | 434 (75.6) | 140 (24.4) |
| D.2 | 34.3 (11.7) | 32 | 18 | 69 | 171 (51.8) | 159 (48.2) | 279 (84.5) | 51 (15.5) | |
| D.3 | 30.3 (12.2) | 29 | 12 | 76 | 158 (40.8) | 229 (59.2) | 329 (85) | 58 (15) | |
| Total | 32.5 (12.2) | 31 | 12 | 76 | 561 (43.5) | 667 (51.7) | 1042 (80.7) | 249 (19.3) | |
| E | E.1 | 70.4 (8.6) | 71 | 29 | 93 | 2515 (68.6) | 1150 (31.4) | 3206 (87.5) | 459 (12.5) |
| E.2 | 66.2 (10.3) | 68 | 25 | 93 | 1132 (38) | 1846 (62) | 2948 (99) | 30 (1) | |
| E.3 | 66 (10.2) | 67 | 20 | 101 | 1109 (37.8) | 1823 (62.2) | 2910 (99.2) | 22 (0.8) | |
| E.4 | 71.9 (9.2) | 73 | 30 | 97 | 3170 (64.9) | 1462 (30) | 4410 (90.4) | 471 (9.6) | |
| E.5 | 68.1 (10.2) | 69.5 | 24 | 94 | 260 (35) | 482 (64.9) | 680 (91.5) | 63 (8.5) | |
| E.6 | 65.6 (11.4) | 66 | 21 | 94 | 507 (42.7) | 681 (57.3) | 1186 (99.8) | 2 (0.2) | |
| E.7 | 64.6 (13) | 67 | 19 | 93 | 960 (46.9) | 1081 (52.9) | 1923 (94) | 122 (6) | |
| E.8 | 68 (9.7) | 69 | 24 | 89 | 1008 (38.6) | 1603 (61.4) | 2493 (95.5) | 118 (4.5) | |
| E.9 | 67 (9.6) | 68 | 26 | 92 | 1012 (36.4) | 1760 (63.3) | 2606 (93.7) | 175 (6.3) | |
| Total | 68.1 (10.3) | 70 | 19 | 101 | 11673 (49) | 11888 (49.9) | 22362 (93.9) | 1462 (6.1) | |
Abbreviation: SD, standard deviation.
Two classes of patients, normal and abnormal, were defined and predictive models were developed to predict the class of individual patients. Normal subjects were those with no elevations of AT, ALP, or TBL, while abnormal subjects had liver signal elevations above the cutoffs derived from the FDA guideline1 at any time point after baseline (see Table 2 for frequencies). Those subjects whose liver chemistry values were elevated above normal ranges, but did not exceed the cutoffs above, were excluded from the dataset. Variables used for predicting the patient class included demographics, concomitant medications, medical history in addition to all laboratory test results. Variables that were considered to be predictive features were analyzed further to determine how they differed between abnormal and normal subjects. This was done by visualizing the differences in distribution between the two classes.
Table 2.
Liver signal frequencies
| Project | Study | Number of subjects | AT | ALP | TBL | Biochemical Hy’s law | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|||||||||
| Total | Normal | >3 | >5 | >8 | >1.5 | >2 | >3 | >1.5 | >2 | |||
| A | A.1 | 1699 | 693 | 42 | 14 | 6 | 50 | 21 | 8 | 65 | 21 | |
| A.2 | 3192 | 1288 | 80 | 20 | 10 | 76 | 35 | 5 | 111 | 37 | 3 | |
| Total | 4891 | 1981 | 122 | 34 | 16 | 126 | 56 | 13 | 176 | 58 | 3 | |
| B | B.1 | 1487 | 841 | 11 | 1 | 13 | 2 | 1 | 10 | 1 | ||
| B.2 | 2767 | 1527 | 20 | 10 | 6 | 37 | 11 | 1 | 21 | 2 | 1 | |
| B.3 | 1573 | 920 | 7 | 1 | 11 | 2 | 11 | 5 | ||||
| B.4 | 568 | 275 | 6 | 2 | ||||||||
| B.5 | 553 | 304 | 2 | 1 | 1 | 7 | 2 | 2 | ||||
| B.6 | 821 | 574 | 3 | 1 | 1 | 14 | 3 | 1 | 5 | 1 | ||
| B.7 | 241 | 160 | 1 | 1 | 1 | 5 | 1 | 2 | ||||
| B.8 | 314 | 243 | 1 | 1 | ||||||||
| Total | 8324 | 4844 | 44 | 15 | 9 | 94 | 22 | 3 | 53 | 9 | 1 | |
| C | C.1 | 1524 | 871 | 13 | 4 | 3 | 22 | 6 | 1 | 13 | 1 | |
| C.2 | 1470 | 921 | 9 | 5 | 5 | 26 | 11 | 3 | 22 | 6 | ||
| Total | 2994 | 1792 | 22 | 9 | 8 | 48 | 17 | 4 | 35 | 7 | 0 | |
| D | D.1 | 574 | 393 | 6 | 3 | 1 | 2 | 15 | 4 | |||
| D.2 | 330 | 117 | 1 | 13 | 2 | 1 | 5 | 1 | ||||
| D.3 | 387 | 234 | 10 | 4 | 8 | 3 | ||||||
| Total | 1291 | 744 | 7 | 3 | 1 | 25 | 6 | 1 | 28 | 8 | 0 | |
| E | E.1 | 3665 | 1860 | 57 | 20 | 5 | 130 | 49 | 15 | 184 | 56 | 3 |
| E.2 | 2978 | 2305 | 7 | 19 | 6 | 2 | 13 | 3 | ||||
| E.3 | 2932 | 1069 | 63 | 10 | 2 | 268 | 123 | 37 | 29 | 3 | ||
| E.4 | 4881 | 2850 | 70 | 25 | 15 | 171 | 66 | 20 | 229 | 60 | 9 | |
| E.5 | 743 | 503 | 3 | 1 | 41 | 20 | 9 | 9 | 3 | |||
| E.6 | 1188 | 684 | 11 | 3 | 2 | 80 | 34 | 2 | 13 | 1 | ||
| E.7 | 2045 | 1144 | 28 | 8 | 1 | 125 | 54 | 14 | 9 | 3 | ||
| E.8 | 2611 | 1712 | 10 | 5 | 1 | 152 | 73 | 18 | 24 | 4 | ||
| E.9 | 2781 | 1840 | 7 | 2 | 1 | 159 | 56 | 16 | 19 | 5 | ||
| Total | 23824 | 13967 | 256 | 74 | 27 | 1145 | 481 | 133 | 529 | 138 | 12 | |
| Total | 41324 | 23328 | 451 | 135 | 61 | 1438 | 582 | 154 | 821 | 220 | 16 | |
Note: Data sets with less than 10 abnormal subjects were indicated with italics and were not used in predictive modeling for the corresponding liver signals.
Abbreviations: ALP, alkaline phosphatase; AT, aminotransferase; TBL, total bilirubin.
Because causality inference, including possible relation to drug treatment, is outside the scope of this analysis, placebo-treated subjects were included in both classes together with drug- and/or comparator-treated subjects. Another reason for this is that we could not find treatment information to separate drug- and placebo-treated subjects for all studies, especially not for some very old studies. Nevertheless, based on data from studies (13 out of the 24 studies) that had individual treatment details, either no or very few abnormal subjects came from placebo-treated arms.
In these trials, normal subjects greatly outnumbered abnormal subjects. In order to avoid bias in models towards the larger class (ie, the normal subjects), 15 balanced subdatasets were created by randomly sampling an equal number of subjects from the normal class as in the respective abnormal class. The results of each dataset are therefore presented with average and standard error across the 15 subdatasets. Datasets that had less than 10 abnormal subjects were discarded due to robustness issues, as indicated by italics in Table 2. Prior to building a random forest model (see Methods) on a dataset, variables with more than 10% missing values in either of the two classes were removed in order to limit the impact of imputation. The remaining missing values were imputed with the imputation function from the random forest package (see Methods). All lab values were normalized with the ULN.
Methods
The random forests method,6 available as an R-project library, was chosen to develop classification models because of the following features: (1) relative high accuracy among current algorithms with an internal unbiased estimate of the generalization error (out-of-bag error), (2) high efficiency on large datasets with thousands of input variables, (3) able to rank the variables that are important in the classification or regression, (4) an effective method for estimating missing data, and (5) can be saved for future predictions on other data.
The random forest models were analyzed in two aspects, their predictive performance and their variable importance (predictive features). The predictive performance was assessed using accuracy and receiver-operating characteristic (ROC) curves, while variable importance was analyzed using the built-in procedure in the random forest method. The variable importance information is a relative measure. It is a score that gives the mean decrease in accuracy when the information of a variable is destroyed using permutation. The importance score should only be interpreted in relation to the rest of the variables. A P-value for the most important variable was derived in a straightforward fashion by simulating a null distribution where the class labels had been permutated. A reasonably correct null distribution for not top-ranked variables is not easily obtained, and consequently a P-value was not calculated for those variables. For a top-ranked variable to be evaluated further, the model accuracy must be above 50%, and the P-value of the variable less than 0.05.
Predictive features of liver signal elevations were evaluated on study, project and cross-project level by pooling datasets accordingly. For each liver signal type, trends of predictive features were evaluated in terms of commonness across all studies within a project, as well as commonness across projects. The purpose with this procedure was twofold; (1) to investigate the degree of predictive generalizability across studies, projects, and therapy areas and (2) collect enough abnormal subjects to distinguish trends for liver signal types of low incidence (eg, Hy’s law cases). A leave-one-project-out (LOPO) validation approach was used to assess predictive performance and predictive features of models across projects. This was done by training a model on four projects and validating it on the fifth project. When pooling data across projects, bias introduced by different population sizes and projectspecific elevation patterns must be considered. To address this bias, two different settings of the LOPO validation approach were tested: (1) using balanced datasets not stratified by the number of subjects in a project, and (2) using balanced datasets, stratified by the number of subjects in a project.
Results
Frequencies of liver signals observed
The frequencies of liver signals from the five projects, including 24 clinical trials, are shown in Table 2. In order of frequency, they were ALP elevations > 1.5 × ULN (3.48%), ALP elevations > 2 × ULN (1.41%), TBL elevations > 1.5 × ULN (1.99%), ALT or AST (AT) elevations > 3 × ULN (1.09%), TBL elevations > 2 × ULN (0.53%), ALT or AST (AT) elevations > 5 × ULN (0.33%), ALT or AST (AT) elevations > 8 × ULN (0.15%), and biochemical Hy’s law cases (0.04%). As would be expected, the use of higher thresholds resulted in lower frequencies of occurrence. The elevations of ALP and bilirubin levels were more common than AT elevations.
The elevation rates of ALT (1.09% > 3 × ULN) and ALP (1.41 > 2 × ULN) in the current analysis were higher than those previously reported from late phase clinical trials.7 This could be due to trial selection criteria. The previous report excluded trials in diseases with a risk of underlying liver abnormalities in addition to trials in which study compounds had liver or renal toxicity. In our analysis, instead, we focused on late phase trails in which study compounds produced liver chemistry signals.
Prediction accuracies of random forest models
In applying random forest models, our goal was to predict retrospectively which patients would develop liver chemistry signals during clinical trials based on their pretreatment (baseline) information. The accuracies of random forest classification models varied across different liver chemistry signals and different projects. The ALP > 1.5 × ULN and TBL > 1.5 × ULN signals were better predicted, compared to AT > 3 × ULN. On datasets pooled by project, these models had an average accuracy of 79% and 78%, respectively for ALP and TBL signals. AT signals (ALT and AST as individual models or merged) had considerable lower accuracy with an average of 65% on pooled datasets. All model accuracies took advantage of the built-in cross-validation mechanism of random forest methodology, which is based on out-of-bag error.6
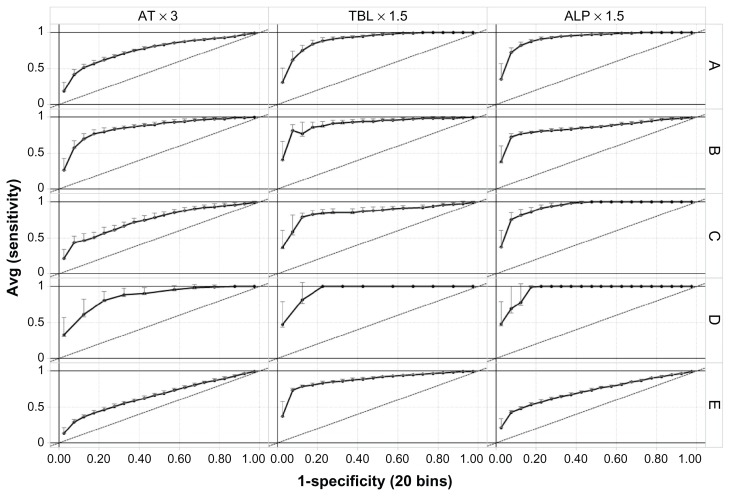
To address directly how applicable the models are across projects, a LOPO validation approach was used to assess the ability of these types of models in predicting liver signals in future studies. The LOPO validation accuracies using balanced and nonstratified datasets (See Methods) are shown in Figure 1. When project E was included in the training set, all models, including the models to predict AT signals, had a high accuracy (both sensitivity and specificity around 80%) when tested on a fifth project. When project E was not included in the training set, but used as a test set instead, all models had a lower accuracy as indicated by a lower area under the curve.
Figure 1.
Receiver-operating characteristic curves showing validation accuracy of random forest models across projects. Average true positive rates (sensitivity) were plotted against false positive rates (1-specificity) of 15 random subdatasets (see Methods) from each of the five projects (rows) for predicting the three liver signals (columns). Standard error is given as error bars. Column headings show the liver signals being predicted, and row headings (A–E) on the right show the projects left out in leave-one-project-out validation, eg, in row A, models were trained on projects (B–E) and tested on project A.
Abbreviations: ALP, alkaline phosphatase; AT, aminotransferase; TBL, total bilirubin.
The observed performances, ranging from low accuracy for models at individual study level (data not shown), to intermediate accuracy for models at individual project level (ie, from data pooled by project), and up to the high accuracy for models at cross-project level (ie, models in Figure 1), suggest that an increase of sample size (in terms of numbers of subjects, studies, and projects) will improve future predictions. This observation was particularly true in the case of predicting AT signals, for which prediction accuracy improved from 65% at individual project level to 80% at cross-project level. This is also consistent with the observed higher model accuracies when project E, which had a large number of patients, was included in the training set. Therefore, the predictability of baseline lab tests and other baseline patient characteristics for liver signals could further improve if more data from other projects is included in the training data.
Predictive baseline variables for individual liver signals
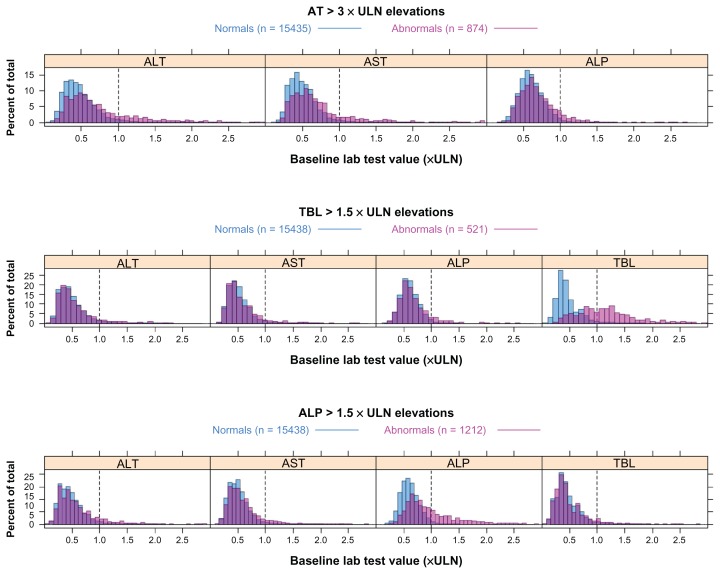
Out of the variables used for predictions (demographics, concomitant medications, medical history, and all laboratory results), the important variables for predicting individual liver signals were identified based on the variable importance scores derived from the random forest classification models (see Methods). Table 3 lists variables that were predictive across multiple projects as well as those that were specific to one project. The predictive variables are listed in the order of importance from high to low for each type of liver signal. The baseline variables that were predictive in multiple projects were further examined and their baseline level distributions are shown in Figure 2.
Table 3.
Important variables for predicting post-baseline liver signals
| Liver signals | Predictive baseline variables |
|---|---|
| AT elevation > 3 × ULN | ALT*, AST*, GGT, ALP |
| TBL elevation > 1.5 × ULN | TBL*, AST, PPC, CRP, HG |
| ALP elevation > 1.5 × ULN | ALP*, ALT*, AST*, GGT, TBL, CRP, LDH |
Note:
Predictive variables common to all projects.
Abbreviations: ALP, alkaline phosphatase; AT, aminotransferase; TBL, total bilirubin; ALT, alanine aminotransferase; AST, aspartate aminotransferase; GGT, γ-glutamyltransferase; PPC, platelet particle count; CRP, C-reactive protein; HG, Mercury; LDH, lactate dehydrogenase.
Figure 2.
Baseline distribution of lab tests important for predicting post-baseline liver signals.
Notes: Predicted liver signal is indicated by the heading. The dashed vertical line indicates the upper limits of normal (ULN) of the predictive lab test. Normal subjects are indicated in blue, abnormal in pink and the overlay of the two distributions is indicated in purple.
Abbreviations: ALP, alkaline phosphatase; AT, aminotransferase; TBL, total bilirubin.
The baseline values of ALT and AST were clearly important for determining a later AT elevation above 3 × ULN during treatment in all five projects (Table 3), while ALP and γ-glutamyltransferase (GGT) were important in only some projects. When investigating the actual values (in multiples of ULN) of AT and ALP at baseline, there is a shift towards higher levels for abnormal subjects (Figure 2). Also, GGT was important in the two projects where it was measured (Table 3). The importance of GGT is also supported when AT and ALP were removed from the training set. It is clear that GGT appears to be correlated with AT, which is consistent with previous publications.8,9 Whether it adds any predictive value to ALT and AST was further investigated below.
The only clear pattern at baseline for predicting TBL elevations above 1.5 × ULN is baseline bilirubin. This is a common feature of all studies and all projects. The distribution of bilirubin values at baseline is shifted towards higher levels for abnormal subjects, and the majority of the abnormal subjects already had a value above the ULN at baseline (Figure 2). Furthermore, AST, ALT, and ALP also seemed to shift slightly towards higher levels. Three other laboratory tests were ranked high in terms of importance (Table 3), two of them, C-reactive protein (CRP) and hepatoglobulin, were project-specific and could not be confirmed when the model was trained without the established liver chemistry tests. The third test, platelet particle count, was confirmed by distribution plots (data not shown) to have a weak association with TBL elevation during treatment.
ALP level at baseline was the most important variable for predicting ALP elevations during the trials (Table 3). This was a common pattern for every study in all five projects, which was also supported by random forest regression models (data not shown). The AT levels also seemed to be important (although less than ALP) in all projects. TBL, which was collected in all studies, was only important in a few studies for predicting ALP elevations. Interestingly, baseline GGT was collected in only two projects, but for all studies in these two projects, it was almost as important as baseline ALP. There is a clear shift of the distribution of ALP values in abnormal subjects towards higher baseline values (Figure 2). A similar shift is seen also in AT and GGT, although not as distinct as for ALP. Whether GGT adds any predictive value is investigated below. A shift of the bilirubin distribution was not observed (not shown). Other potentially predictive variables were CRP and lactate dehydrogenase, but both were only collected in one study.
Did baseline GGT add any predictive value?
It is important to investigate further the importance of baseline GGT in the prediction models of AT and ALP elevation. Also, considering that GGT was not a routine laboratory test in our clinical trials, it is crucial to determine whether GGT adds predictive value to the routine liver chemistry tests (ie, AT, ALP, and TBL). In order to test this, predictive models for liver signals were built on studies in projects B and E where GGT was measured. The accuracies of predictive models, which included GGT in training sets, were compared to those of models that excluded GGT in training sets. However, GGT did not increase predictability on top of already established liver chemistry tests (AT, ALP, and TBL), as measured by average prediction accuracy.
Predicting biochemical Hy’s law cases from baseline
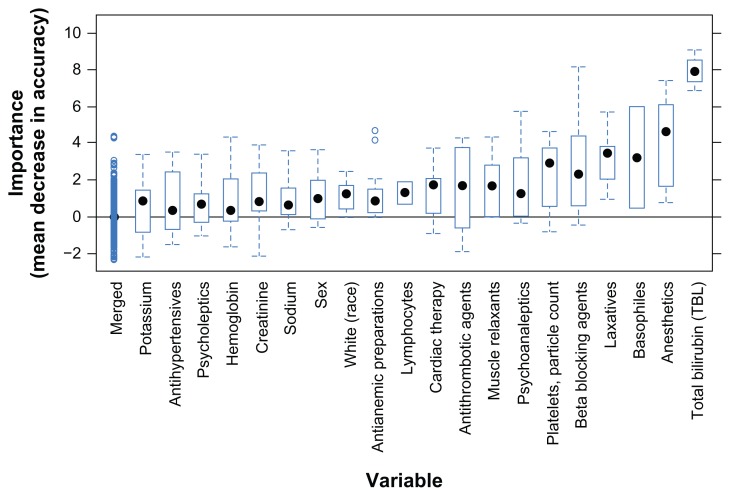
There were only a few biochemical Hy’s law cases (Table 1), therefore, data were pooled from all five projects to build predictive models. Altogether, 16 subjects with the biochemical Hy’s law criteria (AT > 3 × ULN, TBL > 2 × ULN, and ALP < 2 × ULN) were used to develop the random forest models. The models had an average accuracy of 75%. Bilirubin was the most important laboratory test at baseline for predicting biochemical Hy’s law cases (Figure 3). Counts of basophiles, platelets, and lymphocytes were also found somewhat important. Baseline value distribution (data not shown) show that biochemical Hy’s law cases had higher (usually above the normal range) bilirubin levels, but lower platelet, lymphocyte, and basophile counts at baseline. Any interpretation of the small shifts in these cell counts should be done with caution due to the small number of biochemical Hy’s law cases in this analysis.
Figure 3.
Variable importance for predicting biochemical Hy’s law cases from baseline.
Notes: Variable importance is shown as box plots ranked from low to high on x-axis. The most important variable in this dataset is total bilirubin. The importance of the rest lowest ranked variables are grouped together and indicated as “Merged”.
A range of concomitant medications was also identified by variable importance ranking (Figure 3), but remain inconclusive. Anesthetics, laxatives, psychoanaleptics, muscle relaxants, and antianemic preparations were not taken by any of the 16 subjects who had biochemical Hy’s law, but were taken by some patients with normal liver chemistry values (normal subjects). Antithrombotic agents, cardiac therapy, and beta-blocking agents were relatively more frequent among biochemical Hy’s law cases than normal cases. Note that all these observations were made from only 16 abnormal subjects, so no statistical or clinical significance should be inferred. For instance, the concomitant medications found as potential factors here could be simply due to the fact that the majority of biochemical Hy’s law cases came from cardiovascular trials, in which most patients took those medications for their cardiovascular conditions.
Temporal profiles of biochemical Hy’s law cases
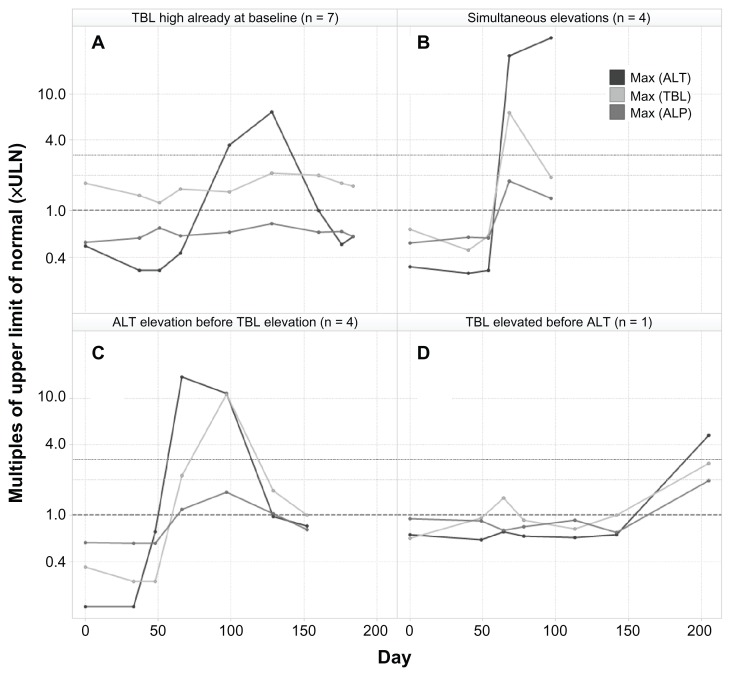
The biochemical definition of Hy’s law was used in the analysis (AT > 3 × ULN, TBL > 2 × ULN, ALP < 2 × ULN during treatment) and hence these cases are referred to as biochemical Hy’s law. The time course of the established liver chemistry tests for all 16 biochemical Hy’s law cases was examined, and a few types of temporal profiles were identified (Figure 4), as following: (A) high TBL (>1 × ULN) at baseline for almost half of the cases (7 of 16), which stayed high or went higher later when AT elevations occurred in the same patients; (B) simultaneous elevations of AT and TBL (4 of 16); (C) AT elevation preceding TBL elevation (4 of 16); and (D) TBL elevation preceding AT elevation (1 of 16), even though bilirubin was normal at baseline.
Figure 4.
Typical examples of different types of temporal profiles of biochemical Hy’s law cases.
Notes: These Hy’s law cases came from different studies, and therefore are of various durations. AST is not shown because it is usually highly correlated to ALT, and ALP is shown to be <2 × ULN.
Abbreviations: ALP, alkaline phosphatase; AST, aspartate aminotransferase; ALT, alanine aminotransferase; TBL, total bilirubin.
Baseline bilirubin levels, biochemical Hy’s law cases, and potential Gilbert’s syndrome
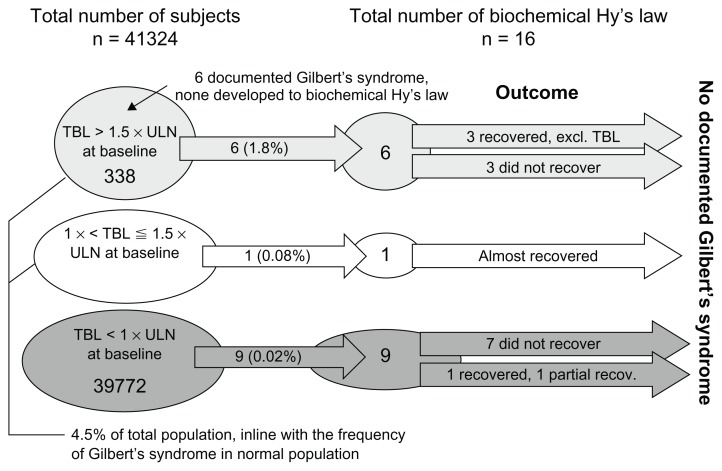
Out of the total of 41,324 patients, 1552 (3.8%) had baseline bilirubin levels > 1 × ULN, and 338 (~1%) had baseline bilirubin levels > 1.5 × ULN. Out of the 338 patients with baseline bilirubin levels > 1.5 × ULN, only six of them had Gilbert’s syndrome recorded in their medical history, but none of the six developed into Hy’s law cases during the trials (Figure 5). The small number of recorded Gilbert’s syndrome can be partially explained by that fact that Gilbert’s syndrome is usually underdiagnosed both in general practice and during the screening stage of clinical trials.
Figure 5.
Number of patients that had high baseline bilirubin levels, biochemical Hy’s law cases, and Gilbert’s syndrome.
Notes: Outcome here refers only to whether the biochemical signals recovered or not. Clinical outcomes of individual cases were not examined here.
Abbreviations: TBL, total bilirubin; ULN upper limit of normal.
Out of the 338 patients, six (1.8%) developed into biochemical Hy’s law cases during the trials, while only nine (0.02%) out of 39,772 patients with normal baseline bilirubin reached the biochemical Hy’s law criteria ( Figure 5). The occurrence of biochemical Hy’s law cases during the trials for subjects with baseline bilirubin > 1.5 × ULN (1.8%) was 78 times more likely than for those with normal baseline bilirubin (0.03%). In other words, the relative risk of a patient with a high baseline bilirubin developing biochemical Hy’s law was 78 times that of patients with a normal baseline bilirubin, even though the absolute risk (1.8%) was low.
The outcome of liver chemistry signals is also shown in Figure 5 for the 16 biochemical Hy’s law cases. In total, 10 of the 16 were not shown to have recovered, since they still met the biochemical criteria of Hy’s law cases at the last visit. It’s interesting to note that three of the six cases (Figure 5) that had bilirubin levels above 1.5 × ULN at baseline recovered (ie, not meeting biochemical criteria of Hy’s law) in later visits, while the majority of (seven of nine) cases that had normal bilirubin levels at baseline did not recover (ie, still meeting biochemical criteria of Hy’s law at the last visit). Also, during the trials, the trajectory of bilirubin changes seemed to be independent of ALT changes in those six cases (Figure 4), which had bilirubin levels above 1.5 × ULN at baseline. With a small number of cases here, however, it is not possible to test the statistical significance of these findings. Also, we didn’t obtain sufficient postelevation data to fully understand the recovery patterns, such as any actions taken on the investigational drug (eg, stopping treatment or changing dose).
Discussion
Retrospectively, predictive models using baseline data were able to predict which patients would develop liver signals during the trials with average validation accuracy around 80% (Figure 1). This finding supports an approach to identify at baseline what patient subpopulation has a high relative risk of developing liver signals during clinical trials. We plan to validate this finding with a prospective analysis in the future, since the predictability of baseline data will further improve when future projects are included in the analysis. Based on our finding that the more diverse data is pooled for analysis the better predictive models perform, we advocate more cross-pharma data sharing and analyses to address common questions regarding DILI signals in drug developments. Two examples are the Predictive Safety Testing Consortium10 and the Safer And Faster Evidence-based Translation Consortium. 11 More generalizable knowledge could be derived from such collaborations to inform decision making in diverse clinical programs.
Specifically, such knowledge might allow more proactive and targeted risk management for individual patients, especially when liver injury is a potential risk of the investigational drug. In addition, such information can make cost-effective clinical sampling possible for novel biomarker research. Collecting frequent clinical samples from all patients in a clinical trial may result in excessive costs and burden to patients, but selective sampling on high-risk patient groups might limit costs while enabling better safety monitoring and prospective research for novel biomarkers.
High bilirubin levels at baseline were found to be not uncommon, highly predictive of subsequent bilirubin elevations in clinical trials, and associated with higher risk of developing biochemical Hy’s law. Whether these high baseline values came from conjugated (direct) or unconjugated (indirect) bilirubin could not be determined because direct bilirubin was not routinely measured in these historical trials. Although none of the six patients who were documented to have Gilbert’s syndrome developed Hy’s law cases during the trials (Figure 5), we cannot totally rule out potential involvement of Gilbert’s syndrome in the category A profile of biochemical Hy’s law (Figure 4), considering that it could be underdiagnosed clinically.12,13 In future clinical trials, if TBL is elevated at baseline or any time during the trial, the sample can be fractionated to test for the contribution of direct and indirect bilirubin in the increase in total bilirubin. If there is an elevation of indirect bilirubin, Gilbert’s syndrome should be considered, though in that case, bilirubin should not rise above 4 mg/dL and AT should not increase. If there is a question of drug association and hemolysis is ruled out, then it is possible to genotype these patients to confirm Gilbert’s syndrome as the source of the increase in indirect bilirubin.
Among the four categories of temporal patterns observed from the biochemical Hy’s law cases, it is arguable whether category A should be even considered for potential Hy’s law. However, the current definition of Hy’s law1 doesn’t explicitly exclude such cases. While category D is a rare case, category C is common and is consistent with the clinical teaching, that transaminase elevation usually precedes bilirubin elevations in hepatocellular damage, the hallmark of a Hy’s law case. In category B, elevations of transaminase and bilirubin cannot be temporally separated, which could be due to infrequent lab testing. Whether or not these biochemical Hy’s law cases were true DILI cases was not determined by this analysis. Nevertheless, it would be helpful to clarify in Hy’s law definition: (1) whether a patient that had a bilirubin elevation before the ALT elevation should be disqualified from being considered a Hy’s law case; (2) whether a patient with a bilirubin elevation at baseline should be always excluded; and (3) if there should be a upper limit of the time interval between the ALT elevation and bilirubin elevation.
This study clearly showed an association between high baseline levels and subsequent elevation in liver chemistry tests, while no patient characteristics, such as demographics, concomitant medications, and medical history, appeared to be consistently associated with liver chemistry signals across different datasets. This suggested that the baseline pretreatment liver condition might be an important factor in determining how the liver reacts to the treatment as reflected by the liver chemistry tests. Related to this finding, there is a common practice to exclude patients from clinical trials because of baseline liver test abnormalities or a history of liver disease, to which the FDA guideline to the industry1 (page 10) commented that “there is no well-established reason to do this, except perhaps to avoid confusion between the previous disease and an effect of the test drug”, and that “patients with acute viral, autoimmune, alcoholic, or other types of hepatitis are unstable and generally not appropriate subjects for clinical trials other than trials of treatments for their acute illness.”
The FDA guideline1 (page 10) advised that “patients with stable liver disease generally should be included in at least some Phase III trials if they are likely to be treated with the drug if it is marketed.” The basis for the advice seems to be that “preexisting liver disease has not been thought to make patients more susceptible to DILI,14,15 but it may be that a diminished liver reserve or the ability to recover could make the consequences of injury worse.” Consistently, if benign conditions like Gilbert’s syndrome can be ruled out, frequent high baseline bilirubin levels (>1.5 × ULN) found here could be an early sign of liver metabolic or excretory function impairment,16 which made those patients more vulnerable to developing biochemical Hy’s law cases during clinical trials. Whether or not these biochemical Hy’s law cases were true DILI cases was not determined by this analysis. Nevertheless, any biochemical Hy’s law cases in clinical trials would trigger close examination of the case to determine if it is a real Hy’s law case by ruling out reasons other than the investigational drug, which is not the purpose of this analysis.
A final point related to our current study is the potential utility of this approach for assessing new biomarkers. The traditional markers for monitoring drug-induced liver injury as described here have been available for over 50 years and, although useful, they have limitations. As a result, many efforts are now underway to discover and develop improved biomarkers for monitoring DILI. Numerous new candidate biomarkers have been described ranging from small molecules to proteins to microRNAs, and commercial vendors are beginning to market these new biomarkers, often in panels containing multiple biomarkers. A significant challenge associated with these new assays, however, is to determine what added value they provide over existing assays. As described here, our analysis of GGT formally demonstrates that while results using this marker can be informative, there is little advantage provided by adding this assay versus using AT and bilirubin alone. As new, exploratory biomarkers make their way into clinical trials, similar approaches could be taken to assess these new biomarkers.
Conclusion
The baseline levels of individual liver chemistry tests were most important for predicting their own elevations during the trials. High bilirubin levels at baseline were not uncommon and were associated with a high risk of developing biochemical Hy’s law cases. Baseline GGT level appeared to have some predictive value, but did not increase predictability beyond using established liver chemistry tests.
In conclusion, it is possible to predict which patients are at a higher risk of developing liver chemistry signals using pretreatment (baseline) data. Derived knowledge from such predictions may allow proactive and targeted risk management, and the type of analysis described here could help determine whether new biomarkers offer improved performance over established ones.
Acknowledgments
We thank the following people for their support or contribution to this study: Harry Southworth, Lars Ståhle, Ina Schuppe-Koistinen, Gerry Kenna, John Pears, Michael Ramaker, Åsa Ström, Anastasia Christianson, and Marianne Keisu.
Footnotes
Disclosure
We were employees of AstraZeneca Pharmaceuticals LP during this study. There are no other potential conflicts of interest.
References
- 1.Food and Drug Administration. Guidance for Industry. Drug-Induced Liver Injury: Premarketing clinical evaluation. [Accessed November 9, 2012]. Available from: http://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/UCM174090.pdf.
- 2.Temple R. Hy’s law. Predicting serious hepatotoxicity. Pharmacoepidemiol Drug Saf. 2006;15(4):241–243. doi: 10.1002/pds.1211. [DOI] [PubMed] [Google Scholar]
- 3.Reuben A. Landmarks in hepatology. Hepatology. 2004;40:1478–1482. doi: 10.1002/hep.20526. [DOI] [PubMed] [Google Scholar]
- 4.Cai Z, Christianson AM, Ståhle L, Keisu M. Reexamining transaminase elevation in Phase I clinical trials: the importance of baseline and change from baseline. Eur J Clin Pharmacol. 2009;65:1025–1035. doi: 10.1007/s00228-009-0684-x. [DOI] [PubMed] [Google Scholar]
- 5.Clinical Data Interchange Standards Consortium. CDISC Study Data Tabulation Model. SDTM Implementation Guide V3.1.1. [Accessed February 12, 2009]. Available from: http://www.cdisc.org/models/sdtm/v1.1/index.html.
- 6.Breiman L. Random forests. Mach Learning. 2001;45:5–32. [Google Scholar]
- 7.Weil JG, Bains C, Linke A, Clark DW, Stirnadel HA, Hunt CM. Background incidence of liver chemistry abnormalities in a clinical trial population without underlying liver disease. Regul Toxicol Pharmacol. 2008;52:85–88. doi: 10.1016/j.yrtph.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 8.Xu Y, Bi YF, Xu M, et al. Cross-sectional and longitudinal association of serum alanine aminotransaminase and γ-glutamyltransferase with metabolic syndrome in middle-aged and elderly Chinese people. J Diabetes. 2011;3:38–47. doi: 10.1111/j.1753-0407.2010.00111.x. [DOI] [PubMed] [Google Scholar]
- 9.George J, Denney-Wilson E, Okely AD, Hardy LL, Aitken R. The population distributions, upper normal limits and correlations between liver tests among Australian adolescents. J Pediatr Child Health. 2008;44:579–585. doi: 10.1111/j.1440-1754.2008.01386.x. [DOI] [PubMed] [Google Scholar]
- 10.Critical Path Institute. Predictive Safety Testing Consortium. [Accessed February 27, 2011]. Available from: http://www.c-path.org/pstc.cfm.
- 11.SAFE-T. Safer And Faster Evidence-based Translation Consortium. [Accessed February 27, 2011]. Available from: http://www.imi-safe-t.eu/
- 12.Kasper DL, Braunwald E, Hauser S, et al. Harrison’s Principles of Internal Medicine. 16th ed. New York, USA: McGraw-Hill; 2005. [Google Scholar]
- 13.Boon NA, Davidson S. Davidson’s Principles and Practice of Medicine. 20th ed. London, UK: Churchill Livingstone; 2006. [Google Scholar]
- 14.Zimmerman HJ. Hepatotoxicity: the Adverse Effects of Drugs and Other Chemicals on the Liver. 1st ed. New York, NY: Appleton-Century-Crofts; 1978. Drug-induced liver disease; pp. 351–353. [Google Scholar]
- 15.Zimmerman HJ. Hepatotoxicity: the Adverse Effects of Drugs and Other Chemicals on the Liver. 2nd ed. Philadelphia, PA: Lippincott Williams & Wilkins; 1999. Drug-induced liver disease; pp. 428–433. [Google Scholar]
- 16.Schmid R. Bilirubin metabolism in man. N Engl J Med. 1972;287:703–709. doi: 10.1056/NEJM197210052871407. [DOI] [PubMed] [Google Scholar]