Abstract
The endocannabinoid 2-arachidonoylglycerol (2-AG) is biosynthesized by diacylglycerol lipases DAGLα and DAGLβ. Chemical probes to perturb DAGLs are needed to characterize endocannabinoid function in biological processes. Here, we report a series of in vivo-active 1,2,3-triazole urea inhibitors, along with paired negative-control and activity-based probes, for the functional analysis of DAGLβ in living systems. Optimized inhibitors showed excellent selectivity for DAGLβ over other serine hydrolases, including DAGLα (~60-fold selectivity), and the limited off-targets, such as ABHD6, were also inhibited by the negative-control probe. Using these agents and Daglb−/− mice, we show that DAGLβ inactivation lowers 2-AG, as well as arachidonic acid and eicosanoids, in mouse peritoneal macrophages in a manner that is distinct and complementary to disruption of cytosolic phospholipase-A2 (PLA2G4A). We observed a corresponding reduction in lipopolysaccharide-induced tumor necrosis factor-α release. These findings indicate that DAGLβ is a key metabolic hub within a lipid network that regulates proinflammatory responses in macrophages.
The endocannabinoids 2-AG1,2 and N-arachidonoyl ethanolamine (anandamide)3 are lipid signaling molecules that activate the G-protein-coupled receptors CB1 and CB24, which are also targets for the psychoactive ingredient in marijuana, Δ9-tetrahydrocannabinol5,6. The characterization and pharmacologic inactivation of key enzymes involved in terminating endocannabinoid signaling, such as the anandamide- and 2-AG-degrading enzymes fatty acid amide hydrolase (FAAH7) and monoacylglycerol lipase (MAGL or MGLL8,9), respectively, have shown that elevating endocannabinoid signaling can affect diverse behavioral processes, including pain, emotionality, and inflammation10–13. Much less is known, however, about the physiological effects of disrupting endocannabinoid production in vivo due, at least in part, to a lack of selective inhibitors for the enzymes that biosynthesize endocannabinoids.
2-AG biosynthesis is regulated by two sequence-related enzymes, diacylglycerol lipase-α and β (DAGLα and DAGLβ, respectively)14. Biochemical studies have provided evidence that these multi-domain, transmembrane serine hydrolases catalyze the sn-1-selective cleavage of arachidonate-containing diglycerides to form 2-AG14. Recent genetic studies with constitutive knockout mice have confirmed that both DAGLα and DAGLβ regulate 2-AG production in vivo, where the relative contribution made by each enzyme depends on tissue type15,16. DAGLα seems to be the principal regulator of 2-AG in the nervous system, where it controls the activity of this endocannabinoid as a retrograde messenger at neuronal synapses15,16 and supports adult neurogenesis15 and pain transmission17. Less is known about the function of DAGLβ, although its genetic disruption in mice leads to a moderate reduction in brain 2-AG and more substantial reductions in 2-AG and arachidonic acid in the liver15. To clarify the impact of DAGLs on 2-AG metabolism and cell and animal physiology, selective and in vivo-active inhibitors of these enzymes would be of value.
Known inhibitors of DAGL enzymes consist mainly of non-selective agents such as RHC80267 and tetrahydrolipstatin, which inactive numerous other mammalian serine hydrolases in proteomes18, as well as tetrahydrolipstatin analogues19 and DAG-like fluorophosphonates20, which have not yet been extensively evaluated for selectivity or in vivo activity. These deficiencies in known DAGL inhibitors have hindered their use as chemical probes of DAGL function in living systems. DAGL substrate assays that are compatible with high-throughput screening have only recently been described21 and have not yet, to our knowledge, been implemented for discovery of new classes of inhibitors. The pursuit of DAGL inhibitors would benefit from the development of assays to directly measure the endogenous activity of DAGL enzymes in proteomes. Finally, determining the selectivity of DAGL inhibitors is important because these enzymes belong to the serine hydrolase class, of which there are ≥200 members in humans that carry out a broad array of functions, including neurotransmitter degradation, peptide hormone processing, proteolysis, and lipid metabolism22.
We recently reported that 1,2,3-triazole ureas (1,2,3-TUs) are a versatile chemotype for the development of selective, irreversible serine hydrolase inhibitors23. Here, we describe screening of DAGL enzymes against a small library of 1,2,3-TUs using a competitive activity-based protein profiling (ABPP) assay24. Optimization of lead hits led to the discovery of two compounds, KT109 and KT172, that potently and selectively inactivated DAGLβ in vitro and in vivo. We confirmed DAGLβ inhibition by KT109 and KT172 in living systems by developing a tailored activity-based probe to report on endogenous DAGL enzymes in complex proteomes. Finally, we also generated a negative-control probe, KT195, that is structurally related to KT109 and KT172 but inactive against DAGLβ. We used this suite of chemical probes and Daglb−/− mice to show that DAGLβ is a principal 2-AG biosynthetic enzyme in peritoneal macrophages and that the enzyme also regulates arachidonic acid, prostaglandins, and TNF-α release in these cells.
RESULTS
Discovery of lead 1,2,3-TU inhibitors for DAGL enzymes
We screened DAGL enzymes against a synthetic library of 1,2,3-TUs, a class of small molecules that has well-suited features for serine hydrolase inhibitor development, including broad reactivity against diverse serine hydrolases, simplified synthetic routes for inhibitor optimization, and an ability to inactivate serine hydrolases in vivo23. To facilitate screening, we established a DAGL activity assay based on competitive ABPP methods using a fluorophosphonate-rhodamine (FP-Rh) probe. We first confirmed that HEK293T cells transfected with cDNAs for mouse DAGLα and DAGLβ enzymes showed higher DAGL activity compared with lysates from mock-transfected HEK293T cells using an LC-MS substrate assay (Supplementary Results, Supplementary Fig. 1). DAGLβ showed much higher activity in transfected cells compared with DAGLα, and this activity was blocked by treatment with FP-Rh with maximal inhibition occurring at ~5 µM (Supplementary Fig. 1). In contrast, the low level of activity observed in DAGLα-transfected cells was insensitive to FP-Rh treatment (Supplementary Fig. 1), consistent with previous findings indicating that DAGLα does not react well with the FP-Rh probe18. On the basis of these results, we established a gel-based competitive ABPP assay for assessing DAGLβ activity with FP-Rh and found that probe labeling was blocked by the non-specific lipase inhibitor tetrahydrolipstatin in a dose-dependent manner (Supplementary Fig. 1).
From a structurally diverse set of 30 1,2,3-TU compounds (1 – 30, Supplementary Fig. 2) screened at 500 nM against DAGLβ, we identified a lead inhibitor KT117 (25) that blocked FP-Rh labeling of DAGLβ > 50% (Supplementary Fig. 3). KT117 possesses a 2-substituted piperidine, a structural feature we have previously found to affect the relative potency of 1,4- and 2,4-regioisomers for 1,2,3-TUs23. We therefore compared the DAGLβ inhibitory activity of KT117 (2,4-regioisomer; Supplementary Fig. 3b) and KT116 (31, 1,4-regioisomer; Supplementary Fig. 3b) and found that the latter compound showed about ten-fold greater potency (Supplementary Fig. 3c, half-maximum inhibitory concentration (IC50) values of 300 and 30 nM, respectively). KT116 and KT117 also showed substantial differences in their selectivity profiles versus serine hydrolases as measured by competitive ABPP assays performed with FP-Rh in the mouse brain proteome (Supplementary Fig. 4). Crystal structures of KT116 and KT117 revealed a more compact structure for the former compound (Supplementary Fig. 3), with potential stacking interactions occurring between the phenyl ring of the 2-benzyl substituted piperidyl group and the triazole ring of the urea. This unusual conformation could explain, at least in part, the differences in activity of KT116 versus KT117, as well as the diminished signal of the triazole-ring proton in the 1H-NMR spectra for KT116 (see Supplementary Methods). These data, taken together, designated KT116 as a suitable lead inhibitor for DAGLβ. However, competitive ABPP experiments revealed that KT116 also inhibited several other serine hydrolases, including PLA2G7, ABHD6, MGLL, and FAAH, with IC50 values from 10 nM – 2 µM (Supplementary Fig. 3d). We therefore next sought to improve the selectivity of KT116 for DAGLβ.
Optimization of DAGLβ inhibitors
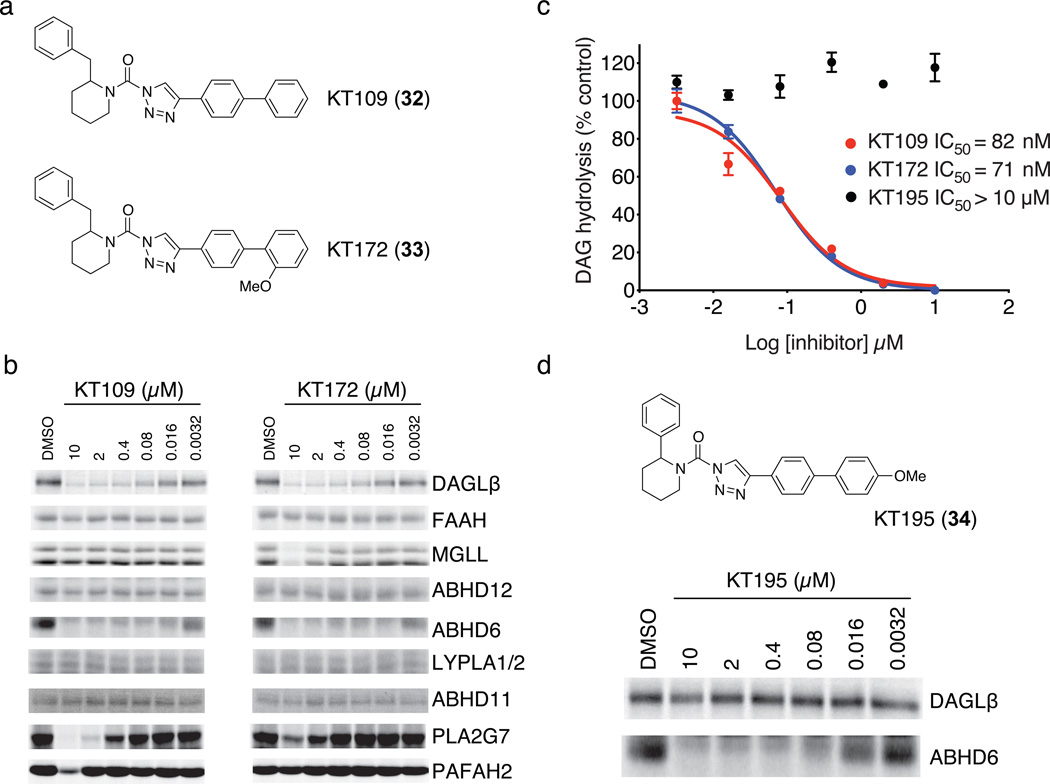
In the course of modifying KT116, we found that the compound KT109 (32), which contains a 4-biphenyl-substituted triazole group (Fig. 1a), showed negligible activity against FAAH, MGLL, and ABHD11 (Fig. 1b) while maintaining good potency against DAGLβ as measured by competitive ABPP (Fig. 1b; IC50 = 42 nM, see Supplementary Fig. 5) and LC-MS substrate (IC50 = 82 nM; Fig. 1c) assays. KT109 still showed some inhibitory activity against PLA2G7 (IC50 = 1 µM), which was mostly eliminated by modifying the distal phenyl ring appended to the triazole with an ortho-methoxy group (compound KT172 (33); Fig. 1a). KT172 showed near-equivalent activity against DAGLβ compared to KT109 (Fig. 1c and Supplementary Fig. 5), but also showed slight cross-reactivity with MGLL (IC50 = 5 µM; Fig. 1b). Neither KT172 nor KT109 showed inhibitory activity against cytosolic phospholipase A2 (cPLA2 or PLA2G4A) (Supplementary Fig. 6). The equivalent potencies shown by KT109 and KT172 for DAGLβ, combined with their complementary selectivity profiles, suggested that they would serve as a useful pair of probes for biological studies.
Figure 1. Optimization of 1,2,3-TU inhibitors for DAGLβ.
(a) Structures of optimized DAGLβ inhibitors KT109 and KT172. (b) Competitive ABPP of KT109 and KT172 against a panel of serine hydrolases detected in mouse brain proteome (FAAH, MGLL, ABHD12, ABHD6, LYPLA1 and LYPLA2 (LYPLA1/2)) or as enzymes recombinantly expressed in HEK293T cells (DAGLβ, ABHD11, PLA2G7, and PAFAH2). Proteomes were analyzed as described in Supplementary Fig. 3. See Supplementary Fig. 5 for IC50 values measured by competitive ABPP for KT109 and KT172 inhibition of DAGLβ. (c) In vitro IC50 values for DAGLβ inhibition by KT109 and KT172 measured with the SAG substrate assay following the protocol described in Supplementary Fig. 3c except SAG substrate was incubated with DAGLβ lysates for only 10 min at 37 °C after pretreatment with inhibitors. Data are mean ± s.e.m. for two independent experiments. 95% confidence intervals for IC50 values: KT109, 50–100 nM; KT172, 50–90 nM. (d) Structure and activity of control probe KT195. KT195 showed negligible cross-reactivity with recombinant DAGLβ (top) and concentration-dependent inhibition of ABHD6 (bottom) as measured by competitive ABPP.
KT109 and KT172 both possessed one remaining off-target, ABHD6 (IC50 values of 16 and 5 nM, respectively), which proved difficult to eliminate despite extensive medicinal chemistry efforts. However, we addressed this problem by generating a structurally related control compound, KT195 (34), that acted as a potent (IC50 = 10 nM) and selective inhibitor of ABHD6 with negligible activity against DAGLβ (Fig. 1d). KT195 also showed a comparable selectivity profile to KT109 and KT172 in competitive ABPP assays against other serine hydrolases (Supplementary Fig. 7). We therefore concluded that KT195 constituted a suitable control probe that could be used in biological studies to assign the pharmacological effects of KT109 and KT172 to DAGLβ versus ABHD6 inhibition.
Development of a DAGL-tailored activity-based probe
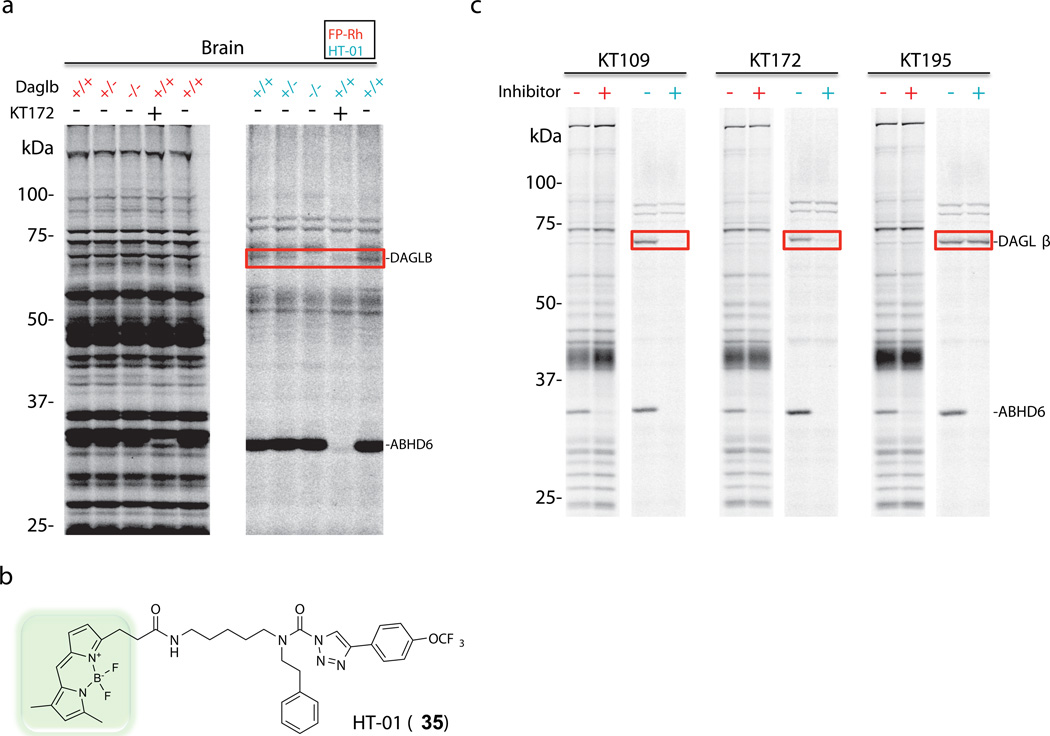
The low expression level of DAGLβ in cells and tissues hindered its detection with broad-spectrum activity-based probes like FP-Rh, which also reacted with more abundant, co-migrating serine hydrolases that masked DAGLβ signals (Fig. 2a, red samples). We surmised that this problem could be addressed by synthesizing an activity-based probe based on the general structure of 1,2,3-TU inhibitors of DAGLβ. In brief, opening the piperidyl ring of DAGLβ inhibitors facilitated attachment of a BODIPY fluorophore to yield probe HT-01 (35) (Fig. 2b), which labeled both DAGLβ and DAGLα (Supplementary Fig. 5). HT-01 was ~five-fold more active against DAGLβ than FP-Rh (Supplementary Fig. 8) and showed a much more restricted labeling profile in mouse brain (Fig. 2a, blue samples), primary neurons (Supplementary Fig. 9), and Neuro2A neuroblastoma cell line (Fig. 2c, blue samples) proteomes that enabled direct detection of DAGLβ activity in these samples. The ~70 kDa, HT-01-reactive protein was confirmed to be DAGLβ by: i) pretreatment of proteomes with our panel inhibitors, which showed that the protein’s reactivity was blocked by pre-treatment with KT109 and KT172, but insensitive to KT195 (Fig. 2a, c and Supplementary Fig. 9), and ii) its absence in brain proteomes from Daglb−/− mice (Fig. 2a). Profiling a larger panel of tissues from Daglb+/+, Daglb+/−, and Daglb−/− mice showed that HT-01 could also detect DAGLβ activity in spinal cord, pancreas, and white adipose tissues (Supplementary Fig. 10).
Figure 2. Development of an activity-based probe tailored for profiling DAGLβ.
(a) Mouse brain proteomes from Daglb+/+, Daglb+/−, and Daglb−/− mice were profiled using the broad-spectrum serine hydrolase probe FP-Rh (1 µM, red samples) or the DAGL-directed probe HT-01 (1 µM, blue samples). Mouse brain proteomes were incubated with probes for 30 min, separated by SDS-PAGE, and serine hydrolase activities detected by in-gel fluorescence scanning. Pretreatment of mouse brain proteome from Daglb+/+ mice with KT172 (2 µM) blocked probe-labeling of the ~70 kDa DAGLβ (red box) and 30-kDa ABHD6 activities. (b) Structure of the DAGL-directed activity-based probe HT-01. (c) Neuro2A membrane proteomes were profiled using FP-Rh (red samples) or HT-01 (blue samples). Cells were treated in situ with KT109 (50 nM), KT172 (25 nM), or KT195 (25 nM) for 4 h and lysed, and membrane proteomes were analyzed by competitive ABPP with either FP-Rh (1 µM) or HT-01 (1 µM). DAGLβ activity, and its inhibition by KT109 and KT172, could only be detected with the HT-01 probe (red box).
The reactivity of HT-01 with DAGLα (Supplementary Fig. 5) enabled us to evaluate the selectivity of inhibitors against this enzyme, which was previously not possible due to its negligible reactivity with FP-Rh (Supplementary Fig. 1). IC50 values of 2.3 and 0.14 µM were determined by competitive ABPP using the HT-01 probe for inhibition of recombinant DAGLα by KT109 and KT172, respectively (Supplementary Fig. 5). Comparison of these IC50 values to those measured in this assay format for inhibition of DAGLβ (IC50 values of 0.04 and 0.06 µM for KT109 and KT172, respectively; Supplementary Fig. 5) indicates that KT109 can be considered an isoform-selective inhibitor of DAGLβ, whereas KT172 exhibits similar activity against both DAGLα and DAGLβ.
DAGLβ regulates 2-AG production in Neuro2A cells
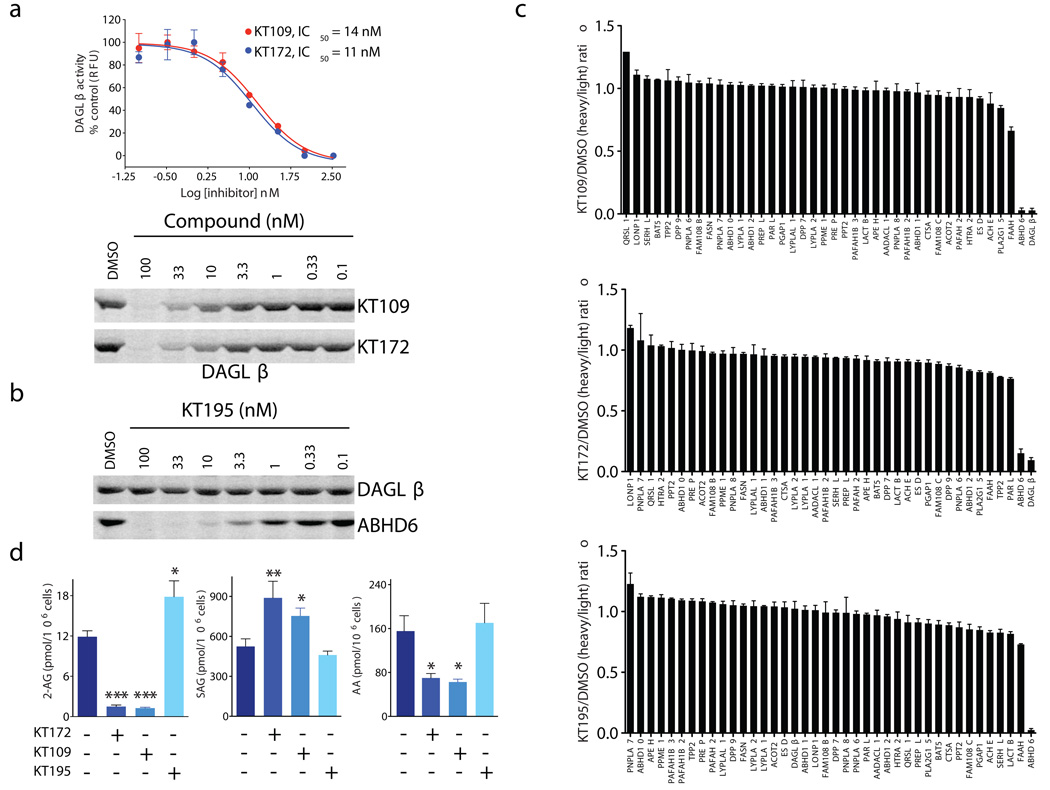
We next asked whether KT109 and KT172 selectively inhibit DAGLβ in living cells. Neuro2A cells were incubated with a range of concentrations of KT109, KT172, or KT195 for 4 h and then lysed and analyzed by competitive ABPP. Both KT109 and KT172 completely inactivated DAGLβ in Neuro2A cells with low-nanomolar potency (IC50 values of 14 and 11 nM, respectively; Fig. 3a). As we expected on the basis of their in vitro selectivity profiles, KT109 and KT172 also inactivated ABHD6 in Neuro2A cells (Supplementary Fig. 11), but did not inhibit any of the other serine hydrolases detected by gel-based ABPP (Fig. 2c, red samples). In contrast, the control-probe KT195 showed negligible activity against DAGLβ while completely inactivating ABHD6 with an IC50 value of ~1 nM (Fig. 3b). To more completely assess the selectivity of DAGLβ inhibitors in cells, we next used the quantitative mass spectrometry-based proteomic method ABPP-SILAC23,25 (Supplementary Fig. 12). We treated cells for 4 h with each compound (25 nM for KT172 and KT195; 50 nM for KT109) prior to analysis following our previously described ABPP-SILAC procedure. We found that each inhibitor showed high selectivity for its respective target; both KT109 and KT172 blocked ≥90% of DAGLβ activity and had negligible activity against other serine hydrolases detected in the Neuro2A proteome with the exception of ABHD6 (Fig. 3c, top). KT195 blocked ABHD6 activity by >90% with no activity against other serine hydrolase detected in the Neuro2A proteome, including DAGLβ (Fig. 3c, bottom). We also performed a control experiment where heavy and light cells were both treated with DMSO, which confirmed that serine hydrolase activities were not changed by treatment with isotopically-labeled media (Supplementary Fig. 13).
Figure 3. In situ activity of DAGLβ inhibitors in Neuro2A cells.
(a) In situ IC50 values for DAGLβ inhibition by KT109 and KT172 in Neuro2A cells as measure by competitive ABPP with the HT-01 probe. 95% confidence intervals for IC50 values: KT109, 9–23 nM; KT172, 6–20 nM). (b) Concentration-dependent inhibition of ABHD6 in Neuro2A cells by KT195. KT195 showed negligible activity against DAGLβ in Neuro2A cells. (c) ABPP-SILAC analysis of serine hydrolase activities from Neuro2A cells treated in situ with KT109 (50 nM), KT172 (25 nM), or KT195 (25 nM) for 4 h. KT109 and KT172 both inhibited DAGLβ and ABHD6 by ≥ 90%, but did not inhibit any of the other detected serine hydrolases. KT195 inhibited ABHD6 by > 90%, but did not affect DAGLβ activity. Error bars represent mean ± s.e.m. of heavy/light ratios for the multiple peptides observed for each enzyme (minimum of two unique peptides per enzyme) in both soluble and membrane fractions. See Supplementary Dataset 1 for complete proteomic data. (d) Inhibition of DAGLβ in Neuro2A cells with KT109 or KT172 (50 and 25 nM, respectively, 4 h) decreased 2-AG and arachidonic acid (AA), and increased SAG compared with DMSO or KT195 (25 nM, 4 h) treated cells. Data are mean ± s.e.m.; n = 5–6 per group. *p < 0.05; **p < 0.01 for inhibitor- versus DMSO-treated cells.
We conducted a metabolomic analysis of Neuro2A cells treated with KT109, KT172, and KT195 to assess the role of DAGLβ in 2-AG biosynthesis. Given the current model for DAGLβ function26,27 and past work showing a reduction in 2-AG and elevations in DAG substrate in Neuro2A cells treated with a DAGLα short hairpin RNA (shRNA) probe28, DAGL enzymes would be expected to convert DAG lipids containing an sn-2 C20:4 (arachidonoyl) acyl chain to 2-AG (Supplementary Fig. 14). Consistent with this model, cells treated for 4 h with KT109 or KT172 (50 and 25 nM, respectively) showed a marked reduction in cellular 2-AG (~90%) compared with untreated or KT195-treated cells (Fig. 3d). Notably, we also observed a significant (KT109, P = 0.018; KT172, P = 0.0098) accumulation of the stearoyl-arachidonoyl (C18:0/C20:4) DAG (SAG, Fig. 3d), designating this lipid species as a probable endogenous DAGLβ substrate. Smaller but still significant elevations in C16:0/C20:4 (KT172, P = 0.0421) and C18:1/C20:4 (KT172, P = 0.0258) DAGs were also observed in DAGLβ-inhibited cells, but no changes were observed in C14:0/C20:4 DAG (Supplementary Fig. 14). These data suggest that DAGLβ prefers DAG substrates with sn-1 acyl chains that are C16 or greater in length. Finally, we also found that DAGLβ inhibition significantly decreased arachidonic acid in Neuro2A cells (Fig. 3d), consistent with previous studies documenting reductions in arachidonic acid in liver tissue from Daglb−/− mice15. These data, combined with past studies using shRNA-mediated knockdown of DAGLα28, indicate that both DAGLβ and DAGLα contribute to 2-AG biosynthesis in Neuro2A cells.
We also found that KT109 and KT172, but not KT195 inhibited human DAGLβ activity with in vitro IC50 values of 580 and 380 nM, respectively (Supplementary Fig. 15). The control probe KT195 did not inhibit human DAGLβ (IC50 > 10 µM; Supplementary Fig. 15). KT109 and KT172 (100 nM, 4 hr) also blocked DAGLβ activity in situ in the human prostate cancer cell line PC3 with high selectivity, showing detectable cross-reactivity against only a single additional target by gel-based competitive ABPP, human ABHD6, which was also inhibited by the control probe KT195 (Supplementary Fig. 15). PC3 cells treated with KT109 and KT172, but not KT195 showed significant elevations in SAG (KT109, P = 0.0004; KT172, P = 0.0078) and reductions in 2-AG (KT109, P < 0.0001; KT172, P < 0.0001) and arachidonic acid (KT109, P = 0.0047; KT172, P = 0.0013) (Supplementary Fig. 15). These data indicate that KT109 and KT172 can be used as selective chemical probes for both the mouse and human orthologues of DAGLβ.
Inactivation of DAGLβ in macrophages in vivo
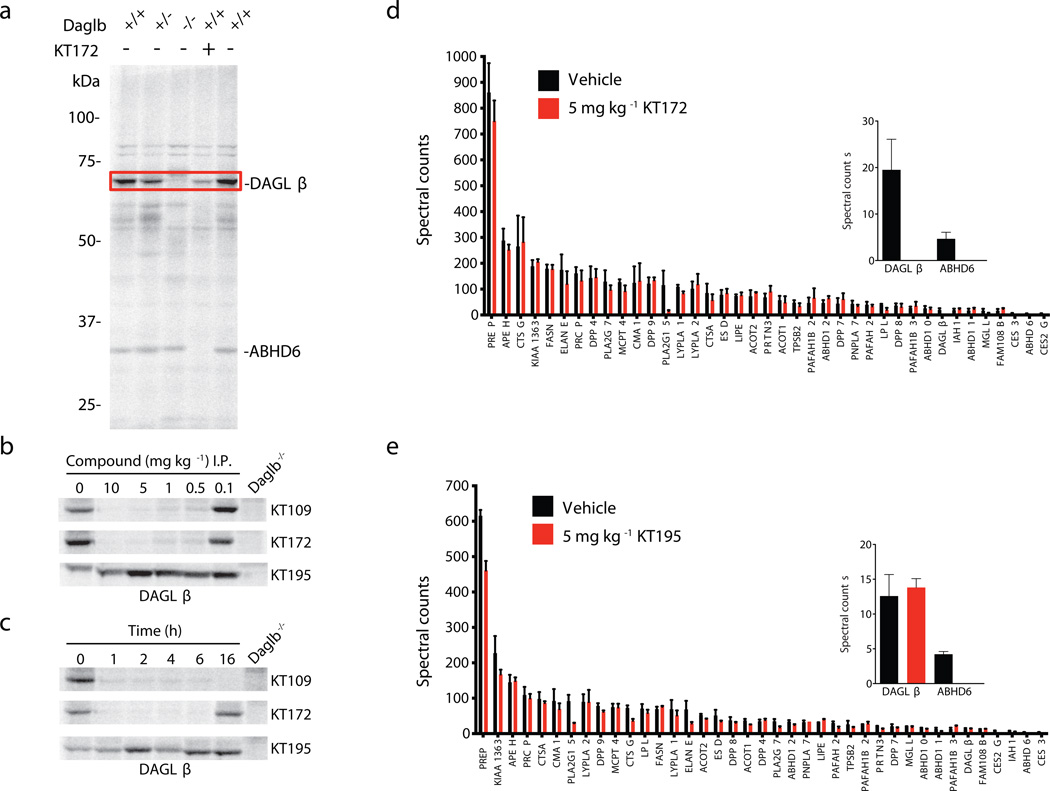
For our in vivo studies, we focused mainly on DAGLβ in peritoneal macrophages from inhibitor-treated mice, as large-scale gene expression (BioGPS, http://biogps.gnf.org/; Supplementary Fig. 16) and our own activity-based proteomic (Fig. 4a and Supplementary Fig. 17) profiles designated these cells as having high DAGLβ expression. Mice were first treated intraperitoneally (i.p.) with 4% (w/v) thioglycollate 4 d before DAGLβ inhibitors to induce recruitment of macrophages to the peritoneal cavity. Mice were treated with KT109, KT172, or KT195 at various doses (0.1 – 10 mg per kg body weight, i.p.) for 4 h, sacrificed, and thioglycollate-elicited peritoneal macrophages were collected, lysed, and analyzed by gel-based competitive ABPP using HT-01. Both KT172 and KT109 completely inactivated DAGLβ at doses as low as 0.5 mg per kg body weight (Fig. 4b, macrophage proteome from Daglb−/− mice is shown as a control). In contrast, KT195 showed no activity against DAGLβ at any dose tested (Fig. 4b). Time-course studies revealed that both KT109 and KT172 produced complete inhibition of macrophage DAGLβ by 1 h after in vivo treatment, and this inhibition was maintained for 6 h with KT172 and > 16 h with KT109 (Fig. 4c). KT195 showed no evidence of DAGLβ inhibition across the entire time-course of treatment (Fig. 4c).
Figure 4. Profiling DAGLβ activity in macrophages from inhibitor-treated mice.
(a) Mouse peritoneal macrophage proteomes from Daglb+/+, Daglb+/−, and Daglb−/− mice were profiled using the HT-01 probe (1 µM). Pretreatment of Daglb+/+ macrophage proteomes with KT172 (2 µM) blocked HT-01-labeling of DAGLβ (red box) and ABHD6 activities. (b) Dose-dependent inhibition of DAGLβ in peritoneal macrophages from mice treated with KT109, KT172, or KT195 (i.p., 4 h) measured by competitive ABPP using HT-01 (1 µM). Macrophage proteomes from Daglb−/− mice are shown as a control. (c) Time-course of inhibition of DAGLβ in peritoneal macrophages from mice treated with KT109, KT172, or KT195 (5 mg per kg body weight, i.p.) for various times (1 – 16 h) measured by competitive ABPP using HT-01 (1 µM). (d, e) ABPP-MudPIT analysis of macrophage serine hydrolase activities from mice treated with (d) KT172 or (e) KT195 (5 mg per kg body weight, i.p., 4 h). Treatment with KT172 completely inactivated DAGLβ and ABHD6, whereas treatment with KT195 inhibited ABHD6, but not DAGLβ (highlighted in inset bar graph). Data are presented as means ± s.e.m.; n = 5 mice per group. See Supplementary Figure 17 for ABPP-MudPIT results for KT109 and Supplementary Dataset 2 for complete proteomic data.
Gel-based ABPP experiments indicated that both KT109 and KT172 maintained good selectivity at doses that completely inactivated DAGLβ (Supplementary Fig. 17). As we expected, both compounds and KT195 also inhibited ABHD6, which is weakly expressed in macrophages and was barely detectable above the signal limits of gel-based ABPP (Supplementary Fig. 17). To more comprehensively profile inhibitor selectivity in vivo, we used the LC/MS method ABPP-MudPIT29 to measure serine hydrolase activities in macrophages from mice treated with KT109, KT172, or KT195 (2 or 5 mg per kg body weight, i.p., 4 h). These analyses identified ~40 serine hydrolases in the peritoneal macrophage proteome and confirmed inhibition of DAGLβ (and ABHD6) by both KT172 and KT109 (Fig. 4d and Supplementary Fig. 17). In contrast, KT195 inhibited ABHD6 but not DAGLβ (Fig. 4e). All three inhibitors also inhibited two carboxylesterases (CES3 and CES2G) that are common targets of mechanism-based serine hydrolase inhibitors in rodents30,31. We also identified an additional potential off-target PLA2G15, which was reduced in activity ~50–80% by all three inhibitors (Fig. 4d, e and Supplementary Fig. 17). We confirmed that recombinantly expressed PLA2G15 is weakly inhibited by KT109, KT172, and KT195 with IC50 values in the low µM range (Supplementary Fig. 18).
These data, taken together, indicate that KT109 and KT172 inhibit DAGLβ in vivo with good potency (< 5 mg per kg body weight) and selectivity, exhibiting cross-reactivity with only a handful of additional serine hydrolases in macrophages that are also inhibited by the control DAGLβ-inactive probe KT195.
DAGLβ regulates a lipid signaling network in macrophages
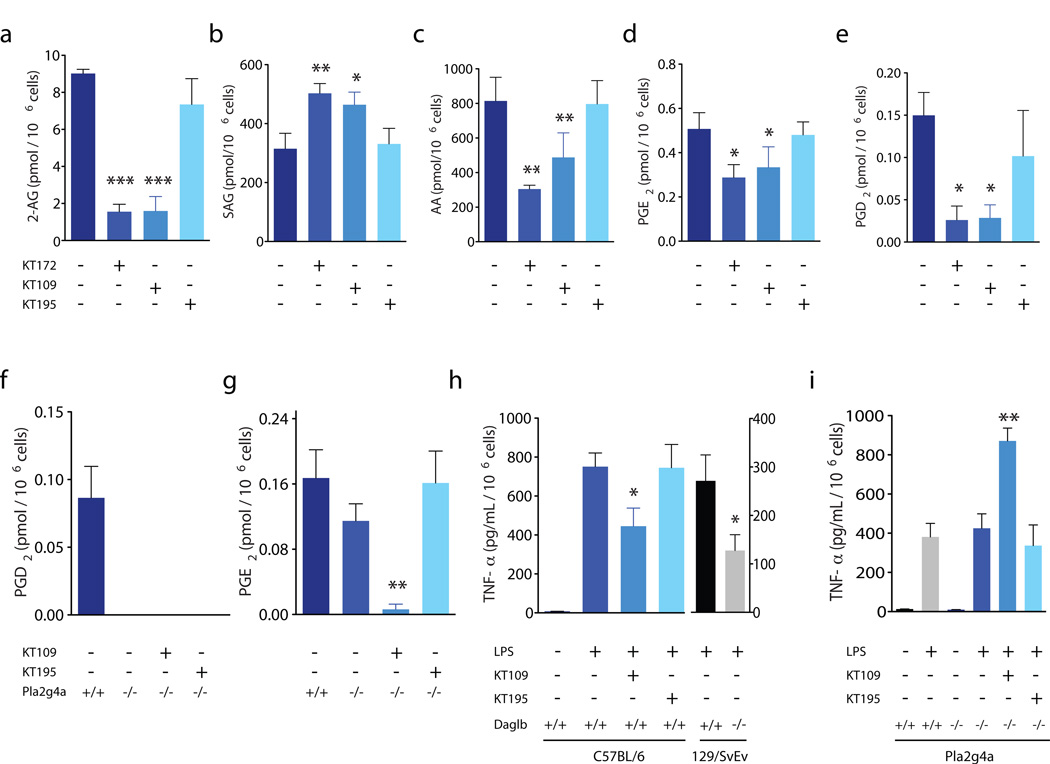
We next examined whether blockade of DAGLβ in peritoneal macrophages would produce similar metabolic effects to those observed in Neuro2A cells. LC/MS analysis showed marked decreases in 2-AG content (Fig. 5a) in macrophages isolated from KT109- and KT172-treated mice (5 mg per kg body weight, 4 h i.p.), whereas no such 2-AG changes were observed in macrophages from KT195-treated mice (5 mg per kg body weight, 4 h, i.p.). Macrophages from mice treated with KT109 and KT172, but not with KT195, also showed significant increases in the DAGLβ substrate SAG (KT109, P = 0.0298; KT172, P = 0.0040) (Fig. 5b) and reductions in arachidonic acid (KT109, P = 0.0012; KT172, P = 0.0050) (Fig. 5c) and in the arachidonic acid-derived prostaglandins PGE2 (KT109, P = 0.0271; KT172, P = 0.0358) and PGD2 (KT109, P = 0.0163; KT172, P = 0.0427) (Fig. 5d and e). Most of these metabolic effects were recapitulated in macrophages from Daglb−/− mice (Supplementary Fig. 19), with the notable exception of SAG, which were paradoxically lower in Daglb−/− macrophages (Supplementary Fig. 19). Chronic genetic disruption of DAGLβ could lead to compensatory pathways that lower cellular SAG in macrophages. We also found that mice treated with KT109 and KT172, but not KT195 (20 mg per kg body weight, 4 h, i.p.), showed reductions in 2-AG and arachidonic acid in liver that was similar in magnitude to the reductions in these lipids in liver tissue from Daglb−/− mice (Supplementary Fig. 20a,b). In contrast, mice treated with KT109 and KT172, or Daglb−/− mice, did not show changes in 2-AG and arachidonic acid in brain tissue (Supplementary Fig. 20c,d), consistent with a principal role for DAGLα in regulating endocannbinoid biosynthesis in the nervous system15,16. We do not fully understand the lack of activity of KT172 against brain DAGLα in vivo (Supplementary Fig. 21), given that this agent can inhibit brain DAGLα in vitro (Supplementary Fig. 21), but KT172 might exhibit insufficient central nervous system penetrance to inactivate DAGLα in vivo.
Figure 5. Metabolic effects of DAGLβ inactivation in macrophages.
(a–e) Peritoneal macrophages from KT109- or KT172-treated, but not KT195-treated mice (5 mg per kg body weight, i.p. 4 h) showed significant reductions in 2-AG (a), AA (c), PGE2 (d), and PGD2 (e) and significant elevations in SAG (b) compared to macrophages isolated from vehicle-treated mice. (f, g) Peritoneal macrophages from Pla2g4a−/− mice showed marked reductions in PGD2 (f) and modest reductions in PGE2 (g) compared with Pla2g4a+/+ mice. Treatment of Pla2g4a−/− mice with KT109, but not KT195 (5 mg per kg body weight, i.p. 4 h) further reduced macrophage PGE2 content. (h) Lipopolysaccharide (LPS)-stimulated TNF-α levels secreted by macrophages from mice treated with vehicle, KT109, or KT195 (5 mg per kg body weight, i.p. 4 h). Macrophages were stimulated with LPS (5 µg mL−1, 90 min) prior to analysis of secreted TNF-α in conditioned media by ELISA. Daglb+/+ mice (C57BL/6) treated with KT109 show significant reductions in TNF-α compared to vehicle-treated or KT195-treated mice. TNF-α levels were also significantly reduced in Daglb−/− compared with Daglb+/+ mice (129/SvEv). (i) LPS-stimulated TNF-α was not significantly different between macrophages from Pla2g4a+/+ and Pla2g4a−/− mice. Treatment of Pla2g4a−/− mice with KT109, however, significantly enhanced TNF-α; KT195 did not produce this effect. Data are presented as means ± s.e.m.; n = 3–5 mice per group. *p < 0.05; **p < 0.01; ***p < 0.001 for all inhibitor-treated groups or Daglb−/− versus vehicle-treated or Daglb+/+ mice.
That DAGLβ regulates not only 2-AG, but also arachidonic acid and prostaglandins in macrophages was notable, considering regulation of the arachidonic acid pools for prostaglandin production in these cells has largely been attributed to PLA2G4A32,33. We were therefore interested in comparing arachidonic acid and prostaglandin content in Pla2g4a−/− mice treated with DAGLβ or control inhibitors. Thioglycollate-elicited macrophages from Pla2g4a+/+ mice or Pla2g4a−/− mice treated with KT109 or KT195 (5 mg per kg body weight, 4 h, i.p.) were collected and subjected to metabolomic analysis. We found that, whereas macrophages from Pla2g4a−/− mice showed a complete loss of PGD2 (Fig. 5f), these cells exhibited only a modest reduction in PGE2 levels (Fig. 5g). PGE2 in Pla2g4a−/− macrophages was strongly decreased by treatment with KT109 but was unaffected by the control probe KT195 (Fig. 5g). That PGE2 was much lower in macrophages from KT109-treated Pla2g4a−/− mice compared with KT109-treated wild-type mice or untreated Pla2g4a−/− mice (Fig. 5g) indicates DAGLβ and PLA2G4A coordinately regulate the production of this prostaglandin in macrophages.
Having discovered that DAGLβ regulates multiple signaling lipids in macrophages (endocannabinoids, diacylglycerols, and prostaglandins), we proceeded to ask whether blocking this enzyme altered macrophage inflammatory responses. Thioglycollate-elicited macrophages from mice treated with KT109 or KT195 (5 mg per kg body weight, 4 h i.p.) were collected, plated, and stimulated with the pro-inflammatory agent lipopolysaccharide. Lipopolysaccharide-stimulated macrophages (5 µg mL−1, 90 min) showed a marked increase in tumor necrosis factor-α (TNF-α) compared with nonstimulated macrophages as measured by ELISAs of their conditioned media (Fig. 5h). Treatment with KT109, but not KT195 significantly reduced secreted TNF-α levels in lipopolysaccharide-stimulated macrophages, and this suppressive effect was also observed in macrophages from Daglb−/− mice (Fig. 5h).
Next, we compared the effects of dual blockade of DAGLβ and PLA2G4A on TNF-α production in macrophages. Peritoneal macrophages from both Pla2g4a+/+ and Pla2g4a−/− mice showed robust secretion of TNF-α upon stimulation with lipopolysaccharide (Fig. 5i). Unexpectedly, however, peritoneal macrophages from KT109-treated, but not KT195-treated Pla2g4a−/− mice showed significantly more secreted TNF-α (P = 0.0022) following lipopolysaccharide stimulation (Fig. 5i). This enhanced secretion of TNF-α resembles the effects previously reported for COX inhibitors, which have been attributed to blocking the production of an autocrine PGE2 signal that attenuates TNF-α release from macrophages34–36. Our data thus indicate that, whereas individually blocking DAGLβ or PLA2G4A is insufficient to enhance TNF-α release from lipopolysaccharide-stimulated macrophages (and, in the case of DAGLβ inhibition, produces the opposite effect of lowering TNF-α), dual disruption of these two arachidonic acid-producing enzymes causes a significant enough reduction in PGE2 (P = 0.0070) (Fig. 5g) to eliminate the suppressive effect of this prostaglandin on TNF-α production (Fig. 5i).
We also performed a complementary set of studies in which thioglycollate-elicited macrophages were isolated, plated for 4 hr, and then treated with inhibitors for an additional 4 hr, followed by treatment with lipopolysaccharide or vehicle for 90 min. We first confirmed that KT109 and KT172, but not KT195 inhibited macrophage DAGLβ activity by competitive ABPP (Supplementary Fig. 22) and then performed lipid and cytokine measurements. Comparison of vehicle versus lipopolysaccharide-treated macrophages (without DAGLβ inhibition) showed variable responses to lipopolysaccharide among the measured lipids, including a reduction in 2-AG, no change in arachidonic acid, and elevations in AEA, SAG, and a variety of eicosanoids, including PGD2, PGE2, thromboxane B2 (TBX2), and leukotriene B4 (LTB4; Supplementary Fig. 23). Similar to the in vivo inhibitor treatment paradigm, we found that KT109 and KT172, but not KT195 caused substantial reductions in 2-AG, arachidonic acid, and all of the measured eicosanoids (PGD2, PGE2, TBX2, LTB4) in lipopolysaccharide-treated macrophages (Supplementary Fig. 23). DAGLβ inhibitors did not cause changes in AEA or SAG in lipopolysaccharide-treated macrophages (Supplementary Fig. 23). KT109, but not KT195 also reduced lipopolysaccharide-stimulated TNF-α from plated macrophages (Supplementary Fig. 24), whereas neither compound affected basal TNF-α secretion (Supplementary Fig. 24). Both KT109 and KT195 also lowered IL1–β secretion from lipopolysaccharide-treated macrophages (Supplementary Fig. 24), suggesting that DAGLβ and ABHD6 may each regulate this cytokine. We did not detect appreciable IL-4 or IL-10 secretion from lipopolysaccharide-stimulated macrophages (Supplementary Fig. 24). A similar overall profile of lipid and cytokine changes was observed in lipopolysaccharide-stimulated macrophages from Daglb−/− mice, including substantial reductions in lipopolysaccharide-stimulated eicosanoids (Supplementary Fig. 25) and cytokines TNF-α and IL1–β (Supplementary Fig. 24).
Discussion
DAGL enzymes occupy a special metabolic hub in mammalian cells that integrate multiple lipid signaling pathways, including DAG37, endocannabinoid38, and eicosanoid39 networks. Understanding the role that DAGLs play in mammalian physiology and disease has, so far been hampered by a lack of selective and in vivo-active inhibitors for these enzymes. Here, we have addressed this problem by creating a versatile set of chemical probes for both profiling and perturbing DAGLβ activity in living systems.
We discovered that the 1,4-regioisomer of our lead 1,2,3-TUs was substantially more potent as a DAGLβ inhibitor compared to the 2,4-regioisomer (Supplementary Fig. 3c). This result contrasts with past work, where we observed nearly equivalent activity for the 1,4- and 2,4-regioisomers for most 1,2,3-TU inhibitors of other serine hydrolases23. The main structural difference found in DAGLβ inhibitors is a 2-benzyl substitution that de-symmetrizes the piperidine ring. Although we do not yet understand how this substitution pattern endows 1,4-regioisomers with preferential activity as DAGLβ inhibitors, small-molecule crystal structures uncovered a marked difference in the relative conformations of the 1,4- and 2,4-regioisomers, KT116 and KT117, respectively (Supplementary Fig. 3b). We speculate that these conformational differences may contribute to the superior activity of 1,4-regioisomeric 1,2,3-TUs as DAGLβ inhibitors.
We used our optimized inhibitors to discover that DAGLβ regulates multiple classes of lipid mediators in primary mouse macrophages, including elevations in DAG substrates and reductions in the endocannabinoid product 2-AG and the downstream lipids arachidonic acid and prostaglandins. Whereas the impact of DAGLβ on DAG and 2-AG are most likely reflective of direct substrate-product relationships, the enzyme’s control over arachidonic acid and prostaglandins is more probably indirect, occurring through the sequential conversion of 2-AG to arachidonic acid by other hydrolases like MGLL13 (Supplementary Fig. 14) and oxidative metabolism of arachidonic acid to prostaglandins by COX enzymes39. Concomitant with these changes in lipid metabolism, DAGLβ inhibitors also substantially reduced the lipopolysaccharide-stimulated production of the proinflammatory cytokine TNF-α. The impact of DAGLβ inhibitors on prostaglandins was initially surprising given a large body of literature supporting a primary role for PLA2G4A in regulating eicosanoid production in macrophages32,33,40. Using a combination of inhibitors and knockout mice, we generated strong evidence that both DAGLβ and PLA2G4A contribute to prostaglandin production in lipopolysaccharide-stimulated macrophages. The distinct and complementary roles played by DAGLβ and PLA2G4A were perhaps best exemplified by the metabolic profile of PGE2, which was modestly reduced by PLA2G4A disruption, more substantially but still partially decreased by DAGLβ inactivation and nearly completely eliminated by dual blockade of both enzymes. We suspect that these metabolic effects are at least partly responsible for the increase in TNF-α release observed following dual inhibition of DAGLβ and PLA2G4A, given that past work with COX inhibitors has shown that PGE2 serves as an autocrine signal to suppress TNF-α production in macrophages34–36. The paradoxical suppression of TNF-α production that we observed upon selective pharmacologic or genetic disruption of DAGLβ is less well understood from a mechanistic perspective, but could relate to changes in endocannabinoids, other arachidonic acid-derived metabolites that are not biosynthesized by COX enzymes, such as LTB4, or a partial (versus a complete) reduction in prostaglandins. Regardless of the precise mechanism by which DAGLβ inhibitors reduce TNF-α release, we believe that this finding could have biomedical relevance given the demonstrated clinical value of TNF-α blockers for treating inflammatory disorders like rheumatoid arthritis41. Macrophages are thought to be a major source of TNF-α production in vivo42 and drugs that impair this process could offer a complementary way to treat inflammatory disorders43. Future studies will be required to assess whether DAGLβ inhibitors have an impact on systemic inflammatory paradigms in vivo.
Finally, we believe that our studies point to many key areas of future research. Could, for instance, 1,2,3-TUs also serve as a scaffold for creating selective inhibitors of DAGLα? Here, improvements not only in potency but also in the central nervous system activity of DAGLα or dual DAGLα and DAGLβ inhibitors may be necessary. Additionally, how is the crosstalk between DAGLβ and cPLA2 regulated at the cellular level, and what is the impact of downstream metabolic enzymes like 2-AG hydrolases? DAGLβ and cPLA2 are activated by distinct and complementary pathways - G-proteins44,45 and calcium influx44–46, respectively - and it is possible that different inflammatory stimuli will preferentially engage one of these two pathways (or both) to produce distinct arachidonic acid pools for prostaglandin biosynthesis in macrophages. In this regard, the profound effect of blocking both DAGLβ and PLA2G4A on cellular PGE2 in macrophages calls to mind a coincidence detection model for the biosynthesis of endocannabinoids, which has been shown to depend, in certain cellular systems, on the coordinated activation of G-protein and calcium-signaling pathways39,44. The inhibitors described here could also prove useful for assessing the relative contribution of DAGLs to orexin signal transduction pathways, which have recently been shown to regulate 2-AG and arachidonic acid release in mammalian cells through a mechanism that involves both PLA2G4A and DAGL enzymes47. Finally, it will also be important to assess the impact of concurrently blocking DAGLβ and downstream 2-AG hydrolases like MGLL, which could further alter the endocannabinoid-eicosanoid signaling network to produce distinct pharmacological effects. Endocannabinoids, for instance, are generally considered anti-inflammatory agents48, which might indicate that downstream reductions in pro-inflammatory eicosanoids due to DAGLβ inhibition override the effect of blocking 2-AG production. Simultaneous disruption of 2-AG degradation could, in principle, mitigate the effect of DAGLβ inhibitors on endocannabinoids, while augmenting reductions in arachidonic acid-derived eicosanoids.
Although we selected macrophages for our initial biological studies because of the high expression level of DAGLβ in these cells and the documented roles that both the endocannabinoid and prostaglandin systems have in macrophage signaling39,49, it is likely that further studies will reveal important functions of DAGLβ in other cell types and organs; this is supported by our findings that DAGLβ inhibitors reduce 2-AG and arachidonic acid in mouse liver (Supplementary Fig. 20) and human prostate cancer cells (Supplementary Fig. 15). More generally, the chemical probes and chemoproteomic methods reported here should find broad utility that extend beyond the study of DAGLβ. KT195, for instance, should prove useful not only as a control probe, but also as a pharmacological tool to study ABHD6, which may function as an alternative endocannabinoid hydrolase9,50. Indeed, we found that KT195 treatment of Neuro2A cells caused significant accumulation of 2-AG (P = 0.0289) (Fig. 3d), supporting a role for ABHD6 in regulating 2-AG in cellular systems that lack detectable MGLL. We also envision an expanding role for next-generation activity-based probes like HT-01, which show tailored selectivity for a subclass of serine hydrolases, enabling the detection of low-abundance enzymes like DAGLβ in proteomes using simple gel-based assay formats as opposed to labor- and sample-intensive MS experiments.
Methods
Synthesis of 1,2,3-triazole urea inhibitors and DAGLβ activity-based probe
See Supplementary Methods for details.
DAGL LC-MS substrate hydrolysis assay
See Supplementary Methods for details of the experiment.
Primary screening of 1,2,3-triazole urea library by gel-based ABPP
Membrane proteome of transiently transfected HEK293T cells overexpressing mouse DAGLβ (50 uL of 0.3 mg/mL; GenBank Accession BC016105) in assay buffer (50 mM HEPES pH 7.2, 100 mM NaCl, 5 mM CaCl2, 0.1% v/v TX-100, 10% v/v DMSO) was treated with test compound (500 nM final concentration) or DMSO for 30 minutes at 37 °C. FP-rhodamine (5 µM final concentration) was added, and the reaction was incubated for 30 minutes at 37 °C, quenched with an equal volume of 2× SDS-PAGE loading buffer (reducing), separated by SDS-PAGE, and visualized by in-gel fluorescent scanning. See Supplementary Methods for details on gel-based competitive ABPP experiments.
In situ treatment of Neuro2A cells with inhibitors
Neuro2A cells were initially grown for 10 passages in either light or heavy SILAC DMEM medium supplemented with 10% v/v dialyzed FCS and 2 mM L-glutamine. Light medium was supplemented with 100 µg mL−1 L-arginine and 100 µg mL−1 L-lysine. Heavy medium was supplemented with 100 µg mL−1 [13C615N4]-L-Arginine and 100 µg mL−1 [13C615N2]-L-Lysine. Heavy cells (in 10 mL medium) were treated with test compound and light cells were treated with DMSO for 4 hours at 37 °C. Cells were washed with DPBS (2×), harvested, and homogenized by sonication in DPBS.
Harvesting thioglycollate-elicited macrophages
Mice were injected with thioglycollate medium (4% w/v, 2.5 mL per mouse) intraperitoneally (i.p.) 4 d prior to proteomics or metabolomics experiments. Mice were anesthetized with isoflurane, euthanized by cervical dislocation, and peritoneal macrophages harvested following protocols available from LIPID MAPS (www.lipidmaps.org). In brief, the peritoneum cavity of mice were exposed, lavaged with 10 mL of sterile PBS, and centrifuged (1400g, 3 min) to pellet cells. To remove red blood cells from the macrophage suspension, the pellet was resuspended and incubated with red blood cell (RBC) lysis buffer (150 mM NH4Cl, 10 mM KHCO3, 0.13 mM EDTA) for 15 min on ice. Macrophages were centrifuged (1400g, 3 min) and RBC lysis buffer removed. Macrophages were then either used immediately or flash frozen in liquid N2 and stored at −80 °C until use.
Competitive ABPP-SILAC
Isotopically light and heavy Neuro2A cells were treated in situ as described above with DMSO or inhibitor, respectively. Cells were lysed, and proteomes were adjusted to a final concentration of 2.0 mg mL−1 and labeled with 10 µM FP-biotin (500 µL total reaction volume) for 2 h at 25 °C. After incubation with probe, samples were prepared for LC/MS/MS analysis as described in Supplementary Methods.
Metabolite measurements in Neuro2A cells and peritoneal macrophages
Lipid measurements were performed as described in Supplementary Methods. In brief, the total cell metabolome was extracted using 2:1:1 chloroform:methanol:1% (w/v) sodium chloride solution. The organic layer was extracted and set aside. The aqueous phase was then acidified and re-extracted with choloroform. The organic phases were pooled, dried under N2, and resuspended in 2:1 chloroform/methanol for LC/MS analysis.
In vivo studies with KT109, KT172, and KT195
Mice were injected with KT109, KT172, or KT195 intraperitoneally i.p. in 18:1:1 (v/v/v) solution of saline:ethanol:PEG40 (ethoxylated castor oil, 10 µL g−1). For dose-response studies, mice were treated with varying doses of compounds for 4 h, anesthetized with isoflurane, and killed by cervical dislocation. For time-course studies, mice were treated with 5 mg per kg body weight of compound and sacrificed after the indicated times. Animal experiments were conducted in accordance with the guidelines of the Institutional Animal Care and Use Committee of The Scripps Research Institute.
Competitive ABPP-MudPIT
Thioglycollate-elicited macrophages from mice treated in vivo with vehicle or compounds were prepared for LC/MS/MS analysis following protocols described in the Supplementary Methods.
Cytokine analysis
Mice were treated with vehicle or compounds (5 mg per kg body weight for 2 h) as described above for in vivo studies. Thioglycollate-elicited peritoneal macrophages were harvested as described above, resuspended in RPMI 1640 media with 10% FCS and 1X penicillin, streptomycin, glutamine solution (Invitrogen), the number of cells determined using a Bio-rad TC10 automated cell counter, and an equal number of cells plated into 6-well plates (2.5 × 106 cells per well). Macrophages were allowed to adhere for 2 h at 37 °C and 5% CO2. Macrophages were then washed 2X with sterile PBS and stimulated with 5 µg mL−1 lipopolysaccharide in serum-free media. After 90 min, the medium was collected and secreted TNF-α was measured using the Mouse Inflammatory Cytokines Single-Analyte ELISArray kit (Qiagen) per the manufacturer’s instructions.
Supplementary Material
Acknowledgments
We thank M. Niphakis, H.-C. Lee, and G. Simon for helpful discussions and C. Joslyn for technical assistance. We thank M. Watanabe (Hokkaido University School of Medicine) for providing the antibody to DAGLα. This work was supported by the US National Institutes of Health (DA009789, DA033760, MH084512) and a Hewitt Foundation Postdoctoral Fellowship (K.-L.H.).
Footnotes
Author contributions. K.-L.H. and B.F.C. designed the experiments; K.-L.H., K.T., and A.A. did the experiments; H.P. and K.M. assisted with experiments; K.-L.H. and B.F.C. analyzed data; K.-L.H. and B.F.C. wrote the manuscript.
Competing financial interests
The authors declare competing financial interests: details accompany the online version of the paper.
References
- 1.Mechoulam R, et al. Identification of an endogenous 2-monoglyceride, present in canine gut, that binds to cannabinoid receptors. Biochem. Pharmacol. 1995;50:83–90. doi: 10.1016/0006-2952(95)00109-d. [DOI] [PubMed] [Google Scholar]
- 2.Sugiura T, et al. 2-Arachidonoylglycerol: a possible endogenous cannabinoid receptor ligand in brain. Biochem. Biophys. Res. Commun. 1995;215:89–97. doi: 10.1006/bbrc.1995.2437. [DOI] [PubMed] [Google Scholar]
- 3.Devane WA, et al. Isolation and structure of a brain constituent that binds to the cannabinoid receptor. Science. 1992;258:1946–1949. doi: 10.1126/science.1470919. [DOI] [PubMed] [Google Scholar]
- 4.Mackie K. Cannabinoid receptors as therapeutic targets. Annu. Rev. Pharmacol. Toxicol. 2006;46:101–122. doi: 10.1146/annurev.pharmtox.46.120604.141254. [DOI] [PubMed] [Google Scholar]
- 5.Ledent C, et al. Unresponsiveness to cannabinoids and reduced addictive effects of opiates in CB1 receptor knockout mice. Science. 1999;283:401–404. doi: 10.1126/science.283.5400.401. [DOI] [PubMed] [Google Scholar]
- 6.Zimmer A, Zimmer AM, Hohmann AG, Herkenham M, Bonner TI. Increased mortality, hypoactivity, and hypoalgesia in cannabinoid CB1 receptor knockout mice. Proc. Natl. Acad. Sci. U S A. 1999;96:5780–5785. doi: 10.1073/pnas.96.10.5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cravatt BF, et al. Molecular characterization of an enzyme that degrades neuromodulatory fatty-acid amides. Nature. 1996;384:83–87. doi: 10.1038/384083a0. [DOI] [PubMed] [Google Scholar]
- 8.Dinh TP, et al. Brain monoglyceride lipase participating in endocannabinoid inactivation. Proc. Natl. Acad. Sci. U S A. 2002;99:10819–10824. doi: 10.1073/pnas.152334899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blankman JL, Simon GM, Cravatt BFA. Comprehensive Profile of Brain Enzymes that Hydrolyze the Endocannabinoid 2-Arachidonoylglycerol. Chem. Biol. 2007;14:1347–1356. doi: 10.1016/j.chembiol.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kathuria S, et al. Modulation of anxiety through blockade of anandamide hydrolysis. Nat. Med. 2003;9:76–81. doi: 10.1038/nm803. [DOI] [PubMed] [Google Scholar]
- 11.Ahn K, et al. Discovery and characterization of a highly selective FAAH inhibitor that reduces inflammatory pain. Chem. Biol. 2009;16:411–420. doi: 10.1016/j.chembiol.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Long JZ, et al. Selective blockade of 2-arachidonoylglycerol hydrolysis produces cannabinoid behavioral effects. Nat. Chem. Biol. 2009;5:37–44. doi: 10.1038/nchembio.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nomura DK, et al. Endocannabinoid hydrolysis generates brain prostaglandins that promote neuroinflammation. Science. 2011;334:809–813. doi: 10.1126/science.1209200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bisogno T, et al. Cloning of the first sn1-DAG lipases points to the spatial and temporal regulation of endocannabinoid signaling in the brain. J. Cell. Biol. 2003;163:463–468. doi: 10.1083/jcb.200305129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao Y, et al. Loss of Retrograde Endocannabinoid Signaling and Reduced Adult Neurogenesis in Diacylglycerol Lipase Knock-out Mice. J. Neurosci. 2010;30:2017–2024. doi: 10.1523/JNEUROSCI.5693-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanimura A, et al. The endocannabinoid 2-arachidonoylglycerol produced by diacylglycerol lipase alpha mediates retrograde suppression of synaptic transmission. Neuron. 2010;65:320–327. doi: 10.1016/j.neuron.2010.01.021. [DOI] [PubMed] [Google Scholar]
- 17.Gregg LC, et al. Activation of Type 5 Metabotropic Glutamate Receptors and Diacylglycerol Lipase-alpha Initiates 2-Arachidonoylglycerol Formation and Endocannabinoid-Mediated Analgesia. J. Neurosci. 2012;32:9457–9468. doi: 10.1523/JNEUROSCI.0013-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoover HS, Blankman JL, Niessen S, Cravatt BF. Selectivity of inhibitors of endocannabinoid biosynthesis evaluated by activity-based protein profiling. Bioorg.Med. Chem. Lett. 2008;18:5838–5841. doi: 10.1016/j.bmcl.2008.06.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ortar G, et al. Tetrahydrolipstatin analogues as modulators of endocannabinoid 2-arachidonoylglycerol metabolism. J. Med. Chem. 2008;51:6970–6979. doi: 10.1021/jm800978m. [DOI] [PubMed] [Google Scholar]
- 20.Bisogno T, et al. Development of the first potent and specific inhibitors of endocannabinoid biosynthesis. Biochim. Biophys. Acta. 2006;1761:205–212. doi: 10.1016/j.bbalip.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 21.Pedicord DL, et al. Molecular characterization and identification of surrogate substrates for diacylglycerol lipase alpha. Biochem. Biophys. Res. Commun. 2011;411:809–814. doi: 10.1016/j.bbrc.2011.07.037. [DOI] [PubMed] [Google Scholar]
- 22.Long JZ, Cravatt BF. The metabolic serine hydrolases and their functions in mammalian physiology and disease. Chem. Rev. 2011;111:6022–6063. doi: 10.1021/cr200075y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adibekian A, et al. Click-generated triazole ureas as ultrapotent in vivo–active serine hydrolase inhibitors. Nat. Chem. Biol. 2011;7:469–478. doi: 10.1038/nchembio.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cravatt BF, Wright AT, Kozarich JW. Activity-based protein profiling: from enzyme chemistry to proteomic chemistry. Annu. Rev. Biochem. 2008;77:383–414. doi: 10.1146/annurev.biochem.75.101304.124125. [DOI] [PubMed] [Google Scholar]
- 25.Bachovchin DA, et al. Academic cross-fertilization by public screening yields a remarkable class of protein phosphatase methylesterase-1 inhibitors. Proc. Natl. Acad. Sci. U S A. 2011;108:6811–6816. doi: 10.1073/pnas.1015248108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oudin MJ, Hobbs C, Doherty P. DAGL-dependent endocannabinoid signalling: roles in axonal pathfinding, synaptic plasticity and adult neurogenesis. Eur. J. Neurosci. 2011;34:1634–1646. doi: 10.1111/j.1460-9568.2011.07831.x. [DOI] [PubMed] [Google Scholar]
- 27.Di Marzo V. Endocannabinoid signaling in the brain: biosynthetic mechanisms in the limelight. Nat. Neurosci. 2011;14:9–15. doi: 10.1038/nn.2720. [DOI] [PubMed] [Google Scholar]
- 28.Jung K-M, et al. A Key Role for Diacylglycerol Lipase-alpha in Metabotropic Glutamate Receptor-Dependent Endocannabinoid Mobilization. Mol. Pharmacol. 2007;72:612–621. doi: 10.1124/mol.107.037796. [DOI] [PubMed] [Google Scholar]
- 29.Jessani N, et al. A streamlined platform for high-content functional proteomics of primary human specimens. Nat. Methods. 2005;2:691–697. doi: 10.1038/nmeth778. [DOI] [PubMed] [Google Scholar]
- 30.Long JZ, Nomura DK, Cravatt BF. Characterization of monoacylglycerol lipase inhibition reveals differences in central and peripheral endocannabinoid metabolism. Chem. Biol. 2009;16:744–753. doi: 10.1016/j.chembiol.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bachovchin DA, et al. Superfamily-wide portrait of serine hydrolase inhibition achieved by library-versus-library screening. Proc. Natl. Acad. Sci. U S A. 2010;107:20941–20946. doi: 10.1073/pnas.1011663107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uozumi N, et al. Role of cytosolic phospholipase A2 in allergic response and parturition. Nature. 1997;390:618–622. doi: 10.1038/37622. [DOI] [PubMed] [Google Scholar]
- 33.Bonventre JV, et al. Reduced fertility and postischaemic brain injury in mice deficient in cytosolic phospholipase A2. Nature. 1997;390:622–625. doi: 10.1038/37635. [DOI] [PubMed] [Google Scholar]
- 34.Rouzer CA, et al. Cyclooxygenase-1-dependent prostaglandin synthesis modulates tumor necrosis factor-alpha secretion in lipopolysaccharidechallenged murine resident peritoneal macrophages. J. Biol. Chem. 2004;279:34256–34268. doi: 10.1074/jbc.M402594200. [DOI] [PubMed] [Google Scholar]
- 35.Watanabe S, Kobayashi T, Okuyama H. Regulation of lipopolysaccharideinduced tumor necrosis factor alpha production by endogenous prostaglandin E2 in rat resident and thioglycollate-elicited macrophages. J. Lipid. Mediat. Cell Signal. 1994;10:283–294. [PubMed] [Google Scholar]
- 36.Rouzer CA, et al. RAW264.7 cells lack prostaglandin-dependent autoregulation of tumor necrosis factor-alpha secretion. J. Lipid. Res. 2005;46:1027–1037. doi: 10.1194/jlr.M500006-JLR200. [DOI] [PubMed] [Google Scholar]
- 37.Carrasco S, Merida I. Diacylglycerol, when simplicity becomes complex. Trends Biochem. Sci. 2007;32:27–36. doi: 10.1016/j.tibs.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 38.Chevaleyre V, Takahashi KA, Castillo PE. Endocannabinoid-mediated synaptic plasticity in the CNS. Annu. Rev. Neurosci. 2006;29:37–76. doi: 10.1146/annurev.neuro.29.051605.112834. [DOI] [PubMed] [Google Scholar]
- 39.Rouzer CA, Marnett LJ. Endocannabinoid oxygenation by cyclooxygenases, lipoxygenases, and cytochromes P450: cross-talk between the eicosanoid and endocannabinoid signaling pathways. Chem. Rev. 2011;111:5899–5921. doi: 10.1021/cr2002799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buczynski MW, Dumlao DS, Dennis EA. Thematic Review Series: Proteomics. An integrated omics analysis of eicosanoid biology. J. Lipid Res. 2009;50:1015–1038. doi: 10.1194/jlr.R900004-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parameswaran N, Patial S. Tumor necrosis factor-alpha signaling in macrophages. Crit. Rev. Eukaryot. Gene Expr. 2010;20:87–103. doi: 10.1615/critreveukargeneexpr.v20.i2.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bondeson J. The mechanisms of action of disease-modifying antirheumatic drugs: a review with emphasis on macrophage signal transduction and the induction of proinflammatory cytokines. Gen. Pharmacol. 1997;29:127–150. doi: 10.1016/s0306-3623(96)00419-3. [DOI] [PubMed] [Google Scholar]
- 43.Lin HI, Chu SJ, Wang D, Feng NH. Pharmacological modulation of TNF production in macrophages. J. Microbiol. Immunol. Infect. 2004;37:8–15. [PubMed] [Google Scholar]
- 44.Hashimotodani Y, et al. Phospholipase Cbeta serves as a coincidence detector through its Ca2+ dependency for triggering retrograde endocannabinoid signal. Neuron. 2005;45:257–268. doi: 10.1016/j.neuron.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 45.Hashimotodani Y, Ohno-Shosaku T, Watanabe M, Kano M. Roles of phospholipase Cbeta and NMDA receptor in activity-dependent endocannabinoid release. J. Physiol. 2007;584:373–380. doi: 10.1113/jphysiol.2007.137497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qiu ZH, Gijon MA, de Carvalho MS, Spencer DM, Leslie CC. The role of calcium and phosphorylation of cytosolic phospholipase A2 in regulating arachidonic acid release in macrophages. J. Biol. Chem. 1998;273:8203–8211. doi: 10.1074/jbc.273.14.8203. [DOI] [PubMed] [Google Scholar]
- 47.Turunen PM, Jantti MH, Kukkonen JP. OX1 Orexin/Hypocretin Receptor Signaling through Arachidonic Acid and Endocannabinoid Release. Mol. Pharmacol. 2012;82:156–167. doi: 10.1124/mol.112.078063. [DOI] [PubMed] [Google Scholar]
- 48.Burstein SH, Zurier RB. Cannabinoids, endocannabinoids, and related analogs in inflammation. AAPS J. 2009;11:109–119. doi: 10.1208/s12248-009-9084-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kunos G, Batkai S. Novel physiologic functions of endocannabinoids as revealed through the use of mutant mice. Neurochem. Res. 2001;26:1015–1021. doi: 10.1023/a:1012301021419. [DOI] [PubMed] [Google Scholar]
- 50.Marrs WR, et al. The serine hydrolase ABHD6 controls the accumulation and efficacy of 2-AG at cannabinoid receptors. Nat. Neurosci. 2010;13:951–957. doi: 10.1038/nn.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.