Abstract
Aims: The BolA protein family is widespread among eukaryotes and bacteria. In Escherichia coli, BolA causes a spherical cell shape and is overexpressed during oxidative stress. Here we aim to elucidate the possible role of its human homolog BOLA1 in mitochondrial morphology and thiol redox potential regulation. Results: We show that BOLA1 is a mitochondrial protein that counterbalances the effect of L-buthionine-(S,R)-sulfoximine (BSO)-induced glutathione (GSH) depletion on the mitochondrial thiol redox potential. Furthermore, overexpression of BOLA1 nullifies the effect of BSO and S-nitrosocysteine on mitochondrial morphology. Conversely, knockdown of the BOLA1 gene increases the oxidation of mitochondrial thiol groups. Supporting a role of BOLA1 in controlling the mitochondrial thiol redox potential is that BOLA1 orthologs only occur in aerobic eukaryotes. A measured interaction of BOLA1 with the mitochondrial monothiol glutaredoxin GLRX5 provides hints for potential mechanisms behind BOLA1's effect on mitochondrial redox potential. Nevertheless, we have no direct evidence for a role of GLRX5 in BOLA1's function. Innovation: We implicate a new protein, BOLA1, in the regulation of the mitochondrial thiol redox potential. Conclusion: BOLA1 is an aerobic, mitochondrial protein that prevents mitochondrial morphology aberrations induced by GSH depletion and reduces the associated oxidative shift of the mitochondrial thiol redox potential. Antioxid. Redox Signal. 18, 129–138.
Introduction
Disbalance of the cellular redox potential has been implicated in normal aging and in aging-related pathologies, such as Alzheimer's disease, Parkinson's disease, and type 2 diabetes. To identify new proteins involved in the defense against such disbalance and/or its consequences, we have analyzed the role of human BolA homolog 1 (BOLA1), a protein whose bacterial homolog (BolA) is upregulated under oxidative stress conditions. The BolA protein family was originally described in Escherichia coli, where overexpression of the protein induces a spherical cell shape (2). In human and yeast, the BolA protein family has three members, BolA homolog 1, 2, and 3. The eukaryotic representatives of the BolA protein family appear derived from the proteobacteria (10) and presumably have resulted from the endosymbiosis of an α-proteobacterium that became the mitochondrion. BolA has been mainly studied in E. coli, where its gene is upregulated under stress (31) and is under control of the transcription regulator RpoS that is induced in the stationary phase and other general stress conditions (28). BolA in E. coli is involved in the regulation of cell wall proteins PBP5, PBP6, and AmpC (32), but its exact molecular function remains unknown. Comparative genomics analyses have pointed to an interaction of BolA with monothiol glutaredoxins, based on gene order conservation (16), phylogenetic distribution (16), and a physical interaction between BolA homolog 2 and Grx3/4 in Saccharomyces cerevisiae (14, 17) and Drosophila melanogaster (11).
BolA homolog 2 has been shown to be cytoplasmic in S. cerevisiae and to interact with the cytoplasmic monothiol glutaredoxins Grx3 and Grx4 (23). In S. cerevisiae, it has been named Fra2 and, together with Grx3/4 and another protein (Fra1), is involved in regulation of the iron regulon via the nucleoplasmatic shuttling of the transcription factor Aft1 (23). BOLA3 has recently been implicated in the biogenesis of [Fe-S] clusters for oxidative phosphorylation complexes and 2-oxo acid dehydrogenase enzymes in mitochondria (6).
Innovation.
The BolA protein family is widespread among eukaryotes and bacteria. There are reports about its role in defense against oxidative stress in bacteria, but its function in mammals is unknown. Furthermore, the mechanism behind its role in defense against oxidative stress has not been elucidated, although it has been predicted to interact with a monothiol glutaredoxin. Here we show that BOLA1, one of the three members of the family in eukaryotes, is mitochondrial in human. Knockdown of the gene increases the level of oxidation of mitochondrial thiol groups. Overexpression of the gene reduces the effect of glutathione (GSH) depletion on the oxidation of thiol groups, and nullifies the effect of GSH depletion on mitochondrial morphology. Consistent with its role in reducing thiol redox potential, we show that BOLA1 orthologs only occur in aerobic species. We supply evidence that BOLA1 interacts with a mitochondrial monothiol glutaredoxin GLRX5, providing a possible mechanism for BOLA1's role in cellular processes, such as mitochondrial morphology regulation.
Little experimental data are available on the function of Bola homolog 1. BOLA1 has been observed to be overexpressed in mononuclear cells of female patients with chronic fatigue syndrome (12), but that observation could not be reproduced in a systematic study (4). Here we set out to examine the function of BOLA1, with specific emphasis on its role in regulating mitochondrial morphology.
Results
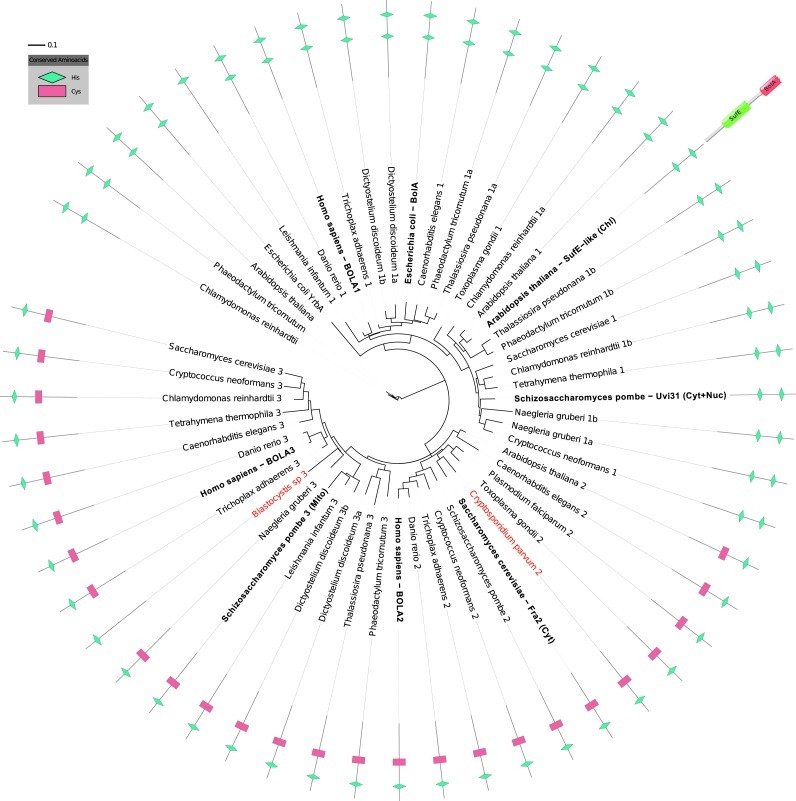
BOLA1 orthologs only occur in aerobic species
In eukaryotes, the BolA protein family comprises three phylogenetically widespread clades (Fig. 1). Notably, none of the genomes of anaerobic eukaryotes sequenced contains a BOLA1 ortholog, whereas orthologs of BOLA2 and BOLA3 are present in the anaerobic genus Cryptosporidium and the anaerobic species Blastocystis hominis, but not in the other anaerobic species (Trichomonas vaginalis, Giardia intestinalis, Encephalitozoon cuniculi, and Entamoeba histolytica). As the last common ancestor of all eukaryotes likely was aerobic (10) and the eukaryotic anaerobes with sequenced genomes belong to five independent evolutionary lineages (T. vaginalis and G. intestinalis belong to the same lineage), BOLA1 orthologs have been lost at least five times in the transition to anaerobic life, and BOLA1's genomic presence strongly correlates with an aerobic metabolism.
FIG. 1.
Phylogenetic distribution of the BolA family among eukaryotes and Escherichia coli and conservation of key residues. A phylogeny was determined for BolA1, 2, and 3 among species that cover the major eukaryotic taxa, including all anaerobic species (indicated in red), and the Bola family members for which experimental knowledge is available (in bold). The phylogeny clearly separates a BolA1 clade (top) from the other two BolA clades (BolA3 on the left, BolA2 on the right-bottom). The C-terminal histidine in the figure (green symbol) represents His-103 in Fra2, an Fe-S cluster ligand in the BolA2-Grx3 complex (25). This histidine is conserved throughout the BolA family in Eukaryotes (Supplementary Fig. S4). A cysteine (Cys-97 in BOLA2, red symbol) that has also been implicated in Fe-S binding in the BolA2-Grx3 complex, although not directly (25), is only conserved in BolA2 and BolA3. In the BolA1 clade, this cysteine appears to be replaced by a conserved histidine (His-67 in BOLA1). The phylogeny indicates that the Arabidopsis thaliana protein that is fused with a SufE domain (40) is a member of the BolA1 clade.
BOLA1 specifically localizes in mitochondria
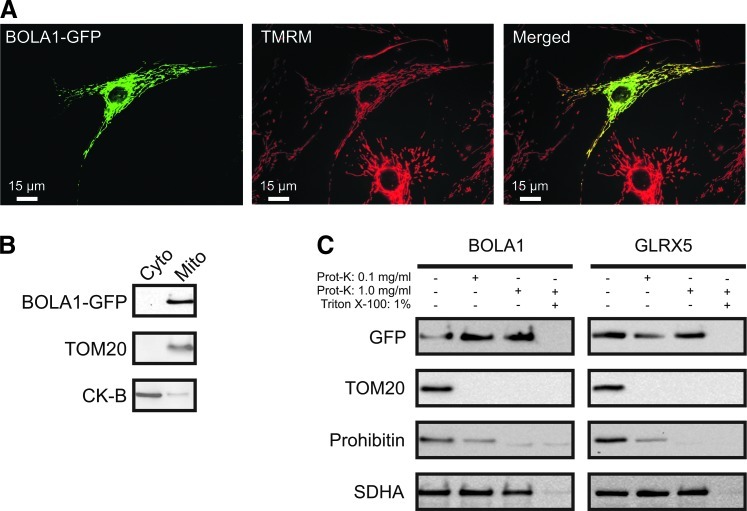
Mouse BolA1 contains a mitochondrial targeting signal [0.97, mitoprot (8)], and its mitochondrial localization was verified by GFP tagging and mass spectrometry (29). The mouse BolA1 protein was found in 13 out of 14 tissues examined (29) and human BOLA1 is ubiquitously expressed, but not overexpressed (at least 10-fold over the median) in any specific tissue (33). Likewise, human BOLA2 and BOLA3 have been found to be ubiquitously expressed, but not overexpressed in any specific tissue examined (33), while mouse Bola3 protein has been detected in mitochondria in 12 out of 14 tissues examined (29). A c-Myc tagged version of BOLA1, overexpressed in African green monkey kidney (Cos-7) cells, was shown to be present in discrete spots that did not encompass the Golgi, but were not further characterized (43). To determine BOLA1's intracellular localization, we transduced primary human skin fibroblasts with a baculovirus containing a fusion of BOLA1 and AcGFP1 (BOLA1-GFP) and loaded them with the mitochondrial membrane potential-sensitive fluorescent dye tetramethylrhodamine methyl ester (TMRM).
Combined digital-imaging microscopy of TMRM and GFP fluorescence revealed that 70% of the fibroblasts expressed BOLA1-GFP. The clear overlap of its fluorescence with that of TMRM demonstrated BOLA1-GFP's mitochondrial localization in human cells (Fig. 2A). BOLA1-GFP overexpression did not detectably alter the mitochondrial appearance (Supplementary Fig. S1A, fifth panel; Supplementary Data are available online at www.liebertpub.com/ars). BOLA2-GFP displayed a cytoplasmic and nuclear distribution, similar to Fra2/BolA2 in S. cerevisiae (23) (Supplementary Fig. S2A), whereas BOLA3-GFP appeared localized to mitochondria, consistent with findings of Cameron et al. (6), aside from cytoplasm and nucleoplasm (Supplementary Fig. S2B). The latter two locations might be a result of overexpression. For BOLA2 and BOLA3, these results are consistent with mitochondrial targeting signal predictions, while for BOLA1 the signal is ambiguous (mitoprot BOLA1: 0.46, BOLA2: 0.27, BOLA3: 0.96).
FIG. 2.
BOLA1 is a mitochondrial protein. (A) Overlap in BOLA1-GFP (left panel) and TMRM (middle panel) fluorescence, demonstrating mitochondrial localization of BOLA1 in human skin fibroblasts (right panel). Note the absence of any TMRM fluorescence bleed through in GFP-negative cells. (B) BOLA1-GFP is exclusively present in the mitochondrial fraction of doxycycline-induced HEK293 cells. (C) Mitochondria-associated BOLA1-GFP and GLRX5-GFP are proteinase K-resistant, suggesting their presence within the limits of the inner-membrane. In (A), images were contrast optimized for visualization purposes.CK-B, cytosolic creatine kinase B; TOM20, import receptor of the outer-membrane translocator; prohibitin 1, mitochondrial inner membrane associated IMS protein; SDHA, A-subunit of the succinate dehydrogenase complex of the mitochondrial matrix; TMRM, tetramethylrhodamine methyl ester.
Fractionation of HEK293 cells induced to express BOLA1-GFP confirmed its mitochondrial localization (Fig. 2B). In mitochondria with digitonin-permeabilized outer membranes, both BOLA1-GFP (Fig. 2C, left panel) and GLRX5-GFP (Fig. 2C, right panel), like subunit A of mitochondrial matrix succinate dehydrogenase (SDHA), appeared resistant to proteinase K, whereas this enzyme readily digested the import receptor (TOM20) of the outer-membrane translocator and prohibitin of the inner membrane, indicating that BOLA1 is located within the limits of the inner-membrane.
BOLA1 is complexed with GLRX5
The BolA-like protein family displays a phylogenetic distribution that is very similar to that of the monothiol glutaredoxins (9, 16), and yeast BolA2 was indeed demonstrated to interact with cytosolic monothiol glutaredoxins Grx3 and Grx4 (23). As human mitochondria contain the monothiol glutaredoxin GLRX5 (30) (Fig. 2C, right panel), we expected BOLA1 to interact with it, although the phylogenetic co-occurrence of BolA1 and Grx5 is not perfect (Supplementary Fig. S3). Supporting an interaction between BOLA1 and GLRX5 is that the S. cerevisiae BolA2 His-103 is conserved in the BolA1 orthologous group (Fig. 1 and Supplementary Fig. S4). This histidine binds the [Fe2S2] cluster that has been implicated in the BolA2-Grx3/4 complex in S. cerevisiae (25).
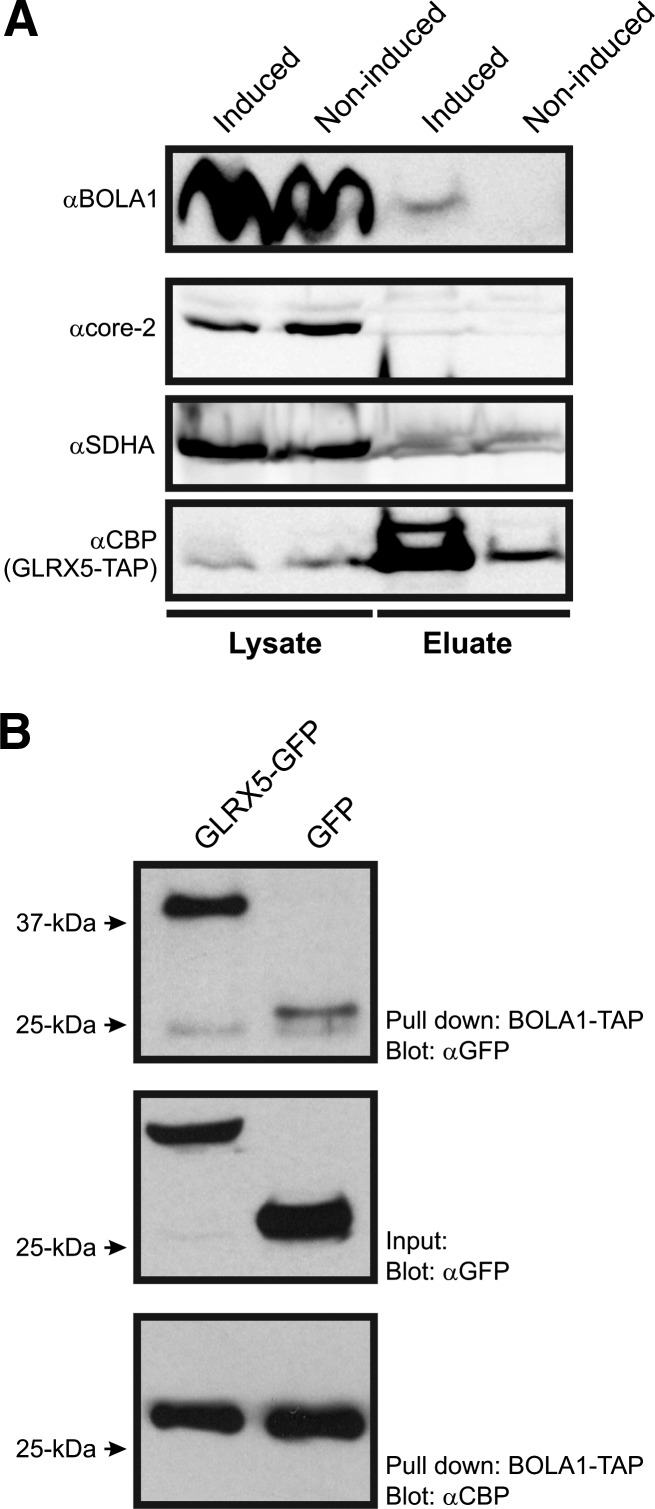
To demonstrate a physical interaction between BOLA1 and GLRX5, we generated HEK293 T-REx cells that inducibly express these proteins with a C-terminal TAP tag. At 24 h after induction, cell lysates were affinity purified and proteins were identified by nanospray ionization liquid chromatography tandem mass spectrometry (nLC-MS/MS). With BOLA1-TAP as a bait, GLRX5 was among the 3 copurified mitochondrial proteins that were specific to BOLA1-TAP, in the sense that they were not also observed in noninduced cells, or in affinity purifications of seven other mitochondrial proteins (Supplementary Table S1a). Conversely, BOLA1 was among the eight copurified mitochondrial proteins that were only copurified with GLRX5-TAP (Supplementary Table S1b). To verify the nLC-MS/MS data, the affinity purifications of BOLA1 and GLRX5 were repeated, and subsequently tested by Western blotting for copurification of GLRX5 and BOLA1, respectively. BOLA1 was copurified with GLRX5-TAP (Fig. 3A). Nevertheless, purification of BOLA1-TAP did not yield detectable amounts of copurified GLRX5 (data not shown). To substantiate the BOLA1-TAP GLRX5 interaction, we mixed lysates of BOLA1-TAP-expressing cells with those of either GLRX5-GFP- or GFP-expressing cells and incubated these with streptactin beads. Possible copurification of GLRX5-GFP by BOLA1-TAP was verified by Western blotting using anti-GFP. This approach resulted in a significant amount of BOLA1-TAP copurified GLRX5-GFP with respect to the GFP control (Fig. 3B, upper panel) strengthening the evidence for an interaction.
FIG. 3.
BOLA1 is complexed to GLRX5. (A) Sodium dodecyl sulfate–polyacrylamide gel electrophoresis and Western blot analysis of a single-step affinity purification performed with doxycycline-induced HEK293 cells expressing GLRX5-TAP. Membranes were probed with antibodies against BOLA1, core 2 protein of mitochondrial complex III, A-subunit of the succinate dehydrogenase complex of the mitochondrial matrix (SDHA), and CBP for detection of the TAP-tag. As a control, noninduced cells were used. (B) Analysis of BOLA1-TAP purification with cell lysates containing GLRX5-GFP or GFP. Membranes were probed with anti-GFP to check for copurification and to determine the input (upper and middle panel, respectively) and anti-CBP to test the efficiency of the pull down. Molecular mass marker (kDa) is shown on the left. CBP, calmodulin-binding protein.
BOLA1 knockdown causes an oxidative shift of the mitochondrial thiol redox potential
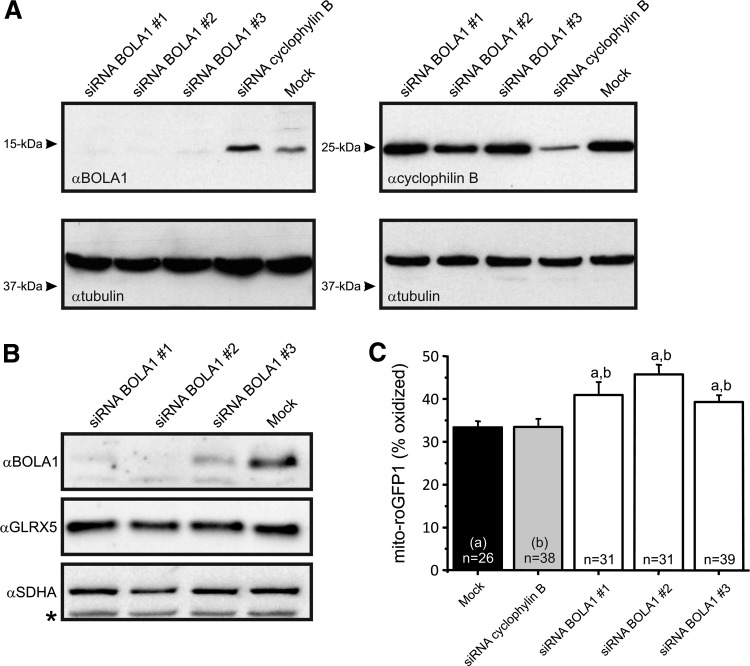
Because a bacterial homolog of BOLA1, BolA, is overexpressed under oxidative stress conditions (31), we investigated whether BOLA1 plays a role in the maintenance of the mitochondrial thiol redox potential. To this end, HeLa cells were transfected with small interfering RNAs (siRNAs) against BOLA1 mRNA. All three siRNAs effectively knocked down BOLA1 (Fig. 4A). BOLA1 knockdown did not alter the amount of GLRX5 (Fig. 4B). Digital-imaging microscopy of cells expressing mito-roGFP1, revealed that BOLA1 knockdown caused an oxidative shift of the mitochondrial thiol/disulfide redox status (Fig. 4C). This shift was not paralleled by a detectable change in the production of hydroethidine (HEt)-specific oxidants as revealed by digital-imaging microscopy of cells loaded with HEt (Supplementary Fig. S5). HEt oxidation measures superoxide production, whereas mito-roGFP1 measures thiol oxidation, processes that have been shown to be rather independent from each other, for example, L-buthionine-(S,R)-sulfoximine (BSO)-induced thiol oxidation can occur in parallel with decreased HEt oxidation (35).
FIG. 4.
Knockdown of BOLA1 increases thiol redox potential in the mitochondrial matrix of HeLa cells. (A) Knockdown of BOLA1 by siRNAs in HeLa cells. Cells were transfected with three different BOLA1 siRNA duplexes (left panel) and a control siRNA targeting cyclophilin B (right panel). The left upper immunoblot probed with anti-BOLA1 demonstrates the effectiveness of the siRNAs. Beta-tubulin was used as protein-loading control. Molecular mass markers (kDa) are indicated on the left. Note that siRNA against cyclophilin B did not alter BOLA1 expression and BOLA1 siRNAs did not affect cyclophilin B expression. (B) BOLA1 knockdown is without effect on GLRX5 expression. (C) Cells, transfected with the indicated siRNA, were transduced with a baculovirus for expression of mito-roGFP. The percent oxidation was calculated after establishment of the maximum and minimum oxidation levels with 1 mM H2O2 and 10 mM DTT, respectively, as described before (35). Numerals indicate the number of cells analyzed. a and b, significantly different from the value of the corresponding bar (p<0.05). DTT, dithiothreitol.
BOLA1 overexpression attenuates the effect of glutathione depletion on the mitochondrial thiol redox potential
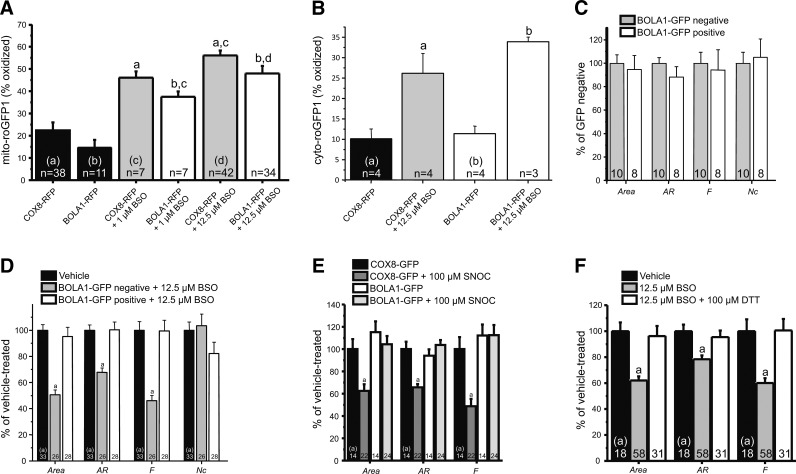
To investigate a putative role of BOLA1 in the maintenance of the mitochondrial thiol redox potential, fibroblasts were cotransduced with a baculovirus for expression of BOLA1-RFP and a baculovirus for expression of mito-roGFP1. Although BOLA1-GFP overexpression tended to cause a reductive shift of the mitochondrial thiol/disulfide redox status, this effect did not reach statistical significance (Fig. 5A).
FIG. 5.
Effect of BOLA1 overexpression on the redox potential of mitochondrial thiol groups and mitochondrial morphology under various (oxidizing) conditions. (A) BOLA1 overexpression attenuates the BSO-induced increase in oxidizing potential of the mitochondrial matrix. Fibroblasts, cotransduced for expression of mito-roGFP1 and either BOLA1-RFP or COX8-RFP, were treated without or with 1 μM or 12.5 μM BSO for 72 h. The percent oxidation in COX8- and BOLA1-RFP-positive cells was calculated as described in the legend to Figure 4. (B) BOLA1 overexpression does not interfere with the BSO-induced increase in oxidizing potential of the cytosol. (C) BOLA1 overexpression has no effect on mitochondrial morphology and number. Fibroblasts, transduced for expression of BOLA1-GFP were loaded with TMRM and subjected to digital-imaging microscopy of GFP and TMRM fluorescence. Quantitative image analysis revealed that compared to BOLA1-GFP-negative cells, BOLA1-GFP-positive cells contained mitochondria that were similar in size (mitochondrial area), length (AR), length and degree of branching (form factor, F), and of mitochondria per cell (Nc), showing that BOLA1 overexpression does not affect mitochondrial morphology and number. (D) BOLA1 prevents the BSO-induced change in mitochondrial morphology. Fibroblasts, transduced for expression of BOLA1-GFP were treated with 12.5 μM BSO for 72 h. Compared to vehicle-treated cells, BSO-treated BOLA1-GFP-negative cells contained mitochondria that were smaller in size, less elongated, and less branched. In sharp contrast, BOLA1-GFP-positive cells, present on the same coverslip, did not display these BSO-induced differences in mitochondrial morphology. The number of mitochondria per cell did not significantly differ between the different conditions. The value obtained with vehicle-treated cells was set at 100%. (E) BOLA1 prevents the SNOC-induced change in mitochondrial morphology. Fibroblasts, transduced for expression of either COX8-GFP or BOLA1-GFP were treated with 100 μM SNOC for 72 h. Subsequent analysis showed that SNOC induced a significant decrease in mitochondrial area, length, and degree of branching in COX8-GFP-positive cells, but not in BOLA1-GFP-positive cells. The value obtained with vehicle-treated COX8-GFP-positive cells was set at 100%. (F) DTT prevents the BSO-induced change in mitochondrial morphology. Fibroblasts were treated with BSO (12.5 μM), alone and in combination with DTT (100 μM), for 72h, and analyzed for mitochondrial morphology. The value obtained with vehicle-treated cells was set at 100%. Numerals indicate the number of cells analyzed. a, b, and c, significantly different (p<0.05) from the corresponding bar. BSO, L-buthionine-(S,R)-sulfoximine; SNOC, S-nitrosocysteine; AR, aspect ratio; COX8, cytochrome c oxidase subunit VIII.
We previously published that 27-h treatment of fibroblasts with 12.5 μM BSO, an inhibitor of the glutathione (GSH)-synthesizing enzyme γ-glutamylcysteine synthetase, caused an oxidative shift of both the cytosolic and mitochondrial thiol/disulfide redox status (35). Here we show that BOLA1-RFP overexpression significantly reduced this effect of BSO in cells treated for 72 h with either 1 μM or 12.5 μM BSO of this inhibitor (Fig. 5A). In agreement with its mitochondrial localization, BOLA1-RFP overexpression did not alter the effect of BSO on the cytosolic thiol redox potential (Fig. 5B), suggesting a compartmentalized regulation of this parameter.
Digital-imaging microscopy of BOLA1-GFP overexpressing fibroblasts loaded with monochlorobimane, a fluorescent indicator of intracellular GSH, revealed that BSO decreased the amount of GSH equally potently in BOLA1-GFP-negative and -positive cells present on the same coverslip, with more than 80% decrease in GSH at 12.5 μM BSO (Supplementary Fig. S6A). This result indicates that BOLA1-GFP overexpression has no effect on BSO-induced GSH depletion.
BOLA1 overexpression prevents the change in mitochondrial shape under GSH depletion
To address a possible effect of BOLA1 on the mitochondrial shape and number, we applied our method for quantitative analysis of these parameters (22) to fibroblasts overexpressing BOLA1-GFP. Mitochondria were visualized by using TMRM. Our analysis revealed that the mitochondrial area, the aspect ratio (AR, a measure of mitochondrial length), the form factor (F, a measure of mitochondrial length and degree of branching) and the number of mitochondria per cell (Nc) were not significantly different between BOLA1-GFP-negative and -positive cells lying next to each other on the same coverslip (Supplementary Fig. S1A and Fig. 5C).
Previous evidence suggests that fibroblast mitochondria become longer and more branched under conditions of increased HEt oxidation (21). Here we show that fibroblast mitochondria become shorter and less branched upon treatment with BSO (Supplementary Fig. S1C and Fig. 5D). The effect of BSO on the mitochondrial area, AR and F was completely absent in BOLA1-GFP-positive cells present on the same coverslip (Supplementary Fig. S1D and Fig. 5D). This result was not observed with BOLA3, the other mitochondria-localized member of the BolA family (6) (Supplementary Fig. S6B). Note that BSO treatment did not alter the amount of mitochondria per cell, suggesting mitochondrial shrinkage rather than fragmentation. BOLA1 overexpression did not counter all the effects of BSO, as it failed to restore NAD(P)H levels, which are significantly increased in BSO-treated cells (Supplementary Fig. S6C) (35).
Similarly to BSO, the NO donor S-nitrosocysteine (SNOC; 100 μM, 72 h) that nitrosylates thiol groups and reduces the amount of GSH in the cell (3), caused a marked decrease in area, AR and F, which was again completely nullified by overexpression of BOLA1-GFP (Fig. 5E). Conversely, and similarly to BOLA1-GFP overexpression, the sulfhydryl-reducing agent dithiothreitol (DTT; 100 μM, 72 h) prevented the BSO-induced change in the mitochondrial shape (Fig. 5F).
Discussion
To date, experimental data on the biological function of BOLA1 are lacking. Here we show that BOLA1 is a mitochondrial protein that, in agreement with comparative genomics analyses, appears to interact with the mitochondrial monothiol glutaredoxin GLRX5. Recent work in human (5) and zebrafish (39) has linked GLRX5 deficiency to decreased haem synthesis in erythroid cells, and it has been linked to protection against oxidative stress in osteoblasts (26). It has a single glutaredoxin domain, and although it can reduce GSH mixed disulfides, it does so at a rate 100 times lower than the dithiol glutaredoxin GLRX2 (18). Furthermore Grx5 in S. cerevisiae is only inefficiently reduced by GSH (34). One explanation for the low catalytic efficiency of Grx5 and GLRX5 is that they interact with a protein that increases the rate at which they reduce GSH mixed disulfides and/or at which they are reduced by GSH. Here we provide first evidence that BOLA1 might exert such a role and that one of the target proteins of the BOLA1/GLRX5 complex is involved in maintaining the normal mitochondrial shape. It should thereby be noted that, although BolA's 3D structure is similar to that of the bacterial reductase OsmC, suggesting a reducing activity of BolA itself, BolA1 does not contain any conserved cysteines, rendering any reducing function dependent on an external source of reducing equivalents (16).
Knockdown of endogenous BOLA1 significantly increased the amount of oxidized mito-roGFP1. Because mito-roGFP1 senses the redox potential of the mitochondrial GSH buffer, this indicates that BOLA1 plays a role in keeping this buffer in a reduced state. As evidence has been provided that glutaredoxins act as mediators of reversible electron flow between GSH and mito-roGFP1 (27), changes in the affinity of a glutaredoxin toward GSH and/or its endogenous protein substrates might lead to alterations in the degree of oxidation of these latter molecules. Conversely, when fibroblasts were treated with BSO to decrease the amount of GSH, BOLA1 overexpression attenuated the BSO-induced increase in mito-roGFP1 oxidation without interfering with the BSO-induced decrease in GSH. These observations are compatible with a model in which the observed physical interaction between BOLA1 and GLRX5 leads to an increase in GLRX5 activity and, therewith, to maintenance of the reduced state of mito-roGFP1 at decreased GSH concentrations. Nevertheless, GLRX5 protein levels do not depend on BOLA1 expression, and we have no direct evidence that BOLA1's effect on mitochondrial morphology and thiol redox potential depends on an interaction with GLRX5.
We provide a first indication for a possible physiological role of BOLA1. Both BSO and SNOC caused a marked decrease in mitochondrial length and degree of branching, which was completely prevented by overexpression of BOLA1. The effect of BOLA1 was mimicked by DTT, indicating that a putative target protein of the BOLA1 has to be in the reduced state to perform its effect. Our observation that BOLA1 could also prevent the SNOC-induced change in the mitochondrial shape can be explained by a SNOC-induced increased oxidation of GSH, thus reducing the amount of GSH available, for example, for GLRX5, to keep the putative target protein in the reduced state. SNOC has been shown to cause a fragmented mitochondrial phenotype, in part, through S-nitrosylation of the mitochondrial fission protein Drp1 (7). Here we show that BSO treatment did not alter the amount of mitochondria per cell, suggesting mitochondrial shrinkage rather than fragmentation. The effect of SNOC on the mitochondrial shape was completely reversed by overexpression of BOLA1. An alternative explanation, that BOLA1 would directly prevent SNOC-induced S-nitrosylation of Drp1, rather than reduce the effect of SNOC-induced increase of GSH oxidation, remains to be established. This might be of critical importance because beta-amyloid protein found in brains of patients with Alzheimer's disease was found to induce S-nitrosylation of Drp1 and S-nitrosylated Drp1 was shown to be increased in brains of these patients, suggesting that S-nitrosylation of Drp1 might contribute to the pathogenesis of neurodegeneration (7).
In bacterial operons, BolA tends to occur not only with a monothiol glutaredoxin, but also with proteins involved in defense against oxidative stress: glutathione-S-transferase and Toluene exporters (16). We found, however, no evidence for upregulation of BOLA1 under conditions of oxidative stress in human gene expression data (Supplementary Table S2). Nor did we detect increased oxidation of HEt, which measures superoxides, in BOLA1 knockdowns, suggesting that BOLA1 is specific for thiol oxidation. Alternatively, to directly affecting the thiol oxidation state of target proteins, BOLA1 could have an indirect effect, for example, by being involved in the assembly of Fe-S clusters, like its mitochondrial paralog BOLA3 (6). Interactions between Grx5 and the Fe-S assembly protein ISA1 have been measured in S. cerevisiae (36) and in Schizosaccharomyces pombe (19), implicating Grx5 in Fe-S assembly, and GLRX5 has been implicated in Fe-S cluster assembly in human erythroblasts (41). The nonassembly of Fe-S clusters in mitochondria will lead to free iron, which via Fenton chemistry can lead to the formation of hydroxyl radicals, and therewith to increased oxidation of biomolecules. The increased thiol redox potential that we observe in BOLA1 knockdowns could thus be an indirect effect of the inability to assemble Fe-S clusters. Also, a fusion protein of BolA1 and the chloroplastic Fe-S assembly protein SufE in plants (40) argue for a role of BolA1 in Fe-S assembly or repair. Nevertheless, mitochondrial Fe-S assembly proteins, including Grx5 (Supplementary Fig. S3) have a wider phylogenetic distribution than only aerobic species. BolA1's strictly aerobic genomic occurrence, its reducing effect on the mitochondrial thiol redox potential, its interaction with Grx5 and the role in Fe-S assembly of the latter, suggest that BolA1 functions by increasing an aspect of Grx5's activity in Fe-S assembly or another process that is specifically required under aerobic, thiol-oxidizing conditions.
Materials and Methods
Sequence analysis and phylogenetic analysis
Eukaryotic orthologs of the BolA protein family were collected using PSI-Blast (default parameters) with BOLA1 as query and iterating until convergence against the NCBI nonredundant protein database in August 2011. To the eukaryotic sequences, E. coli BolA representatives (YrbA and BolA) were added. Alignment for the BolA domain was derived with ClustalX (24) followed by manual adjustments. Representatives from the eukaryotic crown groups, including all the anaerobic species and the proteins with experimental data, were selected for alignment (Supplementary Fig. S4) and phylogeny (Fig. 1). The maximum likelihood phylogeny was computed with PhyML (13), using the LG amino acid substitution model with a Gamma distribution approximated by 4 discrete-rate categories (4G). The proportion of invariant sites (I) and the gamma shape parameter (alpha) were estimated from the data. The Akaike Information Criterion implemented in ProtTest (v2.4) (1) was used to choose the evolutionary model and its parameters. The same procedure was followed for the monothiol glutaredoxins (Supplementary Fig. S3). We refer to the human proteins with BOLA1, BOLA2, BOLA3, and GLRX5, while we use BolA1, BolA2, BolA3, and Grx5 to refer to the orthologous groups that contain these proteins.
Primary human skin fibroblasts and inducible HEK293 and HeLa cell lines
Fibroblasts were obtained according to the relevant Institutional Review Boards from a skin biopsy of a healthy subject and cultured in the HEPES (25 mM)-buffered M199 medium (Invitrogen) supplemented with 10% (v/v) fetal calf serum (FCS), 100 IU/ml penicillin (Gibco), and 100 IU/ml streptomycin (Gibco). Generation of HEK293 cell lines for inducible expression of AcGFP1- and TAP-tagged BOLA1 and TAP-tagged GLRX5 is described in Supplementary Materials and Methods. HEK293 and HeLa cells were cultured in Dulbecco's modified Eagle medium (DMEM; Biowhitaker) with the same additions. T-REx™ Flp-In™ HEK293 cells were cultured in the presence of 5 μg/ml blasticidin (Invitrogen) and 300 μg/ml zeocin (Invitrogen).
Baculoviruses for transient overexpression of proteins of interest in human skin fibroblasts
Baculoviruses for cyto- and mito-roGFP1 are described elsewhere (35). Viruses for AcGFP1-tagged BolA homolog 1 (BOLA1-GFP; BC_063405), 2 (BOLA2-GFP; AF060511), and 3 (BOLA3-GFP; NM_21552) and cytochrome c oxidase subunit VIII (COX8) targeting sequence (COX8-GFP) and for TagRFP-tagged BOLA1 (BOLA1-RFP) and COX8 targeting sequence (COX8-RFP) were generated as reported previously (15). The BOLA protein entry vectors were purchased from Open Biosystems. Briefly, cDNA of the protein of interest was cloned into a modified pFastBacTMDual vector behind a CMV promoter and adjacent to a C-terminal GFP or TagRFP sequence. The obtained constructs were used to generate infectious recombinant baculoviruses by site-specific transposing-mediated insertion into baculovirus genome (barmaid). Isolated recombinant barmaids were used for virus amplification in Spodoptera frugiperda 9 insect cells.
BOLA1 knockdown and analysis of its effect on the cytosolic and mitochondrial thiol redox potential and the rate of HEt oxidation
Three BOLA1 siRNAs were designed using online software provided by the Whitehead Institute for Biomedical Research (42): #1 antisense strand: 5′-UUAACAUGGAACAUCCGGGdTdT, #2 antisense strand: 5′-UUGUUUCCAACUCAUCAGGdTdT, #3 antisense strand: 5′-ACUGUAUCAAAGGGAAGGCdTdT. The siRNAs were synthesized by Biolegio. Transfection of HeLa cells with siRNA as described before (37). Briefly, HeLa cells, plated in DMEM supplemented with 10% (v/v) FCS (without antibiotics), were transfected with siRNA using Dharmafect 1 transfection reagent (Dharmacon) and Optimem (Gibco). Forty-eight hours later, the cells were splitted 1:5, plated on glass bottom dishes (Willco Wells), and incubated overnight. After a second round of transfection, DMEM was replaced with M199 (without antibiotics) containing mito- or cyto-roGFP coding baculovirus (35). Ninety-six hours later, M199 was changed to HEPES–Tris containing 132 mM NaCl, 4.2 mM KCl, 1 mM CaCl2, 1 mM MgCl2, 5.5 mM D-glucose, and 10 mM HEPES (pH 7.4) and cells were subjected to digital-imaging microscopy of roGFP and HEt oxidation products fluorescence (for details, see Supplementary Materials and Methods). Immunoblot analysis (see below) with anti-BOLA1 was used to demonstrate the effectiveness of the siRNAs.
BOLA overexpression and analysis of its intracellular localization and effect on BSO- and SNOC-induced changes in mitochondrial morphology and cytosolic and mitochondrial thiol redox potential
To determine the intracellular localization of BOLA1, 2, and 3 and the effect of BOLA1 on mitochondrial morphology, fibroblasts were seeded on glass bottom dishes, cultured for 24 h, and transduced with the appropriate baculovirus for expression of their GFP-tagged version. At 96 h after transduction, mitochondria were stained with TMRM and cells were subjected to digital-imaging microscopy of GFP and TMRM fluorescence. Quantitative analysis of mitochondrial morphology was performed on TMRM images as described before (20). To assess the effect of BOLA1 on the cytosolic and mitochondrial thiol redox potential, cells were cotransduced with a baculovirus for BOLA1-RFP and a baculovirus for either cyto- or mito-roGFP1. For further details on roGFP measurements, see Supplementary Materials and Methods. BSO and SNOC were added at 24 h after transduction.
Single-step affinity purification and nanospray ionization liquid chromatography tandem mass spectrometry and immunoblot analysis
Single-step affinity purification and nano-LC-MS/MS and immunoblot analysis were performed as described previously (38). For single-step affinity purification of BOLA1-TAP and GLRX5-TAP, cells were resuspended in the lysis buffer provided with the InterPlay TAP Purification Kit (Agilent Technologies) and subjected to three cycles of freeze–thawing followed by 10-min centrifugation at 10,000 g. The supernatant was incubated for 2 h under rotation at 4°C in the presence of Strep-Tactin-Superflow beads (IBA). Next, the beads were washed six times with the streptavidin-binding buffer provided with the kit, supplemented with 0.1% (w/v) lauryl maltoside, and bound proteins were eluted with the Strep-tag elution buffer containing D-Desthiobiotin (IBA). Before being processed for nLC-MS/MS analysis or immunoblotting, eluates were concentrated by passing through a 3 kDa cutoff filter (Millipore). For details on nLC-MS/MS analysis, see Supplementary Materials and Methods. Proteins identified in noninduced cells or in TAP-purifications of background, mitochondrial proteins AURKAIP1, C12orf62, C7orf44, PET100, PET117, ES1, and C10orf65 for which we have no indication that they are functionally or physically linked to BOLA1 or GLRX5, were excluded. Immunoblotting of the concentrated eluates was done as described previously. Sodium dodecyl sulfate–polyacrylamide gel electrophoresis gels were run with 80–300 μg of protein and resolved proteins were transferred onto a nitrocellulose or PVDF membrane for immunostaining with primary antibodies. The GLRX5 antibody was kindly provided by Dr. Wing-Hang Tong (Molecular Medicine Branch, National Institute of Child Health and Human Development, Bethesda).
Supplementary Material
Abbreviations Used
- AR
aspect ratio
- BSO
L-buthionine-(S,R)-sulfoximine
- CBP
calmodulin-binding protein
- CK-B
cytosolic creatine kinase B
- COX8
cytochrome c oxidase subunit VIII
- DMEM
Dulbecco's modified Eagle medium
- DTT
dithiothreitol
- F
form factor
- FCS
fetal calf serum
- GSH
glutathione
- HEt
hydroethidine
- MCB
monochlorobimane
- Nc
number of mitochondria per cell
- nLC-MS/MS
nanospray ionization liquid chromatography tandem mass spectrometry
- RT
room temperature
- SDHA
succinate dehydrogenase subunit A
- SNOC
S-nitrosocysteine
- TMRM
tetramethylrhodamine methyl ester
- TOM20
mitochondrial import receptor subunit TOM20
Acknowledgments
This work was supported by an equipment grant of NWO (Netherlands Organization for Scientific Research, No: 911-02-008). R.S. is supported by CSBR (Centres for Systems Biology Research) from the Netherlands Organisation for Scientific Research (NWO; CSBR09/013V), B.F.J.W. by a Horizon grant (050-71-053), U.K. and J.E. by the Dutch Ministry of Economic Affairs (IOP Grant #IGE05003), I.D. by the Portuguese Foundation for Science and Technology-FCT (SFRH/BD/32959/2006), and by “BolsasRui Tavares 2010.”
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Abascal F. Zardoya R. Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- 2.Aldea M. Hernandez-Chico C. de la Campa AG. Kushner SR. Vicente M. Identification, cloning, and expression of bolA, an ftsZ-dependent morphogene of Escherichia coli. J Bacteriol. 1988;170:5169–5176. doi: 10.1128/jb.170.11.5169-5176.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berendji D. Kolb-Bachofen V. Meyer KL. Kroncke KD. Influence of nitric oxide on the intracellular reduced glutathione pool: different cellular capacities and strategies to encounter nitric oxide-mediated stress. Free Radic Biol Med. 1999;27:773–780. doi: 10.1016/s0891-5849(99)00123-9. [DOI] [PubMed] [Google Scholar]
- 4.Byrnes A. Jacks A. Dahlman-Wright K. Evengard B. Wright FA. Pedersen NL. Sullivan PF. Gene expression in peripheral blood leukocytes in monozygotic twins discordant for chronic fatigue: no evidence of a biomarker. PLoS One. 2009;4:e5805. doi: 10.1371/journal.pone.0005805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Camaschella C. Campanella A. De Falco L. Boschetto L. Merlini R. Silvestri L. Levi S. Iolascon A. The human counterpart of zebrafish shiraz shows sideroblastic-like microcytic anemia and iron overload. Blood. 2007;110:1353–1358. doi: 10.1182/blood-2007-02-072520. [DOI] [PubMed] [Google Scholar]
- 6.Cameron JM. Janer A. Levandovskiy V. Mackay N. Rouault TA. Tong WH. Ogilvie I. Shoubridge EA. Robinson BH. Mutations in iron-sulfur cluster scaffold genes NFU1 and BOLA3 cause a fatal deficiency of multiple respiratory chain and 2-oxoacid dehydrogenase enzymes. Am J Hum Genet. 2011;89:486–495. doi: 10.1016/j.ajhg.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho DH. Nakamura T. Fang J. Cieplak P. Godzik A. Gu Z. Lipton SA. S-nitrosylation of Drp1 mediates beta-amyloid-related mitochondrial fission and neuronal injury. Science. 2009;324:102–105. doi: 10.1126/science.1171091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Claros MG. Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- 9.Couturier J. Jacquot JP. Rouhier N. Evolution and diversity of glutaredoxins in photosynthetic organisms. Cell Mol Life Sci. 2009;66:2539–2557. doi: 10.1007/s00018-009-0054-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gabaldon T. Huynen MA. Reconstruction of the proto-mitochondrial metabolism. Science. 2003;301:609. doi: 10.1126/science.1085463. [DOI] [PubMed] [Google Scholar]
- 11.Giot L, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 12.Grans H. Nilsson P. Evengard B. Gene expression profiling in the chronic fatigue syndrome. J Intern Med. 2005;258:388–390. doi: 10.1111/j.1365-2796.2005.01548.x. [DOI] [PubMed] [Google Scholar]
- 13.Guindon S. Delsuc F. Dufayard JF. Gascuel O. Estimating maximum likelihood phylogenies with PhyML. Methods Mol Biol. 2009;537:113–137. doi: 10.1007/978-1-59745-251-9_6. [DOI] [PubMed] [Google Scholar]
- 14.Ho Y, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 15.Hoefs SJ, et al. NDUFA2 complex I mutation leads to Leigh disease. Am J Hum Genet. 2008;82:1306–1315. doi: 10.1016/j.ajhg.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huynen MA. Spronk CA. Gabaldon T. Snel B. Combining data from genomes, Y2H and 3D structure indicates that BolA is a reductase interacting with a glutaredoxin. FEBS Lett. 2005;579:591–596. doi: 10.1016/j.febslet.2004.11.111. [DOI] [PubMed] [Google Scholar]
- 17.Ito T. Tashiro K. Muta S. Ozawa R. Chiba T. Nishizawa M. Yamamoto K. Kuhara S. Sakaki Y. Toward a protein-protein interaction map of the budding yeast: a comprehensive system to examine two-hybrid interactions in all possible combinations between the yeast proteins. Proc Natl Acad Sci U S A. 2000;97:1143–1147. doi: 10.1073/pnas.97.3.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johansson C, et al. The crystal structure of human GLRX5: iron-sulfur cluster co-ordination, tetrameric assembly and monomer activity. Biochem J. 2010;433:303–311. doi: 10.1042/BJ20101286. [DOI] [PubMed] [Google Scholar]
- 19.Kim KD. Chung WH. Kim HJ. Lee KC. Roe JH. Monothiol glutaredoxin Grx5 interacts with Fe-S scaffold proteins Isa1 and Isa2 and supports Fe-S assembly and DNA integrity in mitochondria of fission yeast. Biochem Biophys Res Commun. 2010;392:467–472. doi: 10.1016/j.bbrc.2010.01.051. [DOI] [PubMed] [Google Scholar]
- 20.Koopman WJ. Distelmaier F. Esseling JJ. Smeitink JA. Willems PH. Computer-assisted live cell analysis of mitochondrial membrane potential, morphology and calcium handling. Methods. 2008;46:304–311. doi: 10.1016/j.ymeth.2008.09.018. [DOI] [PubMed] [Google Scholar]
- 21.Koopman WJ. Verkaart S. Visch HJ. van der Westhuizen FH. Murphy MP. van den Heuvel LW. Smeitink JA. Willems PH. Inhibition of complex I of the electron transport chain causes O2−•-mediated mitochondrial outgrowth. Am J Physiol Cell Physiol. 2005;288:C1440–C1450. doi: 10.1152/ajpcell.00607.2004. [DOI] [PubMed] [Google Scholar]
- 22.Koopman WJ. Visch HJ. Verkaart S. van den Heuvel LW. Smeitink JA. Willems PH. Mitochondrial network complexity and pathological decrease in complex I activity are tightly correlated in isolated human complex I deficiency. Am J Physiol Cell Physiol. 2005;289:C881–C890. doi: 10.1152/ajpcell.00104.2005. [DOI] [PubMed] [Google Scholar]
- 23.Kumanovics A, et al. Identification of FRA1 and FRA2 as genes involved in regulating the yeast iron regulon in response to decreased mitochondrial iron-sulfur cluster synthesis. J Biol Chem. 2008;283:10276–10286. doi: 10.1074/jbc.M801160200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 25.Li H. Mapolelo DT. Dingra NN. Keller G. Riggs-Gelasco PJ. Winge DR. Johnson MK. Outten CE. Histidine 103 in Fra2 is an iron-sulfur cluster ligand in the [2Fe-2S] Fra2-Grx3 complex and is required for in vivo iron signaling in yeast. J Biol Chem. 2010;286:867–876. doi: 10.1074/jbc.M110.184176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Linares GR. Xing W. Govoni KE. Chen ST. Mohan S. Glutaredoxin 5 regulates osteoblast apoptosis by protecting against oxidative stress. Bone. 2009;44:795–804. doi: 10.1016/j.bone.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyer AJ. Brach T. Marty L. Kreye S. Rouhier N. Jacquot JP. Hell R. Redox-sensitive GFP in Arabidopsis thaliana is a quantitative biosensor for the redox potential of the cellular glutathione redox buffer. Plant J. 2007;52:973–986. doi: 10.1111/j.1365-313X.2007.03280.x. [DOI] [PubMed] [Google Scholar]
- 28.Navarro Llorens JM. Tormo A. Martinez-Garcia E. Stationary phase in gram-negative bacteria. FEMS Microbiol Rev. 2010;34:476–495. doi: 10.1111/j.1574-6976.2010.00213.x. [DOI] [PubMed] [Google Scholar]
- 29.Pagliarini DJ, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodriguez-Manzaneque MT. Tamarit J. Belli G. Ros J. Herrero E. Grx5 is a mitochondrial glutaredoxin required for the activity of iron/sulfur enzymes. Mol Biol Cell. 2002;13:1109–1121. doi: 10.1091/mbc.01-10-0517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Santos JM. Freire P. Vicente M. Arraiano CM. The stationary-phase morphogene bolA from Escherichia coli is induced by stress during early stages of growth. Mol Microbiol. 1999;32:789–798. doi: 10.1046/j.1365-2958.1999.01397.x. [DOI] [PubMed] [Google Scholar]
- 32.Santos JM. Lobo M. Matos AP. De Pedro MA. Arraiano CM. The gene bolA regulates dacA (PBP5), dacC (PBP6) and ampC (AmpC), promoting normal morphology in Escherichia coli. Mol Microbiol. 2002;45:1729–1740. doi: 10.1046/j.1365-2958.2002.03131.x. [DOI] [PubMed] [Google Scholar]
- 33.Su AI, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tamarit J. Belli G. Cabiscol E. Herrero E. Ros J. Biochemical characterization of yeast mitochondrial Grx5 monothiol glutaredoxin. J Biol Chem. 2003;278:25745–25751. doi: 10.1074/jbc.M303477200. [DOI] [PubMed] [Google Scholar]
- 35.Verkaart S. Koopman WJ. Cheek J. van Emst-de Vries SE. van den Heuvel LW. Smeitink JA. Willems PH. Mitochondrial and cytosolic thiol redox state are not detectably altered in isolated human NADH: ubiquinone oxidoreductase deficiency. Biochim Biophys Acta. 2007;1772:1041–1051. doi: 10.1016/j.bbadis.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 36.Vilella F. Alves R. Rodriguez-Manzaneque MT. Belli G. Swaminathan S. Sunnerhagen P. Herrero E. Evolution and cellular function of monothiol glutaredoxins: involvement in iron-sulphur cluster assembly. Comp Funct Genomics. 2004;5:328–341. doi: 10.1002/cfg.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vogel RO, et al. Cytosolic signaling protein Ecsit also localizes to mitochondria where it interacts with chaperone NDUFAF1 and functions in complex I assembly. Genes Dev. 2007;21:615–624. doi: 10.1101/gad.408407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wessels HJ. Vogel RO. van den Heuvel L. Smeitink JA. Rodenburg RJ. Nijtmans LG. Farhoud MH. LC-MS/MS as an alternative for SDS-PAGE in blue native analysis of protein complexes. Proteomics. 2009;9:4221–4228. doi: 10.1002/pmic.200900157. [DOI] [PubMed] [Google Scholar]
- 39.Wingert RA, et al. Deficiency of glutaredoxin 5 reveals Fe-S clusters are required for vertebrate haem synthesis. Nature. 2005;436:1035–1039. doi: 10.1038/nature03887. [DOI] [PubMed] [Google Scholar]
- 40.Ye H. Abdel-Ghany SE. Anderson TD. Pilon-Smits EA. Pilon M. CpSufE activates the cysteine desulfurase CpNifS for chloroplastic Fe-S cluster formation. J Biol Chem. 2006;281:8958–8969. doi: 10.1074/jbc.M512737200. [DOI] [PubMed] [Google Scholar]
- 41.Ye H, et al. Glutaredoxin 5 deficiency causes sideroblastic anemia by specifically impairing heme biosynthesis and depleting cytosolic iron in human erythoblasts. J Clin Invest. 2010;120:1749–1761. doi: 10.1172/JCI40372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yuan B. Latek R. Hossbach M. Tuschl T. Lewitter F. siRNA Selection Server: an automated siRNA oligonucleotide prediction server. Nucleic Acids Res. 2004;32:W130–W134. doi: 10.1093/nar/gkh366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou YB, et al. hBolA, novel non-classical secreted proteins, belonging to different BolA family with functional divergence. Mol Cell Biochem. 2008;317:61–68. doi: 10.1007/s11010-008-9809-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.