Abstract
Polyploid cells have genomes that contain multiples of the typical diploid chromosome number and are found in many different organisms. Studies in a variety of animal and plant developmental systems have revealed evolutionarily conserved mechanisms that control the generation of polyploidy and have recently begun to provide clues to its physiological function. These studies demonstrate that cellular polyploidy plays important roles during normal development and also contributes to human disease, particularly cancer.
Keywords: Cancer, Endocycle, Endomitosis, Endoreplication, Genome instability
Introduction
Polyploid cells, which contain multiples of the diploid genome equivalent, have been studied for many years, nearly as long as chromosomes themselves have been studied. Now, over a century after the discovery of polyploidy, we know much about the molecular mechanisms that generate cellular polyploidy, but comparably little regarding the physiological function of the polyploid state. This discrepancy exists despite the frequent occurrence of polyploid cells in most multicellular organisms, as well as in human cancers. An important aspect of deciphering the roles for polyploidy lies in understanding the regulation of endoreplication, a cell cycle variation that generates a polyploid genome by repeated rounds of DNA replication in the absence of cell division. Recent advances show that, although some differences exist in the diverse endoreplication programs found from protists to humans, core principles can be applied. New work in genetic model organisms has identified cases in which programmed endoreplication plays key roles in development. Furthermore, recent evidence indicates that endoreplication can confer genome instability, a major cancer-enabling property. In this Primer (see Box), we review such recent discoveries, focusing on work carried out in the past 3 years, and refer the reader to other comprehensive reviews on this topic (De Veylder et al., 2011; Lee et al., 2009). From these new insights emerge unifying themes regarding endoreplication that hold promise of elucidating the advantages, as well as the potential disadvantages, of polyploidy.
Development: the big picture
This Primer is part of a series entitled ‘Development: the big picture’. This series aims to highlight key developmental systems or processes that have been the subject of intense study because they have broad implications for other developmental, cell and molecular systems, or for disease and therapeutics. Keep an eye out for other articles in this series over the coming months!
Endoreplication and development
Endoreplication is typically studied in the context of endopolyploidy, which refers to the presence of polyploid cells in an otherwise diploid organism. Diverse levels of polyploidy occur throughout nature (Table 1). Although polyploidy is well appreciated in plants (De Veylder et al., 2011), many different animal tissues, including the skin, gut, placenta, liver, brain and blood have polyploid cells (Table 1) (Laird et al., 1980; Corash et al., 1989; Fox et al., 2010; Hedgecock and White, 1985; Melaragno et al., 1993; Sherman, 1972; Unhavaithaya and Orr-Weaver, 2012; Zanet et al., 2010). Interestingly, endoreplication is not limited to multi-cellular organisms, with examples described in ciliated protozoa (Yin et al., 2010) and even bacteria (Mendell et al., 2008).
Table 1.
Examples of the evolutionary diversity of polyploidy
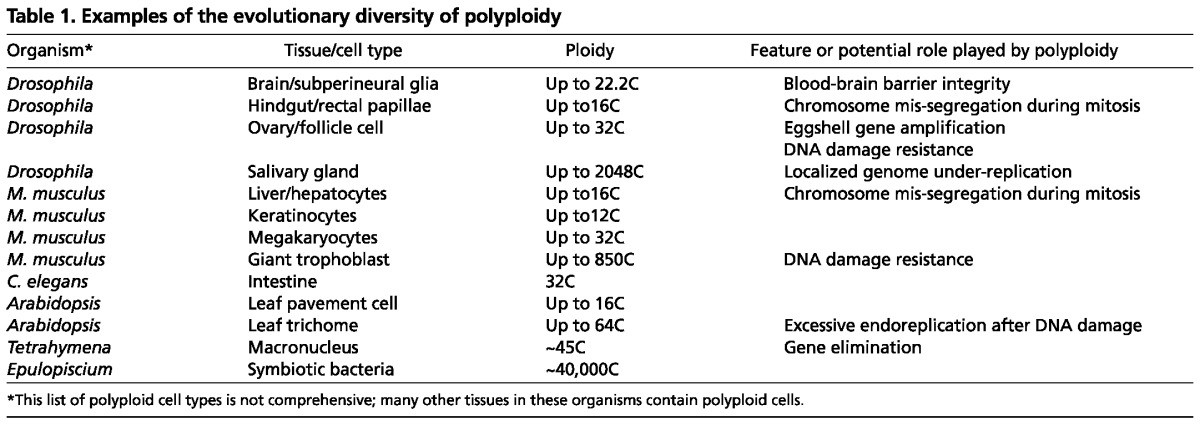
Two primary forms of endoreplication have been described: endocycling and endomitosis (Fig. 1). Endocycles are composed of alternating periods of S phase, during which DNA replication occurs, and gap phase, when cells prepare for the next round of S phase. Endocycling cells do not display any features of mitosis. Cells undergoing endomitosis, by contrast, execute an abortive mitosis that does not result in cell division, followed by subsequent re-entering of S phase. Although some molecular regulators of each cycle type may differ slightly across species, the core regulatory principles are essentially the same. Moreover, given new data showing that both endocycles and endomitosis exist in flies and mammals (Ullah et al., 2009b; Unhavaithaya and Orr-Weaver, 2012), and that varying degrees of both endocycles and endomitosis can occur in the same tissue, the distinction between the two may be insignificant given that they lead to the same outcome (i.e. polyploidy). Because of this similarity, we will refer to all polyploidizing cell cycles as ‘endoreplication’, unless a specific context dictates otherwise.
Fig. 1.
Endoreplication results from cell cycle plasticity during development. (A) A canonical cell division cycle consists of four distinct phases. Chromosomes are duplicated during S phase and segregated to daughter cells during M phase. (B) Endoreplication can occur either by endomitosis, a cell cycle that displays features of mitosis but lacks cytokinesis, or by endocycling in which periods of S and G phase alternate with no cell division.
Cell cycle regulation of endoreplication
With respect to the cell cycle machinery, one of the most interesting features of endoreplication is that the same regulatory proteins that drive a typical cell division cycle are used to drive endoreplication. Developmental signals exploit this inherent plasticity of the cell cycle to convert a typical cell division cycle into an endoreplication cycle. To understand this process, we first need to describe some of the fundamental principles of cell cycle control and DNA replication, and then describe the mechanisms by which cells apply these principles to generate an endoreplication cycle.
A general overview of cell cycle control
Progression through the canonical G1-S-G2-M cell division cycle is controlled by a family of serine/threonine protein kinases, called cyclin-dependent kinases (CDKs). At the appropriate times, CDKs phosphorylate specific substrates to bring about the two major events of the cell division cycle: S phase and mitosis. These two events are coupled such that S phase cannot begin until mitosis is completed. This coupling is enforced by CDK activity, which alternates between relatively low levels (in G1) and high levels (in S-G2-M) during the cell division cycle (Morgan, 2007) (see Box 1). Consequently, two important events must occur in order to convert a mitotic cycle into an endoreplication cycle and achieve polyploidy: (1) cell division must be suppressed; and (2) CDK activity must continue to alternate between low and high levels in order for the genome to be reduplicated in the absence of cell division. As we discuss below, different cell types use different mechanisms to accomplish these tasks.
Box 1. CDK-mediated control of DNA replication
With respect to S phase, low CDK activity permits the assembly of replication initiation proteins into a ‘pre-replicative complex’ (pre-RC) on DNA at places where DNA replication will initiate once S phase begins. Pre-RC assembly during G1 allows DNA to become competent to replicate, which is referred to as ‘replication licensing’. High CDK activity both initiates DNA replication during S phase and prevents DNA from becoming licensed to replicate during S and G2 phases by inhibiting assembly of the pre-RC (Diffley, 2011). Thus, only when CDK activity is abolished at the end of mitosis can the genome become licensed to re-enter S phase. This mechanism ensures that, during a normal cell division cycle, the genome is replicated once and only once – a requisite for maintaining diploidy.
Endoreplication by programmed inhibition of mitosis and cell division
Although transcriptional downregulation of mitotic regulators is one known mechanism for promoting endoreplication (Maqbool et al., 2010; Pandit et al., 2012; Zielke et al., 2008), endoreplicating cells can inhibit mitosis in a number of ways. One of the evolutionarily oldest mechanisms of mitotic inhibition, being conserved between plants and animals, is downregulation of mitotic CDK activity by ubiquitin-mediated proteolysis. Entry into and progression through mitosis requires the function of CDKs that are activated by A- and B-type cyclins. Conversely, exit from mitosis and subsequent cytokinesis requires the inhibition of these CDKs, which is achieved via cyclin proteolysis. This proteolysis is triggered by an E3 ubiquitin ligase called the anaphase-promoting complex/cyclosome (APC/C). An APC/C substrate targeting subunit, called Fzr/Cdh1 (Rap – FlyBase), directs APC/CFzr/Cdh1 activity towards A- and B-type cyclins, beginning in late mitosis and continuing through the subsequent G1 phase, thus helping ensure efficient and irreversible exit from mitosis (Morgan, 2007; Song and Rape, 2011). Some endoreplicating cells take advantage of this mitotic suppression mechanism to achieve polyploidy. For example, Fzr/Cdh1 expression in Drosophila and Arabidopsis is both necessary and sufficient for endoreplication (Larson-Rabin et al., 2009; Sigrist and Lehner, 1997). Drosophila APC/CFzr/Cdh1 activity is continuously required during endocycles to ensure suppression of mitotic regulators (Narbonne-Reveau et al., 2008; Zielke et al., 2008), and genetic evidence in Arabidopsis indicates that destruction of A- and B-type cyclins mediated by the Fzr/Cdh1 homolog CCS52A1 is crucial for endoreplication (Boudolf et al., 2009; Kasili et al., 2010). Moreover, the failure to properly control the timing of inhibition of APC/C activity during Arabidopsis development results in inappropriate endoreplication (Iwata et al., 2011).
CDKs can also be inhibited through direct binding to proteins called cyclin kinase inhibitors (CKIs), and some polyploid cell types use CKIs to inhibit mitosis and trigger endoreplication (Ullah et al., 2009a). This is particularly true for mammalian trophoblast giant cells (TGCs) of the placenta and megakaryocytes, which generate platelets for blood clotting. When trophoblast stem cells differentiate into endoreplicating TGCs, induction of the p57 CKI results in inhibition of Cdk1 (Ullah et al., 2008). The p21 CKI plays a similar role in megakaryocyte endoreplication (Chagraoui et al., 2011; Muñoz-Alonso et al., 2012), as does the SIAMESE gene, which encodes an Arabidopsis CKI required for endoreplication in leaf hairs known as trichomes (Churchman et al., 2006; Morohashi and Grotewold, 2009). CKIs thus play an important part in the differentiation and terminal polyploid phenotype of diverse cell types.
Blocking cytokinesis is another mechanism that can promote endoreplication and polyploidy. For example, horticulturists have long used microtubule poisons, such as colchicine, to inhibit cell division and stimulate polyploid derivatives of important crop species (Hancock, 2005). The small GTPase RhoA is a key regulator of cell division, and multiple mechanisms ensure that RhoA is activated at the correct time and place to initiate cytokinesis (Glotzer, 2005). Megakaryocyte endoreplication requires the downregulation of two guanine nucleotide exchange factors, GEF-H1 and ECT2, that activate RhoA during cytokinesis, and their expression is also sufficient to prevent it (Gao et al., 2012). Endoreplication in rat liver also involves a developmentally regulated block to cytokinesis that occurs during weaning (Celton-Morizur and Desdouets, 2010; Celton-Morizur et al., 2010; Celton-Morizur et al., 2009).
An important concept emerging from these studies is that cells use several different mechanisms to suppress cell division and promote endoreplication. In addition, the combined use of multiple mechanisms to inhibit mitotic CDK activity and cytokinesis (e.g. transcriptional repression, ubiquitin-mediated proteolysis, CKI expression, RhoA inhibition) by a particular cell type ensures robust commitment to endoreplication.
The endoreplication oscillator: toggling between high and low CDK activity
Replication of DNA during S phase requires CDK activity. Indeed, CKI induction during megakaryocyte differentiation must be transient in order not to inhibit the CDK activity required for endoreplication S phase (Muñoz-Alonso et al., 2012). Cdk2 is the important kinase for endoreplication S phase in animal cells, although in the absence of Cdk2 in mammals, Cdk1 can act as a substitute (Ullah et al., 2009b). Cyclin E overexpression increases the ploidy of megakaryocytes (Eliades et al., 2010), suggesting that the Cyclin E/Cdk2 (Cdc2c – FlyBase) complex is the relevant kinase. Likewise, early work suggested that the Cyclin E/Cdk2 complex is the crucial, and perhaps only, CDK required for endoreplication in Drosophila (Lilly and Duronio, 2005). However, Sallé et al. recently showed that Cyclin A regulates endoreplication S phase dynamics in Drosophila mechanosensory organs, although a corresponding CDK was not identified (Sallé et al., 2012). In spite of such important complexities, informative models of endoreplication can be built by considering the S-phase CDK as a single activity.
The alternation of high and low levels of CDK activity that is needed for endoreplication can often be observed cytologically. For example, Cyclin E transcripts and protein are present just prior to and during S phase, but not G phase, in endoreplicating Drosophila cells (Knoblich et al., 1994; Lilly and Spradling, 1996; Weng et al., 2003). Endoreplication is suppressed when such cyclic accumulation of Cyclin E is bypassed by forced continuous transcription of Cyclin E (Follette et al., 1998; Weiss et al., 1998). The Cyclin E/Cdk2 complex also functions to control the cyclic accumulation of replication factors like the pre-RC component Orc1 (Narbonne-Reveau et al., 2008). These observations helped formulate the idea of an endoreplication oscillator that controls periods of high and low S-phase CDK activity.
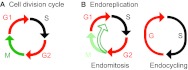
Recently, mathematical modeling of endoreplication oscillations helped guide experiments demonstrating that cyclic accumulation of the transcription factor E2f1 (E2f – FlyBase) is essential for endoreplication in the highly polyploid Drosophila salivary gland (Zielke et al., 2011). E2F transcription factors are potent stimulators of S-phase entry and control the expression of genes required for DNA synthesis, including Cyclin E. Their activity is regulated by the Rb family of tumor suppressors, which bind to and inhibit E2F proteins during G1 phase (van den Heuvel and Dyson, 2008). E2F-Rb interactions regulate endoreplication in plants and worms (Magyar et al., 2012; Ouellet and Roy, 2007). However, Rb-mediated regulation of E2f1 is not essential for endoreplication in Drosophila salivary glands, perhaps because of the action of the E2f2-containing Myb-MuvB complex in repressing Cyclin E expression during G phase (Maqbool et al., 2010; Weng et al., 2003). Zielke et al. (Zielke et al., 2011) provide data in support of a model whereby E2f1 accumulation during G phase results in activation of the Cyclin E/Cdk2 complex, which triggers S phase, which in turn causes the subsequent inactivation of E2f1 via the action of CRL4Cdt2, an E3 ubiquitin ligase that couples proteolysis with DNA replication (Havens and Walter, 2011; Shibutani et al., 2008) (Fig. 2). This model is consistent with earlier observations that Cyclin E activity is required for the downregulation of E2f1 target genes in the endoreplicating embryonic midgut (Duronio and O’Farrell, 1995), and that disruptions to normal oscillations of E2f1 activity in salivary glands suppress endoreplication (Maqbool et al., 2010). Interestingly, CRL4 is also required for endoreplication in Arabidopsis trichomes, although the relevant CRL4 target in trichomes might be CKI proteins (Roodbarkelari et al., 2010). Negative-feedback regulation of E2F activity is also important for endoreplication in hepatocytes (Chen et al., 2012; Pandit et al., 2012), but here it appears to control the transition from cell division to endoreplication, and it is not yet known whether E2F functions as part of a mammalian endoreplication oscillator. Nonetheless, these results emphasize that negative-feedback regulation is a common and important feature of molecular oscillators that control cell cycle progression (Ferrell et al., 2011).
Fig. 2.
The endoreplication oscillator in Drosophila salivary glands. The core regulatory relationships between E2f1, Cyclin E and CRL4Cdt2 (top) give rise to out-of-phase oscillations in the activity of key regulatory proteins controlling S phase (bottom). E2f1, APCFzr/Cdh1 and Cdt1 (red) are active during G phase, whereas the Cyclin E/Cdk2 complex, CRL4Cdt2 and Geminin (black) are active during S phase. Note that CRL4Cdt2 activity is dependent on DNA replication, which is triggered by the Cyclin E/Cdk2 complex, and that Cdt1 is inhibited both by Geminin binding and CRL4Cdt2-mediated destruction.
A central CDK oscillator provides a mechanism through which other important aspects of endoreplication can be controlled (Fig. 2). For example, the oscillation of APC/C activity in Drosophila salivary glands probably results directly from the oscillation of Cyclin E/Cdk2 complex activity, which inhibits the
APC/C (Narbonne-Reveau et al., 2008). Key targets of the APC/C during endoreplication progression are the mitotic cyclins and the protein Geminin, which is an inhibitor of the essential replication licensing protein Cdt1 (Dup – FlyBase). Indeed, the combination of mitotic Cdk1 inhibition and Geminin proteolysis can trigger endoreplication in human cells (Hochegger et al., 2007). By contrast, in Arabidopsis, little or no APC/C activity is required for endoreplication progression (as opposed to endoreplication entry, as described above), perhaps because there is no Geminin protein (Roodbarkelari et al., 2010). In Drosophila salivary glands, Geminin is absent during G phase, when replication licensing occurs, and present during S phase to prevent DNA licensing (Zielke et al., 2008). In this way, the endoreplication S phase is similar to the S phase of a cell division cycle and distinct from the phenomenon of re-replication, in which specific segments of DNA are replicated more than once in a single S phase. In contrast to endoreplication, re-replication is an aberrant situation that causes genomic instability and is observed in some cancer cells (see Box 2).
Box 2. Genome instability
Genome instability refers to an aberrant state of frequent genome alteration. Diverse mechanisms contribute to genome instability. These include chromosome instability, a frequent mis-segregation of chromosomes during cell division (Negrini et al., 2010) or failed or improper repair of DNA damage (Lord and Ashworth, 2012). Because the genome alterations associated with such instability increase the frequency of gene mutation or enable gene overexpression, genome instability is considered the most significant enabling feature of cancer (Hanahan and Weinberg, 2011).
In summary, the central endoreplication oscillator entrains many molecular activities to ensure the cycling of CDK activity and, thus, the cycling of replication licensing. It is important to keep in mind that the mechanism by which cyclic CDK activity is achieved may differ among species or even among different cell types within the same organism. For example, Drosophila nurse cells probably rely more on CKI activity than on E2F activity for CDK control during endoreplication (Hong et al., 2007). Nevertheless, the network of interactions controlling cyclic S phase CDK activity constitutes the central principle of endoreplication that we propose is universally applicable.
Functions of endoreplication and polyploidy
Given the prevalence of polyploid cell types in many different organisms, a reasonable expectation is that endoreplication provides key biological functions during development. Surprisingly, there are relatively few examples of a biological function that is specifically attributed to endopolyploidy. A reason may be that the ideal experiment – converting a tissue composed of polyploid cells into a tissue of the same size composed of diploid cells – is not experimentally trivial and requires a full understanding of endoreplication mechanisms. However, several recent exciting studies have begun to provide evidence that polyploidy is an important functional adaptation in a variety of tissues.
Regulation of the mitosis-to-endoreplication switch
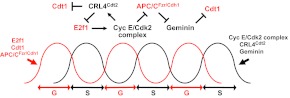
To understand the purpose of endoreplication, we must understand how and when endoreplication initiates. Furthermore, by inhibiting the switch from mitosis to endoreplication, we can learn about the purpose of polyploidy. Perhaps not surprisingly, signal transduction pathways control this switch, and these signals are as variable as the cell types that use them (Fig. 3). These signals control both the decision to make the switch, and the activity of cell cycle regulators that will execute the switch. For example, some of the earliest observations were made in Drosophila follicle cells, which respond to Notch signaling to activate Fzr/Cdh1 expression and suppress Cdk1 activity (Deng et al., 2001; Shcherbata et al., 2004; Sun and Deng, 2007). Recent work in this Drosophila model tissue suggests that chromatin modifications may accompany the Notch-regulated mitosis-to-endoreplication switch (Domanitskaya and Schüpbach, 2012). The involvement of Notch in regulating endoreplication appears conserved, as mammalian megakaryocyte differentiation is regulated in part by Notch, although the nature of this regulation (positive versus negative) may differ between humans and mice (Cornejo et al., 2011; Mercher et al., 2008; Poirault-Chassac et al., 2010).
Fig. 3.
Signals triggering polyploidy. (A-C) Signals that regulate the mitosis-to-endoreplication switch in mammals (A; trophoblast giant cells, TGCs), plant seedlings (B; hypocotyl) and insects (C; Drosophila ovarian follicle cells).
In addition to Notch, recent advances in Arabidopsis and mammalian cell culture systems have identified additional signals that promote the mitosis-to-endoreplication switch. An important principle emerging from these studies is that the developmental signals controlling endoreplication are the very same signals that control other aspects of the cell biology of specific cell types; in other words, endoreplication is just one aspect of the phenotype of differentiated cells. For example, the depletion of fibroblast growth factor 4 (FGF4) from trophoblast stem cells triggers both TGC development and endoreplication (Ullah et al., 2008), as does insulin signaling in hepatocytes (Celton-Morizur et al., 2009).
Plant cells trigger endoreplication using developmental signals that are not relevant to animals, two of the most important of which are light and the plant hormone auxin. Light promotes cell division in hypocotyls, and activates the transcription of DEL1, which encodes an atypical E2F that inhibits endoreplication by repressing the expression of CCA52A1 (Fzr/Cdh1). In the dark, DEL1 expression is extinguished and hypocotyl cells endoreplicate (Berckmans et al., 2011). In the root meristem (which is not exposed to light), auxin signaling regulates the mitosis-to-endoreplication switch (Ishida et al., 2010). Correct timing of this switch is also important, as precocious endoreplication can have drastic consequences. Mutation of the SUMO E3 ligase HPY2 results in stunted root growth because of an increase in cellular ploidy at the expense of cell proliferation (Ishida et al., 2009). Conversely, a failure to switch to endoreplication inhibits trichome cell fate commitment and disrupts normal leaf development (Bramsiepe et al., 2010). A similar connection between cell fate and endoreplication has also been observed in the Drosophila germ line (Lilly and Spradling, 1996).
Consequences of defective endoreplication during development
Polyploid cells are typically large, and endoreplication can thus be considered as a form of tissue growth. Consequently, the pathways regulating cellular growth play important roles in the control of endoreplication (Demontis and Perrimon, 2009; Pierce et al., 2008; Steiger et al., 2008). But what might be the advantages of cell growth mediated via polyploidy? One idea is that more copies of the genome increase the biosynthetic capability of cells. A good example is provided by polyploid Drosophila follicle cells, which require many copies of the chorion genes to produce sufficient protein for eggshell biosynthesis (Calvi and Spradling, 1999). However, increased gene copy number does not always correlate with increased gene expression (Kim et al., 2011). Thus, the idea that polyploidy primarily increases biosynthetic capacity is probably too simplistic.
In addition to increasing gene copy number, endoreplication can alter genome structure in unexpected ways. In Drosophila, many endoreplicating cells fail to complete late S phase, leaving some genome regions dramatically under-replicated, including heterochromatic sequences (Gall et al., 1971; Lilly and Spradling, 1996). Using genomic deep-sequencing and chromatin immunoprecipitation approaches, recent studies of polyploid Drosophila genomes precisely defined a number of under-replicated regions also present in euchromatin (Nordman et al., 2011). Most of these under-replicated regions correlate with marks of transcriptional repression (Sher et al., 2012). Interestingly, the degree and location of endoreplication-induced under-replication is dependent on tissue type, suggesting a role for under-replication in defining tissue identity (Nordman et al., 2011). However, under-replication can be blocked by mutation in a chromatin protein Suppressor of Under-Replication (SuUR) and SuUR mutant flies exhibit no obvious alteration of polyploid tissue function (Belyaeva et al., 1998). Thus, unlike gene amplification, the role of under-replication in polyploid tissue function remains unclear. It remains possible that under conditions of physiological stress, a role for this common yet poorly understood polyploid genome alteration may be uncovered. Nevertheless, coupled with studies of programmed DNA elimination in the polyploid macronucleus of ciliates (Chalker and Yao, 2011), under-replication and amplification all represent examples of ‘genome flexibility’ exhibited by polyploid cells. Further study of such flexibility may lead to novel functions of endoreplication in development.
There are aspects of growth that might be best served by endoreplication rather than cell proliferation. A beautiful example is provided by a recent study of the Drosophila larval nervous system. Here, growth via endoreplication is required for glial cells to maintain tight junctions, and thus the blood-brain barrier, during proliferative expansion of the underlying brain tissue (Unhavaithaya and Orr-Weaver, 2012). Interestingly, these glial cells become polyploid either via endocycles or endomitosis, suggesting that the important outcome is growth via endoreplication rather than cell proliferation, regardless of the cell cycle mechanism. Other cell types regulate endoreplication to control organ shape in addition to growth. This is particularly true for the branched trichome structures of Arabidopsis leaves (Kasili et al., 2011), which can also balance proliferation with endoreplication to regulate size and shape (Li et al., 2009). The emerging theme is that the correct timing and extent of endoreplication is often an essential aspect of tissue growth and morphogenesis in both animals and plants.
In spite of these exciting examples, the function of endoreplication is not always forthcoming. Mammalian livers have a remarkable capacity for regeneration after partial hepatectomy, and regeneration can occur via Cyclin E-mediated regulation of endoreplication rather than proliferation (Miyaoka et al., 2012; Nevzorova et al., 2009). Because many hepatocytes are normally polyploid, a reasonable hypothesis is that endoreplication is important for normal liver function. However, recent studies in mouse show that shifting the hepatocyte population from polyploid cells to diploid cells by manipulating E2F activity does not dramatically affect liver function or regeneration (Chen et al., 2012; Pandit et al., 2012). This was somewhat surprising, and suggests that our understanding of the relationship between polyploidy and normal biological function requires further investigation. The following sections describe emerging evidence suggesting an intriguing and complex relationship between polyploidy, genome stability and cancer.
Endoreplication and disease
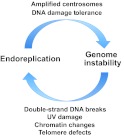
Propagating a stable genome is part of normal cell cycle progression and is required for successful development. It follows that non-canonical cell cycle progression and genome instability (see Box 2) are linked phenomena. Interestingly, recent work from numerous systems suggests a reciprocal connection between genome instability and endoreplication: genome instability promotes endoreplication and, vice versa, endoreplication promotes genome instability (Fig. 4). These findings implicate endoreplication as both an adaptation to genotoxic stress, as well as a potentially novel mechanism for differentiation of polyploid cells. Furthermore, the parallels between normal and cancerous endoreplication cycles highlight that the study of developmentally programmed endoreplication may illuminate mechanisms by which polyploidy promotes precancerous genome alteration.
Fig. 4.
The reciprocal relationship between endoreplication and genome stability. Endoreplication can promote genome instability in the form of amplified centrosomes and an increased tolerance for DNA damage. Conversely, DNA damage, chromatin changes and telomere defects are all examples of genome instability forms that have been demonstrated to initiate endoreplication.
Genome instability promotes endoreplication
A damaged genome presents a challenge for a developing or regenerating tissue. During these dynamic events, tissue mass must be increased, but cell proliferation must be delayed to avoid segregation of damaged chromosomes. Emerging data from multiple systems now suggest a conserved role for endoreplication in providing an alternative to deleterious proliferation in the face of genome instability.
Recent evidence suggests that plants possess endoreplication-promoting mechanisms that enable tissue growth despite genome damage. Following induction of double-stranded DNA breaks in Arabidopsis root tip and sepal cells, normally mitotic cells increase in size via endoreplication. This cell cycle alteration requires the plant-specific transcription factor SUPPRESSOR OF GAMMA RESPONSE 1 (SOG1), which transmits signals from the conserved ATAXIA TELANGIECTASIA MUTATED (ATM) and ATAXIA TELANGIECTASIA-MUTATED AND RAD3-RELATED (ATR) DNA damage sensor kinases. This DNA break response elicits transcriptional changes that are consistent with downregulation of mitotic factors and upregulation of cell cycle genes that can promote endoreplication (Adachi et al., 2011). Similarly, Arabidopsis leaves possess an endoreplication response to genome damage (Radziejwoski et al., 2011). Following UV irradiation, mRNA levels of E2Fe/DEL1, an atypical E2F protein, decrease. Mutant studies showed that loss of E2Fe/DELI elevates transcription of PHOTOLYASE 1 (PHR1), a regulator of UV-induced DNA repair. Plants that lack E2Fe/DELI exhibit enhanced repair of UV-induced breaks and also increased endoreplication (Radziejwoski et al., 2011). Taken together, these recent studies argue that endoreplication is a favorable mode of plant tissue growth following genotoxic stress.
In both plants and animals, genome instability in the form of defective chromatin assembly is also tied to endoreplication. In Arabidopsis, mutations in FASCIATA1 (FAS1), which encodes a subunit of the CHROMATIN ASSEMBLY FACTOR 1 (CAF1) histone chaperone, triggers an inappropriate mitosis to endoreplication switch. These mutations specifically block binding of FAS1 to E2F family members. As a result of this blocked interaction, genes known to regulate the S/G2 checkpoint are upregulated, which is likely to promote endoreplication (Ramirez-Parra and Gutierrez, 2007). Similarly, depletion of Drosophila CAF-1 leads to death in diploid cells but does not block endocycle progression (Klapholz et al., 2009). In both flies and plants, CAF1 loss correlates with DNA damage, providing further evidence of endoreplication as an adaptation to genome damage.
Telomere shortening, which also activates the DNA damage response, appears to be a potent endoreplication-promoting mechanism in many normally proliferative cells. Using an inducible system that activates a persistent DNA damage response at telomeres, Davoli et al. (Davoli et al., 2010) showed that mouse embryonic fibroblasts increase in ploidy via endoreplication. This endoreplication requires ATM/ATR and occurs in the absence of the transcription factor p53, a known suppressor of S phase following polyploidy (reviewed by Aylon and Oren, 2011). These endoreplication cycles resemble developmentally programmed endocycles, exhibiting cyclic Geminin expression and a lack of mitotic CDK activation (Davoli et al., 2010). Inefficient telomere replication can also cause endoreplication. In human mammary epithelial cells, shortened telomeres cause endoreplication through anaphase bridge formation and subsequent cytokinesis failure (Pampalona et al., 2012). Similar to the Davoli et al. study (Davoli et al., 2010), the endoreplication observed in the study by Pampalona et al. (Pampalona et al., 2012) requires aberrant (short) telomeres, but also requires loss of the transcriptional repressor Rb, rather than loss of p53. Telomere replication defects also contribute to endoreplication through failed chromosome segregation in mouse embryonic fibroblasts lacking the chromosome cohesion component SA1 (Remeseiro et al., 2012). These telomere studies provide potential mechanistic insight into how some animal tissues respond to DNA damage. Intriguingly, mice with damaged telomeric DNA regenerate their liver via endoreplication (Lazzerini Denchi et al., 2006). Given the connection between telomere shortening and aging, it is interesting to speculate that endoreplication may be a favored mode of tissue repair in aged humans.
How do cells employ endoreplication to continue the cell cycle despite DNA damage? Regulation of the transcriptional repressor Rb may provide a key mechanistic link. Loss of Rb contributes to tetraploidization following telomere crisis in human fibroblasts and mammary epithelial cells (Davoli and de Lange, 2012). During liver regeneration in normal mice, cell cycling is blocked in hepatocytes with damaged genomes. By contrast, Rb mutant hepatocytes with DNA damage continue to cycle and frequently endoreplicate, as evidenced by the emergence of polyploid cells. Interestingly, this endoreplication phenotype appears specific to Rb loss in the liver and not in other gastrointestinal tissues, suggesting that tissues exhibit differing capacities and requirements for endoreplication (Bourgo et al., 2011). Future work is needed to unravel how diverse genotoxic stresses combine with tissue-specific cell cycle regulation to promote endoreplication.
Endoreplication promotes genome instability
In normal polyploid tissues, programmed endoreplication is usually a permanent departure from a mitotic program. As a result, endoreplication has become synonymous with terminal differentiation. Conversely, polyploidy is associated with mitotic progression of many cancers (Davoli and de Lange, 2011). Experimentally induced endoreplication can enable chromosome instability (CIN), whereby chromosomes mis-segregate during mitosis, and such polyploid instability is associated with tumor growth in mice (reviewed by Ganem and Pellman, 2007). Thus, a simplistic view is that normal endoreplication promotes senescence and differentiation, whereas unscheduled endoreplication promotes tumorigenesis by enabling CIN. In contrast to this view, recent work in both insects and mammals has now shown that polyploid cells can resume mitotic cycles. Surprisingly, these polyploid mitotic cells exhibit CIN, similar to cancerous polyploid cells. This new finding suggests parallels between normal and cancerous endoreplication, but in addition provokes new ideas about apparently ‘unstable’ mitotic divisions contributing to the physiology of normal tissues containing polyploid cells.
During Drosophila larval development, endoreplication occurs frequently in many tissues. The resulting polyploid cells function in the larva but do not proliferate and contribute to the adult fly, with some recently studied exceptions. Fox et al., for example, discovered that intestinal papillar cells, which reclaim up to 90% of ingested water in an insect, form by proliferation of polyploid precursors, which had previously undergone endoreplication (Fox et al., 2010). Thus, the transition from mitotic cycles to endoreplication appears to be reversible in normal animal development. Unexpectedly, these developmentally programmed divisions exhibit frequent signs of CIN, including chromosomal aberrations, anaphase bridging and lagging chromosomes. A similar propensity for CIN was observed in the hind-intestine of mosquitoes (Fox et al., 2010), as well as during endoreplication in Drosophila blood-brain barrier glial cells, which exhibit anaphase bridging during endo-mitotic cycles (Unhavaithaya and Orr-Weaver, 2012). The developmentally tractable nature of these CIN-prone cell cycles provides an accessible entry point into the study of mechanistic connections between endoreplication and genome instability.
The propensity for mitotic instability following endoreplication appears conserved. The liver has long been observed to contain endoreplicating cells, and recent evidence shows that these cells also revert to mitotic cycles that become error prone. Using FACS purification and a marked cell transplantation model, Duncan et al. followed polyploid mouse hepatocytes over time (Duncan et al., 2010). These transplanted hepatocytes frequently produced aneuploid cells (cells with an imbalance in chromosome content). This aneuploidy also occurs in normal mouse and human livers, as verified by karyotyping and in situ hybridization (Duncan et al., 2012a; Duncan et al., 2010). The intriguing observation of polyploid-induced aneuploidy in healthy tissues led the authors of these studies to propose that the transition from endoreplication to polyploidy and finally to aneuploidy represents a means of generating a genetically diverse pool of cells, a subset of which might be amplified under conditions of stress (Duncan et al., 2010). A recent follow up study presented one case in which this model holds true. In a mouse model liver disease model (tyroseinemia-induced injury), cells with a specific aneuploidy containing an extra disease-resistance gene copy can be amplified to suppress liver injury (Duncan et al., 2012b). These recent liver studies present a highly novel role for endoreplication in cellular differentiation.
How does endoreplication drive genome instability? One source is the duplication of the mitotic spindle organizers – the centrosomes. Because centrosome duplication is tied to cell cycle progression, passage through endoreplication could generate multiple centrosomes and aberrant mitotic spindles – a long-hypothesized source of genome instability (Holland and Cleveland, 2009). This appears to be the case in mouse hepatocytes: polyploid hepatocytes with extra centrosomes exhibit cell division errors, owing to apparently improper microtubule attachments (Duncan et al., 2010).
Recent studies in non-mitotic polyploid cells of Drosophila and mice suggest that endoreplication enables tolerance of DNA damage. In flies, endoreplicating cells acquire resistance to DNA damage through a mechanism involving the silencing of cell death genes (Mehrotra et al., 2008). Similarly, endoreplicating mouse trophoblast cells also downregulate the DNA damage response. During the differentiation of trophoblast stem cells into polyploid TGCs, protein levels of the damage-responsive kinase Chk1 diminish, permitting endoreplication. This decrease in Chk1 enables polyploid trophoblast cells to evade apoptosis through suppression of the DNA damage pathway (Ullah et al., 2011; Ullah et al., 2008). Taken together, it is clear that cell cycle alterations tied to endoreplication can encourage genome instability. The consequences of such polyploid genome instability on normal tissue development, as well as in diseases such as cancer, remain to be fully understood.
Endoreplication in cancer
As mentioned above, endoreplication can confer potentially precancerous genome instability but can also represent a permanent block to mitosis. Following endoreplication, a polyploid cell may thus either arrest the cell cycle or regain potentially cancerous proliferation. Recent evidence from diverse models and humans with cancer now suggests that regulation of this decision plays a role in cancer progression.
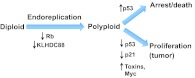
Whether endoreplication suppresses or enables tumorigenesis appears to be dependent on the genetic background and tissue environment (Fig. 5). In mice, Cdk1 deletion results in liver regeneration solely through endoreplication, and correlates with decreased tumor risk (Diril et al., 2012). Similarly, loss of Rb (Rb1 – Mouse Genome Informatics) during mouse liver development promotes increased ploidy but does not promote spontaneous liver cancer, even in combination with p53 (Trp53 – Mouse Genome Informatics) loss (McClendon et al., 2011). However, this p53, Rb double mutant background renders mice highly susceptible to carcinogen-induced liver cancer (McClendon et al., 2011). Furthermore, both ploidy and liver tumorigenesis increase in Rb mutant mice in conjunction with either overexpression of the Myc transcription factor (Saddic et al., 2011) or a liver toxin (Reed et al., 2009). Outside the liver, inappropriate endoreplication may be more tumorigenic. Mouse embryonic cells lacking Rb and p53 undergo endoreplication with de-protected telomeres, and subsequent division of such cells promotes aneuploidy and tumorigenesis (Davoli and de Lange, 2012). Future studies will clarify how genetic background and tissue environment influence whether endoreplication promotes or suppresses tumorigenesis.
Fig. 5.
Potential molecular mechanisms of polyploid tumor formation. Distinct molecular alterations can influence each step of the progression from diploid to polyploid to tumorous proliferation. Shown here are recently identified mechanisms implicated in this progression. Genetic factors such as Rb loss or KLHDC88 mutation can promote endoreplication. Once cells achieve polyploidy, p53 can mediate cell cycle arrest or cell death. Conversely, in the absence of p53 or its target p21, or following administration of toxins such as diethylnitrosamine or aflatoxin B1 (see Reed et al., 2009 and McClendon et al., 2011), or in the presence of increased Myc (see Saddic et al., 2011), tumorigenesis may result.
In parallel to the above mouse studies, recent work has found that, in certain contexts, cancerous cells use endoreplication as a path to drug resistance. Several cancer cell lines treated with growth suppressing drugs enter endoreplication cycles (Sakaue-Sawano et al., 2011; Shen et al., 2008). This switch to endoreplication can be reversed, allowing the now polyploid cancer cells to resume proliferation and acquire resistance to apoptosis-inducing drugs (Shen et al., 2008). This endoreplication and subsequent proliferation requires inhibition of the p53 target p21 (Shen and Maki, 2010; Zheng et al., 2012). Thus, endoreplication may confer additional instability to acquire cancerous proliferative potential.
Many human cancers exhibit polyploidy (reviewed by Davoli and de Lange, 2011; Storchova and Pellman, 2004), but finding direct clinical links to endoreplication is challenging. Recent analysis of multiple related individuals with Hodgkin’s lymphoma led to the identification of a mutation in the cytokinesis regulator KLHDC8B (Salipante et al., 2009). In such individuals, a signature of disease progression is the accumulation of polyploid binucleate Reed-Sternberg cells. Depletion of KLHDC8B generates binucleate cells through endoreplication (failed cytokinesis). This study provides a genetic link between inappropriate, transient endoreplication and cancer progression. Future efforts in personal cancer genome sequencing may be key to identifying additional mutations that enable cancer through endoreplication.
Conclusions
The exciting recent progress highlighted here has re-invigorated efforts to understand the biological functions of endoreplication and polyploidy. Thanks to the development of new tissue models, as well as additional mechanistic insights in familiar systems, it now appears clear that unifying principles apply to all forms of endoreplication, be they ‘endocycles’, ‘endomitosis’ or ‘failed mitosis’. We propose that future efforts should focus on an improved understanding of how tissues benefit from increasing the ploidy of a single cell rather than generating more cells via proliferation. From the recent work highlighted here, we now know that endoreplication can be advantageous to maintain both tissue and genome integrity in cases where proliferation would be disruptive. Furthermore, the continued study of mechanisms connecting endoreplication and genome instability may prove useful in understanding the mysterious role of polyploidy in cancer.
Acknowledgments
We thank David MacAlpine, members of the Fox and Duronio labs for helpful discussions, and the reviewers for their comments on the manuscript.
Footnotes
Funding
D.T.F. is supported by scholar awards from the Whitehead Foundation and the Pew Charitable Trust. R.J.D.’s cell cycle work is supported by the National Institutes of Health. Deposited in PMC for release after 12 months.
Competing interests statement
The authors declare no competing financial interests.
References
- Adachi S., Minamisawa K., Okushima Y., Inagaki S., Yoshiyama K., Kondou Y., Kaminuma E., Kawashima M., Toyoda T., Matsui M., et al. (2011). Programmed induction of endoreduplication by DNA double-strand breaks in Arabidopsis. Proc. Natl. Acad. Sci. USA 108, 10004–10009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylon Y., Oren M. (2011). p53: guardian of ploidy. Mol. Oncol. 5, 315–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyaeva E. S., Zhimulev I. F., Volkova E. I., Alekseyenko A. A., Moshkin Y. M., Koryakov D. E. (1998). Su(UR)ES: a gene suppressing DNA underreplication in intercalary and pericentric heterochromatin of Drosophila melanogaster polytene chromosomes. Proc. Natl. Acad. Sci. USA 95, 7532–7537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berckmans B., Lammens T., Van Den Daele H., Magyar Z., Bögre L., De Veylder L. (2011). Light-dependent regulation of DEL1 is determined by the antagonistic action of E2Fb and E2Fc. Plant Physiol. 157, 1440–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudolf V., Lammens T., Boruc J., Van Leene J., Van Den Daele H., Maes S., Van Isterdael G., Russinova E., Kondorosi E., Witters E., et al. (2009). CDKB1;1 forms a functional complex with CYCA2;3 to suppress endocycle onset. Plant Physiol. 150, 1482–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgo R. J., Ehmer U., Sage J., Knudsen E. S. (2011). RB deletion disrupts coordination between DNA replication licensing and mitotic entry in vivo. Mol. Biol. Cell 22, 931–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramsiepe J., Wester K., Weinl C., Roodbarkelari F., Kasili R., Larkin J. C., Hülskamp M., Schnittger A. (2010). Endoreplication controls cell fate maintenance. PLoS Genet. 6, e1000996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvi B. R., Spradling A. C. (1999). Chorion gene amplification in Drosophila: A model for metazoan origins of DNA replication and S-phase control. Methods 18, 407–417 [DOI] [PubMed] [Google Scholar]
- Celton-Morizur S., Desdouets C. (2010). Polyploidization of liver cells. Adv. Exp. Med. Biol. 676, 123–135 [DOI] [PubMed] [Google Scholar]
- Celton-Morizur S., Merlen G., Couton D., Margall-Ducos G., Desdouets C. (2009). The insulin/Akt pathway controls a specific cell division program that leads to generation of binucleated tetraploid liver cells in rodents. J. Clin. Invest. 119, 1880–1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celton-Morizur S., Merlen G., Couton D., Desdouets C. (2010). Polyploidy and liver proliferation: central role of insulin signaling. Cell Cycle 9, 460–466 [DOI] [PubMed] [Google Scholar]
- Chagraoui H., Kassouf M., Banerjee S., Goardon N., Clark K., Atzberger A., Pearce A. C., Skoda R. C., Ferguson D. J., Watson S. P., et al. (2011). SCL-mediated regulation of the cell-cycle regulator p21 is critical for murine megakaryopoiesis. Blood 118, 723–735 [DOI] [PubMed] [Google Scholar]
- Chalker D. L., Yao M. C. (2011). DNA elimination in ciliates: transposon domestication and genome surveillance. Annu. Rev. Genet. 45, 227–246 [DOI] [PubMed] [Google Scholar]
- Chen H.-Z., Ouseph M. M., Li J., Pécot T., Chokshi V., Kent L., Bae S., Byrne M., Duran C., Comstock G., et al. (2012). Canonical and atypical E2Fs regulate the mammalian endocycle. Nat. Cell Biol. 14, 1192–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman M. L., Brown M. L., Kato N., Kirik V., Hülskamp M., Inzé D., De Veylder L., Walker J. D., Zheng Z., Oppenheimer D. G., et al. (2006). SIAMESE, a plant-specific cell cycle regulator, controls endoreplication onset in Arabidopsis thaliana. Plant Cell 18, 3145–3157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corash L., Levin J., Mok Y., Baker G., McDowell J. (1989). Measurement of megakaryocyte frequency and ploidy distribution in unfractionated murine bone marrow. Exp. Hematol. 17, 278–286 [PubMed] [Google Scholar]
- Cornejo M. G., Mabialah V., Sykes S. M., Khandan T., Lo Celso C., Lopez C. K., Rivera-Muñoz P., Rameau P., Tothova Z., Aster J. C., et al. (2011). Crosstalk between NOTCH and AKT signaling during murine megakaryocyte lineage specification. Blood 118, 1264–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoli T., de Lange T. (2011). The causes and consequences of polyploidy in normal development and cancer. Annu. Rev. Cell Dev. Biol. 27, 585–610 [DOI] [PubMed] [Google Scholar]
- Davoli T., de Lange T. (2012). Telomere-driven tetraploidization occurs in human cells undergoing crisis and promotes transformation of mouse cells. Cancer Cell 21, 765–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoli T., Denchi E. L., de Lange T. (2010). Persistent telomere damage induces bypass of mitosis and tetraploidy. Cell 141, 81–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L., Larkin J. C., Schnittger A. (2011). Molecular control and function of endoreplication in development and physiology. Trends Plant Sci. 16, 624–634 [DOI] [PubMed] [Google Scholar]
- Demontis F., Perrimon N. (2009). Integration of Insulin receptor/Foxo signaling and dMyc activity during muscle growth regulates body size in Drosophila. Development 136, 983–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W. M., Althauser C., Ruohola-Baker H. (2001). Notch-Delta signaling induces a transition from mitotic cell cycle to endocycle in Drosophila follicle cells. Development 128, 4737–4746 [DOI] [PubMed] [Google Scholar]
- Diffley J. F. (2011). Quality control in the initiation of eukaryotic DNA replication. Philos. Trans. R. Soc. B Biol. Sci. 366, 3545–3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diril M. K., Ratnacaram C. K., Padmakumar V. C., Du T., Wasser M., Coppola V., Tessarollo L., Kaldis P. (2012). Cyclin-dependent kinase 1 (Cdk1) is essential for cell division and suppression of DNA re-replication but not for liver regeneration. Proc. Natl. Acad. Sci. USA 109, 3826–3831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domanitskaya E., Schüpbach T. (2012). CoREST acts as a positive regulator of Notch signaling in the follicle cells of Drosophila melanogaster. J. Cell Sci. 125, 399–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan A. W., Taylor M. H., Hickey R. D., Hanlon Newell A. E., Lenzi M. L., Olson S. B., Finegold M. J., Grompe M. (2010). The ploidy conveyor of mature hepatocytes as a source of genetic variation. Nature 467, 707–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan A. W., Hanlon Newell A. E., Smith L., Wilson E. M., Olson S. B., Thayer M. J., Strom S. C., Grompe M. (2012a). Frequent aneuploidy among normal human hepatocytes. Gastroenterology 142, 25–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan A. W., Hanlon Newell A. E., Bi W., Finegold M. J., Olson S. B., Beaudet A. L., Grompe M. (2012b). Aneuploidy as a mechanism for stress-induced liver adaptation. J. Clin. Invest. 122, 3307–3315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duronio R. J., O’Farrell P. H. (1995). Developmental control of the G1 to S transition in Drosophila: cyclin Eis a limiting downstream target of E2F. Genes Dev. 9, 1456–1468 [DOI] [PubMed] [Google Scholar]
- Eliades A., Papadantonakis N., Ravid K. (2010). New roles for cyclin E in megakaryocytic polyploidization. J. Biol. Chem. 285, 18909–18917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrell J. E., Jr, Tsai T. Y., Yang Q. (2011). Modeling the cell cycle: why do certain circuits oscillate? Cell 144, 874–885 [DOI] [PubMed] [Google Scholar]
- Follette P. J., Duronio R. J., O’Farrell P. H. (1998). Fluctuations in cyclin E levels are required for multiple rounds of endocycle S phase in Drosophila. Curr. Biol. 8, 235–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox D. T., Gall J. G., Spradling A. C. (2010). Error-prone polyploid mitosis during normal Drosophila development. Genes Dev. 24, 2294–2302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall J. G., Cohen E. H., Polan M. L. (1971). Reptitive DNA sequences in drosophila. Chromosoma 33, 319–344 [DOI] [PubMed] [Google Scholar]
- Ganem N. J., Pellman D. (2007). Limiting the proliferation of polyploid cells. Cell 131, 437–440 [DOI] [PubMed] [Google Scholar]
- Gao Y., Smith E., Ker E., Campbell P., Cheng E. C., Zou S., Lin S., Wang L., Halene S., Krause D. S. (2012). Role of RhoA-specific guanine exchange factors in regulation of endomitosis in megakaryocytes. Dev. Cell 22, 573–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer M. (2005). The molecular requirements for cytokinesis. Science 307, 1735–1739 [DOI] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R. A. (2011). Hallmarks of cancer: the next generation. Cell 144, 646–674 [DOI] [PubMed] [Google Scholar]
- Hancock J. F. (2005). Contributions of domesticated plant studies to our understanding of plant evolution. Ann. Bot. 96, 953–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havens C. G., Walter J. C. (2011). Mechanism of CRL4(Cdt2), a PCNA-dependent E3 ubiquitin ligase. Genes Dev. 25, 1568–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedgecock E. M., White J. G. (1985). Polyploid tissues in the nematode Caenorhabditis elegans. Dev. Biol. 107, 128–133 [DOI] [PubMed] [Google Scholar]
- Hochegger H., Dejsuphong D., Sonoda E., Saberi A., Rajendra E., Kirk J., Hunt T., Takeda S. (2007). An essential role for Cdk1 in S phase control is revealed via chemical genetics in vertebrate cells. J. Cell Biol. 178, 257–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland A. J., Cleveland D. W. (2009). Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat. Rev. Mol. Cell Biol. 10, 478–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong A., Narbonne-Reveau K., Riesgo-Escovar J., Fu H., Aladjem M. I., Lilly M. A. (2007). The cyclin-dependent kinase inhibitor Dacapo promotes replication licensing during Drosophila endocycles. EMBO J. 26, 2071–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T., Fujiwara S., Miura K., Stacey N., Yoshimura M., Schneider K., Adachi S., Minamisawa K., Umeda M., Sugimoto K. (2009). SUMO E3 ligase HIGH PLOIDY2 regulates endocycle onset and meristem maintenance in Arabidopsis. Plant Cell 21, 2284–2297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T., Adachi S., Yoshimura M., Shimizu K., Umeda M., Sugimoto K. (2010). Auxin modulates the transition from the mitotic cycle to the endocycle in Arabidopsis. Development 137, 63–71 [DOI] [PubMed] [Google Scholar]
- Iwata E., Ikeda S., Matsunaga S., Kurata M., Yoshioka Y., Criqui M. C., Genschik P., Ito M. (2011). GIGAS CELL1, a novel negative regulator of the anaphase-promoting complex/cyclosome, is required for proper mitotic progression and cell fate determination in Arabidopsis. Plant Cell 23, 4382–4393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasili R., Walker J. D., Simmons L. A., Zhou J., De Veylder L., Larkin J. C. (2010). SIAMESE cooperates with the CDH1-like protein CCS52A1 to establish endoreplication in Arabidopsis thaliana trichomes. Genetics 185, 257–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasili R., Huang C. C., Walker J. D., Simmons L. A., Zhou J., Faulk C., Hülskamp M., Larkin J. C. (2011). BRANCHLESS TRICHOMES links cell shape and cell cycle control in Arabidopsis trichomes. Development 138, 2379–2388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. C., Nordman J., Xie F., Kashevsky H., Eng T., Li S., MacAlpine D. M., Orr-Weaver T. L. (2011). Integrative analysis of gene amplification in Drosophila follicle cells: parameters of origin activation and repression. Genes Dev. 25, 1384–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapholz B., Dietrich B. H., Schaffner C., Hérédia F., Quivy J. P., Almouzni G., Dostatni N. (2009). CAF-1 is required for efficient replication of euchromatic DNA in Drosophila larval endocycling cells. Chromosoma 118, 235–248 [DOI] [PubMed] [Google Scholar]
- Knoblich J. A., Sauer K., Jones L., Richardson H., Saint R., Lehner C. F. (1994). Cyclin E controls S phase progression and its down-regulation during Drosophila embryogenesis is required for the arrest of cell proliferation. Cell 77, 107–120 [DOI] [PubMed] [Google Scholar]
- Laird C., Ashburner M., Wilkinson L. (1980). Relationship between relative dry mass and average band width in regions of polytene chromosomes of Drosophila. Chromosoma 76, 175–189 [Google Scholar]
- Larson-Rabin Z., Li Z., Masson P. H., Day C. D. (2009). FZR2/CCS52A1 expression is a determinant of endoreduplication and cell expansion in Arabidopsis. Plant Physiol. 149, 874–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzerini Denchi E., Celli G., de Lange T. (2006). Hepatocytes with extensive telomere deprotection and fusion remain viable and regenerate liver mass through endoreduplication. Genes Dev. 20, 2648–2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. O., Davidson J. M., Duronio R. J. (2009). Endoreplication: polyploidy with purpose. Genes Dev. 23, 2461–2477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Larson-Rabin Z., Masson P. H., Day C. D. (2009). FZR2/CCS52A1 mediated endoreduplication in Arabidopsis development. Plant Signal. Behav. 4, 451–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilly M. A., Spradling A. C. (1996). The Drosophila endocycle is controlled by Cyclin E and lacks a checkpoint ensuring S-phase completion. Genes Dev. 10, 2514–2526 [DOI] [PubMed] [Google Scholar]
- Lilly M. A., Duronio R. J. (2005). New insights into cell cycle control from the Drosophila endocycle. Oncogene 24, 2765–2775 [DOI] [PubMed] [Google Scholar]
- Lord C. J., Ashworth A. (2012). The DNA damage response and cancer therapy. Nature 481, 287–294 [DOI] [PubMed] [Google Scholar]
- Magyar Z., Horváth B., Khan S., Mohammed B., Henriques R., De Veylder L., Bakó L., Scheres B., Bögre L. (2012). Arabidopsis E2FA stimulates proliferation and endocycle separately through RBR-bound and RBR-free complexes. EMBO J. 31, 1480–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maqbool S. B., Mehrotra S., Kolpakas A., Durden C., Zhang B., Zhong H., Calvi B. R. (2010). Dampened activity of E2F1-DP and Myb-MuvB transcription factors in Drosophila endocycling cells. J. Cell Sci. 123, 4095–4106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClendon A. K., Dean J. L., Ertel A., Fu Z., Rivadeneira D. B., Reed C. A., Bourgo R. J., Witkiewicz A., Addya S., Mayhew C. N., et al. (2011). RB and p53 cooperate to prevent liver tumorigenesis in response to tissue damage. Gastroenterology 141, 1439–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrotra S., Maqbool S. B., Kolpakas A., Murnen K., Calvi B. R. (2008). Endocycling cells do not apoptose in response to DNA rereplication genotoxic stress. Genes Dev. 22, 3158–3171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melaragno J. E., Mehrotra B., Coleman A. W. (1993). Relationship between endopolyploidy and cell size in epidermal tissue of Arabidopsis. Plant Cell 5, 1661–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell J. E., Clements K. D., Choat J. H., Angert E. R. (2008). Extreme polyploidy in a large bacterium. Proc. Natl. Acad. Sci. USA 105, 6730–6734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercher T., Cornejo M. G., Sears C., Kindler T., Moore S. A., Maillard I., Pear W. S., Aster J. C., Gilliland D. G. (2008). Notch signaling specifies megakaryocyte development from hematopoietic stem cells. Cell Stem Cell 3, 314–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyaoka Y., Ebato K., Kato H., Arakawa S., Shimizu S., Miyajima A. (2012). Hypertrophy and unconventional cell Ddivision of hepatocytes underlie liver regeneration. Curr. Biol. 22, 1165–1175 [DOI] [PubMed] [Google Scholar]
- Morgan D. O. (2007). The Cell Cycle: Principles of Control. London, UK: New Science Press; [Google Scholar]
- Morohashi K., Grotewold E. (2009). A systems approach reveals regulatory circuitry for Arabidopsis trichome initiation by the GL3 and GL1 selectors. PLoS Genet. 5, e1000396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Alonso M. J., Ceballos L., Bretones G., Frade P., León J., Gandarillas A. (2012). MYC accelerates p21CIP-induced megakaryocytic differentiation involving early mitosis arrest in leukemia cells. J. Cell. Physiol. 227, 2069–2078 [DOI] [PubMed] [Google Scholar]
- Narbonne-Reveau K., Senger S., Pal M., Herr A., Richardson H. E., Asano M., Deak P., Lilly M. A. (2008). APC/CFzr/Cdh1 promotes cell cycle progression during the Drosophila endocycle. Development 135, 1451–1461 [DOI] [PubMed] [Google Scholar]
- Negrini S., Gorgoulis V. G., Halazonetis T. D. (2010). Genomic instability – an evolving hallmark of cancer. Nat. Rev. Mol. Cell Biol. 11, 220–228 [DOI] [PubMed] [Google Scholar]
- Nevzorova Y. A., Tschaharganeh D., Gassler N., Geng Y., Weiskirchen R., Sicinski P., Trautwein C., Liedtke C. (2009) Aberrant cell cycle progression and endoreplication in regenerating livers of mice that lack a single E-type cyclin. Gastroenterology 137, 691–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordman J., Li S., Eng T., Macalpine D., Orr-Weaver T. L. (2011). Developmental control of the DNA replication and transcription programs. Genome Res. 21, 175–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellet J., Roy R. (2007). The lin-35/Rb and RNAi pathways cooperate to regulate a key cell cycle transition in C. elegans. BMC Dev. Biol. 7, 38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pampalona J., Frías C., Genescà A., Tusell L. (2012). Progressive telomere dysfunction causes cytokinesis failure and leads to the accumulation of polyploid cells. PLoS Genet. 8, e1002679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit S. K., Westendorp B., Nantasanti S., van Liere E., Tooten P. C. J., Cornelissen P. W. A., Toussaint M. J. M., Lamers W. H., de Bruin A. (2012). E2F8 is essential for polyploidization in mammalian cells. Nat. Cell Biol. 14, 1181–1191 [DOI] [PubMed] [Google Scholar]
- Pierce S. B., Yost C., Anderson S. A., Flynn E. M., Delrow J., Eisenman R. N. (2008). Drosophila growth and development in the absence of dMyc and dMnt. Dev. Biol. 315, 303–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirault-Chassac S., Six E., Catelain C., Lavergne M., Villeval J.-L., Vainchenker W., Lauret E. (2010). Notch/Delta4 signaling inhibits human megakaryocytic terminal differentiation. Blood 116, 5670–5678 [DOI] [PubMed] [Google Scholar]
- Radziejwoski A., Vlieghe K., Lammens T., Berckmans B., Maes S., Jansen M. A. K., Knappe C., Albert A., Seidlitz H. K., Bahnweg G., et al. (2011). Atypical E2F activity coordinates PHR1 photolyase gene transcription with endoreduplication onset. EMBO J. 30, 355–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Parra E., Gutierrez C. (2007). E2F regulates FASCIATA1, a chromatin assembly gene whose loss switches on the endocycle and activates gene expression by changing the epigenetic status. Plant Physiol. 144, 105–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed C. A., Mayhew C. N., McClendon A. K., Yang X., Witkiewicz A., Knudsen E. S. (2009). RB has a critical role in mediating the in vivo checkpoint response, mitigating secondary DNA damage and suppressing liver tumorigenesis initiated by aflatoxin B1. Oncogene 28, 4434–4443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remeseiro S., Cuadrado A., Carretero M., Martínez P., Drosopoulos W. C., Cañamero M., Schildkraut C. L., Blasco M. A., Losada A. (2012). Cohesin-SA1 deficiency drives aneuploidy and tumourigenesis in mice due to impaired replication of telomeres. EMBO J. 31, 2076–2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roodbarkelari F., Bramsiepe J., Weinl C., Marquardt S., Novák B., Jakoby M. J., Lechner E., Genschik P., Schnittger A. (2010). Cullin 4-ring finger-ligase plays a key role in the control of endoreplication cycles in Arabidopsis trichomes. Proc. Natl. Acad. Sci. USA 107, 15275–15280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saddic L. A., Wirt S., Vogel H., Felsher D. W., Sage J. (2011). Functional interactions between retinoblastoma and c-MYC in a mouse model of hepatocellular carcinoma. PLoS ONE 6, e19758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaue-Sawano A., Kobayashi T., Ohtawa K., Miyawaki A. (2011). Drug-induced cell cycle modulation leading to cell-cycle arrest, nuclear mis-segregation, or endoreplication. BMC Cell Biol. 12, 2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salipante S. J., Mealiffe M. E., Wechsler J., Krem M. M., Liu Y., Namkoong S., Bhagat G., Kirchhoff T., Offit K., Lynch H., et al. (2009). Mutations in a gene encoding a midbody kelch protein in familial and sporadic classical Hodgkin lymphoma lead to binucleated cells. Proc. Natl. Acad. Sci. USA 106, 14920–14925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallé J., Campbell S. D., Gho M., Audibert A. (2012). CycA is involved in the control of endoreplication dynamics in the Drosophila bristle lineage. Development 139, 547–557 [DOI] [PubMed] [Google Scholar]
- Shcherbata H. R., Althauser C., Findley S. D., Ruohola-Baker H. (2004). The mitotic-to-endocycle switch in Drosophila follicle cells is executed by Notch-dependent regulation of G1/S, G2/M and M/G1 cell-cycle transitions. Development 131, 3169–3181 [DOI] [PubMed] [Google Scholar]
- Shen H., Maki C. G. (2010). Persistent p21 expression after Nutlin-3a removal is associated with senescence-like arrest in 4N cells. J. Biol. Chem. 285, 23105–23114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., Moran D. M., Maki C. G. (2008). Transient nutlin-3a treatment promotes endoreduplication and the generation of therapy-resistant tetraploid cells. Cancer Res. 68, 8260–8268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sher N., Bell G. W., Li S., Nordman J., Eng T., Eaton M. L., Macalpine D. M., Orr-Weaver T. L. (2012). Developmental control of gene copy number by repression of replication initiation and fork progression. Genome Res. 22, 64–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman M. I. (1972). The biochemistry of differentiation of mouse trophoblast: alkaline phosphatase. Dev. Biol. 27, 337–350 [DOI] [PubMed] [Google Scholar]
- Shibutani S. T., de la Cruz A. F., Tran V., Turbyfill W. J., 3rd, Reis T., Edgar B. A., Duronio R. J. (2008). Intrinsic negative cell cycle regulation provided by PIP box- and Cul4Cdt2-mediated destruction of E2f1 during S phase. Dev. Cell 15, 890–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist S. J., Lehner C. F. (1997). Drosophila fizzy-related down-regulates mitotic cyclins and is required for cell proliferation arrest and entry into endocycles. Cell 90, 671–681 [DOI] [PubMed] [Google Scholar]
- Song L., Rape M. (2011). Substrate-specific regulation of ubiquitination by the anaphase-promoting complex. Cell Cycle 10, 52–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiger D., Furrer M., Schwinkendorf D., Gallant P. (2008). Max-independent functions of Myc in Drosophila melanogaster. Nat. Genet. 40, 1084–1091 [DOI] [PubMed] [Google Scholar]
- Storchova Z., Pellman D. (2004). From polyploidy to aneuploidy, genome instability and cancer. Nat. Rev. Mol. Cell Biol. 5, 45–54 [DOI] [PubMed] [Google Scholar]
- Sun J., Deng W. M. (2007). Hindsight mediates the role of notch in suppressing hedgehog signaling and cell proliferation. Dev. Cell 12, 431–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah Z., Kohn M. J., Yagi R., Vassilev L. T., DePamphilis M. L. (2008). Differentiation of trophoblast stem cells into giant cells is triggered by p57/Kip2 inhibition of CDK1 activity. Genes Dev. 22, 3024–3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah Z., Lee C. Y., Depamphilis M. L. (2009a). Cip/Kip cyclin-dependent protein kinase inhibitors and the road to polyploidy. Cell Div. 4, 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah Z., Lee C. Y., Lilly M. A., DePamphilis M. L. (2009b). Developmentally programmed endoreduplication in animals. Cell Cycle 8, 1501–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah Z., de Renty C., DePamphilis M. L. (2011). Checkpoint kinase 1 prevents cell cycle exit linked to terminal cell differentiation. Mol. Cell. Biol. 31, 4129–4143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unhavaithaya Y., Orr-Weaver T. L. (2012). Polyploidization of glia in neural development links tissue growth to blood-brain barrier integrity. Genes Dev. 26, 31–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel S., Dyson N. J. (2008). Conserved functions of the pRB and E2F families. Nat. Rev. Mol. Cell Biol. 9, 713–724 [DOI] [PubMed] [Google Scholar]
- Weiss A., Herzig A., Jacobs H., Lehner C. F. (1998). Continuous Cyclin E expression inhibits progression through endoreduplication cycles in Drosophila. Curr. Biol. 8, 239–242 [DOI] [PubMed] [Google Scholar]
- Weng L., Zhu C., Xu J., Du W. (2003). Critical role of active repression by E2F and Rb proteins in endoreplication during Drosophila development. EMBO J. 22, 3865–3875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L., Gater S. T., Karrer K. M. (2010). A developmentally regulated gene, ASI2, is required for endocycling in the macronuclear anlagen of Tetrahymena. Eukaryot. Cell 9, 1343–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanet J., Freije A., Ruiz M., Coulon V., Sanz J. R., Chiesa J., Gandarillas A. (2010). A mitosis block links active cell cycle with human epidermal differentiation and results in endoreplication. PLoS ONE 5, e15701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L., Dai H., Zhou M., Li X., Liu C., Guo Z., Wu X., Wu J., Wang C., Zhong J., et al. (2012). Polyploid cells rewire DNA damage response networks to overcome replication stress-induced barriers for tumour progression. Nat. Commun. 3, 815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielke N., Querings S., Rottig C., Lehner C., Sprenger F. (2008). The anaphase-promoting complex/cyclosome (APC/C) is required for rereplication control in endoreplication cycles. Genes Dev. 22, 1690–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielke N., Kim K. J., Tran V., Shibutani S. T., Bravo M. J., Nagarajan S., van Straaten M., Woods B., von Dassow G., Rottig C., et al. (2011). Control of Drosophila endocycles by E2F and CRL4(CDT2). Nature 480, 123–127 [DOI] [PMC free article] [PubMed] [Google Scholar]