Abstract
In the field of developmental biology, live imaging is a powerful tool for studying, in real time, the dynamic behaviors of tissues and cells during organ formation. Mammals, which develop in utero, have presented a challenge for live imaging. Here, we offer a novel, prolonged and robust live imaging system for visualizing the development of a variety of embryonic tissues in the midgestation mouse embryo. We demonstrate the advantages of this imaging system by following the dynamics of neural tube closure during mouse embryogenesis and reveal extensive movements of the cranial neural tissue that are independent of neural fold zipping.
Keywords: Live imaging, Mouse embryogenesis, Neural tube closure, Shroom3, Non-neural ectoderm
INTRODUCTION
Live imaging of the mouse embryo is very challenging as the embryo develops in utero, unlike other model organisms such as Drosophila, Xenopus, zebrafish and chick. Several techniques have been established to follow the dynamics of different developmental processes during mouse embryogenesis. These take advantage of ex utero embryo culture and fluorescent protein transgenes and have been used for real-time imaging of preimplantation and early postimplantation mouse embryos, including embryonic day (E) 9.5 cranial neural tube closure (Jones et al., 2002; Pyrgaki et al., 2010; Yamaguchi et al., 2011; Xenopoulos et al., 2012).
Although studies using these different imaging techniques have contributed significantly to the understanding of the molecular and cellular mechanisms underlying morphological processes during early mouse embryogenesis, several crucial limitations still hamper the ability to access and manipulate different embryonic tissues. Here, we demonstrate a novel in toto imaging system of the mouse embryo that provides full access to different tissues, a high degree of stabilization of the embryo that allows robust comparisons between wild-type and mutant embryos, and allows manipulation of different tissues followed by extended live imaging to examine the effect of these manipulations.
A particularly challenging stage of mouse embryogenesis is between E8.5 and E9.5, when the heart begins to beat and the embryo undergoes changes in growth and morphology including ‘turning’ of the U-shaped embryo with the ventral surface on the outside to a fetal position with the dorsal surface on the outside. We have created a system that overcomes these challenges to allow real-time imaging as well as robust control of experimental manipulations. Another advantage is the ability to image more than one embryo simultaneously, a crucial parameter in studies of mutant embryos that cannot be distinguished at the onset of the experiment.
To demonstrate the novelty and advantages of our system, we visualized neural tube closure (NTC) in the mouse embryo. Mammalian neural tube (NT) morphogenesis has been dynamically imaged previously but with a focus on the hindbrain area (Pyrgaki et al., 2010; Yamaguchi et al., 2011). Using the novel technique described here, we imaged in real time the complete process of NTC from the face to the primordial brain and spinal cord, and from initiation of neurulation to the meeting and closure of the neural folds. Because of embryo growth and extensive embryo movements, this required following different embryos that were dissected at several developmental stages. Static time points of NTC in mouse embryos and live imaging of other vertebrate embryos, such as chick and Xenopus, have highlighted two major morphological processes: (1) bending or inflection of the neural folds at the medial hinge point and at the two dorsolateral hinge points (DLHPs) to elevate the neural folds and bring them into close apposition and (2) the ‘zipping’ together of the neural folds to form the closed NT (Greene and Copp, 2009; Filla et al., 2004; Ezin et al., 2009; Kieserman and Wallingford, 2009; Schoenwolf and Smith, 1990; Davidson and Keller, 1999; Copp et al., 2003). Our study dynamically follows these two distinct morphological processes during mammalian NTC and we quantify the rate of zipping and distance between the neural folds at different stages of NTC. Moreover, we perform cutting experiments to begin to establish the relationship between neural fold inflection and zipping. Finally, live imaging indicates that DHLP formation is not as fixed in time as previously expected.
MATERIALS AND METHODS
Mice and whole embryo culture and imaging system
Male and female mice transgenic for CAG::myr-venus (Rhee et al., 2006) were mated and embryos dissected at different developmental stages between E8 and E10 in freshly prepared Tyrode’s solution, removing Reichert’s membrane but leaving the yolk sac intact. Animals heterozygous for Shroom3m1Nisw were mated with VenMyr and F1 males, and females positive for Venus and heterozygous for Shroom3m1Nisw were mated to obtain homozygous mutant:Venus embryos. For non-neural ectoderm visualization during NTC, females transgenic for mTomato:mGFP [JAX stock #007576, (Muzumdar et al., 2007)] were mated with males transgenic for Grhl3-Cre (Camerer et al., 2010), and embryos were selected based on GFP expression.
Embryos positive for fluorescence were placed in culture media on the mounting platform for imaging. Culture media was prepared before starting the embryo dissection. Culture media contains three parts rat serum (Harlan #B-4520) preheated at 65°C for 30 minutes and filtered through a 0.2 mm filter, and one part embryonic media (DMEM without Phenol Red and containing 55 U/ml penicillin/streptomycin, 2.2 mM glutamax and 11 mM Hepes buffer). The embryos were mounted on a black Millipore filter paper (catalog #AABG01300). Glass-bottomed dishes were from MatTek (P35G-1.5-20-C).
Time-lapse imaging and post-imaging processing
Mounted embryos were placed in a heat- and humidity-controlled chamber (37°C; 5% O2, 5% CO2 and 90% N2 gas mixture) on a motorized stage of an inverted Zeiss LSM510 Meta confocal microscope. Images were acquired with a 10× lens c-Apochromat NA1.2 with 0.7× digital zoom out or 2× zoom in. Up to five embryos were imaged simultaneously using the multi-time Zeiss Zen software program. For VenMyr embryos, a 448-nm laser (8-15% power) was used. mTomato was excited by a 543-nm laser (50% power) and mGFP by a 448-nm laser (8-15% power). Images were acquired at 512×512 resolution. The thickness of z-slices was 20 μm (total 15-30 slices/each time point, depending on the experiment). No time interval was set. Images were obtained for 8-24 hours, depending on the experiment.
Post-imaging processing and measurements were carried out using LSM image browser (Zeiss). Graphs and statistical analyses were carried out using Microsoft Office Excel. The movies were exported using the LSM image browser and then were flipped using Windows Live Movie Maker. Finally, all movies were reformatted and their size reduced using the software Prism Video File Converter.
RESULTS
A novel in toto imaging technique for studying morphological processes during mouse embryogenesis
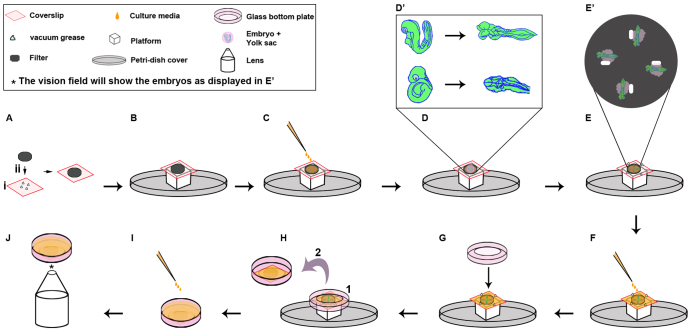
Here we describe a novel live imaging technique that allows dynamic visualization of whole mouse embryos in real time, in this case from E8.0 to E10.5. As shown in Fig. 1 and supplementary material Fig. S1, embryos were removed from the uterus into Tyrode’s solution, the decidua removed and the embryo carefully isolated with yolk sac intact [detailed demonstration of this procedure described by Gray and Ross (Gray and Ross, 2011)]. Embryos were selected for fluorescent protein expression and moved into a droplet of culture media on a paper filter attached to a coverslip with vacuum grease (Fig. 1A-C). As schematized in supplementary material Fig. S1A, starting farthest away from the embryo, the yolk sac was gently peeled open and lightly pressed onto the paper filter, which has adherence qualities, to anchor the embryo to the filter. To expose the tissue of interest, the embryo can be stretched and lightly pressed onto the filter. For example, to expose the neural plate of a 9-somite embryo, the embryo was extended and the ventral sides of the face and the tail adhered to the filter, thus exposing the dorsal side of the embryo such that the neural plate was fully accessible for imaging (Fig. 1D-E′; supplementary material Fig. S1A). After mounting the embryos (up to five embryos on one filter), a glass-bottomed dish was placed over the embryos using vacuum grease drops for spacing and to attach to the coverslip (Fig. 1F,G). This allows flexibility of the distance between the embryos and the glass-bottomed plate (imaging surface). The dish was inverted, filled with culture media and placed in the imaging chamber on an inverted microscope (Fig. 1H-J). As the embryos lie under the filter paper, holes were cut in the filter near the embryos after mounting to ease navigation to their position (supplementary material Fig. S1B). Cuts at the edge of the filter help to identify and correlate each embryo with the final imaging file, especially important when mutants or manipulated embryos are imaged.
Fig. 1.
A novel in toto imaging system for mouse embryogenesis. (A) Four drops of vacuum grease are added to a 24×24 mm coverslip (i). A black filter paper is attached by pressing the filter onto the vacuum grease drops (ii). (B) A mounting platform, such as a Petri dish cover with an OCT-mold taped to the center, is prepared to provide a working height during the mounting procedure. The coverslip with attached filter is placed on top of this platform. (C) On top of the filter paper, 100-200 μl of culture media is added. (D) An embryo with the embryonic yolk sac (YS) intact is transferred to the media drop on the filter. The YS is gently peeled off and adhered to the filter surface. The embryo is extended and lightly pressed against the filter to expose the specific tissue of interest for imaging. To image neural tube closure, embryos were extended to expose the dorsal surface (D′). For a more detailed description of YS and embryo mounting, see supplementary material Fig. S1A. (E) The mounting process is repeated for additional embryos to be imaged. In our experiments, we mounted four embryos for each time-lapse experiment (E′). Supplementary material Fig. S1B demonstrates a filter-cutting method to track different embryos before and after imaging. (F) Four drops of vacuum grease are added to the coverslip, around the filter. The entire surface of the coverslip is covered with 1 ml of culture media. (G) An inverted glass-bottomed dish is attached to the coverslip. (H) (1) Vacuum grease drops provide flexible spacers between the glass bottom dish and the embryos. (2) The plate is flipped. Media will remain around the embryos, which prevents them from drying out. (I) The plate is filled with 2-3 ml culture media. This media mixes with the media surrounding the embryos, as the limited amount of the vacuum grease applied will not seal off the filter with the mounted embryos from the surrounding environment. (J) The plate is placed into a heated and environmentally controlled chamber on the microscope stage for imaging. Asterisk indicates the lens of the microscope visualizing the embryos (as drawn in E′).
Our system overcomes a number of major difficulties faced when following developmental processes during mouse embryogenesis. First, because of the shape of the embryo at different stages, which hampers the ability to image many tissues, our technique provides considerable flexibility in mounting the embryo in different positions, which allows accessibility to the majority of embryonic regions (for examples, see Fig. 2; supplementary material Fig. S2 and Movies 1-7). Second, extensive movement of the embryo due to heartbeat, floatation within the yolk sac, and tissue movements has made live imaging difficult. Our system provides considerable stability of the embryo for long-term imaging. Third, it allows accessibility to different tissues to perform external manipulations (Fig. 3; supplementary material Fig. S3 and Movies 9-12). Fourth, with a programmable motorized microscope stage, multiple embryos can be imaged, which is crucial for experiments with mutants and manipulated embryos (Fig. 4; supplementary material Fig. S4 and Movies 13-17). Finally, live imaging of NTC shows unexpected dynamic behavior during DLHP formation (Fig. 5A-B; supplementary material Movies 18, 19) and extensive cellular activity during NT zipping (Fig. 5C; supplementary material Movie 20).
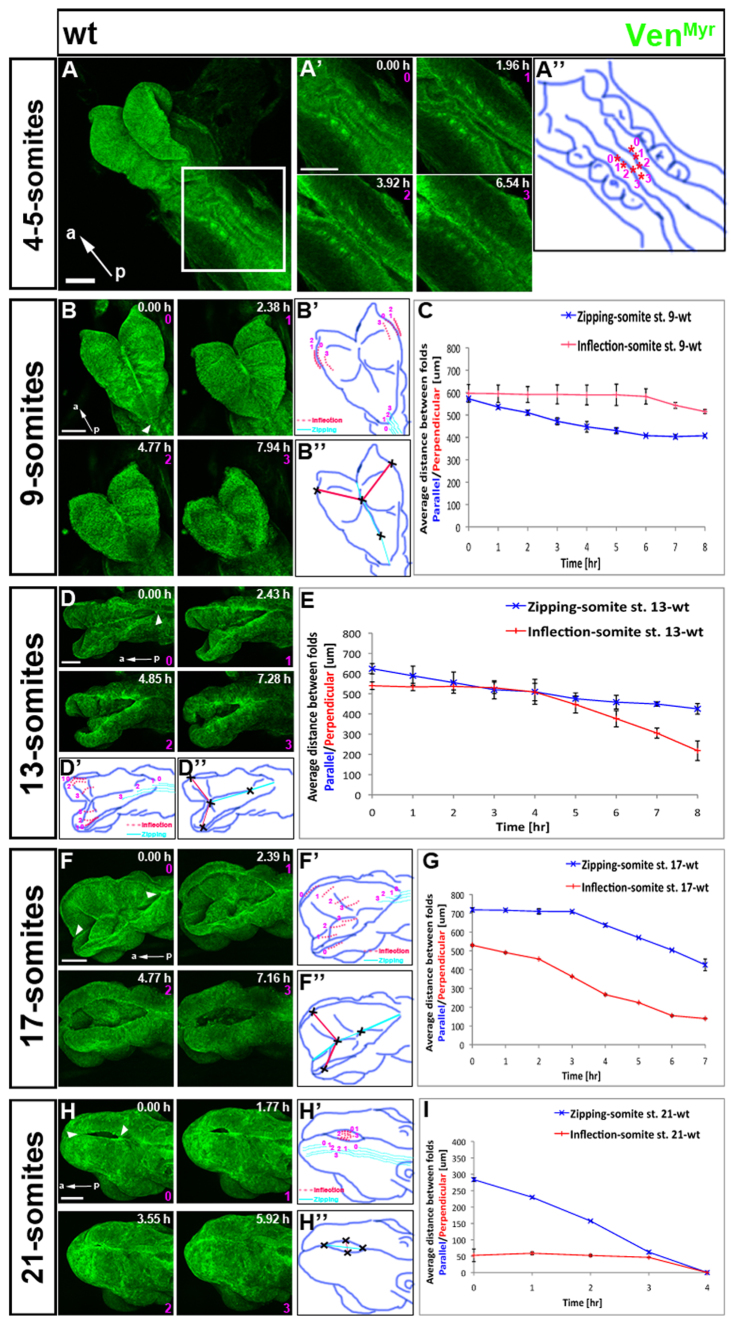
Fig. 2.
Neural tube closure (NTC) in wild-type mouse embryos. Embryos expressing VenMyr constitutively in all tissues were dissected at different developmental stages and mounted to image the dorsal side and NTC. Somite number indicated on the left represents embryo staging at the onset of time-lapse imaging. For still images taken from the corresponding movie, time in hours is shown in each frame and the number in pink below the time refers to frame number. Schematic diagrams depict the progression of NTC and the positions at which measurements of zipping and inflection were taken. (B′,D′,F′,H′) Light blue lines were drawn from the four frames of the corresponding still images to depict NTC due to zipping that proceeds from closure point I (and the rostral closure site if present). Red dotted lines were drawn from the four frames to highlight the extent to which the midbrain neural folds approach one another by inflection. (B″,D″,F″,H″) Red and blue lines and crosses depict where measurements were taken to quantify the distance between the neural folds. Inflection was measured in the midbrain (red lines, perpendicular to the midline, with crosses marking the reference points at the edge of the closing neural folds and midline). Zipping was measured from a fixed point at the rostral midline (or from the rostral closure site if present) to the neural fold closure site proceeding from closure point I (blue lines, parallel to the midline, with crosses marking the reference points). In the graphs, the distance (μm) between folds parallel to the midline (zipping, blue curve) or perpendicular to the midline (inflection, red curve), were measured every hour of the time-lapse movies. (A,A′) Still images from time-lapse supplementary material Movie 1, which starts when the embryo has four to five somites, showing formation of closure point I. Area outlined by white box was magnified and imaged (supplementary material Movie 1). (A″) Drawing of A′, in which each pair of asterisks show the distance between the neural folds in each frame of the still images. Distance between the asterisks decreases with time, until the folds adhere in the last frame. (B) Still images from supplementary material Movie 2 visualizing the hindbrain and midbrain of a 9-somite embryo at onset of imaging. (B′) Light blue lines become longer over time indicating that zipping continues to proceed from closure point I (arrowhead in B). However, the distance between each pair of dotted red lines does not change over the first three frames (0, 1 and 2), and inflection only begins at 6 hours as represented by a shift of line 3 towards the midline. (B″,C) Graph showing progression in time of zipping and inflection during 8 hours of imaging of 9-somite embryos. (D) Still images from supplementary material Movie 4 of the head of a 13-somite embryo. (D′,E) Light blue lines become longer with time indicating progression of zipping from closure point I (arrowhead in D). (D′-E) Distance between each pair of dotted red lines does not change in first two frames (0 and 1; first 4 hours) but inflection progresses rapidly over the last 4 hours of imaging. (F) Still images from supplementary material Movie 5 of the head of 17-somite embryo. (F′-G) Zipping proceeds from both closure point I and II/III (arrowheads in F) and inflection is extensive throughout 7 hours of imaging as highlighted by progressive reduction in distance between each pair of dotted red lines. (H) Still images of supplementary material Movie 6 of 21-somite embryo with a small midbrain opening. (H′-I) Blue lines get longer with time owing to zipping proceeding from closure points I and II/III. Distance between each pair of dotted red lines does not change in the first three frames (0, 1 and 2) and then inflection proceeds dramatically over time of last frame. Error bars represent xxx. Scale bars: 100 μm. a, anterior; p, posterior.
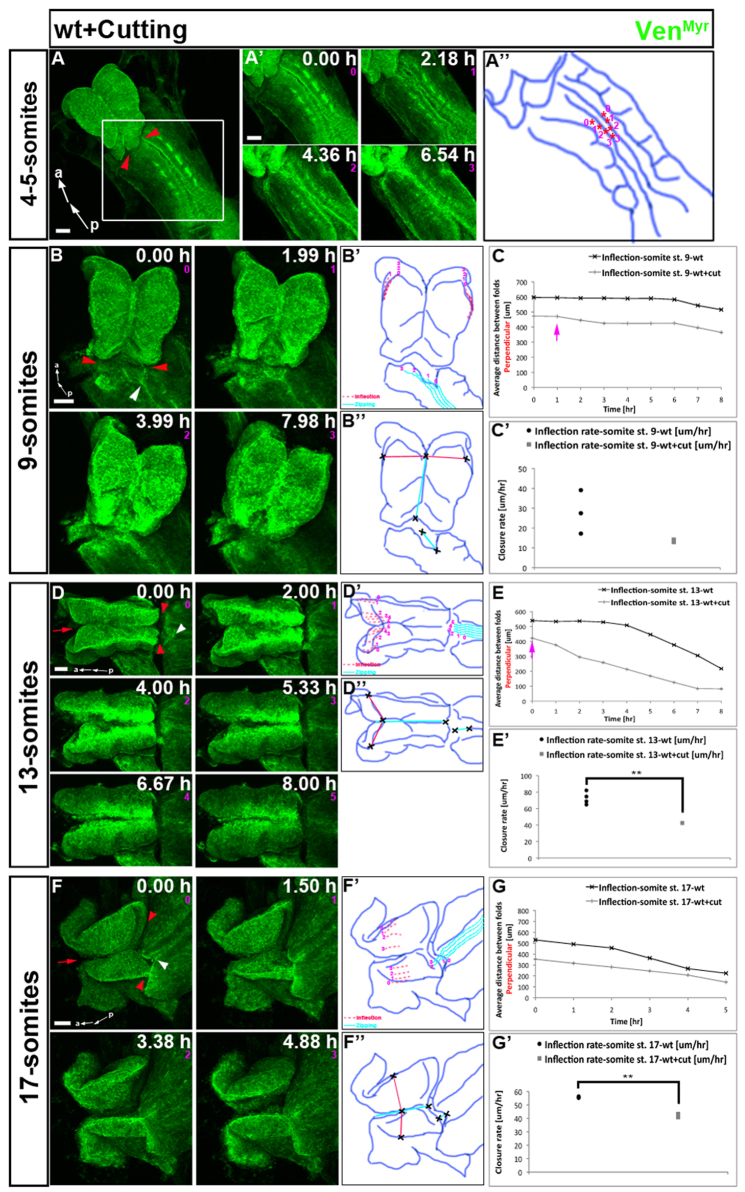
Fig. 3.
Inflection is an active process that can initiate and proceed independently of zipping. Embryos expressing VenMyr constitutively in all tissues were dissected at different developmental stages and mounted to image the dorsal side and NTC. Somite number indicated on the left represents embryo staging at the onset of time-lapse imaging. For still images taken from the corresponding movie, time in hours is shown in each frame and the number in pink below the time refers to frame number. Embryos were cut anterior to closure point I (red arrowheads denote horizontal cut) and in older embryos also at the rostral midline (red arrow) to uncouple zipping and inflection of the neural folds. White arrowheads indicate the position of NTC proceeding from closure point I at the onset of the movie. Quantification of zipping dynamics is presented in supplementary material Fig. S3. (A,A′) Dorsal view of severed 4- to 5-somite embryo and still images of time-lapse supplementary material Movie 9 that shows formation of closure point I. (A″) Distance between the neural folds (asterisks) becomes less with time, until the folds adhere in the last frame. (B) Still images from supplementary material Movie 10 of head of a severed 9-somite embryo. (B′) Light blue lines show progressive zipping from closure point I which ceases at the cut site. (B″-C′) Inflection initiates prematurely (C, gray curve, pink arrow). The rate of inflection (μm/hour) is not significantly different between three uncut (black curve in C, black dots in C′) and three severed (gray curve in C, gray squares in C′) embryos. (D) Still images of supplementary material Movie 11 of a severed 13-somite embryo. (D′) Light blue lines indicate progressive zipping to the cut site over the first two frames. (D″-E′) Inflection initiates prematurely (E, gray curve, pink arrow), although the rate of inflection is significantly slower in severed versus uncut embryos (E′, n=4 each). (F) Still images from supplementary material Movie 12 of a severed 17-somite embryo. (F′-G′) Progressive zipping to the cut site occurs over the first three frames. Distance between each pair of dotted red lines lessens over time indicating inflection occurs independently of zipping but the rate of inflection is significantly less in severed versus uncut embryos (G,G′, n=3 each). For details of schematic diagram labels and measurements taken, see Fig. 2. Scale bars: 100 μm. a, anterior; p, posterior.
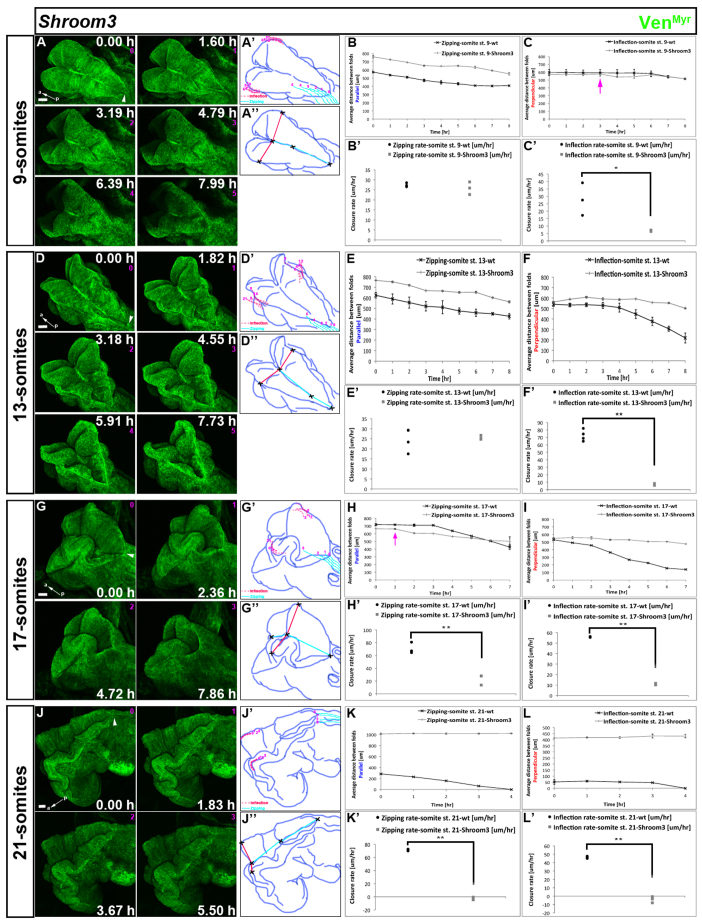
Fig. 4.
Shroom3 mutation disrupts inflection and zipping progression during NTC, resulting in cranial neural tube defects. Embryos expressing VenMyr constitutively in all tissues were dissected at different developmental stages and mounted to image the dorsal side and NTC. Somite number indicated on the left represents embryo staging at the onset of time-lapse imaging. For still images taken from the corresponding movie, time in hours is shown in each frame and the number in pink below the time refers to frame number. Shroom3m1Nisw/m1Nisw embryos expressing VenMyr were imaged during cranial NTC. White arrowheads indicate the position of the closing neural folds proceeding from closure point I at the onset of the movie. Red dotted lines in A′,D′,G′,J′ stay in relatively the same position throughout each movie, indicating that inflection in Shroom3 mutants is disrupted. (A) Still images from supplementary material Movie 14 of a 9-somite Shroom3 mutant embryo. (A′-B′) Light blue lines become longer over time indicating that zipping continues from closure point I and at a similar rate to wild type (gray versus black curve in B, gray squares versus black dots in B′, n=3 each). (C,C′) In mutant embryos, inflection begins prematurely (pink arrow) and its rate was slower compared with wild type (gray and black curves and dots, respectively, n=3 each). (D) Still images of supplementary material Movie 15 of a 13-somite Shroom3 mutant embryo. (D′-E′) Light blue lines become longer over time (D″) indicating that zipping continues and at a similar rate in mutant and in wild type (E,E′, n=3 each). (F,F′) Inflection was significantly perturbed in Shroom3 mutants compared with wild type in both extent and rate (n=3 each). (G) Still images of supplementary material Movie 16 of a 17-somite Shroom3 mutant. (G′-H′) Light blue lines become longer over time (G′), indicating that zipping continues from closure point I, but prematurely and at a significantly slower rate compared with wild type (pink arrow in H,H′, n=3 each). (I,I′) Inflection extent and rate was significantly perturbed in Shroom3 mutants compared with wild type (n=3 each). (J) Still images of supplementary material Movie 17 of a 21-somite Shroom3 mutant embryo. Both zipping and inflection are severely inhibited. Light blue lines do not change over time (J′), indicating a lack of zipping, and the rate of inflection in mutant embryos is negative (K′,L′), indicating that the distance between the cranial neural folds increases with time. For details of schematic diagram labels and measurements taken, see Fig. 2. Error bars represent s.d. Scale bars: 100 μm. a, anterior; p, posterior.
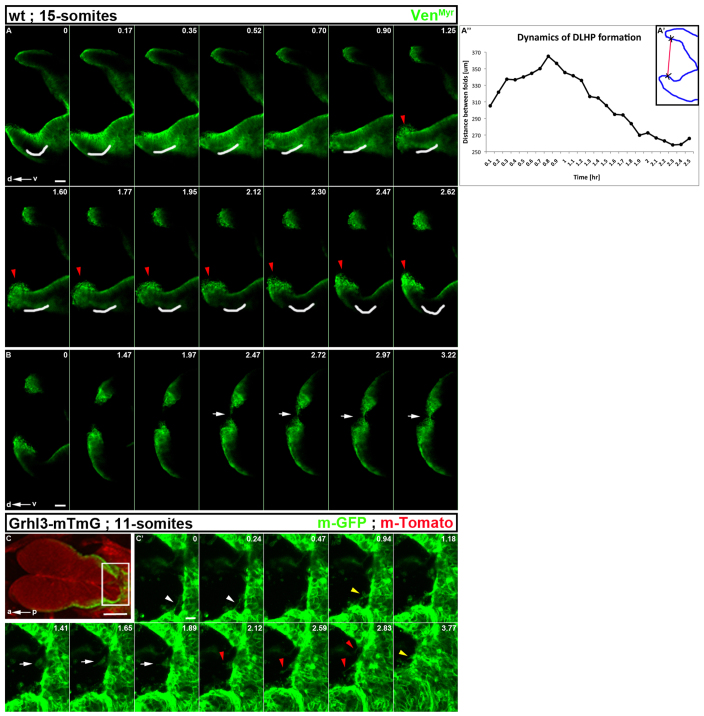
Fig. 5.
Dynamic tissue and cellular activities of the neural folds during inflection and zipping. (A-A″) Still images from supplementary material Movie 18. Optical dorsoventral (d/v) sections showing the midbrain neural folds of a 15-somite embryo. The position of the dorsolateral hinge point (DLHP) of the inflected neural folds is marked by a white line. As time proceeds, the DLHP disappears and the neural folds relax [indicated by straightening of the white line and by increased distance between the neural folds in A″ measured in μm over time (hours)]. Later, the DLHP reforms and inflection brings the neural folds closer. Crosses in A′ schematize the reference points used to measure the distance between the neural folds (red line) at different time points. During the period of DLHP reformation, cells on the edge of each fold become highly active (red arrowheads). Scale bar: 20 μm. (B) Still images of supplementary material Movie 19. After DLHP reformation (A), the neural folds continue to approach one another and the non-neural/neural cells on the edges of the neural folds show dynamic activity and extend long cellular processes (white arrows) towards each other, followed by contact between the folds that completes the closure process. Scale bar: 20 μm. (C) Dorsal side of an 11-somite embryo transgenic for Grhl3-Cre and mTomato-mGFP. The embryo expresses mTomato constitutively in all tissues except for non-neural ectoderm adjacent to the neural folds in which Grhl3-mediated Cre recombination induced mGFP expression. White box indicates region of the still images shown in C′. Scale bar: 100 μm. (C′) Still images from supplementary material Movie 20 (green channel). During zipping of the neural folds proceeding from closure point I, the non-neural ectoderm displays different cellular activities, such as cellular bridges (white arrowheads), cell shape changes (white arrows), filopodia (red arrowheads) and lamellipodia (yellow arrowheads). Scale bar: 20 μm.
Real-time visualization of NTC along the rostral-caudal axis of the mouse embryo
To demonstrate the applicability, novelty and advantages of our system, we focused on NTC in the mouse embryo (Fig. 2; supplementary material Fig. S2A-C and Movies 1-7). NTC is a highly dynamic morphological process that takes place over more than two days (supplementary material Fig. S2A) (Greene and Copp, 2009). At E8.0, initial closure point I is formed at the hindbrain/cervical boundary, and then closure of the NT proceeds both caudally towards the tail and rostrally towards the head (schematic of closure points is shown in supplementary material Fig. S2A). By E9.0, closure point III is formed at the most anterior region of the forebrain, and closure progresses caudally through the brain. In some mouse strains, closure point II is formed at the forebrain/midbrain boundary. Using our technique and a mixed genetic background of 129/SvImJ and C57BL/6J, closure II formation varied and was sometimes observed but sometimes not. Therefore, below we indicate forebrain/midbrain closure as proceeding caudally from closure II/III until it meets with the rostral progression from closure point I to complete cranial NT closure. By E9.5, caudal closure initiated at closure point I proceeds toward the tail, forming the NT of the upper part of the future spinal cord (supplementary material Fig. S2A). At E10, the posterior neuropore at the tail region closes to complete NTC. It is believed that all the closure processes described above proceed in a ‘zippering’ or ‘zipping’ manner. Moreover, to raise the neural folds and bring them close to one another for effective zipping, hinge-point formation is important. A medial hinge point (also called the floor plate) helps to initiate bending of the neural plate and this is followed, depending on the anterior-posterior region, by formation of DHLPs to bring the neural folds in close proximity (Greene and Copp, 2009; Yamaguchi et al., 2011; Copp et al., 2003). Our live imaging system provides the first real-time visualization of all of these steps of neurulation during mammalian embryogenesis, using different embryos dissected at several specific developmental stages (Fig. 2; supplementary material Fig. S2A-C and Movies 1-7).
We used transgenic mice exhibiting widespread expression of myristoylated Venus (VenMyr) fluorescent fusion reporter to highlight the cell membranes (Rhee et al., 2006). To visualize NTC initiation, embryos with four to five somites at dissection were mounted for imaging (Fig. 2A). Closure point I was observed after 4 hours of imaging (Fig. 2A′,A″; supplementary material Movie 1). Closure proceeded both rostrally and caudally in a zipping manner (supplementary material Movie 1). To image closure point III, embryos with eight somites were mounted such that the primordia of the face and forebrain were exposed (supplementary material Fig. S2B). After 6 hours of imaging, closure point III had formed and zipping proceeded caudally, closing the forebrain region (supplementary material Movie 3).
Next, we dissected embryos at five different stages to image NTC proceeding rostrally from closure point I (Fig. 2B,D,F,H; supplementary material Movies 2-6), and at one later stage to image posterior neuropore closure (supplementary material Fig. S2C and Movie 7). We characterized NTC by following and quantifying the dynamics of zipping, which has been visualized in previous imaging studies (Pyrgaki et al., 2010; Yamaguchi et al., 2011), and inflection of the neural ectoderm to bring together the neural folds driven, in part, by DHLPs (Copp et al., 2003). Inflection is most evident in the cranial region, particularly the midbrain, where the neural folds are initially very far apart. Live imaging shows that the cranial neural folds undergo dramatic morphogenesis: elevation, bending and movement towards each other to bring the two folds close together. As the neural folds approach one another, zipping proceeds to complete the formation of the tubular structure of the brain (Greene and Copp, 2009). Zipping closure progress was quantified by measuring the distance between the closure site proceeding rostrally from closure point I to the closure site proceeding caudally from closure point II/III (once these had formed) or to the most rostral position that could be visualized (prior to formation of closure point II/III) over the time course of the movie (distance over time; measurements made parallel to the midline with multiple points of reference to account for curvature of the tissue; Fig. 2B″,D″,F″,H″, light blue lines). Inflection was quantified by measuring the distance between the opposing neural folds at the widest region of the midbrain, perpendicular to the midline (Fig. 2B″,D″,F″,H″, red lines).
Frames from an 8-hour time lapse movie of an embryo at the 9-somite stage at the start of imaging show zipping proceeding rostrally from closure point I, closing the hindbrain region (Fig. 2B, arrowhead). Over the first 6 hours of imaging, zipping closed the cranial neural folds at the rate of 27 μm/hour, whereas no inflection occurred in the midbrain and the folds remained far apart. During the last 2 hours, zipping in the hindbrain paused, whereas inflection of the midbrain started and proceeded at the rate of 28 μm/hour (Fig. 2C; supplementary material Movie 2). Imaging of 13-somite embryos showed zipping proceeding from closure I (Fig. 2D, arrowhead) at a steady rate of 25 μm/hour throughout 8 hours of imaging, whereas inflection was paused during the first 4 hours and then was rapid during the last 4 hours (73 μm/hour; Fig. 2E; supplementary material Movie 4). In 17-somite embryos, zipping from closure points II/III and I (Fig. 2F, arrowheads) paused during the first 3 hours of imaging, and then proceeded rapidly (71 μm/hour) from both closure points. Inflection proceeded throughout the 7 hours of imaging at a rate of 56 μm/hour, bringing the midbrain neural folds considerably closer to one another (Fig. 2G; supplementary material Movie 5). In embryos at the 21-somite stage, all but a small portion of the midbrain was closed (Fig. 2H). Over 4 hours, zipping proceeded at 71 μm/hour, whereas inflection paused for the first 3 hours and then resumed during the last hour, completing cranial NTC (Fig. 2I; supplementary material Movie 6). Closure of the posterior neuropore at E10 (supplementary material Fig. S2C and Movie 7) involved zipping both caudally from closure point I and rostrally from the more posterior region, to complete NT formation. Further study is required to examine whether inflection is a significant aspect of posterior neuropore closure. Thus, using this novel imaging system, we imaged NTC from the first initiation point at E8 to the last step of closure at E10.
The embryo develops normally following mounting, culture and imaging as pairs of somites are added at a normal rate of approximately every 2 hours (supplementary material Fig. S2D and Movie 8). Moreover, the distance between the neural folds of a 9-somite embryo after 4 hours of imaging (which progressed to 11-somites) is similar to that of an 11-somite embryo dissected and measured immediately (supplementary material Fig. S2E,E′,G). Likewise, the distance between the neural folds of a 13-somite embryo after 4 hours of imaging is similar to that of 15-somite embryos dissected and measured immediately (supplementary material Fig. S2F-G).
The relationship between zipping and inflection during NTC in the mouse embryo
The dynamics of zipping and inflection – progression, pausing and continued progression – led us to ask whether inflection is a passive process driven by the zipping forces or an active process that can initiate and proceed independently of zipping. To address this question, embryos were cut between the regions undergoing zipping and inflection to uncouple these two processes, followed by dynamic imaging (Fig. 3; supplementary material Fig. S3). Inflection was quantified as described above in the midbrain (Fig. 3B″,D″,F″, red lines). Zipping was measured over the time of imaging in two places parallel to the midline and these results summed: (1) the distance between the cut site and the posterior neural fold closure site (i.e. zipping proceeding from closure point I) and (2) the distance between a fixed anterior position and the cut site (Fig. 3B″,D″,F″, light blue lines).
First, a 4- to 5-somite embryo was cut anterior to the first somite pair (Fig. 3A, red arrowheads; supplementary material Fig. S3B, red arrowheads; rostral to where closure point I should occur). As in an uncut embryo, closure point I formed after 4 hours of imaging in the severed embryo (Fig. 3A′; supplementary material Movie 9). Next, embryos at 9-, 13- or 17-somite stages were cut anterior to the zipping folds progressing from closure point I, disconnecting zipping from the midbrain neural folds where inflection should take place (Fig. 3B,D,F, red arrowheads; supplementary material Fig. S3D,G,J, red arrowheads). In 13- and 17-somite embryos, a second cut was made along the forebrain midline to prevent zipping from closure point II/III (Fig. 3D,F, red arrows; supplementary material Fig. S3G,J, red arrows). In all severed embryos, the initial distance between the midbrain neural folds, where inflection was measured, was smaller than the initial distance in uncut embryos. This is likely to be because of physical tension that is released by cutting, causing a relaxation of the folds and positioning closer to one another. In embryos cut at 9-, 13- or 17-somite stages, zipping proceeded similarly to uncut embryos up to the position of the cut but did not progress beyond the cut (supplementary material Fig. S3E,H,K, blue arrows). However, in 9- and 13-somite embryos, inflection differed from uncut embryos in two ways: (1) inflection initiated prematurely in severed embryos (Fig. 3C,E, pink arrows; supplementary material Movies 10, 11; within the first hour versus initiation at 6 or 4 hours in uncut 9- or 13-somite embryos) and (2) inflection progressed at a slower rate in cut versus uncut embryos [Fig. 3C′,E′; 14 μm/hour versus 28 μm/hour in 9-somite embryos (P=0.08) and 42 μm/hour versus 73 μm/hour in 13-somite embryos (P=0.005)]. In 17-somite embryos, inflection occurred at the onset of imaging in both cut and uncut embryos (Fig. 3G), but the inflection rate was significantly slower (42 μm/hour) compared with uncut embryos (56 μm/hour) (Fig. 3G′; P=0.002; supplementary material Movie 12).
The fact that inflection in the midbrain can still occur when uncoupled from zipping indicates that inflection is an active process that is not entirely dependent on the forces produced by zipping. However, the slower inflection rate in severed embryos suggests that inflection is facilitated by forces generated by zipping of the neural folds that proceeds from the various closure points. Moreover, alteration of inflection dynamics in cut embryos (premature inflection, disrupted pausing) suggests that zipping and inflection are two distinct morphological processes that are coordinated both in time and in space, throughout NTC in the mouse embryo.
Shroom3 mutation inhibits inflection and affects the dynamics of zipping leading to a neural tube defect
Shroom3 is an actin-binding protein and mutations in Shroom3 lead to cranial neural tube defects (NTDs) (Hildebrand and Soriano, 1999). Shroom3 is thought to induce apical constriction of the neural cells at the DLHPs to promote bending of the neural folds towards each other (Haigo et al., 2003; Lee et al., 2007). We followed the dynamics of zipping and inflection during NTC in Shroom3m1Nisw mutant embryos [an n-ethyl-n-nitrosourea (ENU)-induced allele (Marean et al., 2011)].
Formation of closure point I in Shroom3 mutants was similar to that observed in wild-type embryos (supplementary material Fig. S4B and Movie 13). At the 9-somite stage, the morphology of Shroom3 mutants was slightly different from that of wild type (supplementary material Fig. S4C,D). Imaging starting at this stage showed that the extent of zipping progressing from closure point I was less than that of wild type, although the rate was similar (mutant: 26 μm/hour; wild type: 27 μm/hour) (Fig. 4B,B′). Moreover, the zipping dynamics were altered in Shroom3 mutants as zipping proceeded during the first 3 hours, then paused for 2 hours, and progressed again during the last 3 hours, whereas in wild-type embryos, zipping proceeded during the first 6 hours and paused during the last 2 hours (Fig. 4A-B; supplementary material Movie 14). Inflection was also significantly altered in a number of ways: the inflection rate in Shroom3 mutants was slower (7 μm/hour) than that of wild type (28 μm/hour) (Fig. 4C′; P=0.04) and inflection occurred prematurely (at 3 hours; Fig. 4C, pink arrow) followed by a return to the normal distance between the neural folds during hours 5 and 6 and then continuation of inflection during hours 7 and 8. At the 13-somite stage, Shroom3 embryos were slightly different morphologically from wild type (supplementary material Fig. S4E,F and Movie 15). Over this imaging period, zipping dynamics and rate in mutant embryos were relatively similar to those of wild type (25 μm/hour in both, Fig. 4D,E,E′). However, inflection was severely inhibited in both rate and extent in mutant embryos (7 μm/hour) compared with wild type (73 μm/hour) (Fig. 4F,F′; P=0.0002). By the 17-somite stage, the morphology of the mutant neural folds was highly abnormal and convoluted compared with wild type, which has a relatively smooth and elliptical opening (supplementary material Fig. S4G,H), even though the distances between the folds both parallel and perpendicular to the midline in the mutant were approximately equivalent to those of wild type (Fig. 4H,I). The dynamics of both zipping and inflection were altered: in mutants, zipping occurred prematurely (after 1 hour compared with 3 hours in wild type; Fig. 4G,H, pink arrow; supplementary material Movie 16), yet the zipping rate was significantly slower (23 μm/hour) compared with wild type (71 μm/hour) (Fig. 4H′; P=0.006). Inflection in mutant embryos was dramatically inhibited (Fig. 4I) and the rate was strikingly slower (11 μm/hour) compared with wild type (56 μm/hour) (Fig. 4I′; P=0.00003). By the 21-somite stage, Shroom3 mutant neural tissue was highly abnormal. The forebrain, midbrain and hindbrain remained open in the mutant and the neural tissue was highly convoluted (Fig. 4J, supplementary material Fig. S4I,J). Imaging showed a lack of progression of zipping and inflection (Fig. 4J,K-L′; zipping=–2 μm/hour, P=0.0003; inflection=–4 μm/hour, P=0.001) (supplementary material Movie 17) and the negative rate values indicate that the neural folds become farther away from each other as the tissues grow in the absence of closure.
Our data indicate that the Shroom3 mutation affects both zipping and inflection of the neural folds, which leads to cranial NTDs. Zipping dynamics were altered early during NTC and both the dynamics and rate of zipping were significantly disturbed later. Inflection was highly reduced from the early stages of NTC. Shroom3 protein interacts with actin and regulates actin dynamics (Hildebrand and Soriano, 1999), and it has been suggested that Shroom3 regulates DLHP formation during NTC (Haigo et al., 2003; Lee et al., 2007). DLHP formation is important in bending the neural folds and bringing them closer to each other. Our data, showing a strong effect on inflection in Shroom3 mutants, support previous studies suggesting an important role for Shroom3 during neural fold bending and DLHP formation.
Dynamic formation of DLHPs during inflection of the midbrain neural folds
We followed DLHP formation during inflection of the midbrain neural folds in 15-somite wild-type embryos (Fig. 5A-A″). Our data demonstrate that DLHP formation is not fixed once initiated but instead is dynamic. As highlighted in Fig. 5A by a white line and in supplementary material Movie 18, DLHPs are present but subsequently the DLHP disappears, and then later reforms. This is conceptually different from the ideas derived from static images, which have led to the suggestion that DLHPs once formed are stable and fixed in their position. Fig. 5A″ shows quantification of the distance between the neural folds reflected by inflection, relaxation, re-inflection and continued progression of inflection to bring the opposed midbrain neural folds closer to one another.
During re-formation of the DLHP, the neural/non-neural cells become highly active (Fig. 5A, red arrowheads; supplementary material Movie 18). These cells form long membrane extensions (Fig. 5B, white arrows; supplementary material Movie 19) to connect the opposed neural folds leading to completion of NTC.
Cellular activity of the non-neural ectoderm during zipping of the neural folds
Next, we followed the zipping process at higher magnification and by specific labeling of the non-neural ectoderm cells by mating mGFP transgenic mice (Muzumdar et al., 2007). Embryos carrying both transgenes constitutively express membrane-Tomato in all tissues, and upon Grhl3 expression in non-neural ectoderm cells, Cre is induced and non-neural ectoderm cells begin to express membrane GFP (mGFP). At E8.5, only the non-neural ectoderm adjacent to the neural folds is labeled with mGFP, whereas the rest of the embryo is marked with mTomato (Fig. 5C). As zipping proceeded from closure point I (Fig. 5C′; supplementary material Movie 20; green channel only), different cellular activities were observed including cellular bridges (white arrowheads), cell shape changes (white arrows), filopodia (red arrowheads) and lamellipodia (yellow arrowheads) as the non-neural ectoderm helps to close the neural folds.
DISCUSSION
Live imaging in the mouse embryo has traditionally been highly challenging because the embryo develops in utero. Moreover, the embryo undergoes extensive morphological changes, especially from gastrulation to early neurulation, when the embryo grows and turns around its body axis, which has caused great difficulty for imaging developmental processes in real time, as tissues of interest drift out of the visual field through time. Here, we describe a novel in toto live imaging system applicable to the study of many aspects of mouse embryogenesis, from broad tissue rearrangements to single cell behaviors. The use of transgenic reporters and embryos mounted in different positions allows visualization in real time of tissue- and cellular-level behaviors over prolonged developmental periods in the living mammalian embryo.
A few technologies were previously established for imaging the mouse embryo at preimplantation and early postimplantation stages (Jones et al., 2002; Pyrgaki et al., 2010; Yamaguchi et al., 2011; Xenopoulos et al., 2012). These existing techniques have been important in evaluating a variety of developmental processes; however, they face limitations owing to the challenges mentioned. The live imaging system described here overcomes these limitations and allows more flexibility, control and reproducibility when studying a variety of morphological and developmental processes during mouse embryogenesis, up to E10.5.
The novelty of our system is based upon the following factors. First, more than one embryo can be imaged simultaneously, a crucial factor in studies in which mutant embryos are evaluated (demonstrated in Fig. 4; supplementary material Movies 13-17). Second, our system allows mounting of the embryo in different positions, regardless of the shape of the embryo, and ready access to many embryonic tissues. Third, the embryo is stabilized, preventing drifting out of the imaging field. Finally, stabilization of the embryo allows significant control in performing external manipulations, such as physical methods (cutting as demonstrated in Fig. 2 and supplementary material Movies 9-12, local treatments with drugs, delivery of recombinant proteins on beads, etc.) and optical techniques (photo-bleaching/activation, laser ablation, etc.). Thus, this imaging system should allow the mouse embryo to be manipulated and imaged in much the same way as embryos that develop ex utero. Moreover, the extensive collections of mouse mutants and the molecular insights derived from their study can be probed to reveal how genes influence cellular behaviors. The combined power of genetics and molecular genetic technologies in the mouse, now coupled with this robust imaging strategy should enhance our understanding of mammalian development.
To demonstrate robust characteristics of our imaging system, we focused on NTC. Previous studies have imaged mammalian NTC in real time (Jones et al., 2002; Pyrgaki et al., 2010; Yamaguchi et al., 2011). However, owing to the inverted shape of the embryo and subsequent turning during neurulation, and the extensive movement of the embryo compounded by the heartbeat, imaging of NTC was limited to specific developmental stages and to specific regions along the neural plate. Here, we analyzed the entire process of NTC from the flat neural plate stage through to the completion of neural fold fusion, and from the face through the brain to the posterior neuropore. To do so, we imaged different embryos dissected at different time points between E8 and E10, over the extensive period of NTC (Fig. 1; supplementary material Fig. S1). Our results are similar to the more limited imaging of NTC (Jones et al., 2002; Pyrgaki et al., 2010; Yamaguchi et al., 2011), indicating that our system is reliable and reflective of the process of mammalian NTC as it has been observed so far.
Here, we have dynamically visualized two morphological processes that are active during NTC in the mouse embryo (Figs 2, 3; supplementary material Movies 1-12). In addition to the known zipping process, which proceeds from the different initial closure points to close the neural folds once they are near one another, our data highlights extensive inflection of the neural tissue, especially in the cranial region in which the neural folds begin very far apart. Moreover, by uncoupling zipping from inflection through physical separation, we provide evidence that inflection is an active process. Zipping promotes the progression of inflection, yet inflection, which is due at least in part to DLHP formation, can initiate and proceed independently from zipping. Furthermore, by quantifying the dynamics of zipping and inflection in normal and cut embryos, we suggest that these processes are coordinated both spatially and temporally during NTC. Future studies should help reveal how different intrinsic and extrinsic forces regulate both of these processes in time. In addition, the ability to couple real-time imaging with genetic tools will allow a better understanding of how different cellular machineries are recruited and regulated to facilitate both zipping and inflection at different regions and times along the neural plate during closure. Additionally, our study suggests that dorsolateral hinge-point formation is not as fixed in time as previously expected (Fig. 5A-B; supplementary material Movies 18, 19).
Finally, we used our system to follow the cellular activities of the non-neural ectoderm during zipping of the closing neural folds (Fig. 5C,C′; supplementary material Movie 20). This system also has the potential to follow many other morphological processes during mouse embryogenesis, such as primary palate development (see embryo in supplementary material Fig. S2B mounted with the facial region exposed), heart development and otic placode development. Using the proper combination of transgenic mice expressing different reporter genes in an inducible manner, it will be possible to follow the development and behavior of specific tissues and cells throughout an extended period during mammalian embryogenesis.
Supplementary Material
Acknowledgments
We thank Anna-Katerina Hadjantonakis for CAG::myr-venus transgenic mice; Shaun Coughlin for Grhl3-Cre transgenic mice; and Lori Bulwith and Danielle Marck for technical assistance.
Footnotes
Funding
This work was supported by the Human Frontiers Science Program (postdoctoral fellowship to R.M.) and the Department of Pediatrics. L.N. is an investigator of the Howard Hughes Medical Institute. Deposited in PMC for release after 6 months.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.085001/-/DC1
References
- Camerer E., Barker A., Duong D. N., Ganesan R., Kataoka H., Cornelissen I., Darragh M. R., Hussain A., Zheng Y.-W., Srinivasan Y., et al. (2010). Local protease signaling contributes to neural tube closure in the mouse embryo. Dev. Cell 18, 25–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copp A. J., Greene N. D. E., Murdoch J. N. (2003). The genetic basis of mammalian neurulation. Nat. Rev. Genet. 4, 784–793 [DOI] [PubMed] [Google Scholar]
- Davidson L. A., Keller R. E. (1999). Neural tube closure in Xenopus laevis involves medial migration, directed protrusive activity, cell intercalation and convergent extension. Development 126, 4547–4556 [DOI] [PubMed] [Google Scholar]
- Ezin A. M., Fraser S. E., Bronner-Fraser M. (2009). Fate map and morphogenesis of presumptive neural crest and dorsal neural tube. Dev. Biol. 330, 221–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filla M. B., Czirók A., Zamir E. A., Little C. D., Cheuvront T. J., Rongish B. J. (2004). Dynamic imaging of cell, extracellular matrix, and tissue movements during avian vertebral axis patterning. Birth Defects Res. C Embryo Today 72, 267–276 [DOI] [PubMed] [Google Scholar]
- Gray J., Ross M. E. (2011). Neural tube closure in mouse whole embryo culture. J. Vis. Exp. 56, e3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene N. D. E., Copp A. J. (2009). Development of the vertebrate central nervous system: formation of the neural tube. Prenat. Diagn. 29, 303–311 [DOI] [PubMed] [Google Scholar]
- Haigo S. L., Hildebrand J. D., Harland R. M., Wallingford J. B. (2003). Shroom induces apical constriction and is required for hingepoint formation during neural tube closure. Curr. Biol. 13, 2125–2137 [DOI] [PubMed] [Google Scholar]
- Hildebrand J. D., Soriano P. (1999). Shroom, a PDZ domain-containing actin-binding protein, is required for neural tube morphogenesis in mice. Cell 99, 485–497 [DOI] [PubMed] [Google Scholar]
- Jones E. A. V., Crotty D., Kulesa P. M., Waters C. W., Baron M. H., Fraser S. E., Dickinson M. E. (2002). Dynamic in vivo imaging of postimplantation mammalian embryos using whole embryo culture. Genesis 34, 228–235 [DOI] [PubMed] [Google Scholar]
- Kieserman E. K., Wallingford J. B. (2009). In vivo imaging reveals a role for Cdc42 in spindle positioning and planar orientation of cell divisions during vertebrate neural tube closure. J. Cell Sci. 122, 2481–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C., Scherr H. M., Wallingford J. B. (2007). Shroom family proteins regulate gamma-tubulin distribution and microtubule architecture during epithelial cell shape change. Development 134, 1431–1441 [DOI] [PubMed] [Google Scholar]
- Marean A., Graf A., Zhang Y., Niswander L. (2011). Folic acid supplementation can adversely affect murine neural tube closure and embryonic survival. Hum. Mol. Genet. 20, 3678–3683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar M. D., Tasic B., Miyamichi K., Li L., Luo L. (2007). A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605 [DOI] [PubMed] [Google Scholar]
- Pyrgaki C., Trainor P., Hadjantonakis A.-K., Niswander L. (2010). Dynamic imaging of mammalian neural tube closure. Dev. Biol. 344, 941–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee J. M., Pirity M. K., Lackan C. S., Long J. Z., Kondoh G., Takeda J., Hadjantonakis A.-K. (2006). In vivo imaging and differential localization of lipid-modified GFP-variant fusions in embryonic stem cells and mice. Genesis 44, 202–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenwolf G. C., Smith J. L. (1990). Mechanisms of neurulation: traditional viewpoint and recent advances. Development 109, 243–270 [DOI] [PubMed] [Google Scholar]
- Xenopoulos P., Nowotschin S., Hadjantonakis A.-K. (2012). Live imaging fluorescent proteins in early mouse embryos. Methods Enzymol. 506, 361–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y., Shinotsuka N., Nonomura K., Takemoto K., Kuida K., Yosida H., Miura M. (2011). Live imaging of apoptosis in a novel transgenic mouse highlights its role in neural tube closure. J. Cell Biol. 195, 1047–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.