Abstract
Dipeptidyl aminopeptidase 1 (DPAP1) is an essential food vacuole enzyme with a putative role in hemoglobin catabolism by the erythrocytic malaria parasite. Here, the biochemical properties of DPAP1 have been investigated and compared to those of the human ortholog cathepsin C. To facilitate the characterization of DPAP1, we have developed a method for the production of purified recombinant DPAP1 with properties closely resembling those of the native enzyme. Like cathepsin C, DPAP1 is a chloride-activated enzyme that is most efficient in catalyzing amide bond hydrolysis at acidic pH values. The monomeric quaternary structure of DPAP1 differs from the homotetrameric structure of cathepsin C, which suggests that tetramerization is required for a cathepsin C-specific function. The S1 and S2 subsite preferences of DPAP1 and cathepsin C were profiled with a positional scanning synthetic combinatorial library. The S1 preferences bore close similarity to those of other C1-family cysteine peptidases. The S2 subsites of both DPAP1 and cathepsin C accepted aliphatic hydrophobic residues, proline, and some polar residues, yielding a distinct specificity profile. DPAP1 efficiently catalyzed the hydrolysis of several fluorogenic dipeptide substrates; surprisingly, however, a potential substrate with a P2-phenylalanine residue was instead a competitive inhibitor. Together, our biochemical data suggest that DPAP1 accelerates the production of amino acids from hemoglobin by bridging the gap between the endopeptidase and aminopeptidase activities of the food vacuole. Two reversible cathepsin C inhibitors potently inhibited both recombinant and native DPAP1, thereby validating the use of recombinant DPAP1 for future inhibitor discovery and characterization.
Keywords: malaria, hemoglobin, cathepsin, exopeptidase, vacuole
1. INTRODUCTION
Human malaria is caused by five species of the genus Plasmodium, with most of the 1–2 million annual deaths attributable to P. falciparum. The pathology of malaria arises from infection of host erythrocytes by the parasite. One intriguing feature of the host cell-parasite relationship during this stage is the endocytosis and catabolism of up to 75% of erythrocyte hemoglobin by P. falciparum [1, 2]. Hemoglobin is taken up through the cytostome and delivered to an acidic compartment called the food vacuole or digestive vacuole. Three classes of endopeptidases have been implicated in the hydrolysis of globin into oligopeptides in the food vacuole: aspartic proteases (plasmepsins I, II and IV and the active-site variant histo-aspartic protease), cysteine proteases (falcipain-2, -2′ and -3) and a metalloprotease (falcilysin) [3]. Inhibitors of the first two classes kill parasites and, in the case of cysteine protease inhibitors, lead to the accumulation of undegraded hemoglobin in the food vacuole [4]. In light of this evidence, enzymes that catalyze hemoglobin catabolism are regarded as attractive targets for new anti-malarial drugs.
The endopeptidases described above do not efficiently release small peptides or amino acids from globin oligopeptides; rather, exopeptidases are required for this task. A cysteine exopeptidase termed dipeptidyl aminopeptidase 1 (DPAP1) is located in the P. falciparum food vacuole [5]. We hypothesize that this enzyme reduces the oligopeptide products of endopeptidase cleavage to dipeptides, thereby generating substrates for the vacuolar aminopeptidase PfA-M1 [6]. Attempts to disrupt the DPAP1 gene have been unsuccessful, which suggests that the enzyme makes an important contribution to hemoglobin catabolism during the intraerythrocytic cycle [5]. Although DPAP1 is one of three related DPAP enzymes encoded in the parasite genome, current evidence suggests that only DPAP1 resides in the food vacuole. In contrast to DPAP1, DPAP3 is expressed late in the asexual replication cycle [7]. Abrogation of DPAP3 activity with a specific inhibitor implicates this enzyme in parasite egress [7]. DPAP2 (gene ID PFL2290w) does not appear to be expressed during asexual erythrocytic replication (unpublished observations). Together, these results suggest that the design of inhibitors that block both DPAP1 and DPAP3 could be a desirable anti-malarial strategy as the simultaneous impairment of two distinct, critical pathways would not only kill the parasite but could also delay the onset of resistance.
Mammals possess a single DPAP1 ortholog, termed cathepsin C or dipeptidyl peptidase I (to avoid confusion with DPAP1, the mammalian enzyme is referred to here as cathepsin C). Cathepsin C has two distinct roles. First, it contributes to general protein catabolism within the lysosome. Second, the enzyme is found in secretory granules of immune effector cells (cytotoxic lymphocytes, mast cells and neutrophils) where it activates granular serine proteases by removing an inhibitory N-terminal dipeptide sequence [8–10]. Its role as a mediator of inflammatory processes has made cathepsin C an appealing target for the development of drugs against some inflammation-based pathologies [11, 12].
Cathepsin C and DPAP1 are members of the large C1 family of cysteine endo- and exopeptidases, of which papain is the archetypal member. Papain is comprised of a prodomain, which is proteolytically removed to generate the mature enzyme, and two catalytic domains. In the case of cathepsin C, the polypeptide of the catalytic domains is cleaved into “heavy” and “light” chains. An additional domain, termed the exclusion domain or the residual pro-part, is found in cathepsin C and its orthologs and positions substrates such that peptide bond hydrolysis occurs between the second and third residues [13, 14]. Two other structural features, tetramerization and binding of a monovalent anion in the S2 pocket, differentiate cathepsin C from other C1-family exo- and endopeptidases [13, 14]. (The nomenclature of Schechter and Berger [15] is used here to identify substrate residues (P1, P2 etc.) and the corresponding enzyme subsites (S1, S2 etc.) with which they interact.) While it is reasonable to presume that these latter two variations on the papain scaffold enhance the ability of cathepsin C to carry out its biological roles, their contribution to cathepsin C function remains enigmatic.
The aim of the studies presented here is to elaborate the biochemical properties of DPAP1 for the purpose of further understanding its role in the malaria parasite and of establishing the similarities and differences between this enzyme and its human homolog cathepsin C. Toward this goal, we have compared the oligomerization state, chloride activation and pH-activity relationship of DPAP1 to those of cathepsin C. We have also generated a detailed profile of the subsite specificities of both DPAP1 and cathepsin C for the two substrate residues upstream of the scissile bond. These specificities were then further defined through kinetic characterization of these enzymes with a set of P2-diverse fluorogenic dipeptide substrates and with two established cathepsin C inhibitors. To accomplish these studies, we have developed a method for the recombinant production and in vitro activation of DPAP1.
2. MATERIALS AND METHODS
2.1. Substrates and inhibitors
Phe-Arg-AMC and Arg-Arg-AMC were obtained from Bachem. To synthesize Val-Arg-ACC, Ile-Arg-ACC, Phe-Arg-ACC and Pro-Nle-ACC, N-fluorenylmethyloxycarbonyl-7-amino-4-carbamoylmethylcoumarin (N-Fmoc-ACC; a gift of Jonathan A. Ellman) was linked to Rink Amide AM polystyrene resin and the P1 residue was coupled to the ACC-resin as described [16]. The second Fmoc-protected amino acid was linked using standard N,N-diisopropylcarbodiimide/1-hydroxybenzotriazole coupling conditions. After Fmoc deprotection of the N-terminal amine with 20% piperidine, treatment with trifluoroacetic acid/water/triisopropylsilane (95:2.5:2.5) resulted in the cleavage of fully deprotected dipeptide-ACC substrates. Substrates were purified by high pressure liquid chromatography on a C18 reverse phase column with a gradient of 5 to 60% acetonitrile in water and 0.1 % trifluoroacetic acid. The purity of the collected fractions was assessed by liquid chromatography-mass spectrometry. All substrates were at least 95% pure.
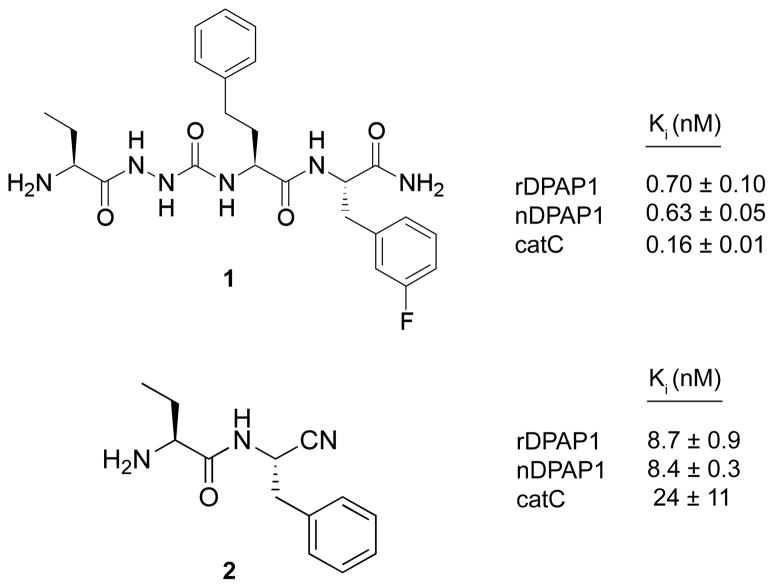
The inhibitor Pro-Arg-fluoromethylketone was custom synthesized by Enzyme Product Systems. Bestatin and trans-epoxysuccinyl-L-leucylamido(4-guanidino)butane (E-64) were obtained from Sigma-Aldrich and 4-(2-Aminoethyl)-benzenesulfonyl fluoride hydrochloride (AEBSF) was obtained from Roche Applied Science. Synthesis of the cathepsin C inhibitors 1-(2S-2-aminobutanoyl)-4-{2S-N-[2S-3-(m-fluorophenyl)propan-2-yl-amide]-4-phenylbutan-2-yl-amide}semicarbazide (compound 1 in Fig. 4) and S-2-aminobutyryl-phenylalanine-nitrile (compound 2 in Fig. 4) has been described previously [17, 18].
Figure 4.
Ki values for inhibition of native DPAP1 (nDPAP1), recombinant DPAP1 (rDPAP1) and cathepsin C (catC) by peptide analogs containing semicarbazide (1) and nitrile (2) pharmacophores.
2.2. Purification of native DPAP1
Native DPAP1 was purified from saponin-treated parasites as previously described [5] with the modification that two rather than three column steps were employed, resulting in higher yield. Briefly, clarified parasite lysate prepared in 20 mM bis-tris·HCl pH 6.0 was loaded onto a MonoQ 5/50 GL column (GE Biosciences) equilibrated in the same buffer. Bound protein was eluted with a linear gradient of 0 – 1 M NaCl. Fractions containing DPAP1 activity were pooled, concentrated and injected onto a Superdex 200 10/300 GL gel filtration column (GE Biosciences) equilibrated in 50 mM Tris-HCl pH 7.5, 200 mM NaCl and 1 mM EDTA. Active fractions were pooled, supplemented with 0.1% Triton X-100, 2 mM dithiothreitol and 10% glycerol, snap frozen, and stored at −80 °C.
2.3. Expression and purification of MBP-TEV protease
The S219V autoinactivation-resistant mutant of tobacco etch virus protease was expressed as a fusion with maltose binding protein from plasmid pRK1043 [19]. The plasmid was transformed into E. coli strain Rosetta2 (EMD Biosciences) and grown in Luria-Bertani broth containing 100 μg/mL ampicillin and 30 μg/mL chloramphenicol to an optical density at 600 nm of about 0.7, at which point the temperature was reduced to 30 °C and 1 mM isopropyl β-D-1-thiogalactopyranoside was added. Cells were harvested 4 hours later by centrifugation. MBP-TEV protease was purified by resuspending the cell pellet in 50 mM HEPES pH 7.5, 200 mM NaCl and 1 mM EDTA, 100 mM AEBSF and 1 mg/mL lysozyme and incubating on ice for 30 minutes. Cells were disrupted by sonication and the lysate was clarified by two rounds of centrifugation at 15 000 g for 10 minutes at 4 °C. The clarified lysate was loaded onto an amylose column (New England Biologicals) equilibrated in 50 mM HEPES pH 7.5, 200 mM NaCl, 1 mM EDTA. MBP-TEV protease was eluted with the same buffer supplemented with 1 M α-methylglucopyranoside. Fractions containing TEV protease were pooled, dialyzed overnight at 4 °C against 50 mM HEPES pH 8.2, 200 mM NaCl, 5 mM dithiothreitol, 1 mM EDTA and 10% glycerol, snap frozen in liquid nitrogen and stored at −80 °C. Because dithiothreitol inhibited DPAP1 activation, MBP-TEV protease was dialyzed against 50 mM HEPES pH 8.2, 200 mM NaCl, 1 mM reduced glutathione, 1 mM EDTA and 10% glycerol prior to addition to MBP-DPAP1-His6.
2.4. Cloning, expression, purification and activation of recombinant DPAP1-His6
The DPAP1 coding sequence was PCR amplified from the genomic DNA of P. falciparum clone 3D7 using primers 5′-GCACGGAATTCGAAAACCTGTATTTTCAGGATTTACCAACCCATGTAGAAAC (EcoRI site is in italics, TEV protease recognition sequence is underlined) and 5′-GCACGCTGCAGTTAATGATGATGATGATGATGATTTCCTAATTCCTTTTGCATTT (PstI site is in italics, hexahistidine tag is underlined). The PCR product was digested with EcoRI and PstI and cloned into the same sites in pMAL-c2x (New England Biolabs) which generated the MBP-DPAP1-His6 chimera. Coding sequences were verified by DNA sequencing.
The MBP-DPAP1-His6 expression plasmid was transformed into a trxB/gor strain of E. coli (Rosetta-gami 2; EMD Biosciences). Bacteria were grown at 37 °C in Luria-Bertani broth containing 100μg/mL ampicillin to an optical density at 600 nm of about 0.7, at which point the temperature was reduced to 25 °C. After 20 minutes, protein expression was induced by adding 0.3 mM isopropyl β-D-1-thiogalactopyranoside and shaking for 6 hours. Bacteria were collected by centrifugation and stored at −80 °C.
To purify MBP-DPAP1-His6, pellets of induced E. coli were resuspended in 15 mL of 20 mM NaH2PO4 pH 7.5, 500 mM NaCl, 30 mM imidazole (immobilized metal affinity chromatography (IMAC) buffer) containing 1 mg/mL hen egg white lysozyme and incubated on ice for 30 minutes. Bacteria were disrupted by sonication and cell debris was removed by two rounds of centrifugation at 20,000 g for 40 minutes at 4 °C. The cleared supernatant was loaded onto an Ni(II)-charged immobilized metal affinity column equilibrated in IMAC buffer. The column was washed extensively and bound protein was eluted with a gradient of 30 – 500 mM imidazole. Fractions containing MBP-DPAP1-His6 were pooled and stored at 4 °C.
Cleavage of MBP from MBP-DPAP1-His6 was achieved by incubating MBP-DPAP1-His6 with purified MBP-TEV protease (100 ng/μL) overnight at room temperature in 10 mM Tris-HCl pH 8, 1 mM reduced glutathione, 0.3 mM oxidized glutathione and 0.5 mM EDTA. To separate DPAP1-His6 from cleaved MBP, uncleaved MBP-DPAP1-His6 and MBP-TEV protease, amylose resin (New England Biolabs) was added, the mixture was rocked gently for 30 minutes at room temperature and the supernatant containing DPAP1-His6 was recovered by centrifugation. To excise the internal DPAP1 proregion, bovine pancreatic trypsin (Sigma, T1426) was added at a concentration of 1.5 μg/mL for 90 minutes at room temperature after which trypsin was inactivated by adding 0.5 mM AEBSF. DPAP1-His6 was separated from trypsin and peptide fragments by IMAC as described above. Active fractions were pooled, concentrated and purified on a Superdex 200 gel filtration column equilibrated with 50 mM Tris-HCl pH 8, 200 mM NaCl and 1 mM EDTA. Fractions with active enzyme were pooled, supplemented with 10% glycerol and 2 mM dithiothreitol, snap frozen in liquid nitrogen and stored at −80 °C. Upon thawing, Triton X-100 was added at 0.1% to preserve full activity. The yield of purified, activated recombinant DPAP1 was about 30 μg per liter of induced E. coli culture. Immunoblot analysis of recombinant DPAP1 was accomplished using a monoclonal antibody that recognizes the exclusion domain (304.4.2.2) and an anti-peptide polyclonal antibody (1502) that recognizes a 14-amino acid peptide sequence in the heavy chain [5].
2.5. Trypsin activity assays
Potential trypsin contamination of purified recombinant DPAP1 was assayed against the endopeptidase substrate N-(carbobenzyloxy)-L-phenylalanyl-L-arginyl-7-amido-4-methylcoumarin (100 μM) in an optimized trypsin assay containing 50 mM Tris-HCl pH 8.0, 20 mM CaCl2 or in the standard DPAP1 assay (section 2.9). Two microliters of recombinant DPAP1 stock were assayed, which represents a ten-fold excess over the amount of enzyme typically added to DPAP1 assays.
2.6. Expression and purification of human cathepsin C
Activated recombinant human cathepsin C was purified from baculovirus-infected insect cell culture medium as previously described [20]. The enzyme was stored in 50% glycerol at −20 °C.
2.7. Protein quantitation
Recombinant DPAP1 was quantified using the fluorescence-based NanoOrange assay (Invitrogen) according to the manufacturer’s instructions. Fluorescence values of samples, matched buffer blanks and bovine serum albumin standards were read on a Spectramax M5e microplate fluorometer (Molecular Devices) with excitation at 470 nm, emission at 590 nm and a 570 nm cutoff filter. The amount of native DPAP1 in a partially purified preparation was estimated by immunoblotting. Known amounts of recombinant protein (20 to 80 ng) were electrophoresed along with native DPAP1 on a 12% SDS-polyacrylamide gel. After blotting to nitrocellulose, DPAP1 was detected using the monoclonal antibody 304.2.4.4 ([5]; 1:100 dilution), an alkaline phosphatase-conjugated secondary antibody and ECL Plus. Fluorescence signal was recorded on a STORM 840 phosphorimager. Peak volumes were quantified using ImageQuant TL version 7.0 software (GE Bioscences). Because the monoclonal antibody recognizes two bands in recombinant DPAP1 (Fig. 1), the volumes of these bands were summed to obtain a final integrated volume. The amount of native enzyme was determined from a standard curve plotted as peak volume vs. amount of rDPAP1. The R2 value for the linear fit was 0.98.
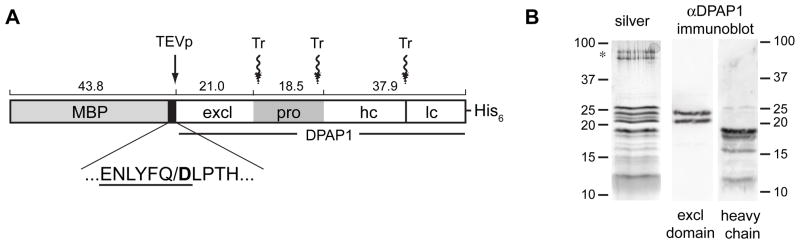
Figure 1.
(A) Schematic diagram of the recombinant MBP-DPAP1-His6 fusion. The white boxes indicate those regions of DPAP1 found in the mature protein, namely the exclusion domain (excl) and catalytic region, the latter being proteolytically cleaved to generate a heavy chain (hc) and a light chain (lc). The gray box within the DPAP1 sequence is the proregion (pro), the boundaries of which are not precisely known but were estimated from a sequence alignment with cathepsin C [5]. The black box represents the linker between MBP and DPAP1. The sequence below shows the TEV protease recognition sequence (underlined) with the cleavage site indicated by “/”. The resulting N-terminal Asp residue is shown in bold. The approximate positions of trypsin (Tr) cleavage are indicated with wavy arrows. Additional/alternate cleavage sites (not shown) give rise to size heterogeneity in the mature DPAP1 polypeptides. The hexahistidine tag is retained in the mature recombinant protein. Predicted polypeptide sizes are shown above the boxes in kDa. (B) Analysis of purified rDPAP1 by reducing SDS-PAGE. Left panel: silver stain. Contaminating proteins (probably keratins) are indicated with an asterisk. Right panels: immunoblots using antibodies that recognize the exclusion domain (“excl domain”) or the heavy chain. Sizes of markers are given in kDa.
2.8. Gel filtration analysis of quaternary structure
Native DPAP1 was purified as described above, dialyzed into 50 mM sodium-MES pH 6.0, 200 mM NaCl, 1 mM EDTA overnight at 4 °C and then injected onto a Superdex 200 gel filtration column equilibrated in the same buffer. Fractions of 0.33 mL were collected and assayed for DPAP1 activity as described in section 2.9. Recombinant cathepsin C was diluted into gel filtration buffer, concentrated and was analyzed as described for DPAP1. Molecular masses were estimated from a calibration curve generated with ferretin (440 kDa), catalase (232 kDa), aldolase (158 kDa), albumin (67 kDa), ovalbumin (43 kDa), chymotrypsinogen A (25 kDa) and ribonuclease A (14 kDa). The void volume was estimated with blue dextran 2000.
2.9. DPAP1 activity assays and effects of ionic strength and chloride
DPAP1 activity was routinely assayed at 25 °C in a volume of 200 μL in 50 mM sodium MES pH 6.0, 30 mM NaCl, 2 mM dithiothreitol, 1 mM EDTA, 0.1% Triton X-100, 10 μM bestatin (to inhibit any remaining aminopeptidase activity in partially purified native DPAP1) and, unless otherwise noted, 100 μM Pro-Arg-AMC. Changes in fluorescence were monitored with a Victor3 microplate fluorometer (PerkinElmer) with an excitation filter of 380 nm (10 nm bandwidth) and an emission filter of 460 nm (25 nm bandwidth).
To evaluate the effect of chloride on DPAP1 activity, chloride was first depleted from native and recombinant DPAP1 by dialysis against 50 mM sodium MES pH 6.0, 1 mM EDTA and 0.1% Triton X-100 in Slide-A-Lyzer MINI dialysis cassettes with a 10,000 molecular weight cutoff (Pierce). Alternately, native DPAP1 was prepared by carrying out the gel filtration step of the purification (section 2.2) in chloride-free buffer (50 mM sodium HEPES pH 7.5, 100 mM Na2SO4 and 1 mM EDTA). DPAP1 assays contained 50 mM sodium MES pH 6.0, 2 mM dithiothreitol, 1 mM EDTA, 0.1% Triton X-100 and 10 μM bestatin. The chloride concentration was varied from 0 – 80 mM by adding NaCl. The ionic strength was held constant by adding Na2SO4 such that the ionic strength contributed by NaCl and Na2SO4 totaled 100 mM in all assays. Prolyl-norleucyl-ACC, which lacks a chloride counterion, was used as the substrate (100 μM).
2.10. S1 and S2 profiling with a positional scanning library
The 40 members of the dipeptide-ACC positional scanning library were assayed at 25 °C in 50 mM sodium MES pH 6.0, 30 mM NaCl, 2 mM dithiothreitol, 1 mM EDTA and 10 μM substrate (diluted from a 1 mM stock in dimethylsulfoxide) in a volume of 100 μL. Since each substrate mixture consisted of 20 compounds, each compound was present in the assay at 0.5 μM. In assays containing partially purified native DPAP1, 1 μM bestatin and 1 μM E-64 were added to suppress aminopeptidase and falcipain activities, respectively. In addition, background rates were assessed using control assays containing native DPAP1 that had been pre-treated for 15 minutes with the irreversible inhibitor Pro-Arg-fluoromethylketone. For recombinant DPAP1 and cathepsin C, no background rate adjustment was made. Assays were conducted in duplicate. Fluorescence values were read in 96 well half-area microplates as described in section 2.9.
2.11. Kinetic analysis of DPAP1 and cathepsin C activity and inhibition
Steady-state kinetic parameters for hydrolysis of fluorogenic dipeptide-AMC or -ACC substrates were determined at 25 °C by assaying DPAP1 and cathepsin C in 50 mM sodium MES pH 6.0, 2 mM dithiothreitol and 1 mM EDTA. The final concentration of chloride, including that carried over with the enzyme and dipeptide-AMC substrates, was maintained at 30 mM by addition of NaCl. Native DPAP1 assays also contained 10 μM bestatin to suppress any contaminating aminopeptidase activity. Substrate concentrations were varied from approximately 0.2Km to 5Km. Enzyme concentrations were 0.3 nM native DPAP1, 0.8 nM rDPAP1 and 0.06 – 0.3 nM cathepsin C. At these concentrations, substrate consumption was less that 10% over the course of the assay. Fluorescence values were converted to AMC concentrations by reference to a standard AMC solution. Rates from dipeptide-ACC substrates were adjusted to reflect the 2.8-fold higher fluorescence of ACC over that of AMC [21]. Assays were conducted in triplicate. Rates were determined from the linear portions of the progress curves. In the case of DPAP1, rates were measured following a ~15 minute non-linear segment that may reflect reductive activation of the enzyme. Rates were fit by non-linear regression to the Michaelis-Menten equation ν = Vs/(Km + s) using Kaleidagraph 4.1 (Synergy Software) where V is the limiting velocity and s is substrate concentration. kcat was calculated from the relationship V = kcat[E]. The Ki for inhibition of DPAP1 by Phe-Arg-ACC was determined by the Dixon method [22]. Methods for the determination of Ki values for the inhibitors in Fig. 4 are provided as Supplementary Methods.
To assess the effects of pH on catalytic efficiency, assays were carried out as described in the above paragraph with PR-AMC (0 – 500 μM) as substrate. Buffering capacity was provided by 50 mM sodium acetate (pH 4.5 and 5.0), sodium MES (pH 5.5, 6.0 and 6.5) or sodium HEPES (pH 7.0). Buffer pKa values were adjusted for an ionic strength of 100 mM and a temperature of 25 °C [23]. Contributions of ionic buffer components to the ionic strength were calculated using the Henderson-Hasselbalch equation. Total ionic strength of the reactions was maintained at 100 mM by adding Na2SO4. Kinetic parameters were obtained from non-linear regression fits to the Michaelis-Menten equation. No substrate inhibition was observed at any pH value.
3. RESULTS
3.1. Purification of native DPAP1
To obtain material for biochemical characterization, we first attempted to purify DPAP1 from saponin-treated trophozoite-stage parasites. As described previously [5], a three-column protocol yielded highly purified native DPAP1; however, yields of enzyme were low and activity was unstable, probably due to the adsorption of the minute quantities of the enzyme to surfaces. By modifying the purification protocol to consist of two (instead of three) chromatographic steps, sufficient native enzyme for the studies described here was obtained; however, the resulting enzyme preparation contained many other polypeptides (data not shown).
3.2. Generation and activation of recombinant DPAP1
To produce greater quantities of DPAP1 at higher purity, we have developed a method for the production of recombinant enzyme by expression in Escherichia coli. We took into consideration three factors that would likely be important for generating recombinant DPAP1 with native-like qualities. First, expression of the protein in soluble form would be required for proteolytic removal of the proregion in vitro. Second, the amino terminus of the exclusion domain would have to begin with a specific sequence (Fig. 1A). Crystal structures of cathepsin C reveal that the amino-terminal Asp residue of the exclusion domain forms a critical interaction through its sidechain carboxylate with the amino group of the substrate [13, 14, 24]. The homologous residue in DPAP1 (Asp28) likely plays an identical role. Thus, the exclusion domain of recombinant DPAP1 should begin at Asp28. Third, the internal proregion (also called the activation peptide) would have to be proteolytically removed to fully activate the enzyme (Fig. 1A). In the case of recombinant cathepsin C, excision of the proregion results in a 2000-fold increase in activity [20].
Prior reports of expression in E. coli of soluble, active P. falciparum peptidases as fusions to the C-terminus of maltose binding protein (MBP) [25, 26] motivated us to attempt expression of an MBP-DPAP1 chimera (Fig. 1). A hexahistidine tag was placed at the C-terminus to permit a second mode of affinity purification. To generate the presumed native N-terminal sequence of the exclusion domain, a modified tobacco etch virus (TEV) protease cleavage site was introduced between MBP and DPAP1 in which the preferred Gly or Ser residue in the last position is replaced with Asp (Fig. 1A). Cleavage of this modified sequence would result in an exclusion domain N-terminus that begins with Asp28. Aspartate is tolerated in the last position of the TEV protease cleavage sequence, although cleavage efficiency is somewhat reduced [27].
MBP-DPAP1-His6 was expressed in soluble form in E. coli and was purified by immobilized metal affinity chromatography (IMAC). Cleavage at the TEV protease recognition site between MBP and DPAP1 was carried out using an MBP-TEV protease fusion [19], which permitted the separation of DPAP1-His6 from MBP-containing species (cleaved MBP, uncleaved MPB-DPAP1-His6 and MBP-TEV protease) by interaction of the latter with amylose resin. Most of the cleaved DPAP1-His6 sedimented upon high-speed centrifugation, a result that is likely due to the formation of aggregates of incorrectly folded protein. The formation of disulfide bonds may be a limiting factor in the folding of DPAP1 in the E. coli cytosol, as soluble enzyme was only obtained when MPB-DPAP1-His6 was produced in a trxB/gor E. coli strain that yields higher levels of disulfide bond formation in the cytosol [28]. Based on a sequence alignment with cathepsin C, three of the five disulfide bonds in the latter protein are expected to be conserved in DPAP1 (not shown).
The identities of the endoproteases that excise the DPAP1 proregion and cleave the catalytic region into heavy and light chains in vivo are unknown. We attempted to mimic the in vivo proteolytic events by treating recombinant DPAP1 with commercially-available peptidases. Trypsin and papain were chosen as one (trypsin) is relatively selective at the P1 position while the other (papain) is not; thus, it was expected that each would produce a distinct spectrum of cleavages in recombinant DPAP1. In addition, papain has been successfully used to activate recombinant cathepsin C [29]. Incubation of DPAP1-His6 with either protease resulted in a moderate (2 to 3-fold) increase in activity. When the kinetics of hydrolysis of the DPAP1 substrate Pro-Arg-AMC [5] by trypsin-treated DPAP1-His6, papain-treated DPAP1-His6 and native DPAP1 was compared, the trypsin-activated enzyme exhibited a hyperbolic relationship between initial velocity and substrate concentration, like that of the native enzyme (Fig. S1). In contrast, the papain-activated enzyme reproducibly exhibited substrate inhibition (Fig. S1). Thus, we selected the trypsin-activated DPAP1-His6 (referred to from here on as “rDPAP1”) for further characterization. rDPAP1 was further purified by metal affinity and gel filtration chromatography.
Highly purified rDPAP1 contains an ensemble of polypeptides with sizes between ~12 and ~27 kDa (Fig. 1B). Immunoblotting with antibodies that recognize the exclusion domain or the heavy chain of the catalytic region revealed heterogeneous processing of rDPAP1 by trypsin (Fig. 1B). The exclusion domain appeared as a doublet of 21 and 23 kDa species. The heavy chain was represented by four major polypeptides ranging in size from 12 to 18 kDa. Other bands appearing in this size range could be light chain polypeptides, for which an antibody is not available. A time course of trypsin activation indicated that, under the conditions employed here, the observed ensemble of polypeptides was stable over time (Fig. S2), which suggests that the size heterogeneity of the exclusion domain and heavy chain is not simply due to incomplete proteolysis. The ~27 kDa band detected by silver staining was not recognized by either antibody (Fig. 1B). This polypeptide is unlikely to correspond to the excised prodomain (predicted size of 18.5 kDa) but could be a C-terminal fragment of DPAP1 (the heavy chain antibody recognizes a peptide epitope 31 kDa from the C-terminus). Since trypsin is predicted to have a molecular mass of ~27 kDa, we assessed whether trypsin activity is present in the rDPAP1 preparation. Using a fluorogenic trypsin substrate and 10-fold more protein than typically included in a DPAP1 assay, no trypsin activity could be detected under the conditions of the standard DPAP1 assay or of an optimized trypsin assay (see Materials and Methods section 2.5). Therefore we conclude that the 27 kDa band corresponds to a C-terminal fragment of DPAP1 or to inactive trypsin.
3.3. DPAP1 quaternary structure
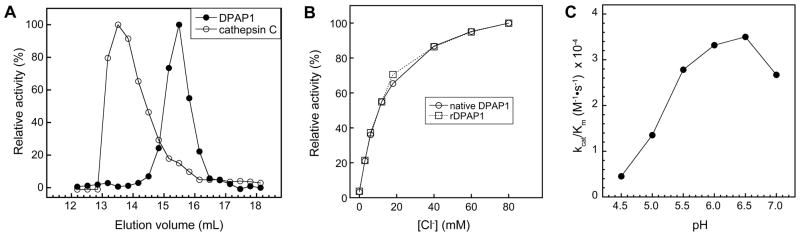
One feature that distinguishes mature mammalian cathepsin C from other C1-family cysteine peptidases is its tetrameric quaternary structure consisting of a dimer of dimers [13, 14]. To determine whether DPAP1 forms a tetramer at physiologically relevant pH values, the apparent molecular mass of the native enzyme was determined by gel filtration chromatography at pH 6 and was found to be 51 kDa (Fig. 2A). Because the sites of in vivo proenzyme cleavage are not known, the predicted molecular mass of the native enzyme cannot be precisely calculated; however, if the boundaries of the proenzyme are assumed to be similar to those of cathepsin C [5], a molecular mass of 59 kDa can be estimated for mature native DPAP1. Thus, the migration of the enzyme on a gel filtration column is consistent with a monomeric rather than a tetrameric structure. In contrast, under the same elution conditions recombinant cathepsin C migrated with an apparent molecular mass of 153 kDa (Fig. 2A), which is close to the value of 140 kDa that was previously reported for this enzyme [20].
Figure 2.
(A) Elution of native DPAP1 (filled circles) and cathepsin C (open circles) from a Superdex 200 gel filtration column in 50 mM Na-MES pH 6, 200 mM NaCl and 1 mM EDTA. For each protein, activities in fractions are shown as percent relative activity with the highest activity set to 100%. (B) Activation of native (circles) and recombinant DPAP1 (squares) by chloride at pH 6.0. Ionic strength was held constant with Na2SO4. Activity values are expressed as relative activity with the highest activity set at 100%. Data points are the average of duplicate assays. (C) The catalytic efficiency (kcat/Km) of recombinant DPAP1 within the pH range 4.5 to 7.0 with the substrate Pro-Arg-AMC. Chloride concentration (30 mM) and total ionic strength (100 mM) were held constant at all pH values.
3.4. Activation of DPAP1 by chloride
Cathepsin C appears to be unique among C1-family cysteine peptidases in exhibiting activation by univalent anions, of which halide ions such as chloride are the most potent activators [30, 31]. To assess whether such activation is a conserved feature of cathepsin C-like dipeptidases, we examined the effects of chloride concentration on DPAP1 activity. Preliminary experiments indicated that DPAP1 activity was sensitive to changes in ionic strength (data not shown); therefore, in the analysis of chloride activation, ionic strength was held constant with sodium sulfate (the divalent sulfate anion does not activate cathepsin C [31]). To deplete chloride from stock solutions of DPAP1, native and recombinant enzyme were extensively dialyzed against chloride-free buffer. In both cases, strong activation was observed upon the addition of millimolar concentrations of chloride to enzyme assays (Fig. 2B). Around 3% activity was observed in the absence of added chloride (relative to activity in 80 mM Cl−). To determine whether this was due to incomplete dialysis of chloride from the DPAP1 stock, native DPAP1 was purified by gel filtration chromatography in chloride-free buffer. This preparation retained a similar level of residual activity (data not shown). These results are most consistent with a hyperbolic activation mechanism for chloride ion in which the enzyme does not exhibit an absolute chloride requirement for either substrate binding or turnover [32]. However, it is formally possible that chloride is essential for binding and/or turnover (i.e., chloride is an essential activator) but has not been completely eliminated from the assay solution or from the enzyme. For practical purposes, a chloride concentration of 30 mM was found to be an optimal balance between the activation of DPAP1 by chloride and the suppression of activity as ionic strength increases. Thus, this concentration was routinely used in DPAP1 assays.
3.5. DPAP1 is an efficient catalyst at acidic pH
If DPAP1 is a catalyst of peptide bond hydrolysis in the food vacuole, it should function efficiently at the acidic lumenal pH of this organelle. The efficiency of rDPAP1 catalysis of Pro-Arg-AMC hydrolysis in the pH range 4.5 to 7.0 was determined at constant chloride concentration and ionic strength (Fig. 2C). The pH-activity profile has an apparent bell shape that resembles those of cathepsin C [33–35]. Catalytic efficiency (kcat/Km) was highest in the pH range 6.0 – 6.5 but was also substantial at pH 5.5, which is within the range of several estimates of food vacuole pH [36–39]. These data are therefore consistent with a catalytic role for DPAP1 in peptide hydrolysis in the malarial food vacuole. The bell-shaped pH-activity profile of C1 family peptidases such as papain has been attributed to protonation of the active site cysteine (acidic arm) and deprotonation of the histidine sidechain that ion pairs with the catalytic cysteine (basic arm) [40]. While that may be the case for DPAP1 and cathepsin C as well, it should be noted that the substrate contains a free α-amino group that ionizes with a pKa of ~8 [41] and that the affinity of chloride for cathepsin C shows a pH dependence, with higher affinity at lower pH [42].
3.6. S1 and S2 subsite specificities of DPAP1 and cathepsin C
The specificities of peptidases are dictated in large part by the interactions of substrate sidechains with cavities or surfaces of the enzyme, which are referred to as subsites. Structures of C1-family cysteine peptidases complexed with inhibitors have revealed that enzyme interactions are most substantial with the P1, P2 and P3 residues on the N-terminal side of the scissile bond and with the P1′ and P2′ residues on the C-terminal side [43, 44]. In the case of the dipeptidyl aminopeptidases DPAP1 and cathepsin C, the S3 subsite is occluded by the exclusion domain; therefore, the S1 and S2 subsites are major determinants of substrate specificity. Using a positional scanning synthetic combinatorial library, we have profiled the S1 and S2 subsite specificities of DPAP1 and human cathepsin C to investigate: i) whether DPAP1 accepts a wide range of amino acid residues in the first two positions of the substrate, which would support a general role for DPAP1 in peptide catabolism in the P. falciparum food vacuole; and ii) whether the subsite preferences of DPAP1 have diverged from those of cathepsin C, perhaps indicating functional specialization of the former as an enzyme dedicated to the catabolism of hemoglobin peptides. The latter point is also clearly of interest if we wish to design inhibitors that are specific for the parasite enzyme over that of the human host.
Positional scanning libraries (PSL) are powerful tools with which to profile the subsite specificities of peptidases [45]. Here, we have employed a fluorogenic dipeptidyl-7-amido-4-carbamoylcoumarin (dipeptide-ACC) PSL (Fig. 3A) to explore the S1 and S2 subsite preferences of native and recombinant DPAP1 and of human cathepsin C. This PSL has been previously used to profile the specificities of the serine dipeptidyl peptidases II, IV and VII [46]. The PSL is divided into two sub-libraries, the structures of which are depicted in Fig. 3A. In each sub-library, one position (P1 or P2) is varied to contain each of twenty amino acids (all of the standard proteinogenic amino acids except cysteine plus norleucine, which is abbreviated Nle here). The other position in the dipeptide is occupied by an equimolar mixture of these same 20 amino acids, which reduces the influence of sidechain interactions between adjacent substrate residues on the observed preferences [45]. We first compared the subsite preferences of native DPAP1 and cathepsin C. Then, to validate the use of rDPAP1 in biochemical studies, its specificity profile was compared to that of the native enzyme.
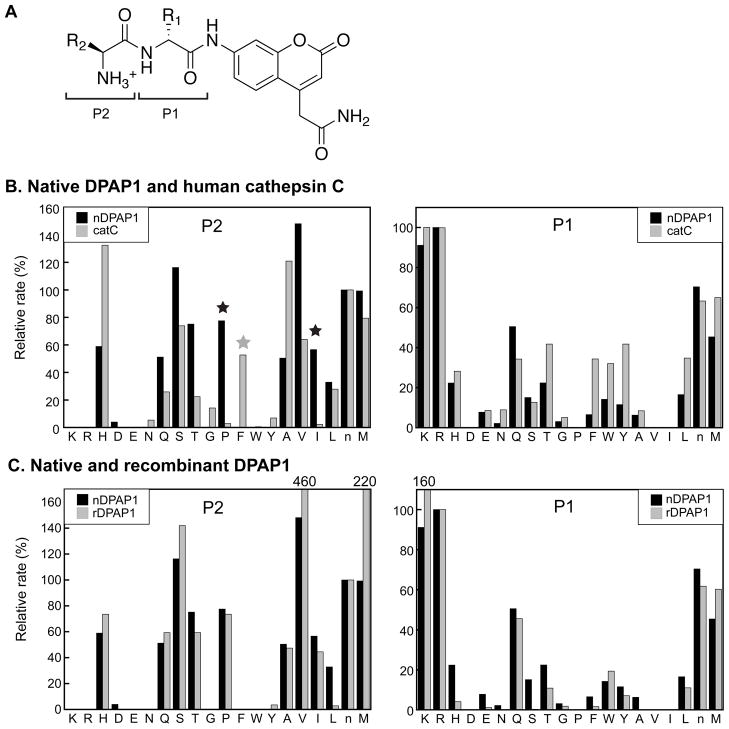
Figure 3.
S1 and S2 specificity profiles of DPAP1 and cathepsin C. (A) Structure of the dipeptidyl-ACC positional scanning library. The P1 and P2 residues are indicated. In the P1 sublibrary, the each member contains a defined residue at the P1 position (the identities of which are indicated in the x-axis labels of the right panels of (B) and (C)) and the P2 position consists of an equimolar mixture of 20 amino acids. In the P2 sublibrary, the P2 position is defined (indicated in the x-axis labels of the left panels of (B) and (C)) and the P1 position is an equimolar mixture. (B) S1 and S2 specificities of native DPAP1 (black bars) and cathepsin C (gray bars). Data are reported as relative rates with those for Arg (P1 dataset) and norleucine (P2 dataset) set to 100%. The P1 or P2 amino acid is indicated with single-letter code, with n representing norleucine. Black stars indicate members of the P2 sublibrary that are substantially better substrates for DPAP1 compared to cathepsin C; a grey star indicates the P2 sublibrary member that is a much better substrate for cathepsin C than DPAP1. Each data point is the average of duplicate assays. nDPAP1, native DPAP1; catC, cathepsin C. (C) Comparison of the S1 and S2 specificities of recombinant DPAP1 with those of the native enzyme. Normalization of rates was as in (B). Values of rates that are off-scale are indicated above the graphs. rDPAP1, recombinant DPAP1. Each data point is the average of duplicate assays.
The S1 subsite specificities of native DPAP1 and cathepsin C are shown in Fig. 3B (right panel). The relative preferences are similar: both enzymes prefer unbranched residues (Arg, Lys, Met and Nle) at this position. Consistent with previous observations with cathepsin C [31, 47], peptides with Ile or Pro in the P1 position were not hydrolyzed. As this is also the case for Val, it appears that β-branched amino acids are not readily accommodated in the S1 subsite of either enzyme. Acidic residues, Ala and Gly in the P1 position also made for poor substrates. The S2 subsite of cysteine peptidases is typically the most restrictive of the subsites upstream of the scissile bond [43, 44].
The S2 preferences of native DPAP1 and cathepsin C are shown in Fig. 3B, left panel. Aliphatic (but not aromatic) hydrophobic residues generally made for good substrates for both enzymes. DPAP1 and cathepsin C also exhibited a preference for the small hydrophilic residues Ser and Thr as well as the larger sidechains of Gln and His at the P2 position. Substrates with the cationic sidechains of Arg and Lys at the P2 position were not detectably hydrolyzed by either DPAP1 or cathepsin C, an observation that is consistent with previous reports for cathepsin C [31, 47, 48]. The specificity profiles for DPAP1 and cathepsin C appear to be distinct from those of other C1-family cysteine proteases for which PSL data are available (see Discussion).
Although the S2 specificities of DPAP1 and cathepsin C were broadly similar, there were a few P2 residues that were much more highly preferred by one enzyme. In the case of DPAP1, Pro and Ile were much more favorable substrates (in a relative sense) than they were with cathepsin C (Fig. 3B, left, black stars). On the other hand, a dipeptide containing a P2 Phe residue made for a good cathepsin C substrate but was not detectably hydrolyzed by DPAP1 (Fig. 3B, left, gray star). These data suggest that there are differences in the accommodation of certain substrate P2 residues within the S2 subsites of the two enzymes. This possibility was further explored with defined substrates (section 3.7).
The S1 and S2 specificities observed with native DPAP1 were reproduced with the recombinant enzyme (Fig. 3C, right and left panels respectively). Importantly, the S2 specificity profile noted above (aliphatic hydrophobic, small hydrophilic, Pro, Gln and His) was observed in the recombinant enzyme. For unknown reasons, a few residues appeared to be much more highly preferred by the recombinant enzyme compared to native DPAP1; for example, relative rates with P2 Val and Met were 3.1- and 2.2-fold higher for recombinant enzyme over native (Fig. 3C). Kinetic analysis of a dipeptide substrate incorporating a P2 Val residue (section 3.7) suggests that these outliers are probably in error.
3.7. Kinetic analysis of DPAP1 and cathepsin C
To further investigate the differences in S2 substrate specificities between DPAP1 and cathepsin C noted above, we determined the kinetics of hydrolysis of a panel of fluorogenic dipeptide substrates with a fixed P1 Arg residue and variable P2 residues. Arg was selected as the P1 residue because it is highly preferred by both enzymes. P2 residues were selected that, based on the PSL results, appeared to be preferred by: i) both enzymes (Val); ii) DPAP1 only (Ile, Pro); or iii) cathepsin C only (Phe), or were disfavored by both enzymes (Arg).
rDPAP1 catalyzed the hydrolysis of the P2-Pro, -Val and -Ile substrates (Table 1). The rank order of catalytic efficiency (kcat/Km) followed that predicted from the PSL data: Val>Pro>Ile. The Km and kcat values varied much more widely for these three substrates than the kcat/Km values; for example, Ile-Arg-ACC exhibited a 5-fold lower Km and a 10-fold lower kcat than Val-Arg-ACC, resulting in a two-fold change in kcat/Km. To validate these results obtained with rDPAP1, the kinetics of hydrolysis of Pro-Arg-AMC and Val-Arg-ACC by partially purified native DPAP1 was examined. Differences in Km values for the two enzymes were within experimental error in both cases (Table 1). The kcat values (and therefore kcat/Km values) were 3- to 5-fold lower for recombinant DPAP1; possible reasons for this are given in the Discussion. Consistent with the PSL data, no hydrolysis was observed for the substrates Phe-Arg-ACC and Arg-Arg-ACC at concentrations up to 500 μM by either recombinant or native DPAP1.
Table 1.
Kinetic parameters for hydrolysis of fluorogenic dipeptide substrates.
| Substrate | Enzyme | Km (μM)a | kcat (s−1)a | kcat/Km (M−1•s−1)a | Ki (μM) |
|---|---|---|---|---|---|
| Pro-Arg-AMC | nDPAP1 | 84 ± 9 | 6.2 ± 0.4 | (7.4 ± 0.2) × 104 | |
| rDPAP1 | 79 ± 2 | 1.8 ± 0.1 | (2.3 ± 0.1) × 104 | ||
| cathepsin C | 130 ± 10 | 490 ± 10 | (3.6 ± 0.1) × 106 | ||
| Val-Arg-ACC | nDPAP1 | 21 ± 2 | 3.5 ± 0.1 | (1.7 ± 0.1) × 105 | |
| rDPAP1 | 20 ± 1 | 0.72 ± 0.02 | (3.7 ± 0.1) × 104 | ||
| cathepsin C | 51 ± 8 | 180 ± 10 | (3.6 ± 0.3) × 106 | ||
| Ile-Arg-ACC | rDPAP1 | 3.9 ± 0.2 | 0.072 ± 0.003 | (1.9 ± 0.1) × 104 | |
| cathepsin C | 28 ± 2 | 17 ± 1 | (6.3 ± 0.4) × 105 | ||
| Phe-Arg-ACC | nDPAP1 | NHb | NH | NH | |
| rDPAP1 | NH | NH | NH | 2.2 | |
| cathepsin C | NDc | ND | ND |
Values are reported as means and standard deviations from triplicate data sets for each substrate/enzyme combination.
NH, no hydrolysis.
ND, kinetic parameters were not determined due to substrate inhibition.
Cathepsin C catalyzed the hydrolysis of substrates with P2-Phe, -Pro, -Val and -Ile substrates. For the latter three, the rank order of catalytic efficiency was Val, Pro > Ile (Table 1). Values of kcat/Km were in the range (106 – 107 M−1•s−1) of those reported previously for similar substrates [47]; these values were up to 50-fold greater than those of DPAP1. Surprisingly, the expected selectivity for a P2-Val over a P2-Pro residue was not apparent in the context of a P1-Arg residue. In the case of Phe-Arg-ACC, substrate inhibition was observed (Fig. S3); therefore, kinetic constants for Phe-Arg-ACC turnover were not determined. As with DPAP1, no turnover of Arg-Arg-AMC was observed.
Given the inhibition of cathepsin C observed with Phe-Arg-ACC, we investigated whether this compound could inhibit DPAP1. Phe-Arg-ACC was a potent inhibitor of the hydrolysis of Pro-Arg-AMC catalyzed by rDPAP1. Dixon analysis yielded a Ki of 2.2 μM and indicated that the inhibition was purely competitive (data not shown). Thus, Phe-Arg-ACC appears to interact with DPAP1 in a conformation that is unproductive for catalysis and that blocks the binding of other substrates.
3.8. Inhibition of DPAP1 by semicarbazide- and nitrile-containing peptide analogs
Peptide analogs containing semicarbazide and nitrile pharmacophores have been shown to be effective reversible inhibitors of C1-family cysteine proteases including cathepsin C [11, 12, 17]. To further validate recombinant DPAP1 as a reagent for small-molecule inhibitor discovery, we have determined the inhibition constants of two established cathepsin C inhibitors (Fig. 4; [11, 17]) for native and recombinant DPAP1. Both were potently inhibited with sub-nanomolar Ki values for the semicarbazide 1 and single-digit nanomolar Ki values for the peptide nitrile 2 (Fig. 4 and Fig. S4). Importantly, for each inhibitor the differences in the Ki values for native and recombinant DPAP1 were within experimental error. We also determined inhibition constants for cathepsin C under the assay conditions used for DPAP1. The semicarbazide 1 was a slow, tight-binding inhibitor of cathepsin C and was analyzed accordingly (see Supplementary Methods and Fig. S4). The Ki values of both inhibitors for cathepsin C (Fig. 4) were within an order of magnitude of the respective Ki values for DPAP1, an observation that underscores the potential difficulty of developing peptide-based compounds that specifically inhibit DPAP1 over its host homolog.
4. DISCUSSION
To obtain biochemical quantities of highly purified DPAP1, we developed a method for the production of recombinant DPAP1 in E. coli and for its processing and activation in vitro. Mature rDPAP1 was obtained by limited cleavage of proDPAP1 with bovine pancreatic trypsin which resulted in removal of the proregion between the exclusion domain and the catalytic domains as well as cleavage of the catalytic domains into heavy and light chains. Heterogeneity of the exclusion domain and heavy chain of rDPAP1 was observed following trypsin treatment. Alternate processing sites have been observed in the heavy chain of native DPAP1 [5] and in the exclusion domain of native cathepsin C [49], the functional significance of which is unknown. It was important to consider whether contaminating trypsin activity could confound the data obtained with rDPAP1. We were unable to detect trypsin activity using a trypsin substrate (which is a more sensitive probe of trypsin activity than the DPAP1 substrates used here) and 10-fold more protein than was typically added to DPAP1 assays. Thus, the presence of active trypsin, if any, in the rDPAP1 preparation appears to be below the limit of detection of our assays.
The biochemical attributes of rDPAP1 were validated against a partially purified preparation of native enzyme. Several key enzymatic properties were recapitulated with the recombinant enzyme, including activation by chloride ion, highly similar S1 and S2 subsite preferences, essentially identical Km values with two fluorogenic dipeptide substrates and nearly identical inhibition constants for two reversible inhibitors with distinct pharmacophores. The one substantive difference between them is the ~4-fold lower kcat values for rDPAP1. Possible explanations for this difference include inaccuracies in the immunoblot-based quantitation of native DPAP1, the presence of inactive enzyme in the recombinant preparation, or subtle structural changes deriving from differences in polypeptide processing in vivo and in vitro. With this one caveat in mind, the results presented here validate the use of recombinant DPAP1 for inhibitor screening and assessment.
A number of characteristics distinguish cathepsin C from its brethren in the C1 cysteine peptidase family: a homotetrameric structure, the presence of the exclusion domain, and activation by chloride. Our comparison of divergent (protozoan and metazoan) homologs sheds light on those features that are conserved (the presence of an exclusion domain and chloride activation) and highlights that which is unique to the mammalian enzyme (a tetrameric quaternary structure). The exclusion domain of cathepsin C is an autonomously-folding β-barrel with structural similarity to a metalloprotease inhibitor from Erwinia chrysanthemi [14]. It defines amino-dipeptidase activity by interacting with the amino terminus of the substrate and thereby preventing endopeptidase activity [13, 14, 24]. In mature DPAP1, the exclusion domain almost certainly serves the same purpose. The exclusion domain of cathepsin C also acts as a chaperone to facilitate folding of the catalytic region [50]. Whether it has a similar chaperone function in DPAP1 remains to be determined. The observation of chloride activation in DPAP1 suggests that this property was present in an ancestral cathepsin C pre-dating divergence of the protozoan and metazoan lineages. The activating chloride ion is most likely that observed at the bottom of the S2 pocket in the x-ray crystal structures of rat and human cathepsin C [13, 14]. Consistent with this notion, the one residue that co-ordinates the chloride ion with its sidechain, Tyr323, is conserved in DPAP1. It has been proposed that the bound chloride ion stabilizes helix 3 of cathepsin C by interacting with the helix dipole [13].
The proenzyme form of mammalian cathepsin C is a dimer that associates to form a dimer of dimers (i.e., a homotetramer) upon maturation [20]. Based on its mobility on a gel filtration column, DPAP1 appears to be a monomeric enzyme, as are other C1-family peptidases; however, we cannot rule out the possibility that DPAP1 forms a tetramer at higher concentration or under different environmental conditions. The reasons for the unusual quaternary organization of cathepsin C have been somewhat of a mystery. It has been suggested that tetramerization stabilizes the association of the exclusion domain with the two catalytic domains [13]. Our results indicate that tetramerization is not a structural requirement for this interaction and that the acquisition of a tetrameric structure probably occurred following divergence of the protozoan and metazoan lineages. One possibility is that the tetrameric structure enhances the affinity and/or specificity of cathepsin C for its protein substrates (the immune cell serine proteases granzyme A and B, mast cell chymase and neutrophil elastase, cathepsin G and proteinase 3 [8–10]) by providing interaction sites separate from the active site.
The S1 subsite preferences of DPAP1 and cathepsin C, as revealed by PSL analysis, are similar to those of other C1 family endo- and exopeptidases for which similar data exist [21, 51, 52]. This is likely due to the paucity of interactions between the P1 residue sidechain and the enzyme domain that defines the S1 site [43], which in turn offers little opportunity to tune the specificity of this site. In contrast, the S2 subsite of C1-family peptidases is the only one that forms a pocket and is generally the most discriminating of the subsites [43, 44]. In many cases, the S2 subsite exhibits a strong preference for hydrophobic sidechains which can be further divided into a preference for aromatic (e.g., cathepsin L) or aliphatic (e.g. cathepsin S) sidechains. The S2 subsite of cathepsin B has a comparatively broad specificity, in part due to the presence of a glutamate sidechain at the bottom of the S2 pocket that can interact with positively charged P2 sidechains [53]. The S2 preferences of DPAP1 and cathepsin C fall in between those of cathepsin S and cathepsin B. While aliphatic hydrophobic P2 sidechains are clearly preferred over aromatic sidechains, polar residues such as His, Ser, Thr and Gln are accepted as readily. However, in contrast to cathepsin B, substrates with Arg or Lys at the P2 position are not cleaved by DPAP1 or cathepsin C. A bias against aromatic and basic P2 sidechains by DPAP1 was also observed with a P2-diverse library of dipeptide vinyl sulfone inhibitors [7]. Crystal structures of cathepsin C have revealed a bound chloride ion at the bottom of the S2 pocket [13, 14] with several water molecules filling the pocket. These water molecules may stabilize the binding of substrates with small polar residues such as Ser and Thr in the S2 pocket through hydrogen bonding. A relatively broad S2 specificity is probably advantageous in enabling the efficient hydrolysis of sequence-diverse substrates in the lysosome (cathepsin C) or the food vacuole (DPAP1).
The ability of DPAP1 and cathepsin C to catalyze the hydrolysis of substrates with a P2 proline residue has been reported previously [5, 31, 47, 54] and was confirmed in our PSL analysis. Substrates with a P2 proline residue are generally not cleaved by C1 family cysteine proteases, prominent exceptions being cathepsin K [51, 55, 56] and cathepsin L2 from the trematode Fasciola hepatica [57]. Mutagenic analysis has revealed that two residues in the S2 pocket of cathepsin K contribute to the acceptance of a P2 proline residue: Tyr67 and Leu205 (papain numbering; [56, 58]). The homologous residues are Phe and Gln in DPAP1 and Phe and Ile in cathepsin C. In contrast, the homologous residues in cathepsin L, which does not efficiently cleave substrates with a P2 proline residue, are Leu and Ala. Thus, these two S2 subsite residues in DPAP1 and cathepsin C are much closer in character to those in cathepsin K. This observation suggests that the S2 subsites of DPAP1/cathepsin C and cathepsin K have evolved in a similar fashion to permit the efficient hydrolysis of substrates with a P2 proline residue. Consistent with this analysis, Arastu-Kapur et al have shown that a dipeptide vinyl sulfone inhibitor with a P2 proline residue exhibits high specificity for DPAP1 over the P. falciparum cysteine peptidases DPAP3 and falcipain-2 and -3 [7].
Despite the overall similarities in the S2 preferences of DPAP1 and cathepsin C, a few pronounced differences stood out. To further explore the S2 preferences of DPAP1 and cathepsin C, we assayed a P2-diverse panel of substrates with a highly-preferred P1 residue (Arg) to define in kinetic terms the differences in specificity observed with the positional scanning library. These experiments supplemented the PSL data by revealing large variations in kcat and Km values that were not apparent from the relative rates of cleavage of the PSL members and by identifying substrate inhibition as a contributor to the observed S2 specificities.
Considering the kcat/Km values for DPAP1 (Table 1), the trends observed with the PSL held up in the kinetic analysis, at least in a qualitative sense. With cathepsin C, a P2-Pro residue was predicted by the PSL data to make for a poor substrate in comparison to P2-Val. However, Pro-Arg-AMC and Val-Arg-ACC were hydrolyzed by cathepsin C with comparable catalytic efficiencies. This finding contrasts with a previous study showing a 10-fold increase in kcat/Km for the hydrolysis of Val-Phe-AMC relative to that of Pro-Phe-AMC [47], which is more in line with expectation from the PSL data. This discrepancy might be explained by the presence of positive co-operativity between P2-Pro and P1-Arg sidechains that does not exist with a P1-Phe or a randomized P1 substrate. It is also possible that differences in the structures of the ACC and AMC fluorophores could affect the relative kcat/Km values.
Cathepsin C was a substantially better catalyst than DPAP1. kcat/Km values for the hydrolysis of Pro-Arg-AMC and Val-Arg-ACC were over an order of magnitude higher for cathepsin C. One trivial explanation for this observation would be that the DPAP1 preparations contain a substantial fraction of catalytically dead enzyme. Given the low concentrations of both native and recombinant DPAP1 in the preparations described here, titration of active enzyme with an inhibitor was not feasible. However, the reasonable concordance of kcat/Km values for native and recombinant DPAP1, which were prepared in very different ways, lends confidence that these are not artifactually low.
Phe-Arg-ACC inhibited both DPAP1 and cathepsin C. DPAP1 activity was competitively inhibited with a low micromolar Ki and no substrate turnover was observed. In the case of cathepsin C, substrate turnover occurred with substrate inhibition at higher concentrations. These results indicate that the apparent “preference” of cathepsin C for a P2 phenylalanine residue (compared with DPAP1) that was observed in the PSL profile cannot simply be attributed to differences in accommodation of this sidechain in the S2 subsites of the two enzymes. The inhibition of cathepsin C by phenylalanine-containing compounds was reported over 50 years ago by Fruton and Mycek [54], who found that phenylalaninamide, DL-Phe-Gly and L-Phe-L-Phe competitively inhibited cathepsin C. Taken together, these observations indicate that a phenylalanyl moiety, in certain contexts, can interact with DPAP1 and cathepsin C in a manner that is unproductive for catalysis.
In conclusion, the relatively broad S1 and S2 specificities of DPAP1 and its high catalytic efficiency at acidic pH are consistent with a key role in peptide turnover in the food vacuole. A chloride concentration inside the parasite of 48 mM [59] is adequate for DPAP1 activation. We envision DPAP1 acting on a wide range of sequence-diverse oligopeptides that are generated by the action of endopeptidases on globin polypeptides. The dipeptides produced through DPAP1 catalysis could then be hydrolyzed into two amino acids by the vacuolar aminopeptidase PfA-M1 [6]. Thus, DPAP1 likely accelerates the rate of production of amino acids from globin by increasing the concentration of PfA-M1 substrates. Conversely, PfA-M1 could remove from the amino termini of oligopeptides those residues that prohibit DPAP1 cleavage (for example, Arg or Lys), thereby preventing the accumulation of such “blocked” peptides. We have proposed a similar role for P. falciparum aminopeptidase P with respect to proline-containing substrates [6]. Our data suggest that inhibitors that block DPAP1 will impede the production of amino acids from hemoglobin and may have anti-malarial activity; however, it appears that the design of an inhibitor that is highly selective for DPAP1 over cathepsin C will be a challenge.
Supplementary Material
Acknowledgments
We wish to thank D. Waugh for the MBP-TEV expression plasmid, J. Ellman for the positional scanning library, D. Goldberg for anti-DPAP1 antibodies, M. Leyva in J. Ellman’s laboratory for Fmoc-ACC, S. Dalal for constructing the DPAP1 expression plasmid and M. Drew for critique of the manuscript.
Abbreviations
- ACC
7-amino-4-carbamoylcoumarin
- AEBSF
4-(2-aminoethyl)benzenesulfonyl fluoride
- AMC
7-amino-4-methylcoumarin
- DPAP
dipeptidyl aminopeptidase
- EDTA
ethylenediaminetetraacetic acid
- IMAC
immobilized metal affinity chromatography
- MBP
maltose binding protein
- MES
4-morpholineethanesulfonic acid
- PSL
positional scanning library
- TEV
tobacco etch virus
APPENDIX A. SUPPLEMENTARY DATA
References
- 1.Krugliak M, Zhang J, Ginsburg H. Intraerythrocytic Plasmodium falciparum utilizes only a fraction of the amino acids derived from the digestion of host cell cytosol for the biosynthesis of its proteins. Mol Biochem Parasitol. 2002;119:249–56. doi: 10.1016/s0166-6851(01)00427-3. [DOI] [PubMed] [Google Scholar]
- 2.Loria P, Miller S, Foley M, Tilley L. Inhibition of the peroxidative degradation of haem as the basis of action of chloroquine and other quinoline antimalarials. Biochem J. 1999;339:363–70. [PMC free article] [PubMed] [Google Scholar]
- 3.Goldberg DE. Hemoglobin degradation. Curr Top Microbiol Immunol. 2005;295:275–91. doi: 10.1007/3-540-29088-5_11. [DOI] [PubMed] [Google Scholar]
- 4.Rosenthal PJ, McKerrow JH, Aikawa M, Nagasawa H, Leech JH. A malarial cysteine proteinase is necessary for hemoglobin degradation by Plasmodium falciparum. J Clin Invest. 1988;82:1560–6. doi: 10.1172/JCI113766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klemba M, Gluzman I, Goldberg DE. A Plasmodium falciparum dipeptidyl aminopeptidase I participates in vacuolar hemoglobin degradation. J Biol Chem. 2004;279:43000–7. doi: 10.1074/jbc.M408123200. [DOI] [PubMed] [Google Scholar]
- 6.Dalal S, Klemba M. Roles for two aminopeptidases in vacuolar hemoglobin catabolism in Plasmodium falciparum. J Biol Chem. 2007;282:35978–87. doi: 10.1074/jbc.M703643200. [DOI] [PubMed] [Google Scholar]
- 7.Arastu-Kapur S, Ponder EL, Fonovic UP, Yeoh S, Garinger M, Yuan F, et al. Chemically mapping protease pathways involved in the regulation of erythrocyte rupture by the human malaria parasite Plasmodium falciparum. Nat Chem Biol. 2008;4:203–13. doi: 10.1038/nchembio.70. [DOI] [PubMed] [Google Scholar]
- 8.Adkison AM, Raptis SZ, Kelley DG, Pham CT. Dipeptidyl peptidase I activates neutrophil-derived serine proteases and regulates the development of acute experimental arthritis. J Clin Invest. 2002;109:363–71. doi: 10.1172/JCI13462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pham CT, Ley TJ. Dipeptidyl peptidase I is required for the processing and activation of granzymes A and B in vivo. Proc Natl Acad Sci USA. 1999;96:8627–32. doi: 10.1073/pnas.96.15.8627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolters PJ, Pham CT, Muilenburg DJ, Ley TJ, Caughey GH. Dipeptidyl peptidase I is essential for activation of mast cell chymases, but not tryptases, in mice. J Biol Chem. 2001;276:18551–6. doi: 10.1074/jbc.M100223200. [DOI] [PubMed] [Google Scholar]
- 11.Bondebjerg J, Fuglsang H, Valeur KR, Pedersen J, Naerum L. Dipeptidyl nitriles as human dipeptidyl peptidase I inhibitors. Bioorg Med Chem Lett. 2006;16:3614–7. doi: 10.1016/j.bmcl.2006.01.102. [DOI] [PubMed] [Google Scholar]
- 12.Methot N, Rubin J, Guay D, Beaulieu C, Ethier D, Reddy TJ, et al. Inhibition of the activation of multiple serine proteases with a cathepsin C inhibitor requires sustained exposure to prevent pro-enzyme processing. J Biol Chem. 2007;282:20836–46. doi: 10.1074/jbc.M702615200. [DOI] [PubMed] [Google Scholar]
- 13.Olsen JG, Kadziola A, Lauritzen C, Pedersen J, Larsen S, Dahl SW. Tetrameric dipeptidyl peptidase I directs substrate specificity by use of the residual pro-part domain. FEBS Lett. 2001;506:201–6. doi: 10.1016/s0014-5793(01)02911-8. [DOI] [PubMed] [Google Scholar]
- 14.Turk D, Janjic V, Stern I, Podobnik M, Lamba D, Dahl SW, et al. Structure of human dipeptidyl peptidase I (cathepsin C): exclusion domain added to an endopeptidase framework creates the machine for activation of granular serine proteases. EMBO J. 2001;20:6570–82. doi: 10.1093/emboj/20.23.6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schechter I, Berger A. On the size of the active site in proteases. I. Papain. Biochem Biophys Res Commun. 1967;27:157–62. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- 16.Maly DJ, Leonetti F, Backes BJ, Dauber DS, Harris JL, Craik CS, et al. Expedient solid-phase synthesis of fluorogenic protease substrates using the 7-amino-4-carbamoylmethylcoumarin (ACC) fluorophore. J Org Chem. 2002;67:910–5. doi: 10.1021/jo016140o. [DOI] [PubMed] [Google Scholar]
- 17.Bondebjerg J, Fuglsang H, Valeur KR, Kaznelson DW, Hansen JA, Pedersen RO, et al. Novel semicarbazide-derived inhibitors of human dipeptidyl peptidase I (hDPPI) Bioorg Med Chem. 2005;13:4408–24. doi: 10.1016/j.bmc.2005.04.048. [DOI] [PubMed] [Google Scholar]
- 18.Bondebjerg J, Fuglsang H, Valeur KR, Pedersen J, Naerum L. Dipeptidyl nitriles as human dipeptidyl peptidase I inhibitors. Bioorg Med Chem Lett. 2006;16:3614–7. doi: 10.1016/j.bmcl.2006.01.102. [DOI] [PubMed] [Google Scholar]
- 19.Kapust RB, Tozser J, Fox JD, Anderson DE, Cherry S, Copeland TD, et al. Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 2001;14:993–1000. doi: 10.1093/protein/14.12.993. [DOI] [PubMed] [Google Scholar]
- 20.Dahl SW, Halkier T, Lauritzen C, Dolenc I, Pedersen J, Turk V, et al. Human recombinant pro-dipeptidyl peptidase I (cathepsin C) can be activated by cathepsins L and S but not by autocatalytic processing. Biochemistry. 2001;40:1671–8. doi: 10.1021/bi001693z. [DOI] [PubMed] [Google Scholar]
- 21.Harris JL, Backes BJ, Leonetti F, Mahrus S, Ellman JA, Craik CS. Rapid and general profiling of protease specificity by using combinatorial fluorogenic substrate libraries. Proc Natl Acad Sci USA. 2000;97:7754–9. doi: 10.1073/pnas.140132697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dixon M. The determination of enzyme inhibitor constants. Biochem J. 1953;55:170–1. doi: 10.1042/bj0550170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ellis KJ, Morrison JF. Buffers of constant ionic strength for studying pH-dependent processes. Methods Enzymol. 1982;87:405–26. doi: 10.1016/s0076-6879(82)87025-0. [DOI] [PubMed] [Google Scholar]
- 24.Molgaard A, Arnau J, Lauritzen C, Larsen S, Petersen G, Pedersen J. The crystal structure of human dipeptidyl peptidase I (cathepsin C) in complex with the inhibitor Gly-Phe-CHN2. Biochem J. 2007;401:645–50. doi: 10.1042/BJ20061389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goh LL, Loke P, Singh M, Sim TS. Soluble expression of a functionally active Plasmodium falciparum falcipain-2 fused to maltose-binding protein in Escherichia coli. Protein Expr Purif. 2003;32:194–201. doi: 10.1016/S1046-5928(03)00225-0. [DOI] [PubMed] [Google Scholar]
- 26.Goh SL, Goh LL, Sim TS. Cysteine protease falcipain 1 in Plasmodium falciparum is biochemically distinct from its isozymes. Parasitol Res. 2005;97:295–301. doi: 10.1007/s00436-005-1430-7. [DOI] [PubMed] [Google Scholar]
- 27.Kapust RB, Tozser J, Copeland TD, Waugh DS. The P1′ specificity of tobacco etch virus protease. Biochem Biophys Res Commun. 2002;294:949–55. doi: 10.1016/S0006-291X(02)00574-0. [DOI] [PubMed] [Google Scholar]
- 28.Bessette PH, Aslund F, Beckwith J, Georgiou G. Efficient folding of proteins with multiple disulfide bonds in the Escherichia coli cytoplasm. Proc Natl Acad Sci USA. 1999;96:13703–8. doi: 10.1073/pnas.96.24.13703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lauritzen C, Pedersen J, Madsen MT, Justesen J, Martensen PM, Dahl SW. Active recombinant rat dipeptidyl aminopeptidase I (cathepsin C) produced using the baculovirus expression system. Protein Expr Purif. 1998;14:434–42. doi: 10.1006/prep.1998.0976. [DOI] [PubMed] [Google Scholar]
- 30.McDonald JK, Reilly TJ, Zeitman BB, Ellis S. Cathepsin C: a chloride-requiring enzyme. Biochem Biophys Res Commun. 1966;22:771–5. doi: 10.1016/0006-291x(66)90392-5. [DOI] [PubMed] [Google Scholar]
- 31.McDonald JK, Zeitman BB, Reilly TJ, Ellis S. New observations on the substrate specificity of cathepsin C (dipeptidyl aminopeptidase I). Including the degradation of beta-corticotropin and other peptide hormones. J Biol Chem. 1969;244:2693–709. [PubMed] [Google Scholar]
- 32.Cornish-Bowden A. Fundamentals of enzyme kinetics. London: Portland Press; 2004. [Google Scholar]
- 33.Dolenc I, Turk B, Pungercic G, Ritonja A, Turk V. Oligomeric structure and substrate induced inhibition of human cathepsin C. J Biol Chem. 1995;270:21626–31. doi: 10.1074/jbc.270.37.21626. [DOI] [PubMed] [Google Scholar]
- 34.Nagler DK, Tam W, Storer AC, Krupa JC, Mort JS, Menard R. Interdependency of sequence and positional specificities for cysteine proteases of the papain family. Biochemistry. 1999;38:4868–74. doi: 10.1021/bi982632s. [DOI] [PubMed] [Google Scholar]
- 35.Schneck JL, Villa JP, McDevitt P, McQueney MS, Thrall SH, Meek TD. Chemical mechanism of a cysteine protease, cathepsin C, as revealed by integration of both steady-state and pre-steady-state solvent kinetic isotope effects. Biochemistry. 2008;47:8697–710. doi: 10.1021/bi8007627. [DOI] [PubMed] [Google Scholar]
- 36.Bennett TN, Kosar AD, Ursos LM, Dzekunov S, Singh Sidhu AB, Fidock DA, et al. Drug resistance-associated pfCRT mutations confer decreased Plasmodium falciparum digestive vacuolar pH. Mol Biochem Parasitol. 2004;133:99–114. doi: 10.1016/j.molbiopara.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 37.Klonis N, Tan O, Jackson K, Goldberg D, Klemba M, Tilley L. Evaluation of pH during cytostomal endocytosis and vacuolar catabolism of hemoglobin in Plasmodium falciparum. Biochem J. 2007;407:343–54. doi: 10.1042/BJ20070934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krogstad DJ, Schlesinger PH, Gluzman IY. Antimalarials increase vesicle pH in Plasmodium falciparum. J Cell Biol. 1985;101:2302–9. doi: 10.1083/jcb.101.6.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuhn Y, Rohrbach P, Lanzer M. Quantitative pH measurements in Plasmodium falciparum- infected erythrocytes using pHluorin. Cell Microbiol. 2007;9:1004–13. doi: 10.1111/j.1462-5822.2006.00847.x. [DOI] [PubMed] [Google Scholar]
- 40.Storer AC, Menard R. Catalytic mechanism in papain family of cysteine peptidases. Methods Enzymol. 1994;244:486–500. doi: 10.1016/0076-6879(94)44035-2. [DOI] [PubMed] [Google Scholar]
- 41.Fersht A. Structure and mechanism in protein science. New York: W.H. Freeman and Company; 1999. [Google Scholar]
- 42.Cigic B, Pain RH. Location of the binding site for chloride ion activation of cathepsin C. Eur J Biochem. 1999;264:944–51. doi: 10.1046/j.1432-1327.1999.00697.x. [DOI] [PubMed] [Google Scholar]
- 43.McGrath ME. The lysosomal cysteine proteases. Annu Rev Biophys Biomol Struct. 1999;28:181–204. doi: 10.1146/annurev.biophys.28.1.181. [DOI] [PubMed] [Google Scholar]
- 44.Turk D, Guncar G, Podobnik M, Turk B. Revised definition of substrate binding sites of papain-like cysteine proteases. Biol Chem. 1998;379:137–47. doi: 10.1515/bchm.1998.379.2.137. [DOI] [PubMed] [Google Scholar]
- 45.Schneider EL, Craik CS. Positional scanning synthetic combinatorial libraries for substrate profiling. Methods Mol Biol. 2009;539:59–78. doi: 10.1007/978-1-60327-003-8_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leiting B, Pryor KD, Wu JK, Marsilio F, Patel RA, Craik CS, et al. Catalytic properties and inhibition of proline-specific dipeptidyl peptidases II, IV and VII. Biochem J. 2003;371:525–32. doi: 10.1042/BJ20021643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tran TV, Ellis KA, Kam CM, Hudig D, Powers JC. Dipeptidyl peptidase I: importance of progranzyme activation sequences, other dipeptide sequences, and the N-terminal amino group of synthetic substrates for enzyme activity. Arch Biochem Biophys. 2002;403:160–70. doi: 10.1016/s0003-9861(02)00217-5. [DOI] [PubMed] [Google Scholar]
- 48.McGuire MJ, Lipsky PE, Thiele DL. Purification and characterization of dipeptidyl peptidase I from human spleen. Arch Biochem Biophys. 1992;295:280–8. doi: 10.1016/0003-9861(92)90519-3. [DOI] [PubMed] [Google Scholar]
- 49.Cigic B, Krizaj I, Kralj B, Turk V, Pain RH. Stoichiometry and heterogeneity of the pro-region chain in tetrameric human cathepsin C. Biochim Biophys Acta. 1998;1382:143–50. doi: 10.1016/s0167-4838(97)00173-8. [DOI] [PubMed] [Google Scholar]
- 50.Cigic B, Dahl SW, Pain RH. The residual pro-part of cathepsin C fulfills the criteria required for an intramolecular chaperone in folding and stabilizing the human proenzyme. Biochemistry. 2000;39:12382–90. doi: 10.1021/bi0008837. [DOI] [PubMed] [Google Scholar]
- 51.Choe Y, Leonetti F, Greenbaum DC, Lecaille F, Bogyo M, Bromme D, et al. Substrate profiling of cysteine proteases using a combinatorial peptide library identifies functionally unique specificities. J Biol Chem. 2006;281:12824–32. doi: 10.1074/jbc.M513331200. [DOI] [PubMed] [Google Scholar]
- 52.Subramanian S, Hardt M, Choe Y, Niles RK, Johansen EB, Legac J, et al. Hemoglobin cleavage site-specificity of the Plasmodium falciparum cysteine proteases falcipain-2 and falcipain-3. PLoS ONE. 2009;4:e5156. doi: 10.1371/journal.pone.0005156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jia Z, Hasnain S, Hirama T, Lee X, Mort JS, To R, et al. Crystal structures of recombinant rat cathepsin B and a cathepsin B-inhibitor complex. Implications for structure-based inhibitor design. J Biol Chem. 1995;270:5527–33. doi: 10.1074/jbc.270.10.5527. [DOI] [PubMed] [Google Scholar]
- 54.Fruton JS, Mycek MJ. Studies on beef spleen cathepsin C. Arch Biochem Biophys. 1956;65:11–20. doi: 10.1016/0003-9861(56)90172-2. [DOI] [PubMed] [Google Scholar]
- 55.Garnero P, Borel O, Byrjalsen I, Ferreras M, Drake FH, McQueney MS, et al. The collagenolytic activity of cathepsin K is unique among mammalian proteinases. J Biol Chem. 1998;273:32347–52. doi: 10.1074/jbc.273.48.32347. [DOI] [PubMed] [Google Scholar]
- 56.Lecaille F, Choe Y, Brandt W, Li Z, Craik CS, Bromme D. Selective inhibition of the collagenolytic activity of human cathepsin K by altering its S2 subsite specificity. Biochemistry. 2002;41:8447–54. doi: 10.1021/bi025638x. [DOI] [PubMed] [Google Scholar]
- 57.Dowd AJ, Smith AM, McGonigle S, Dalton JP. Purification and characterisation of a second cathepsin L proteinase secreted by the parasitic trematode Fasciola hepatica. Eur J Biochem. 1994;223:91–8. doi: 10.1111/j.1432-1033.1994.tb18969.x. [DOI] [PubMed] [Google Scholar]
- 58.Lecaille F, Chowdhury S, Purisima E, Bromme D, Lalmanach G. The S2 subsites of cathepsins K and L and their contribution to collagen degradation. Protein Sci. 2007;16:662–70. doi: 10.1110/ps.062666607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Henry RI, Cobbold SA, Allen RJW, Khan A, Hayward R, Lehane AM, et al. An acid-loading chloride transport pathway in the intraerythrocytic malaria parasite, Plasmodium falciparum. J Biol Chem. 2010;285:18615–26. doi: 10.1074/jbc.M110.120980. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.