Control of cell cycle progression relies on unique regulation of centrosomal cAMP/PKA signals through PKA and PDE4D3 interaction with the A kinase anchoring protein AKAP9.
Abstract
Previous work has shown that the protein kinase A (PKA)–regulated phosphodiesterase (PDE) 4D3 binds to A kinase–anchoring proteins (AKAPs). One such protein, AKAP9, localizes to the centrosome. In this paper, we investigate whether a PKA–PDE4D3–AKAP9 complex can generate spatial compartmentalization of cyclic adenosine monophosphate (cAMP) signaling at the centrosome. Real-time imaging of fluorescence resonance energy transfer reporters shows that centrosomal PDE4D3 modulated a dynamic microdomain within which cAMP concentration selectively changed over the cell cycle. AKAP9-anchored, centrosomal PKA showed a reduced activation threshold as a consequence of increased autophosphorylation of its regulatory subunit at S114. Finally, disruption of the centrosomal cAMP microdomain by local displacement of PDE4D3 impaired cell cycle progression as a result of accumulation of cells in prophase. Our findings describe a novel mechanism of PKA activity regulation that relies on binding to AKAPs and consequent modulation of the enzyme activation threshold rather than on overall changes in cAMP levels. Further, we provide for the first time direct evidence that control of cell cycle progression relies on unique regulation of centrosomal cAMP/PKA signals.
Introduction
The second messenger cAMP mediates the intracellular response to multiple hormones and neurotransmitters and regulates a wide variety of cellular processes, including gene expression, metabolism, and cell growth and division (Stork and Schmitt, 2002). cAMP is generated from ATP by adenylyl cyclases, and phosphodiesterases (PDEs) provide the only means to degrade cAMP (Conti and Beavo, 2007). Therefore, PDEs play a key role in the control of cAMP resting levels as well as in determining the amplitude and duration of cAMP signals in response to extracellular stimuli (Houslay, 2010). The main effector of cAMP is PKA, a tetrameric enzyme that in its inactive form consists of two catalytic subunits (C) and one regulatory subunit (R) dimer. Upon binding of cAMP to the R subunits, the C subunits are released and phosphorylate downstream targets.
A multitude of different stimuli can generate an increase in intracellular cAMP, and active PKA C subunits can potentially phosphorylate a large variety of protein targets within the same cell. However, in order for the cell to execute the appropriate task in response to a specific stimulus, the correct subset of downstream targets must be phosphorylated. To achieve this, spatial confinement (compartmentalization) of the molecular components of the cAMP signaling pathway is critical (Zaccolo, 2009). PKA is tethered to subcellular loci via binding to A kinase–anchoring proteins (AKAPs). AKAPs anchor PKA in proximity to its targets via binding to the amino-terminal dimerization/docking (D/D) domains of PKA R subunits of an amphipathic helix within the AKAP sequence (Wong and Scott, 2004). The cAMP signal is also compartmentalized, with different intracellular subcompartments showing different concentrations of the second messenger (Zaccolo and Pozzan, 2002). Different subsets of anchored PKA are thus exposed to different levels of cAMP, resulting in selective activation and phosphorylation of the appropriate subset of targets (Di Benedetto et al., 2008). PDEs, a large superfamily of enzymes comprised of 11 families (PDE1-11) and >30 isozymes, can also be localized to specific subcellular compartments and, by locally degrading cAMP, play a key role in the spatial control of cAMP signal propagation (Mongillo et al., 2004). Long isoforms of the PDE4 family, including PDE4D3, can be phosphorylated and activated by PKA (Sette and Conti, 1996; MacKenzie et al., 2002), and members of the PDE4D subfamily have been shown to interact with several AKAPs, including AKAP6 (Dodge et al., 2001), AKAP7 (Stefan et al., 2007), and AKAP9 (Taskén et al., 2001). The presence of PKA and PDE4D3 within the same macromolecular complex may thus provide a negative feedback system in which elevated cAMP concentrations trigger PKA to phosphorylate and activate PDE4, reducing local cAMP levels and resetting PKA activity selectively at that site (Dodge et al., 2001).
AKAP9/450/350/CG-NAP (centrosome- and Golgi-localized, PKN-associated protein; hereafter referred to as AKAP450) localizes at the centrosome (Schmidt et al., 1999; Takahashi et al., 1999; Witczak et al., 1999) through a conserved protein interaction module known as the PACT (pericentrin-AKAP350 centrosomal targeting) domain (Gillingham and Munro, 2000). Localization of AKAP450 at the centrosome has been shown to be required for centrosome integrity and centriole duplication (Keryer et al., 2003).
The centrosome plays a key role in cell cycle progression and acts as a scaffold for the accumulation and interaction of different cell cycle regulators (Cuschieri et al., 2007). PKA has been shown to be involved in many aspects of cell cycle regulation, including centrosome duplication, S phase, G2 arrest, mitotic spindle formation, exit from M phase, and cytokinesis (Matyakhina et al., 2002); however, which, if any, of these functions is regulated by a PKA subset targeted at the centrosome remains to be established. In addition, it is not clear how cells achieve appropriate control of cell proliferation while continuously being exposed to hormonal fluctuations and, consequently, to changes in intracellular cAMP levels.
In this study, we use real-time imaging and a combination of fluorescence resonance energy transfer (FRET)–based reporters to explore the hypothesis that anchoring of PKA and PDE4D3 to AKAP450 provides a structural basis for selective regulation of cAMP signals at the centrosome. Our results show, for the first time, that the centrosome is a subcellular compartment undergoing a sophisticated and dynamic control of cAMP signals and PKA activation that relies on PKA anchoring to AKAP450 and on the presence of PDE4D3 and can be independent of overall changes of cAMP levels in the cytosol. In addition, we show that selective disruption of cAMP/PKA signaling at the centrosome has unique effects on cell cycle progression.
Results
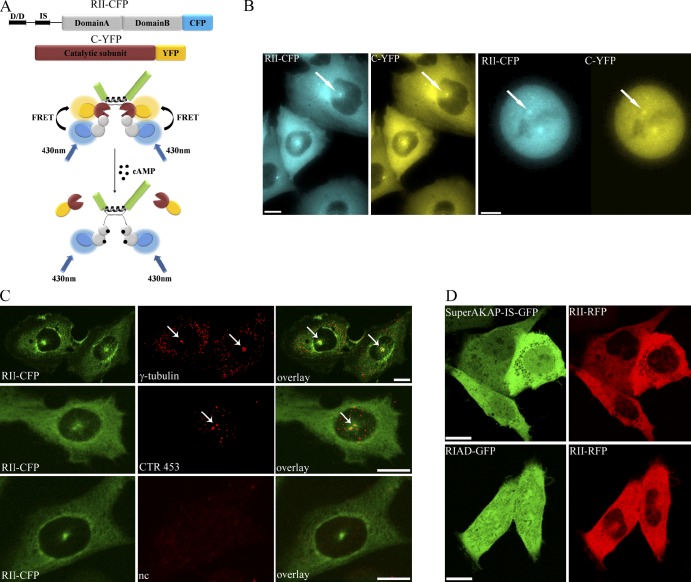
To study cAMP signals at the centrosome, we generated a CHO cell clone that stably expresses a PKA-based FRET sensor, PKA-GFP (Vaasa et al., 2010). The sensor includes the regulatory type II (RII-CFP) and the catalytic (C-YFP) subunits of PKA tagged, at their carboxyl termini, with the cyan and the yellow variants of the GFP, respectively (Fig. 1 A; Zaccolo et al., 2000). In the absence of cAMP, the sensor subunits RII-CFP and C-YFP interact, allowing energy transfer (FRET) from the donor CFP to the acceptor YFP. In the presence of cAMP, the RII-CFP and C-YFP subunits dissociate, and FRET is abolished (Fig. 1 A). We have previously reported that PKA-GFP shows the same cAMP dependence and the same sensitivity to cAMP and ability to phosphorylate substrate as wild-type PKA (Mongillo et al., 2004) and interacts with endogenous AKAPs via the D/D domain of its RII-CFP subunits (Zaccolo and Pozzan, 2002). Expression of PKA-GFP in CHO cells shows a clear localization of the sensor at the centrosome both in interphase cells and in mitotic cells (Fig. 1 B) without affecting centrosome morphology (Fig. S1). Centrosomal localization of the sensor is confirmed by colocalization of RII-CFP with the centrosomal marker γ-tubulin (Figs. 1 C and S1). Immunostaining with CTR453, an mAb specific for AKAP450 (Bailly et al., 1989; Keryer et al., 2003), shows colocalization of PKA-GFP and AKAP450 at the centrosome (Fig. 1 C). To assess whether the centrosomal localization of the sensor is mediated by interaction with centrosomal AKAPs, an RFP-tagged RII subunit was coexpressed with either GFP-tagged SuperAKAP–in silico (IS) or GFP-tagged RI-anchoring disruptor (RIAD). The peptides RIAD (Carlson et al., 2006) and SuperAKAP-IS (Gold et al., 2006) compete selectively with the binding of RI and RII to AKAPs, respectively. As shown in Fig. 1 D, coexpression of SuperAKAP-IS GFP, but not RIAD GFP, completely abolishes the centrosomal localization of RII-RFP.
Figure 1.
The FRET sensor PKA-GFP localizes to the centrosome via binding to endogenous AKAPs. (A, top) Schematic representation of the cAMP sensor PKA-GFP. IS indicates the catalytic inhibitory site and autophosphorylation site. Domains A and B indicate the cAMP-binding domains A and B, respectively. (bottom) Illustration of the interaction of PKA-GFP with an AKAP (in green) and its mechanism of activation upon binding of cAMP. The yellow and blue halos around YFP and CFP indicate fluorescence emission from the fluorophores upon excitation of CFP at 430 nm. (B) Subcellular distribution of the sensor in a CHO cell stably expressing PKA-GFP in interphase (left) and mitosis (right). White arrows point to the centrosome and one of the centrioles. (C) CHO cells stably expressing PKA-GFP and immunostained with a γ-tubulin–specific antibody (top row) and with the AKAP450-specific antibody CTR453 (middle row). A negative control (nc) in which the primary antibody is omitted is shown on the bottom row. The signal from the C-YFP component of the sensor is not shown. Arrows point to the centrosome. (D) CHO cells expressing SuperAKAP-IS-GFP and RIAD-GFP in combination with RII-RFP. Bars, 10 µm.
Basal cAMP levels are lower at the centrosome than in the bulk cytosol in interphase cells
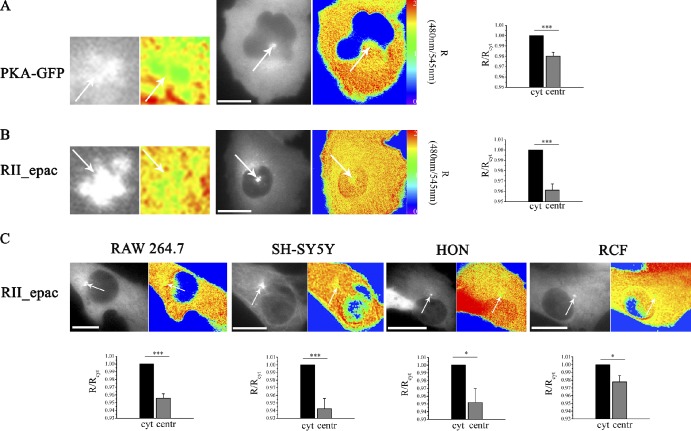
CHO cells expressing the PKA-GFP sensor show a small but highly significant difference in the basal CFP/YFP emission intensity ratio (R) at the centrosome as compared with the bulk cytosol (Fig. 2 A), indicating that in resting, nonstimulated interphase cells, the level of cAMP at the centrosome is lower than the mean cAMP level in the cytosol.
Figure 2.
Basal cAMP in interphase cells is lower at the centrosome than in the cytosol. (A) CHO cell stably expressing the PKA-GFP sensor. The middle image in grayscale is the image acquired at 480 nm upon excitation at 430 nm and shows the subcellular distribution of the sensor. The signal generated by the C-YFP component of the PKA-GFP sensor is not shown. The FRET signal from the same cell, calculated as a 480/540-nm emission intensity ratio upon excitation at 430 nm, is shown in pseudocolor. Images on the left show a higher magnification of the centrosomal region. White arrows point to the centrosome. The image on the right shows the mean basal FRET signal measured in the bulk cytosol (cyt) and at the centrosome (centr) of cells stably expressing PKA-GFP. FRET values are the mean calculated within an ROI drawn to include the entire cytosolic area or the centrosome (an example is provided in Fig. 4, A and B) and are expressed relative to the FRET value measured in the cytosol. n = 34. (B) CHO cells stably expressing the unimolecular sensor RII_epac. Images are as described in A. n = 31. (C) Sensor distribution and FRET pseudocolor images of RAW264.7 cells, SH-SY5Y cells, primary HONs, and primary RCFs expressing RII_epac. For each cell type, bottom images show the mean FRET signal in the cytosol and at the centrosome, calculated as described in A. n ≥ 5. For all experiments, error bars represent SEM. Two-tailed paired t tests were performed (*, P < 0.05; ***, P < 0.001). Bars, 10 µm.
A similar difference between bulk cytosol and centrosome was detected by RII_epac (Fig. S2 A), a unimolecular FRET reporter for cAMP carrying at its amino terminus the D/D domain from the RII subunit of PKA (Di Benedetto et al., 2008) and therefore, similarly to PKA-GFP, able to interact with endogenous centrosomal AKAPs (Fig. 2 B). The RII_epac reporter detects a significant difference between cytosolic and centrosomal basal cAMP levels in several other cell types analyzed, including the macrophage cell line RAW264.7, the human neuroblastoma cell line SH-SY5Y (Biedler et al., 1978), primary human olfactory neurons (HONs), primary rat cardiac fibroblasts (RCFs; Fig. 2 C), and the nontransformed cell line RPE1 (Fig. S3 A).
PDE4D3 is responsible for the low basal cAMP level at the centrosome
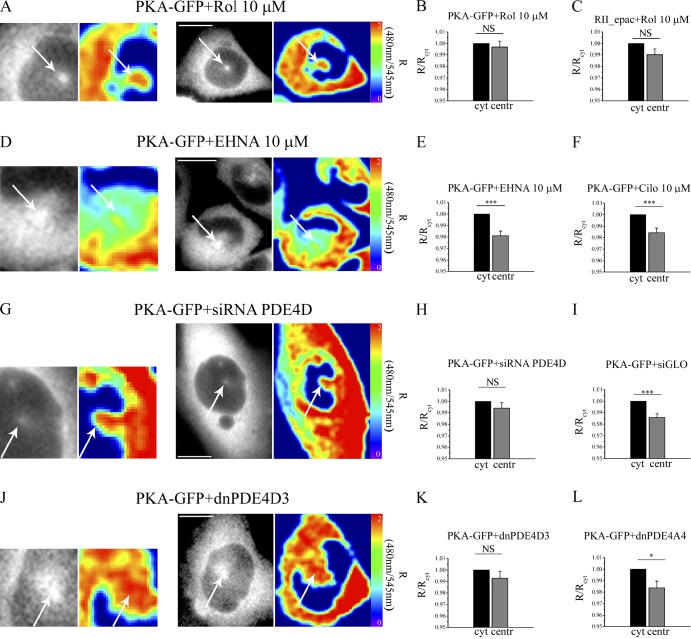
In a variety of cell types, including CHO cells (Fig. S4 A) and RPE1 (Fig. S3 B), PDE4D3 localizes to the centrosome and has been shown to bind to AKAP450 (Taskén et al., 2001; McCahill et al., 2005). Therefore, anchoring of PDE4D3 at the centrosome may explain the observed low cAMP level at this site. In support of this hypothesis, selective inhibition of PDE4 enzymes with 10 µM rolipram completely abolished the difference in cAMP between the centrosome and the bulk cytosol, as detected by both PKA-GFP and RII_epac (Fig. 3, A–C). In contrast, selective inhibition of PDE2 with 10 µM EHNA (erythro-9-(2-hydroxy-3-nonyl)adenine; Fig. 3, D and E) or selective inhibition of PDE3 with 10 µM cilostamide (Fig. 3 F) did not affect the gradient between centrosome and cytosol. In further support of a role of PDE4D3 in maintaining a low basal cAMP level at the centrosome, genetic knockdown of PDE4D using an siRNA approach and resulting in an almost complete ablation of all PDE4D isozymes (Fig. S4 B) completely abolished the differences in cAMP levels between cytosol and centrosome (Fig. 3, G and H), whereas the control oligonucleotide siGLO did not show any effect (Fig. 3 I). Anchoring of active PDE4D3 at the centrosome appears to be necessary to maintain low at the local cAMP concentration, as shown by experiments in which we used a catalytically inactive mutant of PDE4D3 (dnPDE4D3; McCahill et al., 2005). When overexpressed, dnPDE4D3 localizes at the centrosome (Fig. S4 C) and is expected to displace endogenous active PDE4D3 from its centrosomal anchor sites. Overexpression of dnPDE4D3 in CHO cells completely abolished the difference in cAMP concentration between the centrosome and the cytosol, as detected by the coexpressed PKA-GFP (Fig. 3, J and K). In contrast, overexpression of a catalytically inactive mutant of a different PDE4 isozyme (dnPDE4A4; Fig. S4 C; McCahill et al., 2005) did not affect the cAMP gradient between the cytosol and centrosome (Fig. 3 L). Collectively, the aforementioned data strongly indicate that PDE4D3 is responsible for maintaining a microdomain with low cAMP concentration at the centrosome.
Figure 3.
The low cAMP compartment at the centrosome depends on centrosomal PDE4D3. (A) Sensor distribution and FRET pseudocolor image of a representative CHO cell stably expressing PKA-GFP and treated with 10 µM rolipram (Rol). Images on the left are a magnification of the centrosomal region. (B) Mean basal FRET signal calculated in the cytosol (cyt) and at the centrosome (centr) in CHO cells expressing PKA-GFP and treated with the PDE4 inhibitor rolipram (10 µM). n = 44. (C) Mean basal FRET signal calculated in the cytosol and at the centrosome in CHO cells expressing RII_epac and treated with 10 µM rolipram. n = 25. (D) Sensor distribution and FRET pseudocolor image of representative CHO cells stably expressing PKA-GFP and treated with the PDE2 inhibitor EHNA (10 µM). (E) Mean basal FRET signal calculated in the cytosol and at the centrosome in the same cells. n = 46. (F) Mean basal FRET signal calculated in the cytosol and at the centrosome of CHO cells stably expressing PKA-GFP and treated with the PDE3 inhibitor cilostamide (Cilo; 10 µM). n = 39. (G–I) Sensor distribution and FRET pseudocolor image of a representative CHO cell stably expressing PKA-GFP and in which PDE4D isoforms have been knocked down by siRNA treatment (G). The mean FRET signal in the cytosol and centrosome in these cells and in cells expressing the control sequence siGLO are shown in H (n = 40) and I (n = 40), respectively. (J and K) Sensor distribution and FRET pseudocolor image of a representative CHO cell stably expressing PKA-GFP and a catalytically inactive mutant of PDE4D3 (dnPDE4D3); the mean FRET signal in the cytosol and centrosome (n = 31) is shown in K. (A, D, G, and J) Arrows point to the centrosome. (L) Summary of the basal CFP/YFP ratio values recorded in the cytosol and at the centrosome of cells expressing a catalytically inactive mutant of PDE4A4 (n = 21). All error bars represent SEM. Two-tailed paired t tests were performed (*, P < 0.05; ***, P < 0.001). Bars, 10 µm.
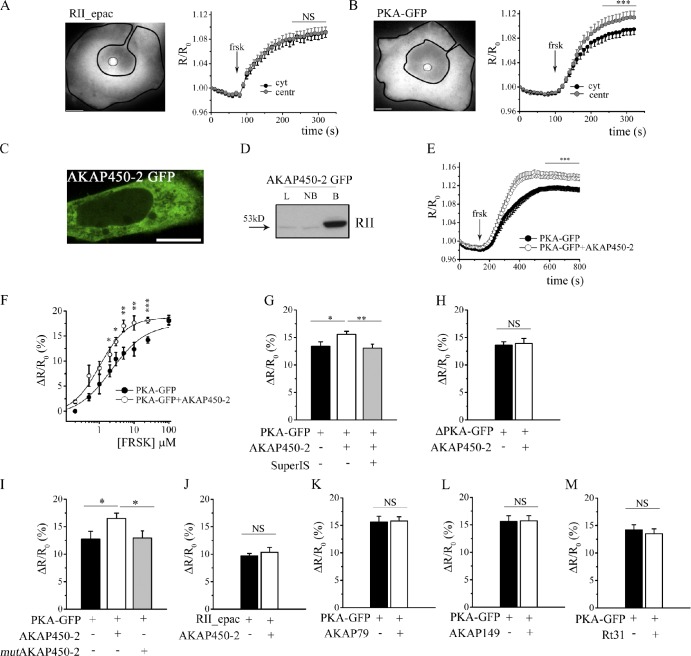
PKA anchored to AKAP450 shows increased sensitivity to cAMP
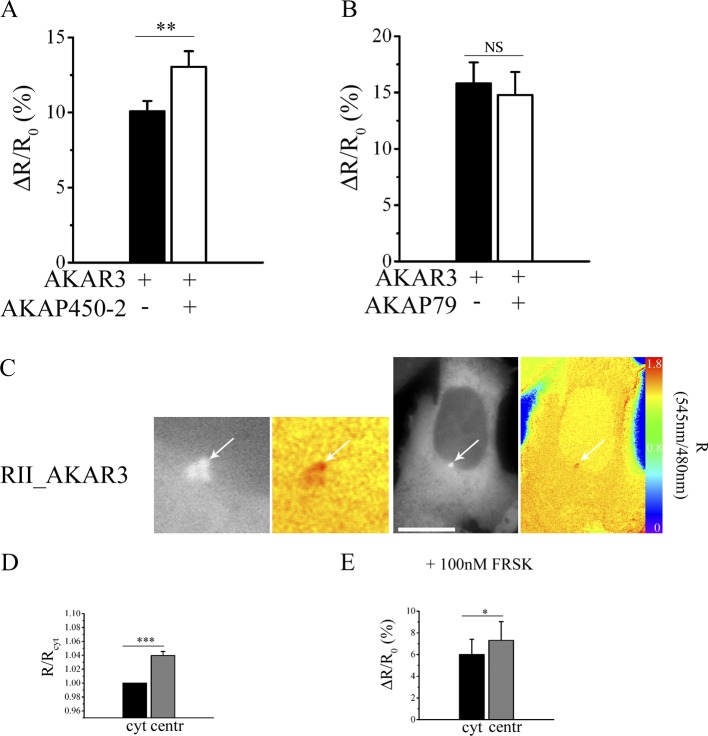
Next, we assessed the cAMP response generated in the bulk cytosol and at the centrosome upon activation of adenylyl cyclases with forskolin, using either the RII_epac or the PKA-GFP reporter. Upon application of 25 µM forskolin, we found that the RII_epac sensor reported an equal FRET change in the cytosol and at the centrosome (Fig. 4 A), whereas, unexpectedly, the PKA-GFP reporter recorded a significantly higher signal at the centrosome than in the bulk cytosol (Fig. 4 B). To explain this discrepancy, we hypothesized that the higher FRET change recorded at the centrosome by PKA-GFP may be the consequence of an increased sensitivity to cAMP of the centrosomal-targeted, PKA-based biosensor. To verify this hypothesis, we expressed the PKA-GFP sensor in CHO cells in combination with a fragment of the centrosomal AKAP450 encompassing amino acids 933–1,804 (AKAP450-2; Witczak et al., 1999) and including the amphipathic helix responsible for binding the RII subunits of PKA. AKAP450-2 lacks the PACT domain responsible for anchoring of AKAP450 to the centrosome (Fig. S2 B; Gillingham and Munro, 2000). Thus, when expressed in cells, AKAP450-2 is a cytosolic polypeptide (Fig. 4 C) that retains its ability to bind to PKA RII subunits (Fig. 4 D). The rationale for this experiment is that if anchoring of PKA to endogenous AKAP450 at the centrosome affects the kinase sensitivity to cAMP, the same effect should result from PKA binding to AKAP450-2 in the cytosol. Fig. 4 E shows that this is the case, and, as reported in Fig. 4 F, the dose-response curve where FRET change is plotted against increasing concentrations of forskolin shows a shift to the left for cells coexpressing the FRET sensor and AKAP450-2, confirming that a lower concentration of cAMP is sufficient to dissociate PKA-GFP when it is bound to AKAP450. The effect of AKAP450-2 on the sensitivity of PKA-GFP to cAMP is completely abolished in the presence of SuperAKAP-IS (Fig. 4 G), confirming that this effect depends on the interaction of PKA-GFP with AKAP450-2. AKAP450-2 has no effect on the FRET change detected by a variant of the PKA-GFP sensor (ΔPKA-GFP) in which the D/D domain of the RII subunit has been deleted (Zaccolo and Pozzan, 2002), thereby resulting in a sensor that cannot bind to AKAPs (Fig. 4 H), or when a serine to proline substitution (S1451P) is introduced in AKAP450-2 (mutAKAP450-2) that disrupts the amphipathic helix and abolishes the ability of PKA-GFP to bind (Fig. 4 I; Feliciello et al., 2001; Alto et al., 2003). As an additional control, we measured the FRET change reported by the sensor RII_epac when coexpressed with the AKAP450-2 fragment, and we found no difference compared with the FRET change recorded in the presence of the sensor alone (Fig. 4 J). The increased sensitivity to cAMP appears to be specific for PKA enzymes anchored to AKAP450, as coexpression of PKA-GFP with fragments from Rt31 (Klussmann et al., 2001), AKAP79 (Herberg et al., 2000), or AKAP149 (Carlson et al., 2003), all including the RII-binding amphipathic helix, did not result in a significant difference in the FRET response compared with control cells expressing the sensor alone (Fig. 4, K–M). Overall, the aforementioned data show that anchoring of PKA to AKAP450 results in an increased sensitivity of the FRET signal to cAMP, which is indicative of a reduced activation threshold of PKA.
Figure 4.
PKA-GFP anchored to AKAP450 shows increased sensitivity to cAMP. (A, left) CHO cell stably expressing the RII_epac sensor. Representative ROIs within which the ratio values are averaged for bulk cytosol (black line) and centrosome (gray line) are shown. (right) Normalized mean kinetics (n = 31) of FRET change induced by 25 µM forskolin (frsk) in CHO cells stably expressing RII_epac and recorded in the cytosol (cyt) and at the centrosome (centr). (B, left) CHO cell stably expressing the PKA-GFP sensor. (right) Normalized mean kinetics of FRET change detected in response to 25 µM forskolin in CHO cells stably expressing PKA-GFP and recorded in the cytosol and at the centrosome (n = 35). (C) Distribution of the GFP-tagged AKAP450-2 fragment in a representative CHO cell. (D) Western blot analysis of lysates from CHO cells overexpressing the GFP-tagged AKAP450-2 fragment. AKAP450-2 was immunoprecipitated using GFP-Trap beads, and the total lysate (L), unbound fraction (NB), and protein-bound fraction (B) to the GFP-Trap beads were immunoblotted with anti-RII antibody. Similar results were obtained in three independent experiments. (E) Normalized mean kinetics (n = 25) of FRET change induced by 25 µM forskolin in CHO cells expressing PKA-GFP in the presence (open circles) or absence (filled circles) of AKAP450-2. (F) Dose-response curve of FRET change at different concentrations of forskolin in CHO cells expressing PKA-GFP in the presence (open circles) or absence (filled circles) of AKAP450-2. For each concentration point, n ≥ 10. (G) FRET change induced by 25 µM forskolin in CHO cells expressing PKA-GFP in the presence or absence of AKAP450-2 fragment and SuperAKAP-IS. n ≥ 18. (H) FRET change induced by 25 µM forskolin in CHO cells expressing the deletion mutant sensor ΔPKA-GFP in the presence or absence of AKAP450-2. n ≥ 23. (I) FRET change induced by 25 µM forskolin in CHO cells expressing PKA-GFP in the presence or absence of the AKAP450-2 and mutAKAP450-2. n ≥ 14. (J) FRET change induced by 25 µM forskolin in CHO cells expressing the RII_epac sensor in the presence or absence of the AKAP450-2. n ≥ 34. (K) FRET change induced by 25 µM forskolin in CHO cells expressing PKA-GFP in the presence or absence of AKAP79 fragment (amino acid 352 to 428). n ≥ 24. (L) FRET change induced by 25 µM forskolin in CHO cells expressing PKA-GFP in the presence or absence of AKAP149 fragment (amino acid 284 to 385). n ≥ 24. (M) FRET change induced by 25 µM forskolin in CHO cells expressing PKA-GFP in the presence or absence of the AKAP Rt31 fragment (amino acid 2 to 1,678). n ≥ 17. All error bars are SEM. Two-tailed unpaired t tests were performed (*, P < 0.05; **, 0.001 < P < 0.01; ***, P < 0.001). Bars, 10 µm.
Binding of PKA to AKAP450 results in increased phosphorylation activity
To establish whether anchoring of PKA to AKAP450 affects PKA-mediated phosphorylation, we measured the activity of endogenous PKA using the cytosolic FRET-based A kinase activity reporter (AKAR) 3 (Allen and Zhang, 2006). As summarized in Fig. 5 A, upon challenge with 25 µM forskolin, cells overexpressing AKAR3 in combination with AKAP450-2 show a significantly higher cytosolic PKA activity than control cells expressing AKAR3 alone. No difference in PKA activity was observed when AKAR3 was coexpressed with the AKAP79 (Fig. 5 B). To establish what is the functional outcome of having at the centrosome a microdomain with low cAMP concentration but a subset of PKA with higher sensitivity for cAMP, we used a variant of AKAR3 that includes the D/D domain at its amino terminus (RII_AKAR3; Stangherlin et al., 2011) and therefore localizes at the centrosome (Fig. 5 C). We found that RII_AKAR3 detects a higher PKA activity at the centrosome both in resting conditions (Fig. 5 D) and in response to forskolin stimulation (Fig. 5 E). The aforementioned results confirm that anchoring of PKA to AKAP450 lowers the activation threshold of PKA, resulting in increased PKA activity at a given cAMP concentration, and show that in interphase cells, the low cAMP concentration at the centrosome is sufficient to maintain a higher basal phosphorylation activity of AKAP450-anchored PKA.
Figure 5.
Binding of PKA to AKAP450-2 increases PKA activity. (A) FRET change measured in response to 25 µM forskolin in CHO cells expressing AKAR3 in the presence or absence of AKAP450-2. n = 53. (B) FRET change measured in response to 25 µM forskolin in CHO cells expressing AKAR3 in the presence or absence of AKAP79 fragment. n = 14. A two-tailed paired t test was performed. (C) Grayscale image acquired at 480 nm upon excitation at 430 nm and showing the subcellular distribution of the RII_AKAR3 sensor. The pseudocolor image shows the FRET signal from the same cell, calculated as a 540/480-nm emission intensity ratio upon excitation at 430 nm. Images on the left show a higher magnification of the centrosomal region. Arrows point to the centrosome. Bar, 10 µm. (D) Mean of basal FRET signal measured in the bulk cytosol (cyt) and at the centrosome (centr) of cells stably expressing the RII_AKAR3 sensor. FRET values are expressed relative to the FRET value measured in the cytosol. n = 30. A two-tailed paired t test was performed. (E) Mean of FRET change elicited by 100 nM forskolin (FRSK) in cells stably expressing the RII_AKAR3 sensor and recorded in the cytosol and at the centrosome. All error bars represent SEM. n = 12. Two-tailed unpaired t tests were performed (*, P < 0.05; **, 0.001 < P < 0.01; ***, P < 0.001).
Anchoring of PKA to AKAP450 enhances RII subunit autophosphorylation
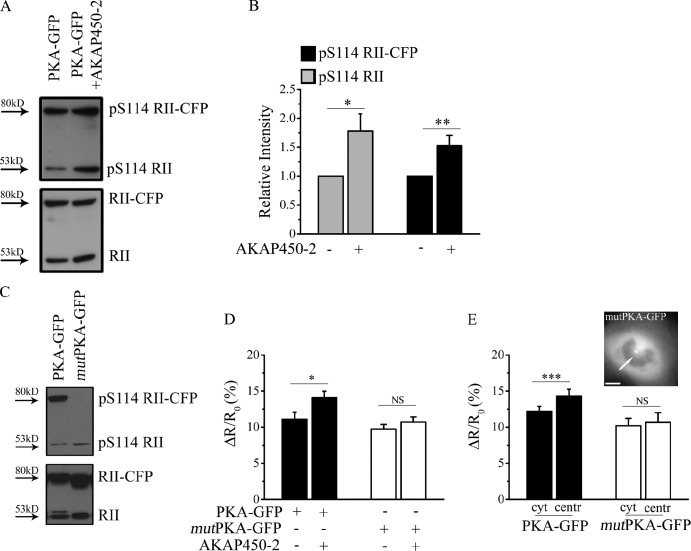
It is well established that autophosphorylation of the RII subunit at Ser114 (S114; Kim et al., 2006) results in a reduced activation threshold for PKA (Taylor et al., 1990, 2008). Therefore, we asked whether anchoring of PKA to AKAP450 may favor autophosphorylation of RII at S114. As shown in Fig. 6 (A and B), we found that phosphorylation at S114 of both endogenous RII subunits and overexpressed recombinant RII-CFP subunits was indeed significantly increased in CHO cells overexpressing AKAP450-2. To further assess whether autophosphorylation of RII is the mechanism responsible for the higher sensitivity to cAMP displayed by the PKA subset anchored to AKAP450, we generated a mutant of the PKA-GFP sensor (mutPKA-GFP) in which the RII subunit contains a S114A substitution, resulting in ablation of RII autophosphorylation (Fig. 6 C; Taylor et al., 1990; Rodríguez-Vilarrupla et al., 2005; Wehrens et al., 2006). When mutPKA-GFP was overexpressed in combination with AKAP450-2 and FRET changes measured in the bulk cytosol, the FRET change in response to 25 µM forskolin was of the same amplitude as the change measured in cells expressing the mutant sensor alone (Fig. 6 D), indicating that autophosphorylation at S114 is necessary for the ability of AKAP450-2 to affect the activation threshold of PKA. mutPKA-GFP maintains an intact D/D domain and can therefore anchor to the centrosome (Fig. 6 E, inset). When CHO cells expressing mutPKA-GFP were challenged with 25 µM forskolin and the FRET change measured in the cytosol and at the centrosome in the same cell, we found no significant difference between the two compartments (Fig. 6 E), confirming that the higher sensitivity to cAMP of the centrosome-anchored PKA requires autophosphorylation of RII at S114.
Figure 6.
AKAP450-bound PKA is more sensitive to cAMP activation as a result of increased autophosphorylation of RII. (A) Representative Western blot analysis of total RII and phospho-RII subunits. Lysates from CHO cells overexpressing PKA-GFP in the presence or absence of AKAP450-2 were blotted and probed for phospho-RII (top blot) and total RII (bottom blot) using specific antibodies. 80 and 53 kD are the expected molecular masses for recombinant RII-CFP and endogenous RII subunit, respectively. (B) Quantification of endogenous phospho-RII (pS114 RII) and recombinant phospho–RII-CFP (pS114 RII-CFP). Data are the mean of five independent experiments. A two-tailed unpaired t test was performed. (C) Western blot analysis of lysates from CHO cells expressing the PKA-GFP sensor or a mutant sensor (mutPKA-GFP) containing an S114A mutation in the RII-CFP subunit. Total and phosphor-RII subunits were detected as in A. (D) FRET changes induced by 25 µM forskolin in CHO cells expressing PKA-GFP (filled bars) and mutPKA-GFP (open bars) in the presence or absence of the AKAP450-2 fragment. n = 28. A two-tailed unpaired t test was performed. (E) Effect of 25 µM forskolin on the FRET signal detected in the cytosol (cyt) and at the centrosome (centr) of CHO cells expressing PKA-GFP (filled bars) or mutPKA-GFP (open bars). n = 16. (inset) Distribution of mutPKA-GFP in a representative CHO cell. The arrow points to the centrosome. Bar, 10 µm. All error bars are SEM. Two-tailed paired t tests were performed (*, P < 0.05; **, 0.001 < P < 0.01; ***, P < 0.001).
The centrosomal microdomain with low cAMP is abrogated in mitosis
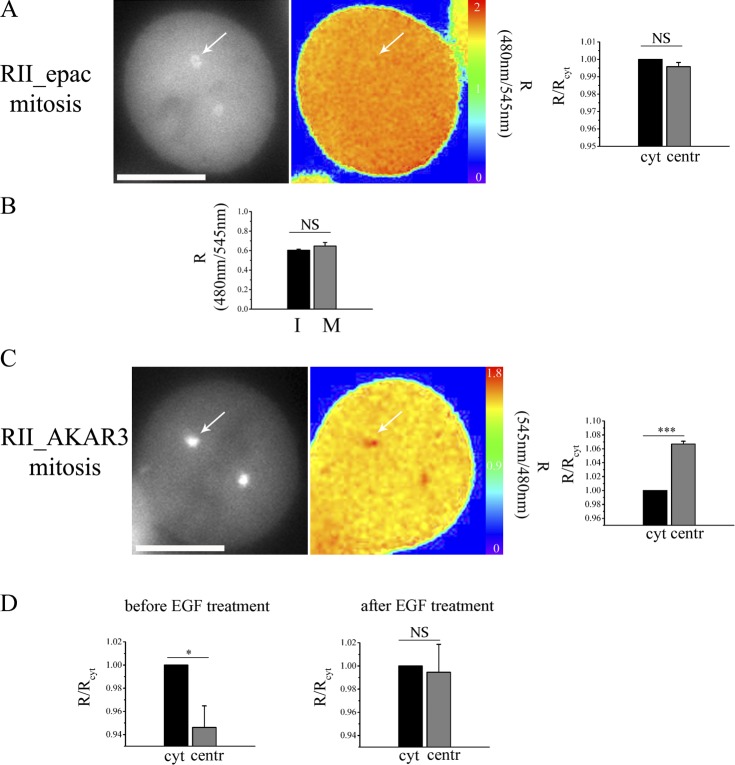
To investigate the possible functional relevance of the centrosomal microdomain with low cAMP, we sought to establish whether the difference between cytosolic and centrosomal cAMP levels changes in different stages of the cell cycle. As shown in Fig. 7 A, mitotic cells expressing RII_Epac show a uniform cAMP level at the centrosome and in the bulk cytosol. The global cAMP concentration in mitotic cells does not appear to be significantly different from the global cAMP concentration in interphase cells (Fig. 7 B), indicating that a selective increase in cAMP concentration occurs at the centrosome site in mitosis. Notably, in agreement with the local increase in cAMP concentration at the centrosome in mitotic cells, PKA phosphorylation activity is further increased selectively at this site, as detected by the RII_AKAR3 reporter (3.98% ± 0.57 and 6.69% ± 0.4 higher FRET signal in the centrosome compared with the cytosol in interphase and mitosis, respectively; P = 0.0005; compare Fig. 7 C and Fig. 5 [C and D]). The aforementioned data indicate that the centrosomal cAMP microdomain is dynamic and is abrogated in mitotic cells.
Figure 7.
The centrosomal microdomain with low cAMP is abrogated in mitosis. (A) Representative mitotic CHO cell stably expressing the RII_epac sensor. The grayscale image shows the subcellular distribution of the RII_epac sensor, and the pseudocolor image shows the FRET signal from the same cell. Arrows point to one of the centrioles. The histogram on the right displays the mean basal FRET signal measured in the bulk cytosol (cyt) and at the centrioles (centr) in n = 30 cells. FRET values are expressed relative to the FRET value measured in the cytosol. (B) Comparison of the FRET signal recorded in the bulk cytosol in interphase (I) and in mitotic (M) CHO cells stably expressing RII_epac. n = 30. A two-tailed unpaired t test was performed. (C) Representative mitotic CHO cell stably expressing the RII_AKAR3 sensor. Images are as described in A. n = 22. A two-tailed paired t test was performed. (D) Mean FRET change measured before and after treatment with 10 nM EGF in serum-depleted CHO cells stably expressing RII_epac. n = 20. All error bars represent SEM. Two-tailed unpaired t tests were performed (*, P < 0.05; ***, P < 0.001). Bars, 10 µm.
As the MAPK extracellular regulation kinase (ERK) has been shown to inhibit PDE4D3 via phosphorylation of its catalytic domain (Baillie et al., 2000), we asked whether mitogenic stimuli that activate the MAPK pathway could affect the centrosomal cAMP microdomain. As shown in Fig. 7 D, in CHO cells stably expressing RII_Epac and treated with 10 nM EGF, the centrosomal microdomain with low cAMP was completely abrogated as a consequence of a selective increase of the FRET signal at this site.
Local manipulation of cAMP signals at the centrosome distinctly affects the cell cycle
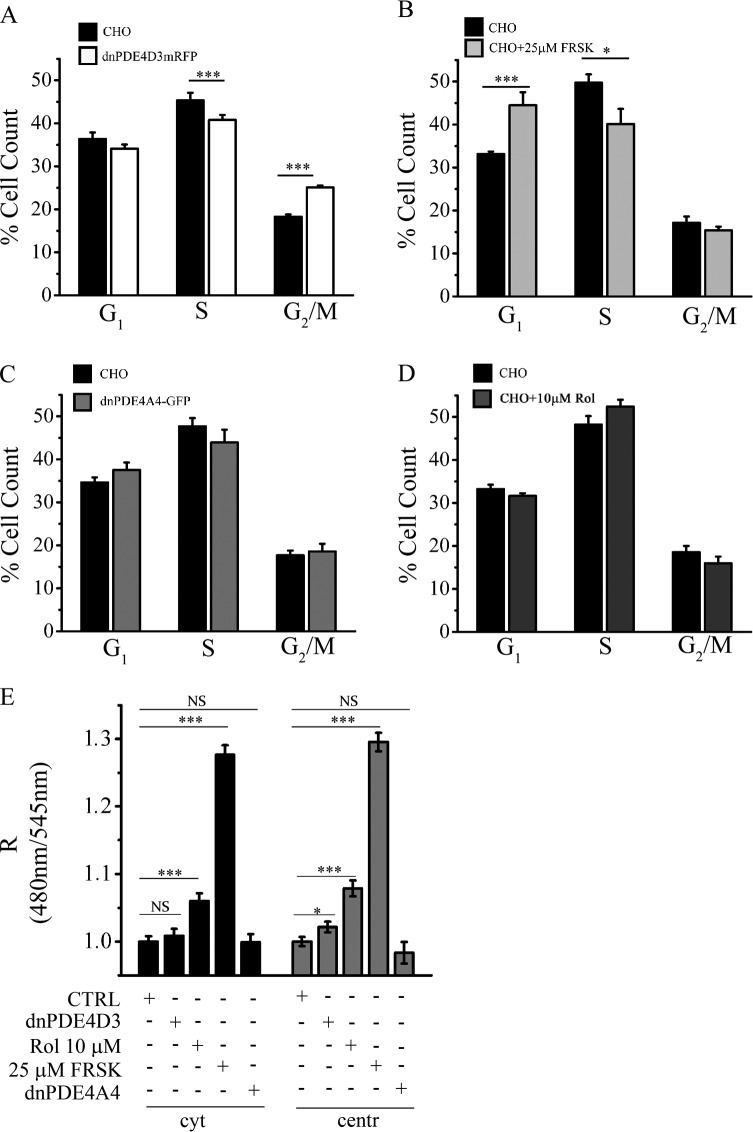
It has been previously reported that displacement of the endogenous centrosomal AKAP450 and the consequent delocalization of centrosomal PKA type II impair cell cycle progression (Keryer et al., 2003), indicating that centrosomal PKA may play an important role in the control of cell division. The data presented in the previous section suggest that the unique handling of cAMP signals at the centrosome at different stages of the cell cycle may be important for the regulation of cell cycle progression. To test this hypothesis, we used flow cytometric analysis to monitor the effects on the cell cycle of displacing endogenous PDE4D3 with dnPDE4D3, a maneuver that results in local increase of cAMP at the centrosome (Fig. 3, J and K; and Fig. 8 E). We found that CHO cells stably expressing dnPDE4D3 show a significantly higher number of cells in G2/M and significantly lower number of cells in S phase, suggestive of a block of the cell cycle in G2/M (Fig. 8 A). Similar results were found in cells synchronized in S phase before analysis (Fig. S5). In contrast, and as previously described (Gützkow et al., 2002), treatment of CHO cells with 25 µM forskolin, which ensues a global increase in cAMP levels (Fig. 8 E), results in a block of the cell cycle in G1, with an increased proportion of cells with 2N DNA content, a significantly reduced number of cells in S phase, but no change in G2/M (Fig. 8 B). Overexpression of a catalytically inactive version of the control enzyme dnPDE4A4 did not show any effect on the cell cycle (Fig. 8 C). Inhibition of all PDE4 isoforms with 10 µM rolipram, a treatment that results in increase of cAMP both at the centrosome and in the cytosol, albeit at a lower level than elicited by forskolin (Fig. 8 E), did not result in a detectable effect on cell cycle progression (Fig. 8 D). The aforementioned findings show that selective local manipulation of cAMP at the centrosome activates a downstream pathway with distinct effects on cell division.
Figure 8.
Displacement of PDE4D3 results in altered cell cycle progression. (A) Quantification of flow cytometry scan analysis of control CHO cells and CHO cells stably expressing the RFP-tagged and catalytically inactive mutant of PDE4D3 (dnPDE4D3mRFP). The same analysis was performed for CHO cells treated with 25 µM forskolin. (B–D) CHO cells stably expressing the catalytic inactive mutant of PDE4A4 (dnPDE4A4-GFP; C) and CHO cells treated with 10 µM rolipram (Rol; D). Histograms indicate the mean percentages of cells in various phases of the cell cycle. Data are the mean of at least six independent experiments. FRSK, forskolin. All error bars are SEM. Two-tailed unpaired t tests were performed (*, P < 0.05; **, 0.05 < P < 0.01; ***, P < 0.001). (E) Effect of overexpression of dnPDE4D3mRFP, 10 µM rolipram, and 25 µM forskolin and overexpression of dnPDE4A4-GFP on cytosolic (cyt) and centrosomal (centr) cAMP compared with untreated and unstimulated CHO control cells (CTRL). All error bars are SEM. Two-tailed unpaired t tests were performed (*, P < 0.05; ***, P < 0.001).
Ablation of the centrosomal microdomain with low cAMP results in accumulation of cells in prophase
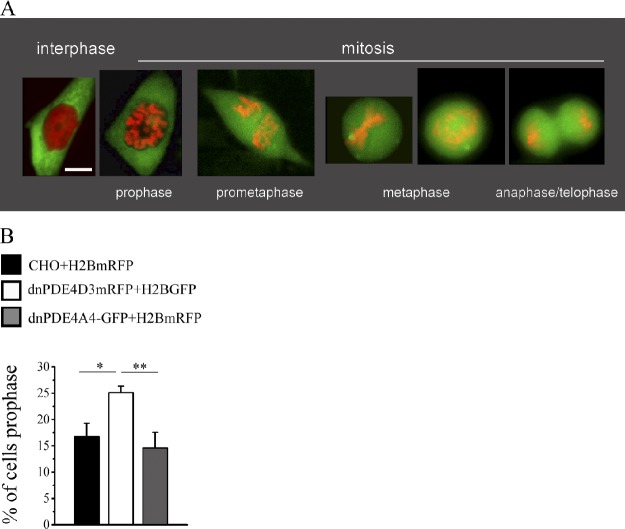
To gain further insight into the mechanism responsible for accumulation of cells in G2/M when the centrosomal cAMP microdomain is perturbed, we generated a stable CHO cell clone expressing an RFP-tagged histone 2B (H2B-RFP) alone or in combination with a GFP-tagged dnPDE4D3 or dnPDE4A4. H2B-RFP labels the chromatin and allows for identification of different phases of the cell cycle. As illustrated in Fig. 9 A, interphase cells show a homogeneous red fluorescence in the nucleus, cells in prophase can be clearly identified by the presence of condensed chromatin and an intact nuclear membrane, and subsequent mitotic phases can be identified by the position of the chromosomes along the mitotic fuse. Using this approach, we assigned cells to one of the following categories: interphase, prophase, metaphase, or anaphase/telophase. We found that CHO cells coexpressing H2B-RFP and dnPDE4D3 have a significantly higher number of cells in prophase compared with CHO cells expressing H2B-RFP alone or H2B-RFP in combination with dnPDE4A4 (Fig. 9 B).
Figure 9.
Overexpression of dnPDE4D3 results in accumulation of cells in prophase. (A) Representative living CHO cells stably expressing dnPDE4A4RFP and the GFP-tagged H2B imaged in interphase and in different phases of the mitotic cycle, as apparent from the analysis of chromosome condensation. Bar, 10 µm. (B) Summary of the results of seven independent experiments in which multiple snapshots of living CHO cells expressing either GFP-tagged H2B and dnPDE4A4RFP or RFP-tagged H2B and dnPDE4D3GFP were acquired and cells assigned to the different stages of the cell cycle, as defined in A. At least 120 mitotic cells were analyzed for each experiment. Data are expressed as the percentage of mitotic cells that appears to be in prophase. All error bars are SEM. *, P < 0.05; **, 0.001 < P < 0.01.
Discussion
The data reported here strongly support the novel proposal that the centrosome in interphase is a subcellular compartment in which basal cAMP levels are lower than in the bulk cytosol as a consequence of centrosomal localization of PDE4D3, anchoring of PKA to centrosomal AKAP450 lowers the activation threshold of PKA as a consequence of increased autophosphorylation of AKAP450-anchored RII subunits at S114, the centrosomal cAMP microdomain is dynamic and is abrogated in mitosis, possibly via activation of the MAPK pathway, and manipulation of cAMP at the centrosome via displacement of PDE4D3 uniquely affects cell cycle progression, resulting in a highly significant increase in the number of cells in G2/M phase with accumulation of cells selectively in prophase. These findings demonstrate that local regulation of cAMP signals at the centrosome is critical for control of cell division.
In addition to a well-established function as a microtubule organizing center, the centrosome has recently been shown to play a role in cell cycle control (Doxsey et al., 2005). For example, active maturation-promoting factor, the key initiator of mitosis, is found at the centrosome during prophase (Jackman et al., 2003), and studies in which the centrosome was removed by microsurgical dissection (Hinchcliffe et al., 2001) or laser ablation (Khodjakov and Rieder, 2001) have provided direct evidence for a role of the centrosome in cell cycle progression. Of particular note, cell cycle arrest in G1 (Gillingham and Munro, 2000; Keryer et al., 2003) as well as a block of cytokinesis (Keryer et al., 2003) were observed when AKAP450 and PKA were selectively displaced from the centrosome, suggesting that a cAMP/PKA signaling module localized at this site may serve a critical role. cAMP/PKA signaling has been shown to be involved in many aspects of cell cycle regulation, including centrosome duplication, S phase, G2 arrest, mitotic spindle formation, exit from M phase, and cytokinesis (Matyakhina et al., 2002), and it is possible that different cAMP/PKA signaling modules may be responsible for the regulation of specific cell cycle–related events. In line with this view, our results show that whereas a global increase in cAMP levels, as generated by forskolin stimulation, results in an accumulation of cells in G1, the local increase of cAMP generated by displacing PDE4D3 from the centrosome has a completely different effect, resulting in accumulation of cells in G2/M. Further investigations will be necessary to identify the specific targets downstream of AKAP450-anchored PKA. However, in agreement with our findings, cAMP/PKA-dependent reduction of histone H3 phosphorylation (Rodriguez-Collazo et al., 2008b), an event that results in disruption of G2 progression in adenocarcinoma cells, has been shown to require a concentration of cAMP that is significantly lower than the amount of cAMP necessary for PKA-mediated phosphorylation of cAMP response element–binding protein (Rodriguez-Collazo et al., 2008a), suggesting that the pool of PKA responsible for control of G2 progression is more sensitive to cAMP than the pool of PKA that regulates gene transcription.
The data reported here clearly show that cAMP signals are uniquely processed at the centrosome, where a high-sensitivity PKA subset is associated with a PKA-activatable and ERK-inhibitable PDE. Our findings are compatible with a model whereby the lower activation threshold of PKA tethered to AKAP450 allows for local activation of PKA at a concentration of cAMP that is insufficient to activate PKA subsets at other subcellular locations. Mitogenic stimuli selectively increase cAMP at the centrosome, resulting in further activation of PKA at this site in the absence of global increase of cAMP levels. In agreement with this model, overexpression of a catalytically inactive mutant of PDE4D3 in COS1 cells was shown to result in its PKA-dependent hyperphosphorylation selectively at the centrosome even at resting levels of cAMP (McCahill et al., 2005), suggesting that the centrosomal PDE4D3 modulates activation of the local pool of PKA at basal cAMP concentrations. The feedback loop mechanism described here and involving a high-sensitivity subset of PKA coupled with a PKA-activatable and ERK-inhibitable PDE at the centrosome not only allows tight temporal control of centrosomal cAMP signals but also provides a potential basis for autonomous regulation of centrosomal cAMP/PKA-dependent events, independently of global increase in cAMP and therefore of Gs protein–coupled receptor activation.
The functional relevance of the centrosomal cAMP microdomain that we have identified is illustrated by the disruption of cell cycle progression in CHO cells in which cAMP levels are selectively elevated at the centrosome via overexpression of a catalytically inactive PDE4D3. We found that in these conditions, cell cycle progression is disrupted, and cells accumulate in prophase. Of note, we also found that overexpression of dnPDE4D3 is not tolerated in RPE1 cells (Fig. S3), indicating that the effect of manipulating the centrosomal cAMP microdomain may be incompatible with cell cycle progression in nontransformed cells.
The data presented here show that during interphase, although the cAMP level at the centrosome is lower than in the bulk cytosol, it is sufficient to maintain a tonic activity of PKA at this site as a consequence of the reduced activation threshold of the local PKA subset. This finding is in agreement with the established notion that PKA activity is required to maintain cells in interphase (Bombik and Burger, 1973; Lamb et al., 1991). Our analysis shows that in mitosis, there is a further increase in PKA activity at the centrosome, raising the question of how PKA activity is tuned temporally to allow progression from interphase to mitosis. Further studies with higher temporal resolution will be necessary to dissect cAMP signals and PKA activity at the centrosome within the same cell as it progresses from interphase and through mitosis to establish whether there is a short temporal window within which PKA activity is reduced to allow the interphase/mitosis transition. In this context, the activity of phosphatases may also be critical, as it may counterbalance substrate phosphorylation by a tonically active centrosomal PKA subset.
Our study uncovers a completely novel mechanism of PKA activity regulation. Such regulation relies on binding of PKA to AKAP450 and the consequent reduction of the kinase activation threshold rather than on changes in the level of cAMP and is therefore effective only at the sites where AKAP450 is localized. Noncentrosomal splice variants of AKAP450 localize at the sarcolemma of cardiac myocytes in a complex with the slowly activating potassium channels IKs (Walsh and Kass, 1988) and to NR1 subunits of glutamate receptors at postsynaptic sites in neurons (Lin et al., 1998), and it will be interesting to establish whether the regulation described here at the centrosome also operates at these other sites. The novel mechanism we describe defines a new function for AKAPs and introduces a further level of complexity to the already sophisticated regulation of cAMP/PKA signaling and may have implications that extend beyond the control of cell cycle progression.
Materials and methods
Reagent
Forskolin (catalog no. F6886), rolipram (catalog no. R6520), cilostamide (catalog no. C7971), EHNA (catalog no. E114), H-89 (catalog no. B1427), and EGF (catalog no. E9644) were purchased from Sigma-Aldrich.
Generation of fluorescent chimeras
The CFP-tagged R subunit of PKA (Lissandron et al., 2005) was subcloned into pCDNA3.1/Zeo (+) as an NheI–XbaI fragment. For the generation of mutRII-CFP, the S114A mutation was introduced using the QuikChange site-directed mutagenesis kit (Agilent Technologies). The AKAP450-2 fragment (provided by K. Taskén, University of Oslo, Oslo, Norway) from amino acid 933 to amino acid 1,804 encoded by the AKAP450 cDNA (DDBJ/EMBL/GenBank accession no. AJ131693; Witczak et al., 1999) was amplified by PCR and subcloned as an NheI–BamHI fragment in pcDNA3.1/Hygro (+). For mutAKAP450-2, the S1451P mutation was introduced using the QuikChange site-directed mutagenesis kit. The Rt31 fragment from amino acid 2 to amino acid 1,678 (GenBank/EMBL/DDBJ accession no. AF387102; Klussmann et al., 2001) was PCR amplified and subcloned as an EcoRI–XhoI insert into pIRES vector (catalog no. 631605; Takara Bio Inc.). Generation of dnPDE4D3mRFP, the sequence encoding for dnPDE4D3 (McCahill et al., 2005), was amplified by PCR and subcloned into the BstXI site of the multiple cloning site of pcDNA3.1/Hygro (+). The monomeric RFP was then inserted as an XhoI–XbaI fragment in frame at the C terminus of dnPDE4D3. A schematic of the sensors and AKAP fragments used in this study is shown in Fig. S2. AKAR3 was a gift from J. Zhang (Johns Hopkins Institute, Baltimore, MA), AKAP-IS-GFP was a gift from J.D. Scott (Howard Hughes Medical Institute, University of Washington, Seattle, WA), and RIAD-GFP was a gift from K. Taskén.
Cell culture and transfection
CHO-K1 cells from CHO (Puck et al., 1958) were grown in Ham’s F12 medium (catalog no. 21765-029; Invitrogen) supplemented with 10% (vol/vol) FBS (catalog no. 10270-106, Invitrogen), 2 mM L-Glutamine (catalog no. 25030-024; Invitrogen), 100 U/ml penicillin, and 100 µg/ml streptomycin (catalog no. 15070063; Invitrogen) at 37°C in a humidified atmosphere containing 5% CO2. SH-SY5Y cells from human neuroblastoma (Biedler et al., 1978) were grown in Ham’s F12/DME (1:1; catalog no. 42430-025; Invitrogen) supplemented with 10% (vol/vol) FBS, 2 mM L-Glutamine, 1% (vol/vol) nonessential amino acids (catalog no. 11140-035; Invitrogen), 100 U/ml penicillin, and 100 µg/ml streptomycin at 37°C in a humidified atmosphere containing 5% CO2. RAW264.7 cells (Raschke et al., 1978) were grown in DME (catalog no. 41966; Invitrogen) supplemented with 10% (vol/vol) FBS (catalog no. F9665; Sigma-Aldrich), 2 mM L-Glutamine, 100 U/ml penicillin, and 100 µg/ml streptomycin at 37°C in a humidified atmosphere containing 5% CO2. Enriched primary cultures of neonatal ventricular heart fibroblasts (RCF) were obtained from 1–3-d-old Sprague Dawley rats, as described in Mongillo et al. (2006) . In brief, rats were killed by cervical dislocation, and ventricular tissue was enzymatically digested with a mixture of collagenase (Roche) and pancreatin (Sigma-Aldrich). The isolated cell suspension was preplated for 2 h in DME high glucose (catalog no. 42430025; Invitrogen) supplemented with 20% (vol/vol) Medium 199 (catalog no. 31150-022; Invitrogen), 5% (vol/vol) horse serum, 0.5% (vol/vol) newborn calf serum, 2 mM L-Glutamine, 10 U/ml penicillin, and 10 µg/ml streptomycin at 37°C in a humidified atmosphere containing 5% CO2. The plastic-adherent nonmyocyte cells obtained are fibroblasts. These were trypsinized and plated on coverslips for further analysis. HONs were grown in DME/F12 (catalog no. 11330-032; Invitrogen) supplemented with 10% (vol/vol) FBS, 100 U/ml penicillin, and 100 µg/ml streptomycin at 37°C in a humidified atmosphere containing 5% CO2. The retinal pigment epithelial RPE1 cells (Bodnar et al., 1998) were grown in DME/F-12 medium (catalog no. 11320–074; Invitrogen) supplemented with 10% (vol/vol) FBS (catalog no. 10270-106; Invitrogen), 2 mM L-Glutamine, 100 U/ml penicillin, and 100 µg/ml streptomycin at 37°C in a humidified atmosphere containing 5% CO2.
For transient expression, cells were seeded onto 24-mm glass coverslips in complete medium and grown for 24 h, as described in Terrin et al. (2006). Transfections were performed at 50–70% of confluence. All cell types were transfected with TransIT-LT1 reagent (catalog no. 2300; Mirus Bio LLC) following the supplier’s instructions and using 2–4 µg DNA per coverslip. Experiments were performed 24–48 h after transfection.
Knockdown of PDE4D was achieved by using an siRNA oligonucleotide targeting the PDE4D gene (125 nM final concentration, 5′-GAACUUGCCUUGAUGUACA-3′ sequence; Thermo Fisher Scientific), as previously described (Lynch et al., 2005). Control experiments were performed using siGLO red transfection indicator (125 nM; catalog no. D-001630-02-20; Thermo Fisher Scientific).
Generation of stable clones
Stable clones expressing the PKA-GFP sensor have been previously described (Vaasa et al., 2010). In brief, a CHO clone stably expressing RII-CFP was selected with 300 µg/ml Zeocine and successively used to select a stable clone expressing the C-YFP subunit by using 800 µg/ml Geneticin (Promega). CHO clones stably expressing either RII_epac or RII_AKAR3 sensor (Di Benedetto et al., 2008) were obtained by selection with 800 µg/ml Geneticin. CHO and RPE1 clones stably expressing dnPDE4D3mRFP1 were selected using 700 µg/ml Hygromycin B (Invitrogen). CHO clone stably expressing pcDNA3dnPDE4A4-GFP was selected using 800 µg/ml Geneticin. Stable clones expressing either the GFP- or the RFP-tagged histone H2B were obtain by selection with 5 µg/ml Blasticidin S (Invitrogen). In all cases, after 12 d of treatment with the antibiotic, cells were seeded in a 96-well plate at 0.8 cells/well, and single clones growing in individual wells were selected for further expansion.
RT-PCR
Total mRNA was extracted with TRIzol reagent (catalog no. 15596-026; Invitrogen) from cells transfected with dnPDE4D3mRFP before or after selection with Geneticin. An aliquot of total mRNA was reversed transcribed with 1 µl SuperScript II Reverse Transcriptase (2,000 U/µl; catalog no. 18064-022; Invitrogen) to generate cDNA. Amplification of the coding regions of dnPDE4D3mRFP was performed by using specific primers annealing on the D484A mutation (McCahill et al., 2005). The primers used are as follows: 5′-GGTAACCGGCCCTTGACTG-3′ and 5′-GGTTCTTCAGAATATGGTGCACTGTGCAGAT-3′ for amplification of PDE4D3 wild type and 5′-GGTAACCGGCCCTTGACTG-3′ and 5′-GGTTCTTCAGAATATGGTGCACTGTGCAGCA-3′ for amplification of dnPDE4D3.
FRET imaging
Cells stably or transiently expressing a FRET-based cAMP sensor were imaged 24 h after transfection, as described in Monterisi et al. (2012). In brief, cells were imaged on an inverted microscope (IX81; Olympus) equipped with a Plan Apochromat N 60×/1.42 oil immersion objective, a charge-coupled device camera (ORCA-AG; Hamamatsu Photonics), and a custom-made beam splitter including the specific set of emission filters for CFP and YFP acquisition (dichroic mirror 505DCLP, YFP emission of 545 nm, and CFP emission of 480 nm; Chroma Technology Corp.). During FRET experiments, cells were bathed with 37°C prewarmed PBS. Images were acquired using CellR software (Olympus) and processed using ImageJ (National Institutes of Health). FRET changes were measured in different cell compartments by drawing a region of interest (ROI) around a specific compartment (centrosome or cytosol). FRET changes of all the cAMP sensors were measured as changes in the 480/545-nm fluorescence emission intensities after background subtraction on excitation at 430 nm. For AKAR3 and RII_AKAR3 sensors, FRET changes were measured as changes in the ratio between 545/480-nm fluorescent emission intensities after background subtraction upon excitation at 430 nm. For dynamic FRET changes, the kinetic of the 480/545-nm emission intensity ratio is plotted against time, and the mean FRET response is expressed as the percentage of ΔR/R0, in which ΔR = R − R0. R0 is the ratio at time = 0 s, and R is the ratio at time = t seconds. For steady-state (or basal) FRET, 480/545-nm emission intensity values measured in the cytosol (Rcyt) and at the centrosome (Rcentr) are expressed as normalized values with respect to the basal FRET ratio value measured in the cytosol. Ratiometric images are displayed in pseudocolor, according to a user-defined lookup table that assigns a different color to each ratio value, as indicated.
Western blotting and immunoprecipitation
Untransfected CHO cells or CHO cells stably expressing PKA-GFP were seeded on 10-cm tissue culture dishes, treated as indicated, and washed twice with ice-cold D-PBS before cell lysis. Cell lysates were prepared in lysis buffer containing 25 mM Hepes, pH 7.5, 2.5 mM EDTA, 50 mM NaCl, 30 mM sodium pyrophosphate, 10% (vol/vol) glycerol, 1% (vol/vol) Triton X-100 (catalog no. T8532; Sigma-Aldrich), and cOmplete EDTA-free protease inhibitor cocktail tablets (catalog no. 11836170001; Roche). AKAP450-GFP was isolated from cell lysates via immunoprecipitation with GFP-Trap beads (catalog no. gta-100; ChromoTek) following the manufacturer’s instructions. Protein concentration was determined using the Bradford protein assay (Bio-Rad Laboratories). Proteins were separated by gradient gel electrophoresis on NuPAGE Novex 4–12% Bis-Tris gels (Invitrogen) and transferred to polyvinylidene fluoride membranes (EMD Millipore). Membranes were then blocked either with protein-free T20 (TBS) blocking buffer (Thermo Fisher Scientific) or 5% (wt/vol) skimmed milk in TBS-T for 1 h at room temperature. The following antibodies were used to probe the membranes: mouse anti-PKARII (BD), mouse anti-PKARII (pS114; BD), goat pan-PDE4D (a gift from M. Houslay), and goat anti–γ-tubulin (C-20; Santa Cruz Biotechnology, Inc.). Results, representing the mean of at least three independent experiments, were normalized to the amount of γ-tubulin.
Cells synchronization
G1/S synchrony was obtained by double block with thymidine (Sigma-Aldrich). In brief, cells were treated with 5 mM thymidine in FBS-free medium for 16 h, released to cycle in medium supplemented with FBS for 8 h, and blocked again for an additional 16 h. Cells were allowed to recover for 24 h in completed medium with or without 10 µM rolipram before FACS analysis.
Immunostaining and confocal imaging
Cells transiently or stably expressing the PKA-GFP sensor were washed three times with ice-cold D-PBS. The centrosome was exposed by treatment with PHEM solution (45 mM Pipes, 45 mM Hepes, 10 mM EGTA, 5 mM MgCl2, 1 mM PMSF, and 0.1% (vol/vol) Triton X-100, pH 6.9) for 30 s at room temperature. Cells were then fixed with ice-cold methanol for 5 min at −20°C, washed twice in D-PBS, and saturated in 3% BSA for 30 min at room temperature. Primary antibodies were diluted in 3% BSA and incubated overnight in a wet chamber. CTR453 (provided by G. Keryer, Institut Curie, Orsay, France; Bailly et al., 1989) was used at a 1:5 dilution, rabbit anti-PDE4D3 (a gift from M. Houslay, University of Glasgow, Scotland, UK) was used at a 1:500 dilution, and goat anti–γ-tubulin (C-20) was used at 1:2,000. Goat anti–mouse Alexa Fluor 568 (Invitrogen), goat anti–rabbit Alexa Fluor 568 (Invitrogen), and donkey anti–goat Alexa Fluor 488 (Invitrogen) were used as secondary antibodies. Secondary antibody alone was used for controls. Confocal images were acquired with an inverted microscope (Eclipse TE300; Nikon) equipped with a spinning-disk confocal system (Ultraview Live Cell Imager; PerkinElmer), a 60× 1.4 NA Plan Apochromat objective (Nikon), and a 12-bit charge-coupled device camera (Orca-ER; Hamamatsu Photonics). Cells were excited at a 568-nm laser line of a 643-series argon krypton laser (643-Ryb-A02; CVI Melles Griot) for imaging of the Alexa Fluor 568 fluorophore and the 405-nm line of a diode laser (iFLEX2000; Point Source) for imaging CFP. The emission filters were 607/45 for the red emission and 480/30 for the cyan emission.
Flow cytometry scan analysis
Approximately 106 of cells were treated as indicated and grown in a T75 flask for 48 h. After 48 h, exponentially growing cells were trypsinized, washed twice with D-PBS, and resuspended in 300 µl D-PBS. 700 µl of ice-cold 70% (vol/vol) EtOH/PBS was added dropwise, and the samples were incubated at 4°C for 1 h. After incubation, cells were spun down, washed with 1 ml D-PBS, resuspended in 250 µl D-PBS containing 5 µl of 10 mg/ml RNAaseA (Sigma-Aldrich), and incubated for 1 h at 37°C. Samples were stained with 5 µl of 1 mg/ml of propidium iodide and kept in the dark at 4°C until analysis. Flow cytometric analysis was performed using a FACSCalibur flow cytometer (BD), and data collected were analyzed with FlowJo software and computed using the Dean-Jett-Fox model.
Statistics
Data are presented as mean ± SEM. Two-tailed paired and unpaired Student’s t tests were used to determine significance between groups, as indicated. The number of replicates and the type of Student’s t test used are indicated in the text. Asterisks are used to indicate levels of significance based on P values: *, P < 0.05; **, 0.001 < P < 0.01; ***, P < 0.001.
Online supplemental material
Fig. S1 shows that PKA-GFP localizes at the centrosome and that its overexpression does not affect the centrosome morphology. Fig. S2 shows a schematic representation of the RII_epac sensor, AKAP450, and AKAP450-2 fragment. A schematic of the interaction between the FRET-based sensors and AKAP constructs used in this study is also shown. Fig. S3 shows the localization of the PKA-GFP sensor and PDE4D3 in the nontransformed cell line RPE1 and also shows analysis of the basal FRET signal at the centrosome and in the bulk cytosol in the same cell line. Fig. S4 shows the localization of endogenous PDE4D3 in CHO cells, the efficiency of its knockdown, and the localization of the overexpressed catalytically dead PDE4D3 isoform in CHO cells. Fig. S5 shows the FACS analysis of cell cycle progression in CHO, CHO treated with rolipram, and CHO stably expressing the catalytically dead PDE4D3 after synchronization in G1/S phase. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201201059/DC1.
Supplementary Material
Acknowledgments
The authors would like to thank Miles Houslay for dnPDE4D3, dnPDE4A4-GFP, and goat anti–pan-PDE4D and rabbit anti-PDE4D3 antibodies; John D. Scott for SuperAKAP-IS-GFP; Kjetil Taskén for AKAP450-2, AKAP79, AKAP149, and RIAD-GFP; Enno Klussmann (Max-Delbrück-Centrum für Molekulare Medizin, Berlin, Germany) for Rt31; Jin Zhang for AKAR3; and Guy Keryer for the CTR453 AKAP450–specific antibody.
This work was supported by the Fondation Leducq (O6 CVD 02), the British Heart Foundation (PG/07/091/23698), and the National Science Foundation–National Institutes of Health Collaborative Research in Computational Neuroscience program (National Institutes of Health R01 AA18060) to M. Zaccolo.
Footnotes
Abbreviations used in this paper:
- AKAP
- A kinase–anchoring protein
- AKAR
- A kinase activity reporter
- D/D
- dimerization/docking
- ERK
- extracellular regulation kinase
- FRET
- fluorescence resonance energy transfer
- IS
- in silico
- PDE
- phosphodiesterase
- RCF
- rat cardiac fibroblast
- RIAD
- RI-anchoring disruptor
- ROI
- region of interest
References
- Allen M.D., Zhang J. 2006. Subcellular dynamics of protein kinase A activity visualized by FRET-based reporters. Biochem. Biophys. Res. Commun. 348:716–721 10.1016/j.bbrc.2006.07.136 [DOI] [PubMed] [Google Scholar]
- Alto N.M., Soderling S.H., Hoshi N., Langeberg L.K., Fayos R., Jennings P.A., Scott J.D. 2003. Bioinformatic design of A-kinase anchoring protein-in silico: A potent and selective peptide antagonist of type II protein kinase A anchoring. Proc. Natl. Acad. Sci. USA. 100:4445–4450 10.1073/pnas.0330734100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillie G.S., MacKenzie S.J., McPhee I., Houslay M.D. 2000. Sub-family selective actions in the ability of Erk2 MAP kinase to phosphorylate and regulate the activity of PDE4 cyclic AMP-specific phosphodiesterases. Br. J. Pharmacol. 131:811–819 10.1038/sj.bjp.0703636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly E., Dorée M., Nurse P., Bornens M. 1989. p34cdc2 is located in both nucleus and cytoplasm; part is centrosomally associated at G2/M and enters vesicles at anaphase. EMBO J. 8:3985–3995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biedler J.L., Roffler-Tarlov S., Schachner M., Freedman L.S. 1978. Multiple neurotransmitter synthesis by human neuroblastoma cell lines and clones. Cancer Res. 38:3751–3757 [PubMed] [Google Scholar]
- Bodnar A.G., Ouellette M., Frolkis M., Holt S.E., Chiu C.P., Morin G.B., Harley C.B., Shay J.W., Lichtsteiner S., Wright W.E. 1998. Extension of life-span by introduction of telomerase into normal human cells. Science. 279:349–352 10.1126/science.279.5349.349 [DOI] [PubMed] [Google Scholar]
- Bombik B.M., Burger M.M. 1973. c-AMP and the cell cycle: Inhibition of growth stimulation. Exp. Cell Res. 80:88–94 10.1016/0014-4827(73)90278-4 [DOI] [PubMed] [Google Scholar]
- Carlson C.R., Ruppelt A., Taskén K. 2003. A kinase anchoring protein (AKAP) interaction and dimerization of the RIalpha and RIbeta regulatory subunits of protein kinase a in vivo by the yeast two hybrid system. J. Mol. Biol. 327:609–618 10.1016/S0022-2836(03)00093-7 [DOI] [PubMed] [Google Scholar]
- Carlson C.R., Lygren B., Berge T., Hoshi N., Wong W., Taskén K., Scott J.D. 2006. Delineation of type I protein kinase A-selective signaling events using an RI anchoring disruptor. J. Biol. Chem. 281:21535–21545 10.1074/jbc.M603223200 [DOI] [PubMed] [Google Scholar]
- Conti M., Beavo J. 2007. Biochemistry and physiology of cyclic nucleotide phosphodiesterases: Essential components in cyclic nucleotide signaling. Annu. Rev. Biochem. 76:481–511 10.1146/annurev.biochem.76.060305.150444 [DOI] [PubMed] [Google Scholar]
- Cuschieri L., Nguyen T., Vogel J. 2007. Control at the cell center: The role of spindle poles in cytoskeletal organization and cell cycle regulation. Cell Cycle. 6:2788–2794 10.4161/cc.6.22.4941 [DOI] [PubMed] [Google Scholar]
- Di Benedetto G., Zoccarato A., Lissandron V., Terrin A., Li X., Houslay M.D., Baillie G.S., Zaccolo M. 2008. Protein kinase A type I and type II define distinct intracellular signaling compartments. Circ. Res. 103:836–844 10.1161/CIRCRESAHA.108.174813 [DOI] [PubMed] [Google Scholar]
- Dodge K.L., Khouangsathiene S., Kapiloff M.S., Mouton R., Hill E.V., Houslay M.D., Langeberg L.K., Scott J.D. 2001. mAKAP assembles a protein kinase A/PDE4 phosphodiesterase cAMP signaling module. EMBO J. 20:1921–1930 10.1093/emboj/20.8.1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxsey S., Zimmerman W., Mikule K. 2005. Centrosome control of the cell cycle. Trends Cell Biol. 15:303–311 10.1016/j.tcb.2005.04.008 [DOI] [PubMed] [Google Scholar]
- Feliciello A., Gottesman M.E., Avvedimento E.V. 2001. The biological functions of A-kinase anchor proteins. J. Mol. Biol. 308:99–114 10.1006/jmbi.2001.4585 [DOI] [PubMed] [Google Scholar]
- Gillingham A.K., Munro S. 2000. The PACT domain, a conserved centrosomal targeting motif in the coiled-coil proteins AKAP450 and pericentrin. EMBO Rep. 1:524–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold M.G., Lygren B., Dokurno P., Hoshi N., McConnachie G., Taskén K., Carlson C.R., Scott J.D., Barford D. 2006. Molecular basis of AKAP specificity for PKA regulatory subunits. Mol. Cell. 24:383–395 10.1016/j.molcel.2006.09.006 [DOI] [PubMed] [Google Scholar]
- Gützkow K.B., Naderi S., Blomhoff H.K. 2002. Forskolin-mediated G1 arrest in acute lymphoblastic leukaemia cells: Phosphorylated pRB sequesters E2Fs. J. Cell Sci. 115:1073–1082 [DOI] [PubMed] [Google Scholar]
- Herberg F.W., Maleszka A., Eide T., Vossebein L., Tasken K. 2000. Analysis of A-kinase anchoring protein (AKAP) interaction with protein kinase A (PKA) regulatory subunits: PKA isoform specificity in AKAP binding. J. Mol. Biol. 298:329–339 10.1006/jmbi.2000.3662 [DOI] [PubMed] [Google Scholar]
- Hinchcliffe E.H., Miller F.J., Cham M., Khodjakov A., Sluder G. 2001. Requirement of a centrosomal activity for cell cycle progression through G1 into S phase. Science. 291:1547–1550 10.1126/science.1056866 [DOI] [PubMed] [Google Scholar]
- Houslay M.D. 2010. Underpinning compartmentalised cAMP signalling through targeted cAMP breakdown. Trends Biochem. Sci. 35:91–100 10.1016/j.tibs.2009.09.007 [DOI] [PubMed] [Google Scholar]
- Jackman M., Lindon C., Nigg E.A., Pines J. 2003. Active cyclin B1-Cdk1 first appears on centrosomes in prophase. Nat. Cell Biol. 5:143–148 10.1038/ncb918 [DOI] [PubMed] [Google Scholar]
- Keryer G., Witczak O., Delouvée A., Kemmner W.A., Rouillard D., Tasken K., Bornens M. 2003. Dissociating the centrosomal matrix protein AKAP450 from centrioles impairs centriole duplication and cell cycle progression. Mol. Biol. Cell. 14:2436–2446 10.1091/mbc.E02-09-0614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A., Rieder C.L. 2001. Centrosomes enhance the fidelity of cytokinesis in vertebrates and are required for cell cycle progression. J. Cell Biol. 153:237–242 10.1083/jcb.153.1.237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C., Vigil D., Anand G., Taylor S.S. 2006. Structure and dynamics of PKA signaling proteins. Eur. J. Cell Biol. 85:651–654 10.1016/j.ejcb.2006.02.004 [DOI] [PubMed] [Google Scholar]
- Klussmann E., Edemir B., Pepperle B., Tamma G., Henn V., Klauschenz E., Hundsrucker C., Maric K., Rosenthal W. 2001. Ht31: The first protein kinase A anchoring protein to integrate protein kinase A and Rho signaling. FEBS Lett. 507:264–268 10.1016/S0014-5793(01)02995-7 [DOI] [PubMed] [Google Scholar]
- Lamb N.J., Cavadore J.C., Labbe J.C., Maurer R.A., Fernandez A. 1991. Inhibition of cAMP-dependent protein kinase plays a key role in the induction of mitosis and nuclear envelope breakdown in mammalian cells. EMBO J. 10:1523–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J.W., Wyszynski M., Madhavan R., Sealock R., Kim J.U., Sheng M. 1998. Yotiao, a novel protein of neuromuscular junction and brain that interacts with specific splice variants of NMDA receptor subunit NR1. J. Neurosci. 18:2017–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lissandron V., Terrin A., Collini M., D’alfonso L., Chirico G., Pantano S., Zaccolo M. 2005. Improvement of a FRET-based indicator for cAMP by linker design and stabilization of donor-acceptor interaction. J. Mol. Biol. 354:546–555 10.1016/j.jmb.2005.09.089 [DOI] [PubMed] [Google Scholar]
- Lynch M.J., Baillie G.S., Mohamed A., Li X., Maisonneuve C., Klussmann E., van Heeke G., Houslay M.D. 2005. RNA silencing identifies PDE4D5 as the functionally relevant cAMP phosphodiesterase interacting with beta arrestin to control the protein kinase A/AKAP79-mediated switching of the beta2-adrenergic receptor to activation of ERK in HEK293B2 cells. J. Biol. Chem. 280:33178–33189 10.1074/jbc.M414316200 [DOI] [PubMed] [Google Scholar]
- MacKenzie S.J., Baillie G.S., McPhee I., MacKenzie C., Seamons R., McSorley T., Millen J., Beard M.B., van Heeke G., Houslay M.D. 2002. Long PDE4 cAMP specific phosphodiesterases are activated by protein kinase A-mediated phosphorylation of a single serine residue in Upstream Conserved Region 1 (UCR1). Br. J. Pharmacol. 136:421–433 10.1038/sj.bjp.0704743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matyakhina L., Lenherr S.M., Stratakis C.A. 2002. Protein kinase A and chromosomal stability. Ann. N. Y. Acad. Sci. 968:148–157 10.1111/j.1749-6632.2002.tb04333.x [DOI] [PubMed] [Google Scholar]
- McCahill A., McSorley T., Huston E., Hill E.V., Lynch M.J., Gall I., Keryer G., Lygren B., Tasken K., van Heeke G., Houslay M.D. 2005. In resting COS1 cells a dominant negative approach shows that specific, anchored PDE4 cAMP phosphodiesterase isoforms gate the activation, by basal cyclic AMP production, of AKAP-tethered protein kinase A type II located in the centrosomal region. Cell. Signal. 17:1158–1173 10.1016/j.cellsig.2005.04.003 [DOI] [PubMed] [Google Scholar]
- Mongillo M., McSorley T., Evellin S., Sood A., Lissandron V., Terrin A., Huston E., Hannawacker A., Lohse M.J., Pozzan T., et al. 2004. Fluorescence resonance energy transfer-based analysis of cAMP dynamics in live neonatal rat cardiac myocytes reveals distinct functions of compartmentalized phosphodiesterases. Circ. Res. 95:67–75 10.1161/01.RES.0000134629.84732.11 [DOI] [PubMed] [Google Scholar]
- Mongillo M., Tocchetti C.G., Terrin A., Lissandron V., Cheung Y.F., Dostmann W.R., Pozzan T., Kass D.A., Paolocci N., Houslay M.D., Zaccolo M. 2006. Compartmentalized phosphodiesterase-2 activity blunts beta-adrenergic cardiac inotropy via an NO/cGMP-dependent pathway. Circ. Res. 98:226–234 10.1161/01.RES.0000200178.34179.93 [DOI] [PubMed] [Google Scholar]
- Monterisi S., Favia M., Guerra L., Cardone R.A., Marzulli D., Reshkin S.J., Casavola V., Zaccolo M. 2012. CFTR regulation in human airway epithelial cells requires integrity of the actin cytoskeleton and compartmentalized cAMP and PKA activity. J. Cell Sci. 125:1106–1117 10.1242/jcs.089086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puck T.T., Cieciura S.J., Robinson A. 1958. Genetics of somatic mammalian cells. III. Long-term cultivation of euploid cells from human and animal subjects. J. Exp. Med. 108:945–956 10.1084/jem.108.6.945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raschke W.C., Baird S., Ralph P., Nakoinz I. 1978. Functional macrophage cell lines transformed by Abelson leukemia virus. Cell. 15:261–267 10.1016/0092-8674(78)90101-0 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Collazo P., Snyder S.K., Chiffer R.C., Bressler E.A., Voss T.C., Anderson E.P., Genieser H.G., Smith C.L. 2008a. cAMP signaling regulates histone H3 phosphorylation and mitotic entry through a disruption of G2 progression. Exp. Cell Res. 314:2855–2869 10.1016/j.yexcr.2008.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Collazo P., Snyder S.K., Chiffer R.C., Zlatanova J., Leuba S.H., Smith C.L. 2008b. cAMP signaling induces rapid loss of histone H3 phosphorylation in mammary adenocarcinoma-derived cell lines. Exp. Cell Res. 314:1–10 10.1016/j.yexcr.2007.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Vilarrupla A., Jaumot M., Abella N., Canela N., Brun S., Díaz C., Estanyol J.M., Bachs O., Agell N. 2005. Binding of calmodulin to the carboxy-terminal region of p21 induces nuclear accumulation via inhibition of protein kinase C-mediated phosphorylation of Ser153. Mol. Cell. Biol. 25:7364–7374 10.1128/MCB.25.16.7364-7374.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt P.H., Dransfield D.T., Claudio J.O., Hawley R.G., Trotter K.W., Milgram S.L., Goldenring J.R. 1999. AKAP350, a multiply spliced protein kinase A-anchoring protein associated with centrosomes. J. Biol. Chem. 274:3055–3066 10.1074/jbc.274.5.3055 [DOI] [PubMed] [Google Scholar]
- Sette C., Conti M. 1996. Phosphorylation and activation of a cAMP-specific phosphodiesterase by the cAMP-dependent protein kinase. Involvement of serine 54 in the enzyme activation. J. Biol. Chem. 271:16526–16534 10.1074/jbc.271.28.16526 [DOI] [PubMed] [Google Scholar]
- Stangherlin A., Gesellchen F., Zoccarato A., Terrin A., Fields L.A., Berrera M., Surdo N.C., Craig M.A., Smith G., Hamilton G., Zaccolo M. 2011. cGMP signals modulate cAMP levels in a compartment-specific manner to regulate catecholamine-dependent signaling in cardiac myocytes. Circ. Res. 108:929–939 10.1161/CIRCRESAHA.110.230698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefan E., Wiesner B., Baillie G.S., Mollajew R., Henn V., Lorenz D., Furkert J., Santamaria K., Nedvetsky P., Hundsrucker C., et al. 2007. Compartmentalization of cAMP-dependent signaling by phosphodiesterase-4D is involved in the regulation of vasopressin-mediated water reabsorption in renal principal cells. J. Am. Soc. Nephrol. 18:199–212 10.1681/ASN.2006020132 [DOI] [PubMed] [Google Scholar]
- Stork P.J., Schmitt J.M. 2002. Crosstalk between cAMP and MAP kinase signaling in the regulation of cell proliferation. Trends Cell Biol. 12:258–266 10.1016/S0962-8924(02)02294-8 [DOI] [PubMed] [Google Scholar]
- Takahashi M., Shibata H., Shimakawa M., Miyamoto M., Mukai H., Ono Y. 1999. Characterization of a novel giant scaffolding protein, CG-NAP, that anchors multiple signaling enzymes to centrosome and the golgi apparatus. J. Biol. Chem. 274:17267–17274 10.1074/jbc.274.24.17267 [DOI] [PubMed] [Google Scholar]
- Taskén K.A., Collas P., Kemmner W.A., Witczak O., Conti M., Taskén K. 2001. Phosphodiesterase 4D and protein kinase a type II constitute a signaling unit in the centrosomal area. J. Biol. Chem. 276:21999–22002 10.1074/jbc.C000911200 [DOI] [PubMed] [Google Scholar]
- Taylor S.S., Buechler J.A., Yonemoto W. 1990. cAMP-dependent protein kinase: Framework for a diverse family of regulatory enzymes. Annu. Rev. Biochem. 59:971–1005 10.1146/annurev.bi.59.070190.004543 [DOI] [PubMed] [Google Scholar]
- Taylor S.S., Kim C., Cheng C.Y., Brown S.H., Wu J., Kannan N. 2008. Signaling through cAMP and cAMP-dependent protein kinase: Diverse strategies for drug design. Biochim. Biophys. Acta. 1784:16–26 10.1016/j.bbapap.2007.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrin A., Di Benedetto G., Pertegato V., Cheung Y.F., Baillie G., Lynch M.J., Elvassore N., Prinz A., Herberg F.W., Houslay M.D., Zaccolo M. 2006. PGE1 stimulation of HEK293 cells generates multiple contiguous domains with different [cAMP]: Role of compartmentalized phosphodiesterases. J. Cell Biol. 175:441–451 10.1083/jcb.200605050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaasa A., Lust M., Terrin A., Uri A., Zaccolo M. 2010. Small-molecule FRET probes for protein kinase activity monitoring in living cells. Biochem. Biophys. Res. Commun. 397:750–755 10.1016/j.bbrc.2010.06.026 [DOI] [PubMed] [Google Scholar]
- Walsh K.B., Kass R.S. 1988. Regulation of a heart potassium channel by protein kinase A and C. Science. 242:67–69 10.1126/science.2845575 [DOI] [PubMed] [Google Scholar]
- Wehrens X.H., Lehnart S.E., Reiken S., Vest J.A., Wronska A., Marks A.R. 2006. Ryanodine receptor/calcium release channel PKA phosphorylation: A critical mediator of heart failure progression. Proc. Natl. Acad. Sci. USA. 103:511–518 10.1073/pnas.0510113103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witczak O., Skålhegg B.S., Keryer G., Bornens M., Taskén K., Jahnsen T., Orstavik S. 1999. Cloning and characterization of a cDNA encoding an A-kinase anchoring protein located in the centrosome, AKAP450. EMBO J. 18:1858–1868 10.1093/emboj/18.7.1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong W., Scott J.D. 2004. AKAP signalling complexes: Focal points in space and time. Nat. Rev. Mol. Cell Biol. 5:959–970 10.1038/nrm1527 [DOI] [PubMed] [Google Scholar]
- Zaccolo M. 2009. cAMP signal transduction in the heart: Understanding spatial control for the development of novel therapeutic strategies. Br. J. Pharmacol. 158:50–60 10.1111/j.1476-5381.2009.00185.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaccolo M., Pozzan T. 2002. Discrete microdomains with high concentration of cAMP in stimulated rat neonatal cardiac myocytes. Science. 295:1711–1715 10.1126/science.1069982 [DOI] [PubMed] [Google Scholar]
- Zaccolo M., De Giorgi F., Cho C.Y., Feng L., Knapp T., Negulescu P.A., Taylor S.S., Tsien R.Y., Pozzan T. 2000. A genetically encoded, fluorescent indicator for cyclic AMP in living cells. Nat. Cell Biol. 2:25–29 10.1038/71345 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.