Localization of the miRNA-induced silencing complex to GW/P bodies by GW220/TNGW1 may regulate the fate of target mRNAs.
Abstract
The microRNA (miRNA)-induced silencing complex (miRISC) controls gene expression by a posttranscriptional mechanism involving translational repression and/or promoting messenger RNA (mRNA) deadenylation and degradation. The GW182/TNRC6 (GW) family proteins are core components of the miRISC and are essential for miRNA function. We show that mammalian GW proteins have distinctive functions in the miRNA pathway, with GW220/TNGW1 being essential for the formation of GW/P bodies containing the miRISC. miRISC aggregation and formation of GW/P bodies sequestered and stabilized translationally repressed target mRNA. Depletion of GW220 led to the loss of GW/P bodies and destabilization of miRNA-targeted mRNA. These findings support a model in which the cellular localization of the miRISC regulates the fate of the target mRNA.
Introduction
MicroRNAs (miRNAs) represent a large class of noncoding small RNAs that are predicted to regulate the expression of over half of the genes encoded in the human genome (Bartel, 2004). They have emerged as major regulators of important developmental processes. Additionally, deregulation of miRNAs has been implicated in various diseases, including cancer (Ambros, 2004). Generally, miRNAs base pair imperfectly with the 3′ untranslated region (UTR) of target mRNAs and down-regulate gene expression through a posttranscriptional mechanism that remains poorly understood (Carthew and Sontheimer, 2009; Fabian et al., 2010).
Initial studies proposed that miRNAs mediate gene silencing through translational inhibition of the target mRNA (Lee et al., 1993; Wightman et al., 1993; Olsen and Ambros, 1999). How this translational repression is achieved at the molecular level still remains unclear (Humphreys et al., 2005; Pillai et al., 2005; Maroney et al., 2006; Nottrott et al., 2006; Petersen et al., 2006). Recent studies have shown that miRNAs are also capable of promoting deadenylation and subsequent degradation of target mRNAs (Bagga et al., 2005; Lim et al., 2005; Giraldez et al., 2006; Wu et al., 2006). Using large-scale quantitative experiments in mammalian cells, it was demonstrated that the effects of miRNAs on target protein expression are typically mirrored by changes in the levels of their cognate mRNAs (Baek et al., 2008; Selbach et al., 2008). Also, a recent genome-wide ribosome-profiling study argued that miRNAs predominantly elicit gene silencing in mammalian cells by regulating the mRNA levels of their endogenous targets (Guo et al., 2010). These results support a model by which miRNAs, in addition to inhibiting translation, are capable of target mRNA destabilization. Both of these processes contribute toward gene silencing. The modest magnitudes of miRNA-mediated repression of endogenous targets in cells make it difficult to conclusively determine the molecular mechanisms behind these processes. A recent ribosome-profiling study in zebrafish and a kinetics study in Drosophila melanogaster S2 cells suggest that a translational repression event, mostly likely an inhibition of translation initiation, occurs before mRNA deadenylation and decay (Bazzini et al., 2012; Djuranovic et al., 2012). However, how miRNAs coordinate the regulation of translational repression and mRNA stability is still unclear.
The miRNA-induced silencing complex (miRISC) is a multimeric protein complex, which elicits the posttranscriptional silencing mediated by miRNAs. Two highly conserved families of proteins, Argonaute (Ago) and GW182/TNRC6 (GW), represent the core components of the miRISC (Eulalio et al., 2009b). Ago proteins directly associate with miRNA and recruit GW proteins to the target mRNA. GW proteins are essential for miRNA-mediated gene silencing (Jakymiw et al., 2005; Liu et al., 2005a; Behm-Ansmant et al., 2006; Eulalio et al., 2008). Recent studies have shown that the N-terminal WG/GW motif of GW proteins interacts with Ago, whereas the C-terminal domain of GW proteins is essential and sufficient for the gene-silencing function (Chekulaeva et al., 2009; Eulalio et al., 2009a; Lazzaretti et al., 2009; Zipprich et al., 2009). The C-terminal silencing domain of GW proteins has been shown to associate with poly(A)-binding protein (PABP), PAN2/PAN3, and CNOT1/CCR4/CAF1 cytoplasmic deadenylase complexes (Chen et al., 2009; Fabian et al., 2009, 2011; Zekri et al., 2009; Piao et al., 2010; Braun et al., 2011; Chekulaeva et al., 2011). The recruitment of these proteins activates miRNA-induced mRNA deadenylation and subsequent destabilization.
Both GW and Ago proteins accumulate in specific cytoplasmic foci known as processing bodies (P bodies or GW bodies) in metazoa (Jakymiw et al., 2005; Liu et al., 2005a,b; Pillai et al., 2005; Sen and Blau, 2005; Behm-Ansmant et al., 2006; Leung et al., 2006). P bodies are heterogeneous messenger RNP (mRNP) granules that are implicated in both mRNA degradation and storage (Eulalio et al., 2007a; Parker and Sheth, 2007; Franks and Lykke-Andersen, 2008). However, miRNAs are fully functional in gene silencing in the absence of microscopically visible P bodies (Chu and Rana, 2006; Eulalio et al., 2007b). Therefore, the functional significance of miRISC aggregation and localization to the P bodies remains unknown.
We identified a unique GW family member, GW220/TNGW1, as being crucial to the process that regulates the localization of the miRISC in mammalian cells. We showed that GW220 promotes aggregation and sequestration of the miRISC into GW/P bodies. These GW/P bodies are more stable aggregates than the classically unstable and dynamic P bodies. This aggregation of the miRISC stabilized the associated mRNA. Depletion of GW220 led to the loss of GW/P bodies, concomitant release of the retained miRISC into the cytosol, and subsequent destabilization of the target mRNA. These results suggest that the aggregation and localization of miRISC into GW/P bodies could regulate the stability of the mRNAs that are translationally repressed by miRNAs and that the subcellular localization of the miRISC could play an important role in the regulation of the fate of the target mRNA.
Results
GW220/TNGW1 is a unique GW family protein capable of nucleating the de novo formation of mammalian GW/P bodies
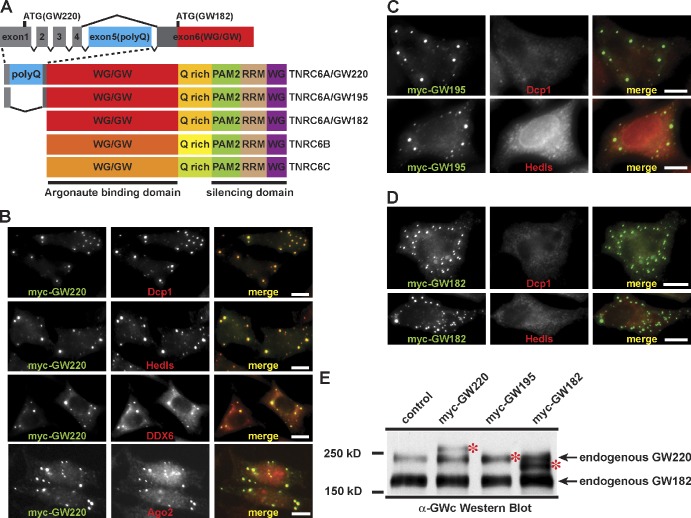
To understand how the miRISC elicits miRNA-mediated gene silencing, we focused on the function and role of the GW family of proteins. In mammals, there are three paralogues of GW182 (TNRC6A, TNRC6B, and TNRC6C), which share highly conserved domain structures (Fig. 1 A). During work to generate the expression constructs for the GW family proteins, we isolated two alternatively spliced TNRC6A transcripts, GW220 and GW195, named after their respective predicted molecular masses (220 and 195 kD). These three TNRC6A isoforms (GW220, GW195, and GW182) differ only in their N-terminal regions through alternative splicing (Fig. 1 A). The largest isoform, GW220, contains a unique N-terminal extension, containing an expanded polyglutamine (polyQ) repeat motif. This motif is mammalian specific and is encoded in additional exons upstream of the canonical start codon of GW182. The GW220 isoform was previously reported and referred to as TNGW1 (Li et al., 2008). The second novel alternative isoform, GW195, also contains an N-terminal extension but lacks the polyQ domain.
Figure 1.
GW220 is a unique GW family member capable of nucleating the de novo formation of GW/P bodies. (A) Schematic representation of the domain structure of GW family proteins. RRM, RNA recognition motif. (B) GW220 recruits all the known GW/P-body components into the cytoplasmic granules termed GW/P bodies. (C and D) Other GW family proteins aggregate into cytoplasmic granules that are distinct from GW/P bodies, as shown in GW195 (C) and GW182 (D). The presence of epitope-tagged GW proteins and endogenous components of GW/P body was determined using the indicated antibodies. (E) The expression levels of the endogenous GW proteins and myc-tagged overexpressed GW proteins (indicated by asterisks) were determined by Western blot analysis using an antibody that recognized the common C terminus of the TNRC6A/GW proteins. Bars, 10 µm.
Given the essential role of GW proteins in P-body formation, we examined the ability of different GW proteins to nucleate the de novo formation of P bodies in mammalian cells. We chose to use HeLa Tet-OFF cells, a cell line that displayed very few endogenous P bodies. We transiently transfected myc-tagged TNRC6A expression constructs into HeLa Tet-OFF cells and examined the localization of the proteins by indirect immunofluorescence microscopy, using the well-characterized P body markers Dcp1, Hedls, DDX6, and Ago2. This assay showed that only GW220 was capable of recruiting endogenous components of the P body, with ∼95% of the transfected cells now containing detectable P bodies (Fig. 1 B). GW220 was also capable of recruiting endogenous TNRC6B into P bodies (Fig. S1 A). When GW195 and GW182 were transiently expressed in HeLa Tet-OFF cells, both isoforms exhibited the tendency to form large cytoplasmic foci. However, these foci were distinct from the granules generated by GW220, with <10% of these foci containing endogenous Ago2 and none containing other endogenous P-body components, such as Dcp1 and Hedls (Fig. 1, C and D; and Fig. S1 B). When we transiently transfected FLAG-tagged TNRC6B and TNRC6C into the cells, both proteins were able to form cytoplasmic foci. However, these foci also lacked Dcp1, Hedls, and DDX6, and <5% of the foci contained endogenous Ago2 (Fig. S1 C and not depicted). Similar aggregation patterns were observed in the osteosarcoma U2OS cell line (Fig. S1 E).
Next we generated a polyclonal antibody against the common C terminus of the TNRC6A/GW proteins (Fig. S2 A) and examined the expression levels of the endogenous and ectopically expressed GW proteins. The level of endogenous GW182 was approximately two- to threefold higher than the level of endogenous GW220 in HeLa and U2OS cell lines (Fig. S2 C). These cells expressed a much lower relative level of endogenous GW195. The expression levels of the myc-tagged TNRC6A/GW proteins were close to the endogenous levels of GW proteins in HeLa Tet-OFF cells and were approximately two- to threefold higher than those of the endogenous GW proteins in U2OS cells (Fig. 1 E and Fig. S1 E).
All three GW paralogues (TNRC6A, TNRC6B, and TNRC6C) have previously been shown to localize to the mammalian P bodies (Meister et al., 2005; Lazzaretti et al., 2009). However, our findings that TNRC6A/GW195, TNRC6A/GW182, TNRC6B, and TNRC6C are unable to recruit other endogenous P body markers suggest that the colocalization noted in previous studies is most likely a result of Ago protein coexpression. Ectopic expression of myc-Ago2 is sufficient to trigger GW/P-body formation and recruitment of endogenous TNRC6A/GW and TNRC6B proteins (Fig. S1 D). Overexpressed GW protein has also been reported to localize to the multivesicular bodies in mammalian cells (Gibbings et al., 2009). The different cell lines used in these studies and differing levels of overexpression could also affect the localization and the aggregation patterns of the GW proteins. Our results suggest that mammalian GW proteins have distinctive roles in the formation of P bodies that contain the miRISC. The unique and essential role of GW220 in the formation and maintenance of mammalian GW/P bodies is further demonstrated when endogenous GW220 is depleted from the cells (see Fig. 9 and Fig. S5).
Figure 9.
GW220 is essential for the accumulation of miRISC in the GW/P bodies, and maintenance of GW/P bodies is essential to protect mRNA targeted by miRNA from degradation. (A) GW220, endogenous or stably expressed, can be repressed by specific siRNAs, as determined by Western blot analysis. Depletion of GW220 did not affect Ago2 expression levels. (B and C) Depletion of GW220 led to loss of the GW/P bodies, as determined by immunofluorescence using GW220 (B) and Ago2 (C) antibodies. (D) Depletion of GW220 led to destabilization of the miRNA target mRNA, as determined by qRT-PCR analysis in wild-type U2OS cells. (E) Depletion of GW220 led to reduced half-lives of the miRNA target mRNA in wild-type U2OS cells, as determined by transcriptional shutoff (addition of doxycycline) and qRT-PCR analysis. GAPDH, glyceraldehyde 3-phosphate dehydrogenase. Mean values ± standard deviations from three independent experiments are shown. Bars, 10 µm.
The middle Q-rich domain of GW220 is not required for GW/P-body formation
All the GW family member proteins contain a middle glutamine-rich (Q rich) domain between the N-terminal Ago-binding domain and the C-terminal silencing domain (Fig. 1 A). Previous studies suggested that the Q-rich domain plays an essential role in the P-body localization of the GW proteins (Behm-Ansmant et al., 2006; Lazzaretti et al., 2009). We systematically examined the various domains within GW220 to determine their role, if any, in GW/P-body formation. Our experiments demonstrated that neither the N-terminal polyQ domain, the middle Q-rich domain, nor the C-terminal silencing domain are sufficient to trigger the formation of GW/P bodies, despite the high expression levels of these small truncations (Fig. 2). Interestingly, deletion of the middle Q-rich domain from GW220 has no effect on the ability of GW220 to nucleate the GW/P-body formation and recruit P-body marker proteins. The expression level of GW(ΔQ rich) protein is similar to the endogenous GW proteins (Fig. 2).
Figure 2.
The middle Q-rich domain is not required for GW/P-body formation. The N-terminal polyQ domain (1–253 aa), the middle Q-rich domain (1,151–1,490 aa), and the C-terminal silencing domain (1,491–1,962 aa) cannot nucleate the de novo formation of GW/P bodies. Deletion of the middle Q-rich domain does not affect the ability of GW220 (GWΔQ rich) to nucleate the formation of GW/P bodies. The presence of myc-tagged GW proteins and endogenous components of GW/P body were determined using the indicated antibodies. The expression levels of the myc-tagged truncated domains of the GW220 proteins (indicated by asterisks) were determined by Western blot analysis using the myc antibody. The expression levels of full-length GW220 and GW(ΔQ rich; indicated by asterisks) were also determined by Western blot analysis using an antibody that recognizes the C terminus of the TNRC6A/GW proteins. RRM, RNA recognition motif. Bars, 10 µm.
GW/P bodies contain the miRISC and are different from the classical P bodies
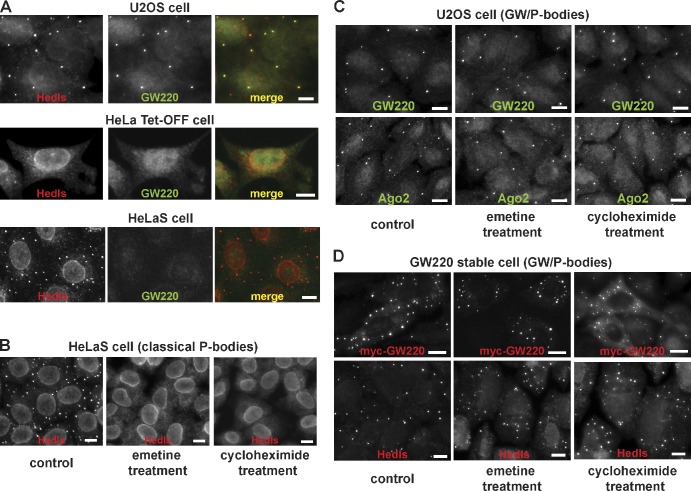
Next, we generated a polyclonal antibody against GW220 and confirmed that purified anti-GW220 serum was specific for the GW220 isoform using tagged GW constructs (Fig. S2 A). Using this GW220 antibody along with other P-body markers, we screened several cell lines to detect the presence of endogenous GW220-positive P bodies. From this screen, we detected GW220-positive P bodies present in the U2OS cells, with GW220-positive P bodies accumulating in between 60 and 75% of these cells as detected by antibodies against Hedls and GW220 (Fig. 3 A). Because <1% of the HeLa Tet-OFF cells contained microscopically visible P bodies (Fig. 3 A), we examined the presence of endogenous GW220 in the classical P bodies of HeLaS cells instead. This subtype of the HeLa cell line contains a high number of visible P bodies in ∼80–90% of the cells as determined by staining with Hedls (Fig. 3 A). Examination of these P bodies showed that >95% of these foci did not contain detectable GW220 or Ago2 (Fig. 3 A and Fig. S2 B). We also could not detect a positive signal in these P bodies using the GW antibody that recognizes the C terminus of GW/TNRC6A proteins. These results suggest that GW182 protein did not accumulate in these classical P bodies (Fig. S2 B). Given that not all P bodies contain GW220 and Ago2, these findings suggest that GW220/Ago-positive P bodies represent a unique class of P bodies that contain the miRISC. We classified them as GW/P bodies (Moser and Fritzler, 2010). The absence of GW/P bodies in the HeLa cell lines was not the result of an absence of expression of endogenous GW220. We observed comparable protein expression levels of the TNRC6A/GW proteins in HeLaS, HeLa Tet-OFF, and U2OS cells by Western blotting (Fig. S2 C). These results argue that endogenous GW220 in the HeLa cells is not sufficient for GW/P-body formation and indicate that other factors, including additional protein/RNA cofactors or posttranslational modifications of GW220, might play a role in regulating the formation of endogenous GW/P bodies.
Figure 3.
GW/P bodies are a unique class of cytoplasmic P bodies in mammalian cells and are different from classical P bodies. (A) U2OS cells accumulate microscopically visible GW/P bodies, whereas HeLa Tet-OFF cells lack microscopically visible P bodies. The majority of the P bodies in HeLaS cells do not contain GW220. The presence of GW/P bodies and classical P bodies was determined by immunofluorescence using antibodies against Hedls and GW220. (B–D) GW/P bodies are more stable aggregates and are different from the classically unstable and dynamic P bodies. (B) GW220-negative P bodies in HeLaS cells are highly sensitive to the emetine or cycloheximide treatment, whereas GW/P bodies in the U2OS cells (C) and GW220 stable cells (D) are resistant to the emetine or cycloheximide treatment. Bars, 10 µm.
Classical P-body formation and maintenance is dependent on the presence of mRNA. The treatment of cells with drugs such as cycloheximide or emetine, which block translation elongation, has the effect of trapping mRNA in association with the ribosomes, thereby reducing the supply of nontranslating mRNA and leading to disassembly of P bodies (Teixeira et al., 2005). We treated the HeLaS and U2OS cells with either emetine or cycloheximide and assayed the behavior of the classical P bodies and GW/P bodies in the respective cell lines. When the HeLaS cells were treated with emetine or cycloheximide, all of the classical P bodies disappeared within 30 min of the treatment (Fig. 3 B), likely reflecting the degradation and/or release of the mRNA in these P bodies. However, after the same treatment, the GW/P bodies remained largely unaffected in the U2OS cells (Fig. 3 C) and in the GW220 stable cells (which overexpress a myc-tagged GW220; Fig. 3 D and Fig. S2 D), as determined by Hedls, Ago2, and GW220 staining.
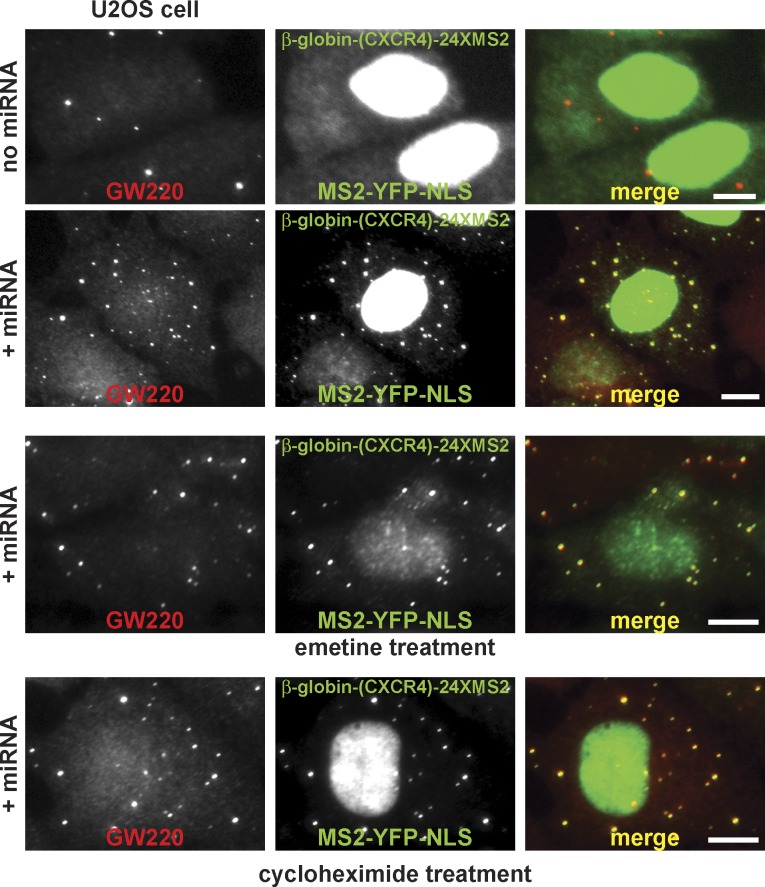
Next, we examined whether miRNA-targeted mRNA would accumulate within these GW/P bodies. We generated a β-globin reporter construct with a 3′ UTR containing both the miRNA target sites and 24 binding sites for the MS2 coat protein, allowing the tracking of the reporter mRNA by coexpression of the MS2-YFP-NLS fusion protein as previously described (Liu et al., 2005b). In the absence of miRNA, the MS2-YFP protein mostly localized to the nucleus because of the presence of an NLS. However, in the presence of miRNA, MS2-YFP protein became concentrated into the foci marked by endogenous GW220, signifying that the miRNA-targeted mRNA had been sequestered into the endogenous GW/P bodies in U2OS cells (Fig. 4). The same result was obtained in the GW220 stable cells (Fig. S2 E). Importantly, the β-globin reporter mRNA targeted by miRNA was retained in the GW/P bodies after treatment with emetine or cycloheximide (Fig. 4). The resistance of the GW/P bodies to the cycloheximide or emetine exposure demonstrates the unique nature of GW/P bodies and suggests GW/P bodies may function in the storage of sequestered mRNA.
Figure 4.
mRNA targeted by miRNA accumulates in endogenous GW/P bodies in the U2OS cells and is retained in the endogenous GW/P bodies after either emetine or cycloheximide treatment. A β-globin reporter construct with a 3′ UTR containing both the miRNA target sites and 24 binding sites for the MS2 coat protein allowed the tracking of the reporter mRNA by coexpression of the MS2-YFP-NLS fusion protein. In the absence of miRNA, the MS2-YFP protein mostly localized to the nucleus as a result of the presence of an NLS. In the presence of an miRNA, MS2-YFP protein became concentrated into the foci marked by endogenous GW220. Bars, 10 µm.
PABP, an miRISC cofactor, is excluded from classical P bodies (Kedersha et al., 2005). We examined whether PABP is recruited to GW/P bodies. We found that endogenous PABP protein localizes largely in the cytoplasm in both HeLa and U2OS cells and does not concentrate in endogenous GW/P bodies, classical P bodies, or in the granules formed by the overexpression of GW proteins (Fig. S3 A).
The formation of classical P bodies has been shown to be regulated by deadenylation in mammalian cells. Overexpression of catalytically inactive CAF1 mutant impairs deadenylation and blocks the formation of P bodies (Zheng et al., 2008). Therefore, we examined the effect of CAF1 mutant on the formation of endogenous GW/P bodies. The classical P bodies disappeared in the HeLaS cells overexpressing the CAF1 mutant, confirming the previous study. However, the GW/P bodies in U2OS cells were not affected by the expression of CAF1 mutant (Fig. S3 B). These data suggest that the formation of GW/P bodies is regulated by a different mechanism than the formation of classical P bodies.
We established a method to assay for the presence of GW/P bodies in the cell extract using an in vitro immunostaining method. We prepared miRISC-containing extract as previously described (Liu et al., 2004) and examined the lysate from U2OS cells by centrifugal sedimentation of the extract through a glycerol cushion onto microscope coverslips. The coverslips were fixed and examined for the presence of GW/P bodies. The GW/P bodies were clearly present in the extract, as determined by Ago2 and Hedls staining, GW220 and Hedls staining, or DDX6 and Hedls staining (Fig. 5 A).
Figure 5.
GW/P bodies are stable mRNP granules in vitro and are insensitive to RNase treatment. (A) Stable GW/P bodies were present in the in vitro RISC extract, determined by immunostaining using antibodies against Hedls, GW220, Ago2, and DDX6. (B) GW/P bodies extracted from U2OS cells were insensitive to in vitro RNase treatment, determined by immunostaining using antibodies against Hedls, Ago2, and GW220. (C) Classical P bodies extracted from HeLaS cells were unstable and disappeared completely after in vitro RNase treatment, determined by immunostaining using antibodies against DDX6 and Hedls. Bars, 10 µm.
A previous study showed that the classical P bodies in yeast were sensitive to RNase treatment in vitro (Teixeira et al., 2005). Therefore, we examined the stability of GW/P bodies in vitro after RNase treatment. After a 30-min incubation of the RISC extract with RNase, both 28S and 18S ribosomal RNA were completely degraded (not depicted). The GW/P bodies from U2OS cells remained largely unchanged as determined by Hedls and Ago2 staining or Hedls and GW220 staining (Fig. 5 B). On the other hand, the classical P bodies prepared from HeLaS cells were unstable and disappeared completely after the same RNase treatment as determined by DDX6 and Hedls staining (Fig. 5 C). Although we cannot determine whether mRNA remained intact in the GW/P bodies, these results further highlighted the differences between GW/P bodies and classical P bodies.
The fate of miRNA-targeted mRNA correlates with the aggregation of the miRISC into GW/P bodies
To examine the functional consequence of miRISC aggregation into GW/P bodies, we used a well-established β-globin reporter construct containing different miRNA target sites in the 3′ UTR to examine the fate of the target mRNA (Fig. 6 A). We assayed this construct in HeLa Tet-OFF and U2OS cell lines. We used a perfect siRNA (siCXCR4p) targeting the reporter as a positive control for the degradation-mediated gene-silencing pathway (Fig. 6 B). In the HeLa Tet-OFF cell line, which contains very few visible P bodies, we observed a significant destabilization of β-globin mRNA targeted by miRNA (let-7) or miRNA mimics (CXCR4). Overall, the level of β-globin protein repression correlated well with mRNA degradation (Fig. 6 B). This result is in agreement with previous studies, which showed that deadenylation leads to the decay of miRNA-targeted mRNA in mammalian cells (Wu et al., 2006; Chen et al., 2009). Unlike the widely used luciferase reporter system, the β-globin system we used revealed a much more significant role of mRNA destabilization in miRNA-mediated gene silencing in mammalian cells. The effects observed in the HeLa Tet-OFF cell line are consistent with a dispensable role of P bodies in miRNA-mediated silencing (Chu and Rana, 2006; Eulalio et al., 2007b), supporting the model that the accumulation of microscopically visible P bodies is not required for miRNA-mediated mRNA deadenylation and degradation.
Figure 6.
Cell type–specific silencing mechanisms mediated by miRNA correlated with the miRISC aggregation into GW/P bodies. (A) Schematic representation of the FLAG–β-globin miRNA reporter. (B) miRNA target mRNA for degradation in HeLa Tet-OFF cells. Western blot analysis confirmed repression of the β-globin reporter at the protein level, whereas qRT-PCR confirmed concomitant degradation of the β-globin mRNA. The protein level of β-globin was determined by α-FLAG Western blot analysis normalized to tubulin. The mRNA level of β-globin was determined by qRT-PCR analysis, normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH). A perfect siRNA (siCXCR4p) targeting the CXCR4 reporter was used as a control. (C) mRNA targeted by miRNA is partially protected in U2OS cells, correlating with the accumulation of microscopically visible GW/P bodies. (D–F) Elevated expression of GW220 promotes miRISC aggregation and GW/P-body formation and protects mRNA targeted by miRNA from deadenylation and degradation. (E) Schematic representation of the oligo(dT) RNaseH/Northern assay used to determine the poly(A) status of the miRNA targeted mRNA. (F) The distribution of the miRNA target β-globin mRNA poly(A) tail in HeLaTet-OFF and GW220 stable cells. (left) The β-globin mRNAs were destabilized in HeLaTet-OFF cells that lacked visible GW/P bodies. (right) The β-globin mRNAs were protected and retained the poly(A) tail in cells stably expressing GW220 and visible GW/P bodies. Mean values ± standard deviations from three independent experiments are shown.
Next, we assayed the same reporter construct in the U2OS cell line and observed far less mRNA destabilization, even though protein expression was equally well repressed as in the HeLa Tet-OFF cell line (Fig. 6 C). The disproportional change in protein repression versus mRNA destabilization revealed a modest degree of translational repression and suggested that an alternative silencing mechanism was used to silence the target mRNA. These results show that the stability of miRNA-targeted mRNA correlated with the presence of miRISC in the GW/P bodies.
To minimize the heterogeneity of GW220/Ago-containing P bodies in the U2OS cells, we established a U2OS cell line stably overexpressing a myc-tagged GW220 protein. More than 95% of the P bodies present in this stable cell line were positive for GW220 and Ago2 (Fig. S2 D). We then examined the effect of elevated GW220 expression and increased miRISC aggregation and localization to the GW/P body on miRNA-mediated gene silencing. The miRNA-mediated β-globin protein repression was comparable with that observed in the HeLa Tet-OFF and U2OS cell lines. However, there was only a modest decrease in the β-globin mRNA levels, as confirmed by quantitative RT-PCR (qRT-PCR) analysis (Fig. 6 D). The target mRNA stabilization resembled the situation we observed in the wild-type U2OS cell line; however, in the GW220 stable cell line, there was greater protection of the target mRNA, correlating with increased miRISC aggregation into the GW/P bodies. This result suggests that increased GW220 expression could protect the miRNA-targeted mRNA from degradation. Given the general exclusion of the translational machinery from P bodies (Kedersha et al., 2005; Anderson and Kedersha, 2006; Souquere et al., 2009), the sequestering of the miRISC and the target mRNA into GW/P bodies would maintain the gene silencing through translational repression.
The ability of miRNAs to promote deadenylation of target mRNAs led us to examine the effect of miRISC localization to the GW/P body on the poly(A) tail of the stabilized β-globin miRNA reporter mRNA. We applied a gene specific oligonucleotide (oligo), with or without oligo(dT), coupled to an RNaseH/Northern assay, to determine the length of the poly(A) tail as previously described (Fig. 6 E; Wu et al., 2006). In HeLa Tet-OFF cells lacking microscopically visible GW/P bodies and miRISC aggregations, the poly(A) tail length distribution of the detectable mRNA targeted by miRNA showed similar patterns to that of the mRNA not targeted by miRNA (Fig. 6 F, left). We did not observe any obvious accumulation of mRNA containing a short poly(A) tail, consistent with the model that miRNA-mediated deadenylation is normally coupled with mRNA degradation. In the GW220 stable cell line, the stability of the β-globin mRNA was not significantly affected by miRNA. We again did not observe any accumulation of mRNA containing a short poly(A) tail (Fig. 6 F, right). This result suggests that increased GW220 expression could mediate repression without activating the rapid deadenylation and decay of the miRNA-targeted mRNA.
GW220 provides partial protection to a tethered mRNA target
The tethering of GW/TNRC6 proteins to mRNA bypasses the requirement of Ago/miRNA for target mRNA recognition and is sufficient to trigger deadenylation and silencing of the target mRNA (Behm-Ansmant et al., 2006; Chen et al., 2009). We applied a β-globin–tethering reporter to determine the effect of various GW proteins and the C-terminal silencing domain of GW proteins on the target mRNA levels. The λN-HA–tagged GW proteins were expressed in HeLa Tet-OFF cells, together with a β-globin reporter that contained five boxB sites in the 3′ UTR (Fig. 7 A). The C-terminal silencing domain of GW/TNRC6A, GWSD(1,491C), did not form aggregates and localized to both the nucleus and the cytoplasm in the cell. The λN-HA–tagged GW220 formed GW/P bodies, whereas GW195 and GW182 formed abnormal aggregates that were not GW/P bodies, similar to myc-tagged GW proteins (Fig. 7, B and C). Tethering of GWSD(1,491C) and GW220 led to a similar degree of target protein repression as determined by Western blotting (Fig. 7 B, bottom left). Although tethering of GWSD(1,491C) led to significant mRNA destabilization that correlated well with the protein repression, tethering of GW220 provided a partial protection to the repressed β-globin mRNA, as determined by Northern blotting (Fig. 7 B, bottom right). Tethering of GW182 and GW195 also led to target protein repression that correlated well with the mRNA destabilization (Fig. 7 C). These results suggest that GW220 could mediate repression without activating the rapid degradation of the associated mRNA.
Figure 7.
GW220 provides partial protection to a tethered mRNA target. (A) Schematic representation of the λN/boxB FLAG–β-globin–tethering reporter system. (B) The λN-HA–tagged C-terminal silencing domain of GW, GWSD(1,491C), localized in the nucleus and cytoplasm, whereas the λN-HA–tagged GW220 formed GW/P bodies in HeLa Tet-OFF cells. Tethering of GWSD(1,491C) led to significant mRNA destabilization that correlated well with protein repression. Tethering of GW220 caused less mRNA destabilization even though the protein was equally well repressed as tethering of GWSD(1,491C). (C) The λN-HA–tagged GW195 and GW182 formed cytoplasmic aggregates that were distinct from the GW/P bodies in the HeLa Tet-OFF cells. Tethering of GW195 and GW182 led to mRNA destabilization that correlated well with protein repression. The protein repression and mRNA destabilization were quantified by ImageJ software and Quantity One 1-D Analysis Software and confirmed by qRT-PCR analysis. GAPDH, glyceraldehyde 3-phosphate dehydrogenase. Mean values ± standard deviations from three independent experiments are shown. Bars, 10 µm.
Next, we generated a λN-tagged Hedls expression construct and examined the effect of classical P-body formation on the tethered mRNA target (Fenger-Grøn et al., 2005). The λN-tagged Hedls formed aggregates similar to classical P bodies in the mammalian cells as determined by costaining with Hedls, DDX6, Dcp1, and Lsm1 (Fig. S4 A and not depicted). These granules did not contain GW220 and Ago2 (Fig. S4 A); therefore, they were different from the GW/P bodies nucleated by GW220. Tethering of Hedls to the 3′ UTR of the β-globin reporter did not repress the β-globin protein expression (Fig. S4 B, top) nor did it trigger the destabilization of the β-globin mRNA (Fig. S4 B, bottom). A recent study showed that the expression of PAT1b, another classical P-body component, also led to the formation of classical P bodies but resulted in significant destabilization and repression of the tethered mRNA (Ozgur et al., 2010). Considered together, these results suggest that the protection of the target mRNA provided by GW220 is unique.
GW220 provides partial protection of a miRNA-targeted mRNA
As previously shown, the miRNA-targeted β-globin reporter mRNA was significantly destabilized in HeLa Tet-OFF cells (Fig. 6 B). We examined the effect of overexpression of different GW proteins on the target mRNA level in these cells. We first applied a mixture of three siRNAs (siTNRC6) targeting all human TNRC6A/6B/6C proteins to deplete the endogenous GW proteins. Western blot analysis of the endogenous TNRC6A/GW and the overexpressed myc-tagged TNRC6B and TNRC6C confirmed the efficiency of the knockdowns (Fig. 8 A). The levels of the endogenous Ago proteins were not affected by the depletion of all three TNRC6 proteins, as determined by an antibody that recognizes all four human Ago proteins (Fig. 8 A). Repression of the β-globin miRNA reporter was significantly impaired upon depletion of endogenous TNRC6 proteins (Fig. 8 C, lane 2).
Figure 8.
GW220 provides partial protection to a miRNA-targeted mRNA. (A) The endogenous TNRC6 proteins were depleted by a mixture of siRNAs (siTNRC6) targeting all human GW proteins. The endogenous TNRC6A/GW proteins were examined by Western blot analysis using an antibody that recognized the C terminus of the proteins (GWc). The efficiencies of the knockdown of myc-tagged TNRC6B and TNRC6C proteins were confirmed by Western blotting using the myc antibody. The expressions of siRNA-resistant forms of the myc-tagged TNRC6A/GW proteins were confirmed by the GWc antibody (indicated by asterisks). (B) The myc-tagged siResistant GW220 formed GW/P bodies in HeLa Tet-OFF cells, whereas myc-tagged siResistant GW195 and GW182 formed different granules. (C) Depletion of all endogenous TNRC6 proteins impaired miRNA-mediated repression of the β-globin reporter. Expression of siRNA-resistant TNRC6A/GW proteins rescued the repression. The expression of GW195 and GW182 led to significant mRNA destabilization that correlated well with protein repression, whereas GW220 provided partial protection to the same target mRNA. The protein repression and mRNA destabilization were quantified by Quantity One 1-D Analysis Software and confirmed by qRT-PCR analysis. Mean values ± standard deviations from three independent experiments are shown. Bars, 10 µm.
Next, we expressed siRNA-resistant forms of myc-tagged TNRC6A/GW to rescue the miRNA-mediated repression (Fig. 8, A and C). myc-GW220siResistant triggered the formation of GW/P bodies in HeLa Tet-OFF cells, whereas myc-GW195siResistant and myc-GW182siResistant formed granules that were different from the GW/P bodies (Fig. 8 B). All three TNRC6A/GWsiResistant proteins were able to rescue the repression of the β-globin reporter protein to approximately fivefold (Fig. 8 C), despite all having different expression levels (Fig. 8 A). We observed a partial protection of the β-globin mRNA by GW220 rescue but not by GW195 and GW182 rescues (Fig. 8 C), similar to the effect we observed in the tethering assay. We emphasize that the protection of the target mRNA was not complete in both assays, and mRNA destabilization remained a significant contributing factor to the repression in both systems.
Depletion of GW220 causes disassembly of the GW/P bodies and concomitant mRNA destabilization
To directly test the hypothesis that the GW/P-body formation sequestered and protected miRNA-targeted mRNA from degradation, we examined the consequences of removing GW220. We depleted the GW220 isoform by using two specific siRNAs targeting the GW220-specific polyQ region (siGW220#A and siGW220#B). A previously described siRNA (siGW) targeting all three isoforms of GW proteins was used as a control (Liu et al., 2005a). Western blot analysis confirmed the specificity and efficiency of these siRNAs (Fig. 9 A). More importantly, the significant reduction in the levels of GW220 did not affect the levels of endogenous Ago2 (Fig. 9 A). We observed a small degree of reduction in the levels of GW182 protein upon GW220 depletion, suggesting that miRISC aggregation might increase the stability of GW proteins retained within the foci.
Next, we examined the effect of GW220 depletion on GW/P-body formation. Although P bodies remained readily detectable, as determined by Hedls staining, GW220 and Ago2 were no longer present in these foci, with Ago2 becoming diffusely distributed throughout the cytoplasm (Fig. 9, B and C). Moreover, the endogenous TNRC6B was excluded from the P bodies when GW220 is depleted (Fig. S5, A–C). These results further support an essential role for GW220 in the formation and maintenance of the GW/P bodies and the retention of Ago2 and miRISC in these distinctive foci.
We next examined the fate of the β-globin miRNA-targeted mRNA in U2OS cells depleted of GW220 and GW/P bodies. Although β-globin target protein levels were similarly reduced, there was now a significant destabilization of the target mRNA in these cells (Fig. 9 D). Similar destabilization of the target mRNA was observed when we depleted GW220 in the myc-GW220 stable cells (Fig. S5 D). We further determined the half-life of the reporter mRNA upon transcriptional shutoff. The half-life of the nontargeted control β-globin mRNA was ∼6 h, whereas the half-life of the miRNA-targeted β-globin mRNA was reduced to ∼4 h. Depletion of GW220 and GW/P bodies further destabilized the β-globin mRNA and reduced its half-life to ∼2 h (Fig. 9 E). Furthermore, depletion of GW220 led to a significant loss of miRNA-targeted mRNA in the remaining miRISC-negative P bodies (Fig. S5 E).
Overall, these results support the model that miRISC aggregation and GW/P-body formation protect miRNA-targeted mRNA from degradation. Preventing aggregation allows the miRISC to efficiently recruit the cytoplasmic deadenylase complex and destabilize the associated mRNA targets.
Discussion
The GW/TNRC6 family proteins are essential components of the miRISC and are critical for miRNA function (Eulalio et al., 2009b). Recruitment of the cytoplasmic deadenylase complex to the mRNA is a critical regulatory step in posttranscriptional gene regulation. Recent studies have shown that the C-terminal silencing domain of the GW/TNRC6 proteins interacts with PABP, PAN2/PAN3, and CNOT1/CCR4/CAF1 (Fabian et al., 2009, 2011; Zekri et al., 2009; Braun et al., 2011; Chekulaeva et al., 2011). Therefore, the cytosolic miRISC serves as an adaptor complex to recruit the cytoplasmic deadenylases and control the target mRNA deadenylation and stability in mammalian cells.
Our results show that mammalian miRISC can also accumulate into GW/P bodies in certain cells. GW/P-body formation stabilizes the target mRNA associated with these unique granules. The exact mechanism of mRNA stabilization by the GW/P body remains to be determined. However, given that the stabilized miRNA-targeted mRNA retains its poly(A) tail, the protection provided by the aggregation and sequestration of miRISC into GW/P body might involve either blocking of the deadenylation machinery from accessing the target mRNA or inhibition of the activity of cytoplasmic deadenylases.
The cytoplasmic compartmentalization of the miRISC could play an important role in the choice between an irreversible silencing process (deadenylation and degradation of the target mRNA) and a reversible silencing process (protection and translational repression of the target mRNA). In mammalian cells, GW220, an alternative isoform of GW182/TNRC6A, plays an essential role in this control process. GW220 is capable of nucleating the de novo formation of GW/P bodies and is essential for their maintenance in mammalian cells. Its unique N-terminal polyQ motif may influence protein–protein interactions that are required for the miRISC sequestration. GW220 is necessary but not always sufficient for GW/P-body formation. The additional miRISC cofactors involved in the aggregation and formation of GW/P body remain to be identified. In our experiments, overexpression of GW195 and GW182 proteins in U2OS cells often led to the disassembly of endogenous GW/P bodies (Fig. S1 E). It is possible that these ectopically expressed GW proteins compete with endogenous GW220 for the additional protein and/or mRNA cofactors required for GW/P-body formation.
Posttranslational modifications of GW proteins could also play important roles in regulating GW/P-body formation. A recent study suggested that the assembly and disassembly of germline P granules in Caenorhabditis elegans is regulated not only by the concentration of critical components but also through an additional mechanism involving posttranslational modifications of unknown components (Gallo et al., 2010). These modifications and the regulatory mechanisms are yet to be identified.
It is still unclear how repression by the miRISC is initiated. Recent studies suggest that a translation initiation block occurs before deadenylation (Fabian et al., 2010; Bazzini et al., 2012; Djuranovic et al., 2012). A recent study in Xenopus laevis suggests that the deadenylase CAF1 could inhibit translation independent of deadenylation (Cooke et al., 2010). The recruitment of a large complex containing some or all of the GW-associated proteins could potentially disrupt the closed-loop mRNA conformation. This conformational change might be sufficient to trigger the initial translational repression. Further studies are required to dissect the processes that occur when miRISC associates with the target mRNA. Our results suggest that the cellular localization of the miRISC plays an important role in coordinating the translational repression, degradation, and storage of miRNA-targeted mRNA. It is possible that GW220 and GW/P-body formation could enhance the initial translational repression without activating the rapid decay of the translationally repressed mRNA. Therefore, we propose a dual level of control involving the miRISC. Cytosol-localized miRISCs are capable of recruiting the deadenylase complex and initiating mRNA degradation. When miRISCs are aggregated into GW/P bodies, mRNA is sequestered, protected from degradation, and silenced through exclusion from the translational machinery (Fig. 10).
Figure 10.
A model for the cytoplasmic miRISC action and the functional implication for GW/P-body formation. The cytosolic miRISCs are capable of recruiting the deadenylase complex and initiating mRNA degradation. In the situation where miRISCs are aggregated into GW/P bodies, mRNA is sequestered, protected from degradation, and silenced through exclusion from the translational machinery. GW220 is essential for the formation of GW/P bodies. GW220 and GW/P-body formation enhance the translational repression without activating rapid decay of the translationally repressed mRNA. Unknown miRISC cofactors could play a role in initiating the miRISC aggregation and GW/P-body formation. These cofactors are likely involved in the initial translational repression by the miRISC. The presence of endogenous Ago1/3/4 and TNRC6C in the GW/P body has not been confirmed because of the lack of suitable antibodies. The endogenous mRNA targets of GW/P bodies remain to be determined.
The formation of microscopically visible P bodies is not a requirement for mRNA degradation (Decker et al., 2007; Hu et al., 2009). Accumulation of microscopically visible foci is more consistent with a role in mRNA storage or inhibition of mRNA degradation. Recent studies suggest that P body–like structures present during the oogenesis of C. elegans (Boag et al., 2008) and during the sexual development of the protozoan Plasmodium berghei (Mair et al., 2006) function by sequestering a cohort of specific mRNA species, protecting them from degradation and, at the same time, promoting translational repression. It has been demonstrated in yeast that mRNA stored in P bodies is able to return to translation once released from the foci (Brengues et al., 2005). The same possible scenario could apply to the miRNA-targeted mRNA sequestered in the GW/P body (Bhattacharyya et al., 2006). The mRNAs sequestered in the GW/P bodies could potentially return to translation when GW/P bodies disassemble. Therefore, a function of the GW/P bodies could be to provide spatial and temporal control of translation for miRNA-targeted mRNA.
Recently, Han et al. (2012) and Kato et al. (2012) established a cell-free system to assemble RNA granules. The aggregation and formation of these RNA granules in vitro was triggered by incubation of the lysates either with a biotinylated isoxazole chemical or with the overexpressed domains of certain RNA-binding proteins that contain low complexity sequences. Proteomic analysis of these granules identified a large number of RNA-binding proteins, including TNRC6B and TNRC6C. Both Ago2 and TNRC6A were absent in these granules. These findings raise an interesting possibility that different TNRC6 proteins have distinct functions in different types of RNA granules. The RNA-binding proteins that contain low complexity sequences could potentially play key roles in regulating GW/P-body formation. Here, we described a method to extract and assay classical P bodies and GW/P bodies in vitro. Additional purification is required to separate these granules from the cytosolic fraction for proteomic analysis or biochemical assay. Further characterization of GW/P bodies and identification of endogenous mRNAs that are regulated by these GW/P bodies will provide better understanding of the role of these unique cytoplasmic mRNP granules in the miRNA pathway.
Materials and methods
DNA constructs
cDNAs encoding full-length human GW220, GW195, and GW182 were generated by RT-PCR from RNAs extracted from either HEK293T, U2OS, or HeLa cells. Plasmids expressing various GW/TNRC6 proteins were made by cloning the cDNAs into pcDNA3 (Invitrogen)-based mammalian expression vectors. Various truncations were generated by PCR subcloning. The β-globin expression construct was provided by A.-B. Shyu (The University of Texas Medical School, Houston, TX). The β-globin miRNA reporters were constructed into a tetracycline response elements–Tight vector (Takara Bio Inc.). The FLAG epitope tag was fused to the reporter to facilitate the analysis of protein levels by Western blot analysis. miRNA-responsive elements (containing six CXCR4 sites or three let-7 sites) were inserted into the 3′ UTR of the reporter constructs (Doench et al., 2003; Pillai et al., 2005). The 24 MS2 repeat–containing β-globin reporter was constructed in a similar fashion to the previously reported 24 MS2 repeat–containing luciferase reporters (Liu et al., 2005b).
Cell culture and transfection
Human HEK293T, U2OS, or HeLa cells were cultured in DME (10% FBS) in a 37°C incubator with 5% CO2. Plasmid transfections were performed using TransIT-LT1 or TransIT-HeLaMONSTER transfection reagent (Mirus Bio LLC) as per the manufacturer’s instructions. siCXCR4, micro–let-7, and other siRNAs were purchased from Thermo Fisher Scientific. siRNA-targeted sequences used in this study were as follows: GW, 5′-GAAAUGCUCUGGUCCGCUA-3′; GW220, 5′-TCGGTATCCTCGTGAAGTA-3′ and 5′-CAGATAAAGCCCAGTGTAA-3′; TNRC6B, 5′-GCACTGCCCTGATCCGATA-3′; and TNRC6C, 5′-CTATTAACCTCGCCAATTA-3′. siRNA transfections were performed using Oligofectamine reagent obtained from Invitrogen as per the manufacturer’s instructions. To generate the GW220 stable cell line, a full-length GW220 expression construct containing an N-terminal c-myc tag and a neomycin selectable cassette was transfected into the wild-type U2OS cells using TransIT-LT1 reagent. After 2 wk in selection with 500 µg/ml Geneticin G418 (Invitrogen), individual clones were expanded for analysis by both Western blotting and immunofluorescence to confirm the expression of GW220. Clones were further selected by single-cell dilution and expansion. The miRNA reporter assay and the tethering assay were performed as described previously (Liu et al., 2005a). In brief, exponentially growing cells were plated into 6-well plates at a density of 2.5 × 105 cells/well on the day before transfection. The cells were transfected with 2.5 µg of total DNA (β-globin reporter construct, pRevTet-Off-IN vector expressing tetracycline-controlled transactivator, and various GW expression constructs) and/or 100 nM siRNA. The transfection efficiency of HeLa Tet-OFF and U2OS cells ranges from 30 to 60%, monitored by GFP fluorescence assay. Protein and RNA analyses were performed 48 h after the transfection.
Protein analysis
Procedures for cell lysis, immunoprecipitation, and immunoblotting were performed as previously described (Liu et al., 2004). Lysis buffer contained 0.5% NP-40, 150 mM NaCl, 2 mM MgCl2, and 20 mM Tris HCl, pH 7.5. EDTA-free protease inhibitor (Roche) and DTT (final of 1.5 mM) were added immediately before lysis. The cell lysates were precleared by centrifugation at 15,000 g for 10 min at 4°C.
100-µg cell lysates were examined by Western blot analysis. For immunoprecipitation, a 10-mg cell lysate was incubated with 10 µg antibody and 30 µl protein A or G agarose or Dynabeads (Life Technologies) for 3 h at 4°C. For the in vitro GW/P-body assay, we modified the well-characterized RISC extract preparation method (Liu et al., 2004). Hypotonic extracts were prepared in buffer A (10 mM Hepes, pH 7.9, 1.5 mM MgCl2, and 10 mM NaCl). Two rounds of low speed clarification steps (1,000 g for 10 min) were used to obtain the cytoplasm extract. After adding 0.11 vol buffer B (0.3 M Hepes, pH 7.9, 1.4 M NaCl, and 0.03 M MgCl2), the extract was examined for the presence of miRISC and GW/P bodies. The lysates from low speed RISC extract were centrifuged onto microscope coverslips through a glycerol cushion. The coverslips were fixed and examined for the presence of GW/P bodies following the same immunofluorescence protocol (see Immunofluorescence section). Western blots of the straight cell lysates or of the immunoprecipitations were probed with the indicated antibodies. Antibodies to c-myc and HA were purchased from Roche, and antibodies to FLAG were purchased from Sigma-Aldrich. Specific antibodies against GW family proteins, Ago2 protein, and DDX6 protein were generated by immunizing rabbits with various synthetic peptides from either the N terminus or the C terminus of the corresponding proteins. Human IC-6 serum that recognizes Hedls is a gift from M.J. Fritzler (University of Calgary, Calgary, Alberta, Canada), and the rabbit polyclonal antibody against Dcp1 was a gift from J. Lykke-Andersen (University of California, San Diego, La Jolla, CA).
RNA analysis
RNA extractions were performed as previously described (Liu et al., 2004) using TRIZOL reagent (Invitrogen). Two-step qRT-PCR was performed using SuperScript III First-Strand Synthesis kit (Invitrogen), SYBR green supermix (iQ; Bio-Rad Laboratories), and iQ5 Real-Time PCR system (Bio-Rad Laboratories) as per the manufacturers’ instructions. Northern blotting and RNaseH/Northern analysis was performed as previously described (Wu et al., 2006). In brief, 10 µg of total RNA was annealed at 75°C for 5 min with a gene-specific oligo (500 ng), with or without the addition of an oligo(dT) oligo (500 ng). RNA/oligo hybrids were then digested with RNaseH (Invitrogen) for 1 h at 37°C. Samples were precipitated and resuspended in loading buffer (Ambion), separated on a 6% urea denaturing polyacrylamide gel, transferred to nitrocellose (Hybond-N+; GE Healthcare), and cross-linked using a UV stratalinker (Spectronics). The probe was labeled with [32P]CTP using the random labeling kit (Roche) as per the manufacturer’s instructions, and membranes were hybridized overnight at 42°C. Detection and analysis of the membrane after hybridization was carried out using a phosphoimager (Personal Molecular Imager; Bio-Rad Laboratories) and Quantity One 1-D Analysis Software (Bio-Rad Laboratories) or ImageJ software (National Institutes of Health).
Immunofluorescence
Cultured cells grown on a glass chamber slide were fixed using 4% paraformaldehyde (in PBS) for 12 min at room temperature. The cells were then permeabilized in PBS containing 0.2% Triton X-100 for 6 min at 4°C. Alexa Fluor (488 or 594)–conjugated secondary antibodies were purchased from Invitrogen. Antibody incubation was performed at room temperature in PBS containing 0.5% BSA as a blocking agent. The slides were mounted with the fluorescence mounting medium (Dako). The cells were examined using objective EC Plan-Neofluar 40×/0.75 NA phase contrast optics lens and fluorescent microscope (Axio Imager.Z1; Carl Zeiss) at room temperature. The images were acquired using a high resolution microscopy camera (AxioCam MRm Rev. 3 FireWire; Carl Zeiss) and AxioVision Rel. 4.6 software (Carl Zeiss). AxioVision image files were then exported to the TIFF file format. In the target mRNA localization assay, longer exposure was required to detect the cytoplasmic MS2-YFP signal in the control cells. We observed that ∼30–40% of the cells were transfected with the MS2-YFP protein, and >80% of these cells showed GW/P-body accumulation of the target mRNA in the presence of miRNA in five independent experiments.
Online supplemental material
Fig. S1 shows that different TNRC6 proteins form different mRNP granules in both HeLa Tet-OFF cells and U2OS cells. Fig. S2 shows the characterization of the GW/TNRC6A antibodies and the myc-GW220 stable cells. Fig. S3 shows that endogenous PABP protein does not accumulate in the GW/P bodies and that the formation of GW/P bodies in U2OS cells is not affected by the overexpression of catalytically inactive CAF1 mutant. Fig. S4 shows that Hedls promotes classical P-body formation in mammalian cells and does not silence the tethered mRNA target. Fig. S5 shows that depletion of GW220 leads to loss of the GW/P bodies and significant loss of the miRNA-targeted mRNA in the remaining miRISC-negative Hedls-positive P bodies. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201201153/DC1.
Supplementary Material
Acknowledgments
We thank Alan Hall, Christopher Lima, and Ian Ganley for critical reading of the manuscript, Roy Parker, Ed Chan, and Marvin J. Fritzler for helpful discussions, Marvin J. Fritzler for providing human index serum, Jens Lykke-Andersen for providing the Dcp1 antibody, and Ann-Bin Shyu for providing the β-globin expression vector. We thank Angelica Schreyer for characterization of various GW isoforms and editing the manuscript.
This work is supported by Sloan-Kettering Institute, Memorial Sloan-Kettering Cancer Center, and in part by a Special Fellow award from the Leukemia and Lymphoma Society (to J. Liu).
Footnotes
Abbreviations used in this paper:
- Ago
- Argonaute
- GW
- GW182/TNRC6
- miRNA
- microRNA
- mRNP
- messenger RNP
- PABP
- poly(A)-binding protein
- qRT-PCR
- quantitative RT-PCR
- RISC
- RNA-induced silencing complex
- UTR
- untranslated region
References
- Ambros V. 2004. The functions of animal microRNAs. Nature. 431:350–355 10.1038/nature02871 [DOI] [PubMed] [Google Scholar]
- Anderson P., Kedersha N. 2006. RNA granules. J. Cell Biol. 172:803–808 10.1083/jcb.200512082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D., Villén J., Shin C., Camargo F.D., Gygi S.P., Bartel D.P. 2008. The impact of microRNAs on protein output. Nature. 455:64–71 10.1038/nature07242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagga S., Bracht J., Hunter S., Massirer K., Holtz J., Eachus R., Pasquinelli A.E. 2005. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 122:553–563 10.1016/j.cell.2005.07.031 [DOI] [PubMed] [Google Scholar]
- Bartel D.P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 116:281–297 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- Bazzini A.A., Lee M.T., Giraldez A.J. 2012. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science. 336:233–237 10.1126/science.1215704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm-Ansmant I., Rehwinkel J., Doerks T., Stark A., Bork P., Izaurralde E. 2006. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 20:1885–1898 10.1101/gad.1424106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya S.N., Habermacher R., Martine U., Closs E.I., Filipowicz W. 2006. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 125:1111–1124 10.1016/j.cell.2006.04.031 [DOI] [PubMed] [Google Scholar]
- Boag P.R., Atalay A., Robida S., Reinke V., Blackwell T.K. 2008. Protection of specific maternal messenger RNAs by the P body protein CGH-1 (Dhh1/RCK) during Caenorhabditis elegans oogenesis. J. Cell Biol. 182:543–557 10.1083/jcb.200801183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun J.E., Huntzinger E., Fauser M., Izaurralde E. 2011. GW182 proteins directly recruit cytoplasmic deadenylase complexes to miRNA targets. Mol. Cell. 44:120–133 10.1016/j.molcel.2011.09.007 [DOI] [PubMed] [Google Scholar]
- Brengues M., Teixeira D., Parker R. 2005. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 310:486–489 10.1126/science.1115791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew R.W., Sontheimer E.J. 2009. Origins and mechanisms of miRNAs and siRNAs. Cell. 136:642–655 10.1016/j.cell.2009.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekulaeva M., Filipowicz W., Parker R. 2009. Multiple independent domains of dGW182 function in miRNA-mediated repression in Drosophila. RNA. 15:794–803 10.1261/rna.1364909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekulaeva M., Mathys H., Zipprich J.T., Attig J., Colic M., Parker R., Filipowicz W. 2011. miRNA repression involves GW182-mediated recruitment of CCR4-NOT through conserved W-containing motifs. Nat. Struct. Mol. Biol. 18:1218–1226 10.1038/nsmb.2166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.Y., Zheng D., Xia Z., Shyu A.B. 2009. Ago-TNRC6 triggers microRNA-mediated decay by promoting two deadenylation steps. Nat. Struct. Mol. Biol. 16:1160–1166 10.1038/nsmb.1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C.Y., Rana T.M. 2006. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol. 4:e210 10.1371/journal.pbio.0040210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke A., Prigge A., Wickens M. 2010. Translational repression by deadenylases. J. Biol. Chem. 285:28506–28513 10.1074/jbc.M110.150763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker C.J., Teixeira D., Parker R. 2007. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J. Cell Biol. 179:437–449 10.1083/jcb.200704147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djuranovic S., Nahvi A., Green R. 2012. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science. 336:237–240 10.1126/science.1215691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench J.G., Petersen C.P., Sharp P.A. 2003. siRNAs can function as miRNAs. Genes Dev. 17:438–442 10.1101/gad.1064703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A., Behm-Ansmant I., Izaurralde E. 2007a. P bodies: at the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol. 8:9–22 10.1038/nrm2080 [DOI] [PubMed] [Google Scholar]
- Eulalio A., Behm-Ansmant I., Schweizer D., Izaurralde E. 2007b. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol. Cell. Biol. 27:3970–3981 10.1128/MCB.00128-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A., Huntzinger E., Izaurralde E. 2008. GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat. Struct. Mol. Biol. 15:346–353 10.1038/nsmb.1405 [DOI] [PubMed] [Google Scholar]
- Eulalio A., Helms S., Fritzsch C., Fauser M., Izaurralde E. 2009a. A C-terminal silencing domain in GW182 is essential for miRNA function. RNA. 15:1067–1077 10.1261/rna.1605509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A., Tritschler F., Izaurralde E. 2009b. The GW182 protein family in animal cells: new insights into domains required for miRNA-mediated gene silencing. RNA. 15:1433–1442 10.1261/rna.1703809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian M.R., Mathonnet G., Sundermeier T., Mathys H., Zipprich J.T., Svitkin Y.V., Rivas F., Jinek M., Wohlschlegel J., Doudna J.A., et al. 2009. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol. Cell. 35:868–880 10.1016/j.molcel.2009.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian M.R., Sonenberg N., Filipowicz W. 2010. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 79:351–379 10.1146/annurev-biochem-060308-103103 [DOI] [PubMed] [Google Scholar]
- Fabian M.R., Cieplak M.K., Frank F., Morita M., Green J., Srikumar T., Nagar B., Yamamoto T., Raught B., Duchaine T.F., Sonenberg N. 2011. miRNA-mediated deadenylation is orchestrated by GW182 through two conserved motifs that interact with CCR4-NOT. Nat. Struct. Mol. Biol. 18:1211–1217 10.1038/nsmb.2149 [DOI] [PubMed] [Google Scholar]
- Fenger-Grøn M., Fillman C., Norrild B., Lykke-Andersen J. 2005. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol. Cell. 20:905–915 10.1016/j.molcel.2005.10.031 [DOI] [PubMed] [Google Scholar]
- Franks T.M., Lykke-Andersen J. 2008. The control of mRNA decapping and P-body formation. Mol. Cell. 32:605–615 10.1016/j.molcel.2008.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo C.M., Wang J.T., Motegi F., Seydoux G. 2010. Cytoplasmic partitioning of P granule components is not required to specify the germline in C. elegans. Science. 330:1685–1689 10.1126/science.1193697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbings D.J., Ciaudo C., Erhardt M., Voinnet O. 2009. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat. Cell Biol. 11:1143–1149 10.1038/ncb1929 [DOI] [PubMed] [Google Scholar]
- Giraldez A.J., Mishima Y., Rihel J., Grocock R.J., Van Dongen S., Inoue K., Enright A.J., Schier A.F. 2006. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 312:75–79 10.1126/science.1122689 [DOI] [PubMed] [Google Scholar]
- Guo H., Ingolia N.T., Weissman J.S., Bartel D.P. 2010. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 466:835–840 10.1038/nature09267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han T.W., Kato M., Xie S., Wu L.C., Mirzaei H., Pei J., Chen M., Xie Y., Allen J., Xiao G., McKnight S.L. 2012. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell. 149:768–779 10.1016/j.cell.2012.04.016 [DOI] [PubMed] [Google Scholar]
- Hu W., Sweet T.J., Chamnongpol S., Baker K.E., Coller J. 2009. Co-translational mRNA decay in Saccharomyces cerevisiae. Nature. 461:225–229 10.1038/nature08265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys D.T., Westman B.J., Martin D.I., Preiss T. 2005. MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc. Natl. Acad. Sci. USA. 102:16961–16966 10.1073/pnas.0506482102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakymiw A., Lian S., Eystathioy T., Li S., Satoh M., Hamel J.C., Fritzler M.J., Chan E.K. 2005. Disruption of GW bodies impairs mammalian RNA interference. Nat. Cell Biol. 7:1267–1274 10.1038/ncb1334 [DOI] [PubMed] [Google Scholar]
- Kato M., Han T.W., Xie S., Shi K., Du X., Wu L.C., Mirzaei H., Goldsmith E.J., Longgood J., Pei J., et al. 2012. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 149:753–767 10.1016/j.cell.2012.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Stoecklin G., Ayodele M., Yacono P., Lykke-Andersen J., Fritzler M.J., Scheuner D., Kaufman R.J., Golan D.E., Anderson P. 2005. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169:871–884 10.1083/jcb.200502088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzaretti D., Tournier I., Izaurralde E. 2009. The C-terminal domains of human TNRC6A, TNRC6B, and TNRC6C silence bound transcripts independently of Argonaute proteins. RNA. 15:1059–1066 10.1261/rna.1606309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R.C., Feinbaum R.L., Ambros V. 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 75:843–854 10.1016/0092-8674(93)90529-Y [DOI] [PubMed] [Google Scholar]
- Leung A.K., Calabrese J.M., Sharp P.A. 2006. Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proc. Natl. Acad. Sci. USA. 103:18125–18130 10.1073/pnas.0608845103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Lian S.L., Moser J.J., Fritzler M.L., Fritzler M.J., Satoh M., Chan E.K. 2008. Identification of GW182 and its novel isoform TNGW1 as translational repressors in Ago2-mediated silencing. J. Cell Sci. 121:4134–4144 10.1242/jcs.036905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim L.P., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M. 2005. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 433:769–773 10.1038/nature03315 [DOI] [PubMed] [Google Scholar]
- Liu J., Carmell M.A., Rivas F.V., Marsden C.G., Thomson J.M., Song J.J., Hammond S.M., Joshua-Tor L., Hannon G.J. 2004. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 305:1437–1441 10.1126/science.1102513 [DOI] [PubMed] [Google Scholar]
- Liu J., Rivas F.V., Wohlschlegel J., Yates J.R., III, Parker R., Hannon G.J. 2005a. A role for the P-body component GW182 in microRNA function. Nat. Cell Biol. 7:1261–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Valencia-Sanchez M.A., Hannon G.J., Parker R. 2005b. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 7:719–723 10.1038/ncb1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair G.R., Braks J.A., Garver L.S., Wiegant J.C., Hall N., Dirks R.W., Khan S.M., Dimopoulos G., Janse C.J., Waters A.P. 2006. Regulation of sexual development of Plasmodium by translational repression. Science. 313:667–669 10.1126/science.1125129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroney P.A., Yu Y., Fisher J., Nilsen T.W. 2006. Evidence that microRNAs are associated with translating messenger RNAs in human cells. Nat. Struct. Mol. Biol. 13:1102–1107 10.1038/nsmb1174 [DOI] [PubMed] [Google Scholar]
- Meister G., Landthaler M., Peters L., Chen P.Y., Urlaub H., Lührmann R., Tuschl T. 2005. Identification of novel argonaute-associated proteins. Curr. Biol. 15:2149–2155 10.1016/j.cub.2005.10.048 [DOI] [PubMed] [Google Scholar]
- Moser J.J., Fritzler M.J. 2010. Cytoplasmic ribonucleoprotein (RNP) bodies and their relationship to GW/P bodies. Int. J. Biochem. Cell Biol. 42:828–843 10.1016/j.biocel.2009.11.018 [DOI] [PubMed] [Google Scholar]
- Nottrott S., Simard M.J., Richter J.D. 2006. Human let-7a miRNA blocks protein production on actively translating polyribosomes. Nat. Struct. Mol. Biol. 13:1108–1114 10.1038/nsmb1173 [DOI] [PubMed] [Google Scholar]
- Olsen P.H., Ambros V. 1999. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev. Biol. 216:671–680 10.1006/dbio.1999.9523 [DOI] [PubMed] [Google Scholar]
- Ozgur S., Chekulaeva M., Stoecklin G. 2010. Human Pat1b connects deadenylation with mRNA decapping and controls the assembly of processing bodies. Mol. Cell. Biol. 30:4308–4323 10.1128/MCB.00429-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R., Sheth U. 2007. P bodies and the control of mRNA translation and degradation. Mol. Cell. 25:635–646 10.1016/j.molcel.2007.02.011 [DOI] [PubMed] [Google Scholar]
- Petersen C.P., Bordeleau M.E., Pelletier J., Sharp P.A. 2006. Short RNAs repress translation after initiation in mammalian cells. Mol. Cell. 21:533–542 10.1016/j.molcel.2006.01.031 [DOI] [PubMed] [Google Scholar]
- Piao X., Zhang X., Wu L., Belasco J.G. 2010. CCR4-NOT deadenylates mRNA associated with RNA-induced silencing complexes in human cells. Mol. Cell. Biol. 30:1486–1494 10.1128/MCB.01481-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai R.S., Bhattacharyya S.N., Artus C.G., Zoller T., Cougot N., Basyuk E., Bertrand E., Filipowicz W. 2005. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 309:1573–1576 10.1126/science.1115079 [DOI] [PubMed] [Google Scholar]
- Selbach M., Schwanhäusser B., Thierfelder N., Fang Z., Khanin R., Rajewsky N. 2008. Widespread changes in protein synthesis induced by microRNAs. Nature. 455:58–63 10.1038/nature07228 [DOI] [PubMed] [Google Scholar]
- Sen G.L., Blau H.M. 2005. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat. Cell Biol. 7:633–636 10.1038/ncb1265 [DOI] [PubMed] [Google Scholar]
- Souquere S., Mollet S., Kress M., Dautry F., Pierron G., Weil D. 2009. Unravelling the ultrastructure of stress granules and associated P-bodies in human cells. J. Cell Sci. 122:3619–3626 10.1242/jcs.054437 [DOI] [PubMed] [Google Scholar]
- Teixeira D., Sheth U., Valencia-Sanchez M.A., Brengues M., Parker R. 2005. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 11:371–382 10.1261/rna.7258505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman B., Ha I., Ruvkun G. 1993. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 75:855–862 10.1016/0092-8674(93)90530-4 [DOI] [PubMed] [Google Scholar]
- Wu L., Fan J., Belasco J.G. 2006. MicroRNAs direct rapid deadenylation of mRNA. Proc. Natl. Acad. Sci. USA. 103:4034–4039 10.1073/pnas.0510928103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zekri L., Huntzinger E., Heimstädt S., Izaurralde E. 2009. The silencing domain of GW182 interacts with PABPC1 to promote translational repression and degradation of microRNA targets and is required for target release. Mol. Cell. Biol. 29:6220–6231 10.1128/MCB.01081-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D., Ezzeddine N., Chen C.Y., Zhu W., He X., Shyu A.B. 2008. Deadenylation is prerequisite for P-body formation and mRNA decay in mammalian cells. J. Cell Biol. 182:89–101 10.1083/jcb.200801196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipprich J.T., Bhattacharyya S., Mathys H., Filipowicz W. 2009. Importance of the C-terminal domain of the human GW182 protein TNRC6C for translational repression. RNA. 15:781–793 10.1261/rna.1448009 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.