In the absence of SCAR, Dictyostelium WASP is relocalized from coated pits to the leading edge of the cell and activated to drive pseudopod formation.
Abstract
Under normal conditions, the Arp2/3 complex activator SCAR/WAVE controls actin polymerization in pseudopods, whereas Wiskott–Aldrich syndrome protein (WASP) assembles actin at clathrin-coated pits. We show that, unexpectedly, Dictyostelium discoideum SCAR knockouts could still spread, migrate, and chemotax using pseudopods driven by the Arp2/3 complex. In the absence of SCAR, some WASP relocated from the coated pits to the leading edge, where it behaved with similar dynamics to normal SCAR, forming split pseudopods and traveling waves. Pseudopods colocalized with active Rac, whether driven by WASP or SCAR, though Rac was activated to a higher level in SCAR mutants. Members of the SCAR regulatory complex, in particular PIR121, were not required for WASP regulation. We thus show that WASP is able to respond to all core upstream signals and that regulators coupled through the other members of SCAR’s regulatory complex are not essential for pseudopod formation. We conclude that WASP and SCAR can regulate pseudopod actin using similar mechanisms.
Introduction
Eukaryotic cells migrate using pseudopods that are made of actin filaments (Insall and Machesky, 2009). These actin filaments are initiated by the seven-member Arp2/3 complex. The key step controlling the formation of new pseudopods occurs when SCAR/WAVE proteins activate the Arp2/3 complex. SCAR/WAVE thus acts as the central regulator of cell movement. Other actin nucleation promotion factors that activate the Arp2/3 complex include Wiskott–Aldrich syndrome proteins (WASPs) and WASH. WASPs are typically found at the plasma membrane in punctate structures associated with vesicle traffic, for example, clathrin-coated pits (Taylor et al., 2011; Veltman et al., 2011) and invadopods (Yamaguchi et al., 2005). WASH, on the other hand, acts intracellularly on vesicles such as recycling endosomes (Derivery et al., 2009; Gomez and Billadeau, 2009) and lysosomes (Carnell et al., 2011).
Arp2/3 activators’ activation and recruitment are controlled by signaling proteins. Both SCAR/WAVE and WASP are activated by small GTPases of the Rho family (Higgs and Pollard, 2001), though numerous other signaling pathways have been described. WASPs bind directly to Cdc42 (Symons et al., 1996) and (in the case of neural WASP [N-WASP]) Rac (Tomasevic et al., 2007) through their Cdc42- and Rac-interacting and binding (CRIB) domains. SCAR/WAVEs do not possess any GTPase-binding domains but are activated by Rac as part of a five-member complex including PIR121, Nap1, Abelson kinase (Abl) interactor (Abi), and HSPC300 (Eden et al., 2002). PIR121 binds directly to Rac, whereas Abi has also been found to bind to WASP (Innocenti et al., 2005), thus providing a connection between WASP and SCAR/WAVE regulation. The detailed roles of the complex members are not well understood; however, Nap1 has been associated with adhesion (Ibarra et al., 2006; Weiner et al., 2006), but the direct regulators are not known, and HSPC300 remains a mystery.
SCAR was first discovered in Dictyostelium discoideum (Bear et al., 1998), a widely used target of cell migration research. SCAR, its regulatory complex, and WASP are all well conserved relative to the metazoan proteins (Veltman and Insall, 2010) but are encoded by single genes, making D. discoideum an ideal organism for analysis of the control of the Arp2/3 complex (King and Insall, 2009). However, D. discoideum is unusual in one respect. Mutants in SCAR or its complex have multiple, strong phenotypes. Cell size and speed, pseudopod size, and actin polymerization in response to signals are all decreased (Blagg et al., 2003; Ura et al., 2012). In qualitative terms, though, cells move and chemotax surprisingly well, considering the principal pseudopod pathway has been deleted. D. discoideum has a strong bleb pathway (Yoshida and Soldati, 2006), which can generate protrusions independently of the Arp2/3 complex using hydrostatic pressure. Loss of WASP is believed to be lethal (Myers et al., 2005).
In this paper, we show that pseudopods in SCAR mutant cells are generated after WASP relocalizes to the leading edge. This shows unexpected plasticity in the regulation of actin polymerization but, more surprisingly, that WASP is able to respond to all the signals that are necessary for pseudopod generation and chemotaxis.
Results and discussion
Separate roles of SCAR and WASP in wild-type cells
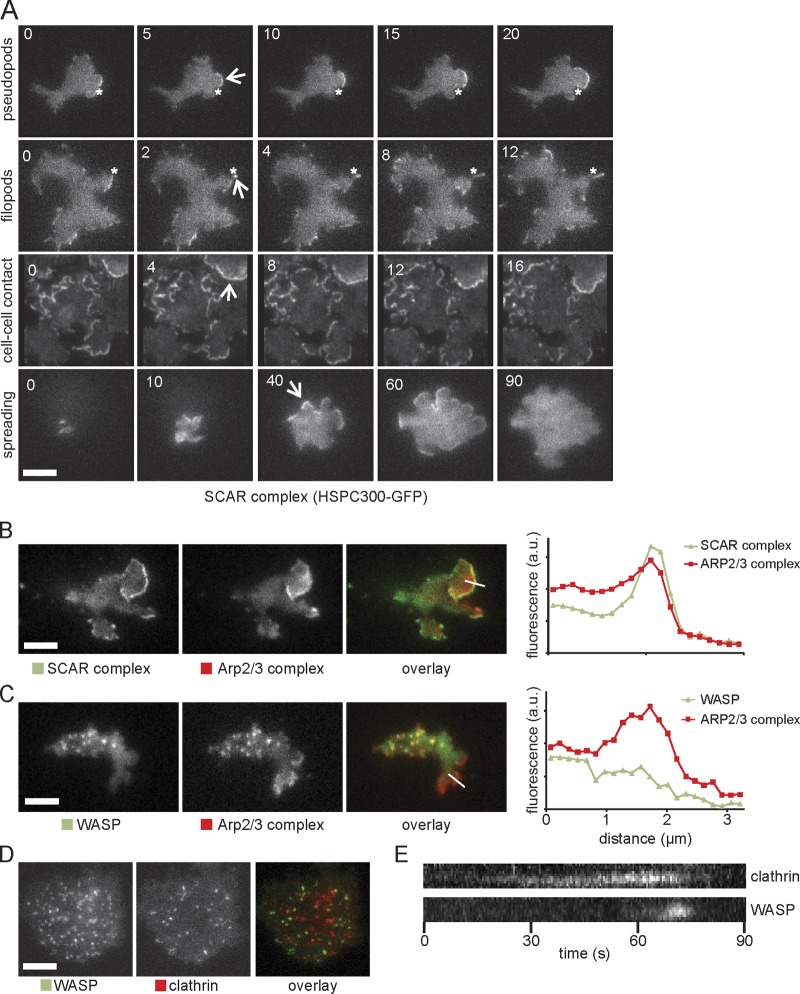
We examined the localization of the SCAR complex in living cells by total internal reflection fluorescence (TIRF) microscopy. Several different GFP fusions all behaved similarly, indicating that the GFP tag was not causing SCAR mislocalization (Fig. S1 A); GFP-HSPC300 gave a clear signal and was experimentally easy, so we used it as a marker for the complete SCAR complex. We saw an extremely dynamic pattern of localization (Video 1), as has been seen in cultured human neutrophils (Weiner et al., 2007). There were the expected localizations at the extreme front of pseudopods and the tips of filopods (Fig. 1 A); essentially all actin pseudopods in wild-type cells have a bright region of localized SCAR. This contrasts starkly with fixed cells (Pollitt et al., 2006) in which SCAR shows almost no distinct localization, presumably because the protein is too dynamic to fix. More surprisingly, two conditions revealed particularly strong SCAR localization. Cell–cell contacts were bright in vegetative cells and throughout development, and recruitment was particularly bright all around the edge when cells first spread after adhering (Fig. 1 A). Once cells were fully spread, the SCAR localization became more fleeting and restricted to the leading edge.
Figure 1.
SCAR complex and WASP localization in wild-type D. discoideum cells. (A) TIRF microscopy of SCAR complex (labeled with HSPC300-GFP) in pseudopods, filopods, cell–cell contact, and cell spreading of wild-type cells. Numbers indicate the time in seconds. The position of the asterisks is fixed across different time points. Arrows show SCAR-rich protrusions. (B and C) Colocalization of GFP-tagged SCAR complex (B) and WASP (C) with RFP-tagged Arp2/3 complex (subunit ARPC4) during cell migration. Quantitations of the fluorescence intensity along the indicated lines through the pseudopod are displayed on the right. Images are representative of ≥50 cells observed. (D) Colocalization of GFP-WASP and RFP-clathrin light chain. (E) Kymograph from a video similar to D showing arrival and disappearance of clathrin and WASP during a single clathrin-mediated endocytosis event. a.u., arbitrary unit. Bars, 5 µm.
The Arp2/3 complex is principally seen in two sites, leading pseudopods (Hahne et al., 2001) and puncta at the base of the cell that colocalize with clathrin (Kaksonen et al., 2006; Taylor et al., 2011). We examined the colocalization in TIRF of each structure with SCAR and the D. discoideum WASP orthologue. In pseudopods, only SCAR was normally visible (Fig. 1 B), in a narrow strip marginally in front of the start of the broader Arp2/3 band. WASP was recruited to clathrin puncta as previously described (Veltman et al., 2011), but SCAR was not. Detailed examination of the leading edge (Fig. 1 C) showed no discernible increase in WASP levels at the leading edge or throughout the Arp2/3 complex band. This conflicts with a previous study (Myers et al., 2005). We did not observe the broad bands of WASP reported by Myers et al. (2005) by TIRF, confocal, or wide-field microscopy (Fig. S1 B), and quantitative analysis (Fig. 1 C) does not show significant WASP localization in pseudopods. Yeast WASP (Sun et al., 2006) and human N-WASP (Taylor et al., 2011) in clathrin puncta are connected with the final stage of endocytosis, and the same is true for D. discoideum (Veltman et al., 2011). Each clathrin spot acquires a spot of WASP shortly before it vanishes from the TIRF field (which occurs as coated pits bud off into the cell; Fig. 1 D and Video 1), and every WASP spot is associated with a clathrin punctum. There is thus a clear separation of function in wild-type cells, with WASP connected with clathrin-mediated endocytosis and SCAR with leading edge protrusion.
Redeployment of WASP in SCAR mutants
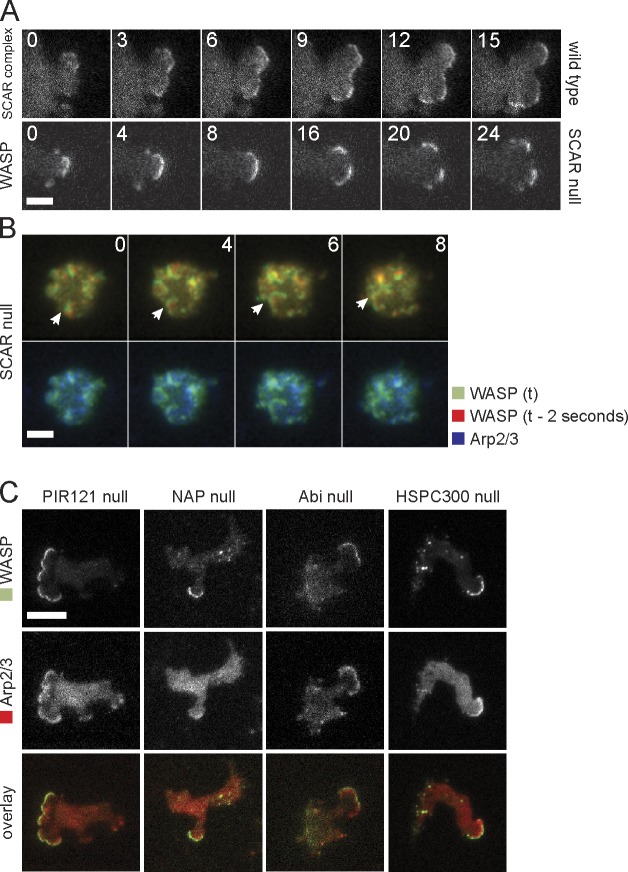
The relatively normal migration of SCAR mutants (Blagg et al., 2003) has been a puzzle for several years. We previously suspected that SCAR knockouts moved using blebs. Although SCAR mutants generate substantially more blebs than wild type, they still generate true actin pseudopods with normal Arp2/3 complex localization. We therefore transfected SCAR mutants with RFP-Arp2/3 and GFP-WASP and examined them by TIRF microscopy. Surprisingly, Arp2/3-positive pseudopods all contained WASP at the extreme leading edge in the position normally filled by SCAR (Fig. 2 A and Video 2). WASP did not lose its normal localization to puncta (Fig. 2, A and F, last image) but took on both functions simultaneously.
Figure 2.
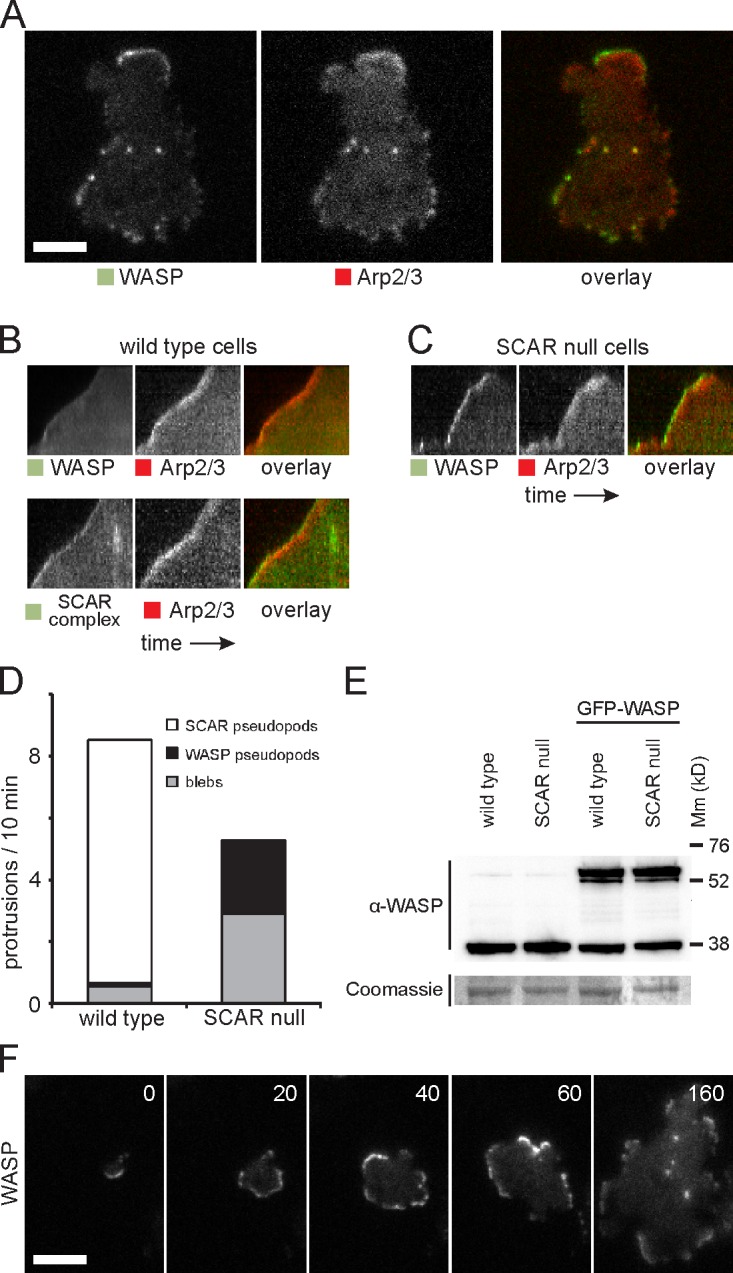
WASP compensates for the loss of SCAR. (A) Colocalization of WASP and Arp2/3 complex in SCAR-null cells. (B) Kymograph showing distribution of GFP-tagged WASP and SCAR complex with RFP-tagged Arp2/3 complex in a wild-type cell protrusion. The protrusion extends upwards, and time is along the horizontal axis. (C) Kymograph of WASP and the Arp2/3 complex in a protrusion of a SCAR-null cell. (D) Quantification of the number of pseudopods with SCAR complex and with WASP and the number of blebs in wild-type and SCAR-null cells. Over 200 pseudopods/blebs were counted in a total of 40 cells that were recorded with time-lapse microscopy for a length of 10 min each. (E) Western blot of whole-cell lysates of the indicated strains with an anti-WASP antibody. After immunodetection, the blot was stained with Coomassie brilliant blue. Molecular mass (Mm) size markers are indicated on the right in kilodaltons. (F) TIRF microscopy image of GFP-WASP in a SCAR-null cell that is dropping out of solution and spreading on the glass substratum. Time is indicated in seconds. Bars, 5 µm.
The cells were less polar, and fewer pseudopods were generated (Fig. 2 D), but they behaved normally. The WASP patch also migrated with the leading edge and drove protrusion at normal rates (Fig. 2, B and C, kymographs). Thus, WASP was redeployed to replace the function lost in SCAR knockouts. WASP expression levels remained unaltered in SCAR knockout cells (Fig. 2 E). WASP replaced SCAR in other locations besides pseudopods (Fig. 2 F), particularly spreading edges (Video 3). Overall, the only difference between localizations of SCAR in normal cells and WASP in SCAR knockouts was in the number of events. As far as we can tell, every structure that usually contains SCAR is still present in SCAR knockouts but with WASP in the place of SCAR.
Evolution of WASP-driven pseudopods
We examined the evolution of pseudopods using TIRF microscopy. SCAR localization is highly dynamic and typically spreads laterally along the cell edge. SCAR patches can become unstable and divide into two subregions (Fig. 3 A), causing the pseudopod to split (Andrew and Insall, 2007). Again, WASP behaved very similarly in SCAR mutants (Fig. 3 A). In neutrophils, the SCAR complex is recruited in expanding waves that underpin pseudopod formation (Weiner et al., 2006). We do not usually see expanding waves of SCAR complex in D. discoideum, but interestingly, the WASP in SCAR mutants can sometimes form expanding waves that resemble SCAR in neutrophils (Fig. 3 B and Video 4).
Figure 3.
WASP dynamics in SCAR complex mutants. (A) TIRF image series of splitting pseudopods in a wild-type cell labeled with HSPC300-GFP and a SCAR knockout cell labeled with GFP-WASP. Cells are moving to the right, and time is indicated in seconds. (B) TIRF image series of WASP waves in a SCAR-null cell. In the top images, this image is overlaid with the localization of WASP 2 s before the current frame, and in the bottom images, this image is overlaid with the Arp2/3 complex. A single-pass Gaussian blur was applied to the full image to smooth background fluorescence. The arrow tracks a single WASP wave. (C) TIRF images of GFP-WASP and RFP-tagged Arp2/3 complex in strains deleted for PIR121, Nap1, Abi, and HSPC300. Bars, 5 µm.
Other SCAR complex subunits are not required
The unexpected replacement of SCAR by WASP raises fundamental questions about the regulation of Arp2/3 complex in pseudopods. SCAR is thought to couple to most signaling pathways through its regulatory complex (Chen et al., 2010), in particular by PIR121 binding to activated Rac (Kobayashi et al., 1998) and Abi-coupling protein kinases, such as Src (Ardern et al., 2006) and Abl (Dai and Pendergast, 1995), to actin dynamics. Furthermore, Abi has been shown to interact with mammalian WASPs and connect SCAR and N-WASP function (Innocenti et al., 2005). We therefore examined WASP dynamics in SCAR complex mutants. Actin pseudopods still contained both WASP and the Arp2/3 complex in mutants lacking each of the SCAR complex subunits (Fig. 3 B). Thus, the connection between signaling pathways and WASP is not mediated by cross talk through the SCAR complex.
Rac activation and WASP localization
If WASP is not recruited through the SCAR regulatory complex, how is its localization controlled? It is not simply carried by F-actin because wild-type pseudopods have plenty of F-actin but no WASP. The most likely shared component is Rac, which binds to the PIR121 subunit of the SCAR complex. Mammalian WASPs contain a small GTPase-binding CRIB domain. It is usually reported that this is specific for Cdc42 binding (Aspenström et al., 1996), but several workers have found that Rac1 is also effective (Aspenström et al., 1996; Kolluri et al., 1996; Tomasevic et al., 2007). In D. discoideum, an unusual Rac (RacC) normally mediates WASP activation, but GTP-bound Rac1 isoforms also bind (Han et al., 2006).
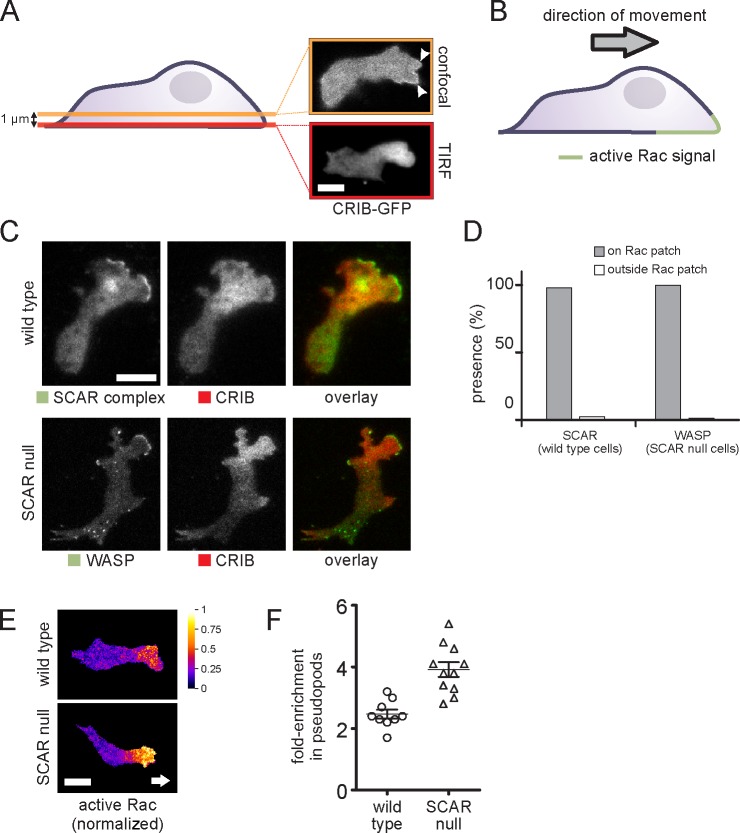
We examined the location of active Rac in living cells. We initially examined FRET-based probes based on the Raichu format (Mochizuki et al., 2001), but they caused an unacceptable dominant Rac1 activation effect, causing even vegetative cells to become hyperpolar and adhesive. However, GFP and monomeric RFP (mRFP) variant, mRFPmars fusions of the CRIB domain from PakB (de la Roche et al., 2005), which binds strongly to Racs, in particular Rac1a–c, yielded good localization. When CRIB-GFP was examined by confocal microscopy, a small peak was visible in planes above the base of the cell (Fig. 4 A). In TIRF microscopy, however, the localization was far clearer, but the active Rac was present in a patch of membrane around the leading edge. Thus, active Rac is found in the pattern shown in Fig. 4 B, defining the front portion of the cell. This closely resembles the localization seen in mammalian cells.
Figure 4.
Correlation between WASP, SCAR, and active Rac. (A) Visualization of the active Rac marker CRIB-GFP in a migrating cell in a confocal slice at a position 1 µm above the basal membrane (confocal) and directly on the basal membrane (TIRF). Arrowheads mark active protrusions. (B) Schematic drawing of the distribution of active Rac in a migrating cell. (C) TIRF image of a migrating cell coexpressing the active Rac marker CRIB-RFP and a marker for the SCAR complex (HSPC300-GFP) or GFP-WASP. (D) Quantification of the number of visible SCAR and WASP protrusions that are located on active Rac patches and outside of active Rac patches, respectively (n = 130 pseudopods from two experiments). (E) CRIB-GFP and free RFP were coexpressed in the indicated cells and visualized using TIRF microscopy. Active Rac distribution was normalized by dividing the CRIB-GFP signal by the free RFP signal. Background signal outside of the cell was masked. Arrow indicates cell’s direction. (F) Short videos of cells coexpressing CRIB-GFP and free RFP were recorded. Active Rac levels were normalized as described in E. The fold enrichment of active Rac in pseudopods in the normalized image was calculated by dividing the mean pixel value of the pseudopod by the mean pixel value of the cytosol. For each cell, the fold enrichment in pseudopods of each cell is plotted. Each data point is the mean of at least three pseudopods. The difference between the means is significant (Student’s t test, P < 0.001). Lines and error bars indicate means ± SEM. Bars, 5 µm.
When SCAR and active Rac reporter were colocalized in living cells, it became clear that SCAR recruitment only occurred at sites of Rac activation (Fig. 4 C). In SCAR mutants, the pattern of Rac activation was not greatly altered, but WASP was recruited to the areas of active Rac. A detailed analysis shows that SCAR recruitment is totally restricted to the patches of the membrane in which Rac was activated (Fig. 4 D); in SCAR− cells, WASP behaves in exactly the same way. Thus, in each case, Rac activation is a principal determinant of Arp2/3 complex activation and pseudopod formation.
Why does WASP relocalize in SCAR− mutants?
While examining the colocalization of WASP and active Rac, we observed that SCAR mutants consistently showed a stronger CRIB-GFP localization in pseudopods. We quantified this ratiometrically to normalize for uneven TIRF illumination. CRIB-GFP was coexpressed with free RFP, and Rac activation was measured by the green/red ratio (Fig. 4 E). Quantification of the data (Fig. 4 F) confirms that the enrichment of active Rac in pseudopods is consistently substantially higher in the mutants.
This suggests that WASP can be recruited to pseudopods by higher than usual levels of activated Rac1. To test this, we inducibly expressed dominant-active (G12V) Rac1A in wild-type cells and dominant-negative Rac1A (S17N) in SCAR mutants (Fig. S2). The dominant-active Rac1 caused strong recruitment of GFP-WASP all the way round the periphery of the wild-type cells, confirming that Rac1 activation is sufficient to recruit WASP. A complete ring, which is never normally seen, was observed in 11/100 cells. The dominant-negative Rac1 caused almost all WASP localization and pseudopod formation to be lost in 100% of observed cells (n > 100).
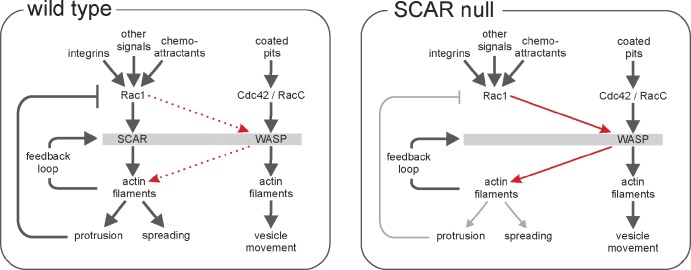
This allows us to propose a consistent and convincing model for the relocalization of WASP when SCAR function is lost (Fig. 5). In normal cells, SCAR recruitment occurs in patches of activated Rac. This causes Arp2/3 recruitment and pseudopod formation but also (directly or indirectly) results in Rac1 inactivation. When SCAR function is lost, by deletion of the genes for SCAR or any members of its regulatory complex, this Rac inactivation no longer occurs, and the level of activated Rac increases. When the level of activated Rac becomes high enough, WASP is recruited through its CRIB domain. In normal cells, SCAR is recruited by lower levels of active Rac1, and this recruitment maintains a low level Rac1 activation, so WASP is not normally recruited.
Figure 5.
Model of protrusion formation by SCAR and WASP in wild-type cells. Rac1 activation of the SCAR complex leads to new actin filament formation. We predict the following: (a) Feedback through actin filaments results in excitable behavior and a normal pseudopod cycle. (b) The Rac signal is turned off through negative feedback as the pseudopod matures. (c) In the absence of SCAR, protrusion formation is diminished, and active Rac levels increase until they engage WASP, which then replaces SCAR in essentially all respects. (d) In SCAR-null cells, as in wild-type cells, evolving pseudopods are formed through feedback, and normal protrusions are formed, though at a reduced frequency.
Implications
Our conclusion that WASP can replace the functions of SCAR has surprising implications for the control of actin polymerization and cell movement. Most authors currently consider WASP and SCAR to be parts of completely separate pathways (Stradal et al., 2004). They perform different roles within the cell and have different regulators. Even in mammalian cells, there are data contradicting this point—Rac1, the upstream regulator of mammalian SCAR/WAVE, can also bind and activate WASP and (particularly) N-WASP (Tomasevic et al., 2007). Our data go much further—in SCAR knockout cells, we find that WASP acquires SCAR’s localization, and the data suggest WASP replaces its fundamental functions. Of course, the global proteomes of SCAR mutants will be altered in complex and unforeseeable ways, but the obvious conclusion is that all essential upstream regulatory pathways and downstream effector pathways are shared between WASP and SCAR.
The recapitulation of the traveling waves is particularly striking. Such waves, which have been seen in other cells like neutrophils, require the existence of a positive feedback loop including SCAR and actin. We had previously presumed that this was mediated by the other four members of the SCAR regulatory complex (Nap1, PIR121, HSPC300, and Abi). However, because WASP does not bind to these proteins and because waves propagate effectively without them (Fig. 3), none of them is needed to make a positive feedback loop. They may take part in the wave-generating loop in normal cells, but none is essential.
One other form of feedback is also implied by our data. The increased Rac activation in SCAR knockouts implies that SCAR inhibits its upstream regulators. Rac activates SCAR, but SCAR activation feeds back negatively to Rac. This type of self-limitation is an important feature of robust systems (Brandman and Meyer, 2008) that also use positive feedback. It could be mediated by either SCAR itself, a member of the SCAR regulatory complex, or a downstream effector, such as actin or the Arp2/3 complex. Such feedback would have a key role in limiting the extent of pseudopod generation and thus in cell polarization. It could act through direct inhibition of any of the huge family of RacGEFs, activation of the similarly huge family of RacGAPs, or through more complex indirect pathways. One possible candidate is the WAVE-associated RacGAP (Soderling et al., 2002); though in D. discoideum, we did not find a strong association between the WAVE-associated RacGAP homologues (MEGAPs 1 and 2) and SCAR (Heath and Insall, 2008).
Overall, our data provide important and very unexpected limits on the mechanisms that generate pseudopods and actin regulation. Future work should focus on those interactors that are shared between SCAR and WASP (Rac and the Arp2/3 complex, in particular) as well as the specific components of each pathway.
Materials and methods
Cell strains
The axenic D. discoideum strain AX3 was used as wild type. Knockout cell strains for SCAR (IR46), PIR121 (SB3), NAP (SB12), Abi (AP5b), and HSPC300 (AP1) are previously described elsewhere (Blagg et al., 2003; Ibarra et al., 2006; Pollitt and Insall, 2008, 2009). In each strain, the gene was disrupted through homologous recombination by inserting a blasticidin resistance cassette into the open reading frame of the gene. Knockouts were confirmed by PCR. The marked cells expressing dominant Rac mutants were made as follows: A single vector expressing both GFP-WASP and PakB CRIB-mRFP (pDM982) was integrated into the genome of wild-type and SCAR-null cells. Transformants with appropriate expression levels of both GFP and mRFP were selected after cloning and then transfected with an extrachromosomal, inducible vector expressing constitutively active Rac1 G12V (pDM987) and dominant-negative Rac1 T17N (pDM985). WASP polyclonal antibody was a gift from T. Soldati (University of Geneva, Geneva, Switzerland).
DNA constructs
Selected genes were amplified from cDNA. Primers were designed using the genome data on Dictybase as a guideline. BglII–SpeI-compatible restriction sites were added before the start codon and after the last codon of the open reading frame. The CRIB motif was amplified from cDNA using primers 5′-ATGTCAATTTTCACTAATAAGAAAAAG-3′ and 5′-TTGAGCTTGTTGTTGTTGTTGTTGCATATG-3′ encoding residues 339–420 of D. discoideum PakB. PCR fragments were cloned in the PCR ligation vector pDM368 and fully sequenced. Correct clones were selected, and the genes were subcloned into a GFP expression vector (pDM448/pDM450 for N-terminal or C-terminal fusions, respectively; Veltman et al., 2009). For coexpression, the second gene was cloned into an mRFP shuttle vector (pDM411/pDM413 for N-terminal or C-terminal fusions, respectively). The entire expression cassette was excised with NgoMIV and cloned into the single NgoMIV site of the GFP expression vector that contains the first gene.
Microscopy and image analysis
Cells were incubated overnight under LoFlo medium (ForMedium) to remove autofluorescence in preparation for fluorescence microscopy. Cells were washed with development buffer (DB; 10 mM Na/K phosphate buffer, pH 6.5, 2 mM MgCl2, and 1 mM CaCl2) and starved as a monolayer in a Petri dish under DB until the onset of streaming to obtain developed cells. Vegetative cells were imaged in LoFlo medium, and developed cells were imaged in DB.
TIRF microscopy was performed at 21°C on a microscope (Eclipse TE2000-U; Nikon) that was fitted with a custom TIRF condenser and a 1.45 NA, 100× Plan Apochromat TIRF objective (Nikon). Red and green fluorescent signals were separated using a beam splitter and projected onto an EM charge-coupled device camera (Cascade II:512; Photometrics). MetaMorph software (Molecular Devices) was used to control the camera, shutters, and light sources. Wide-field fluorescence microscopy was performed on the same microscope but with a mercury lamp (X-Cite 120PC Q; Lumen Dynamics) as a light source. Confocal microscopy was performed on a microscope (A1R; Nikon) using a 1.4 NA, 60× Plan Apochromat objective.
Microscopy data were analyzed using ImageJ (National Institutes of Health). For Video 1, a windowed-sinc filter was applied to reduce noise. Active Rac levels were quantified as follows. A single-pass Gaussian blur was first applied to the image to remove potential zero-value pixels. Then, pixel values of the green channel (CRIB-GFP) were divided by the pixel values of the red channel (free RFP) to compensate for potential illumination differences originating from local differences in the distance between the membrane and the glass surface. The fold enrichment of the active Rac signal in the pseudopod was calculated by dividing the mean pixel value in the pseudopod by the mean pixel value of the cell body.
Online supplemental material
Fig. S1 shows the normal location of SCAR complex and WASP. Fig. S2 shows how dominant Rac mutants affect WASP localization in SCAR knockout cells. Videos 1 and 2 show the change in WASP distribution in pseudopods of wild-type and SCAR knockout cells, respectively. Video 3 shows cell spreading of SCAR in wild-type cells and WASP in SCAR knockout cells. Video 4 shows self-propagating waves of WASP in SCAR-null cells that mimic the waves of SCAR seen elsewhere. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201205058/DC1.
Supplementary Material
Acknowledgments
We are very grateful to Dr. David Knecht for critical comments on the manuscript, to Dr. Thierry Soldati for the anti-WASP antibody, and to Drs. Kurt Anderson and Juliana Schwarz and the Beatson Advanced Imaging Resource for assistance with microscopy.
We are grateful to Cancer Research UK for central support to R.H. Insall, J.S. King, D.M. Veltman, and L.M. Machesky.
Footnotes
Abbreviations used in this paper:
- Abi
- Abl interactor
- Abl
- Abelson kinase
- CRIB
- Cdc42 and Rac interacting and binding
- DB
- development buffer
- mRFP
- monomeric RFP
- N-WASP
- neural WASP
- TIRF
- total internal reflection fluorescence
- WASP
- Wiskott–Aldrich syndrome protein
References
- Andrew N., Insall R.H. 2007. Chemotaxis in shallow gradients is mediated independently of PtdIns 3-kinase by biased choices between random protrusions. Nat. Cell Biol. 9:193–200 10.1038/ncb1536 [DOI] [PubMed] [Google Scholar]
- Ardern H., Sandilands E., Machesky L.M., Timpson P., Frame M.C., Brunton V.G. 2006. Src-dependent phosphorylation of Scar1 promotes its association with the Arp2/3 complex. Cell Motil. Cytoskeleton. 63:6–13 10.1002/cm.20101 [DOI] [PubMed] [Google Scholar]
- Aspenström P., Lindberg U., Hall A. 1996. Two GTPases, Cdc42 and Rac, bind directly to a protein implicated in the immunodeficiency disorder Wiskott-Aldrich syndrome. Curr. Biol. 6:70–75 10.1016/S0960-9822(02)00423-2 [DOI] [PubMed] [Google Scholar]
- Bear J.E., Rawls J.F., Saxe C.L., III 1998. SCAR, a WASP-related protein, isolated as a suppressor of receptor defects in late Dictyostelium development. J. Cell Biol. 142:1325–1335 10.1083/jcb.142.5.1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagg S.L., Stewart M., Sambles C., Insall R.H. 2003. PIR121 regulates pseudopod dynamics and SCAR activity in Dictyostelium. Curr. Biol. 13:1480–1487 10.1016/S0960-9822(03)00580-3 [DOI] [PubMed] [Google Scholar]
- Brandman O., Meyer T. 2008. Feedback loops shape cellular signals in space and time. Science. 322:390–395 10.1126/science.1160617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnell M., Zech T., Calaminus S.D., Ura S., Hagedorn M., Johnston S.A., May R.C., Soldati T., Machesky L.M., Insall R.H. 2011. Actin polymerization driven by WASH causes V-ATPase retrieval and vesicle neutralization before exocytosis. J. Cell Biol. 193:831–839 10.1083/jcb.201009119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Borek D., Padrick S.B., Gomez T.S., Metlagel Z., Ismail A.M., Umetani J., Billadeau D.D., Otwinowski Z., Rosen M.K. 2010. Structure and control of the actin regulatory WAVE complex. Nature. 468:533–538 10.1038/nature09623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Z., Pendergast A.M. 1995. Abi-2, a novel SH3-containing protein interacts with the c-Abl tyrosine kinase and modulates c-Abl transforming activity. Genes Dev. 9:2569–2582 10.1101/gad.9.21.2569 [DOI] [PubMed] [Google Scholar]
- de la Roche M., Mahasneh A., Lee S.F., Rivero F., Côté G.P. 2005. Cellular distribution and functions of wild-type and constitutively activated Dictyostelium PakB. Mol. Biol. Cell. 16:238–247 10.1091/mbc.E04-06-0534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derivery E., Sousa C., Gautier J.J., Lombard B., Loew D., Gautreau A. 2009. The Arp2/3 activator WASH controls the fission of endosomes through a large multiprotein complex. Dev. Cell. 17:712–723 10.1016/j.devcel.2009.09.010 [DOI] [PubMed] [Google Scholar]
- Eden S., Rohatgi R., Podtelejnikov A.V., Mann M., Kirschner M.W. 2002. Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature. 418:790–793 10.1038/nature00859 [DOI] [PubMed] [Google Scholar]
- Gomez T.S., Billadeau D.D. 2009. A FAM21-containing WASH complex regulates retromer-dependent sorting. Dev. Cell. 17:699–711 10.1016/j.devcel.2009.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahne P., Sechi A., Benesch S., Small J.V. 2001. Scar/WAVE is localised at the tips of protruding lamellipodia in living cells. FEBS Lett. 492:215–220 10.1016/S0014-5793(01)02239-6 [DOI] [PubMed] [Google Scholar]
- Han J.W., Leeper L., Rivero F., Chung C.Y. 2006. Role of RacC for the regulation of WASP and phosphatidylinositol 3-kinase during chemotaxis of Dictyostelium. J. Biol. Chem. 281:35224–35234 10.1074/jbc.M605997200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath R.J., Insall R.H. 2008. Dictyostelium MEGAPs: F-BAR domain proteins that regulate motility and membrane tubulation in contractile vacuoles. J. Cell Sci. 121:1054–1064 10.1242/jcs.021113 [DOI] [PubMed] [Google Scholar]
- Higgs H.N., Pollard T.D. 2001. Regulation of actin filament network formation through ARP2/3 complex: activation by a diverse array of proteins. Annu. Rev. Biochem. 70:649–676 10.1146/annurev.biochem.70.1.649 [DOI] [PubMed] [Google Scholar]
- Ibarra N., Blagg S.L., Vazquez F., Insall R.H. 2006. Nap1 regulates Dictyostelium cell motility and adhesion through SCAR-dependent and -independent pathways. Curr. Biol. 16:717–722 10.1016/j.cub.2006.02.068 [DOI] [PubMed] [Google Scholar]
- Innocenti M., Gerboth S., Rottner K., Lai F.P., Hertzog M., Stradal T.E., Frittoli E., Didry D., Polo S., Disanza A., et al. 2005. Abi1 regulates the activity of N-WASP and WAVE in distinct actin-based processes. Nat. Cell Biol. 7:969–976 10.1038/ncb1304 [DOI] [PubMed] [Google Scholar]
- Insall R.H., Machesky L.M. 2009. Actin dynamics at the leading edge: from simple machinery to complex networks. Dev. Cell. 17:310–322 10.1016/j.devcel.2009.08.012 [DOI] [PubMed] [Google Scholar]
- Kaksonen M., Toret C.P., Drubin D.G. 2006. Harnessing actin dynamics for clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 7:404–414 10.1038/nrm1940 [DOI] [PubMed] [Google Scholar]
- King J.S., Insall R.H. 2009. Chemotaxis: finding the way forward with Dictyostelium. Trends Cell Biol. 19:523–530 10.1016/j.tcb.2009.07.004 [DOI] [PubMed] [Google Scholar]
- Kobayashi K., Kuroda S., Fukata M., Nakamura T., Nagase T., Nomura N., Matsuura Y., Yoshida-Kubomura N., Iwamatsu A., Kaibuchi K. 1998. p140Sra-1 (specifically Rac1-associated protein) is a novel specific target for Rac1 small GTPase. J. Biol. Chem. 273:291–295 10.1074/jbc.273.1.291 [DOI] [PubMed] [Google Scholar]
- Kolluri R., Tolias K.F., Carpenter C.L., Rosen F.S., Kirchhausen T. 1996. Direct interaction of the Wiskott-Aldrich syndrome protein with the GTPase Cdc42. Proc. Natl. Acad. Sci. USA. 93:5615–5618 10.1073/pnas.93.11.5615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki N., Yamashita S., Kurokawa K., Ohba Y., Nagai T., Miyawaki A., Matsuda M. 2001. Spatio-temporal images of growth-factor-induced activation of Ras and Rap1. Nature. 411:1065–1068 10.1038/35082594 [DOI] [PubMed] [Google Scholar]
- Myers S.A., Han J.W., Lee Y., Firtel R.A., Chung C.Y. 2005. A Dictyostelium homologue of WASP is required for polarized F-actin assembly during chemotaxis. Mol. Biol. Cell. 16:2191–2206 10.1091/mbc.E04-09-0844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollitt A.Y., Insall R.H. 2008. Abi mutants in Dictyostelium reveal specific roles for the SCAR/WAVE complex in cytokinesis. Curr. Biol. 18:203–210 10.1016/j.cub.2008.01.026 [DOI] [PubMed] [Google Scholar]
- Pollitt A.Y., Insall R.H. 2009. Loss of Dictyostelium HSPC300 causes a scar-like phenotype and loss of SCAR protein. BMC Cell Biol. 10:13 10.1186/1471-2121-10-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollitt A.Y., Blagg S.L., Ibarra N., Insall R.H. 2006. Cell motility and SCAR localisation in axenically growing Dictyostelium cells. Eur. J. Cell Biol. 85:1091–1098 10.1016/j.ejcb.2006.05.014 [DOI] [PubMed] [Google Scholar]
- Soderling S.H., Binns K.L., Wayman G.A., Davee S.M., Ong S.H., Pawson T., Scott J.D. 2002. The WRP component of the WAVE-1 complex attenuates Rac-mediated signalling. Nat. Cell Biol. 4:970–975 10.1038/ncb886 [DOI] [PubMed] [Google Scholar]
- Stradal T.E., Rottner K., Disanza A., Confalonieri S., Innocenti M., Scita G. 2004. Regulation of actin dynamics by WASP and WAVE family proteins. Trends Cell Biol. 14:303–311 10.1016/j.tcb.2004.04.007 [DOI] [PubMed] [Google Scholar]
- Sun Y., Martin A.C., Drubin D.G. 2006. Endocytic internalization in budding yeast requires coordinated actin nucleation and myosin motor activity. Dev. Cell. 11:33–46 10.1016/j.devcel.2006.05.008 [DOI] [PubMed] [Google Scholar]
- Symons M., Derry J.M., Karlak B., Jiang S., Lemahieu V., Mccormick F., Francke U., Abo A. 1996. Wiskott-Aldrich syndrome protein, a novel effector for the GTPase CDC42Hs, is implicated in actin polymerization. Cell. 84:723–734 10.1016/S0092-8674(00)81050-8 [DOI] [PubMed] [Google Scholar]
- Taylor M.J., Perrais D., Merrifield C.J. 2011. A high precision survey of the molecular dynamics of mammalian clathrin-mediated endocytosis. PLoS Biol. 9:e1000604 10.1371/journal.pbio.1000604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasevic N., Jia Z., Russell A., Fujii T., Hartman J.J., Clancy S., Wang M., Beraud C., Wood K.W., Sakowicz R. 2007. Differential regulation of WASP and N-WASP by Cdc42, Rac1, Nck, and PI(4,5)P2. Biochemistry. 46:3494–3502 10.1021/bi062152y [DOI] [PubMed] [Google Scholar]
- Ura S., Pollitt A.Y., Veltman D.M., Morrice N.A., Machesky L.M., Insall R.H. 2012. Pseudopod growth and evolution during cell movement is controlled through SCAR/WAVE dephosphorylation. Curr. Biol. 22:553–561 10.1016/j.cub.2012.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltman D.M., Insall R.H. 2010. WASP family proteins: their evolution and its physiological implications. Mol. Biol. Cell. 21:2880–2893 10.1091/mbc.E10-04-0372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltman D.M., Akar G., Bosgraaf L., Van Haastert P.J. 2009. A new set of small, extrachromosomal expression vectors for Dictyostelium discoideum. Plasmid. 61:110–118 10.1016/j.plasmid.2008.11.003 [DOI] [PubMed] [Google Scholar]
- Veltman D.M., Auciello G., Spence H.J., Machesky L.M., Rappoport J.Z., Insall R.H. 2011. Functional analysis of Dictyostelium IBARa reveals a conserved role of the I-BAR domain in endocytosis. Biochem. J. 436:45–52 10.1042/BJ20101684 [DOI] [PubMed] [Google Scholar]
- Weiner O.D., Rentel M.C., Ott A., Brown G.E., Jedrychowski M., Yaffe M.B., Gygi S.P., Cantley L.C., Bourne H.R., Kirschner M.W. 2006. Hem-1 complexes are essential for Rac activation, actin polymerization, and myosin regulation during neutrophil chemotaxis. PLoS Biol. 4:e38 10.1371/journal.pbio.0040038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner O.D., Marganski W.A., Wu L.F., Altschuler S.J., Kirschner M.W. 2007. An actin-based wave generator organizes cell motility. PLoS Biol. 5:e221 10.1371/journal.pbio.0050221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H., Lorenz M., Kempiak S., Sarmiento C., Coniglio S., Symons M., Segall J., Eddy R., Miki H., Takenawa T., Condeelis J. 2005. Molecular mechanisms of invadopodium formation: the role of the N-WASP–Arp2/3 complex pathway and cofilin. J. Cell Biol. 168:441–452 10.1083/jcb.200407076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K., Soldati T. 2006. Dissection of amoeboid movement into two mechanically distinct modes. J. Cell Sci. 119:3833–3844 10.1242/jcs.03152 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.