Abstract
To understand the regulation and expression of pyrimidine biosynthesis in plants, we have examined the effect of the metabolic inhibitor 5-fluoroorotic acid (FOA) on uridine-5′-monophosphate synthase (UMPSase) expression in cell cultures of Nicotiana plumbaginifolia. UMPSase is the rate-limiting step of pyrimidine biosynthesis in plants. Addition of FOA causes an up-regulation of UMPSase enzyme activity in cell cultures after a lag phase of several days. Western-blot analysis demonstrated that the up-regulation in enzyme activity was caused by increased expression of the UMPSase protein. Northern-blot analysis demonstrated a higher level of UMPSase mRNA in the FOA-induced tissues than in control tissues. Run-on transcriptional assays showed that the UMPSase gene was transcriptionally activated after FOA treatment. The mechanism of toxicity of FOA is through thymine starvation. We found that addition of thymine abrogated the FOA-mediated up-regulation of UMPSase. In addition, methotrexate and aminopterin, which affect thymine levels by inhibiting dihydrofolate reductase, also up-regulate UMPSase in N. plumbaginifolia cells.
Pyrimidines play a central role in cellular regulation and metabolism. They are substrates for DNA/RNA biosynthesis, regulators of the biosynthesis of some amino acids, and cofactors in the biosynthesis of phospholipids, glycolipids, sugars, and polysaccharides. The classical de novo pyrimidine biosynthetic pathway ends with the synthesis of UMP, and other divergent pathways lead to the formation of CTP and TTP (Neuhard and Nygaard, 1987). Also, several salvage pathways exist that allow cells to utilize preformed nucleotides, thereby avoiding the high metabolic cost of biosynthesis (Jones and Hahn, 1979; Neuhard and Nygaard, 1987).
The enzymatic activities of the de novo pyrimidine biosynthetic pathway are well known and invariant in all examined organisms; however, the gene organization of these several steps varies among organisms. Plants differ from most higher eukaryotes in that the first three steps of the de novo pathway are carried out by separate enzymes that are individually encoded (Williamson and Slocum, 1994; Williamson et al., 1996; Zhou et al., 1997). In mammals, some fungi, and insects, however, the genes encoding these enzymes have been rearranged during evolution (van den Hoff et al., 1995) into a single transcriptional unit that encodes a single polyprotein termed CAD (Kim et al., 1992). This polyprotein has three enzymatic activities, carbamoylphosphate synthase, aspartyl transcarbamoylase, and dihydroorotase.
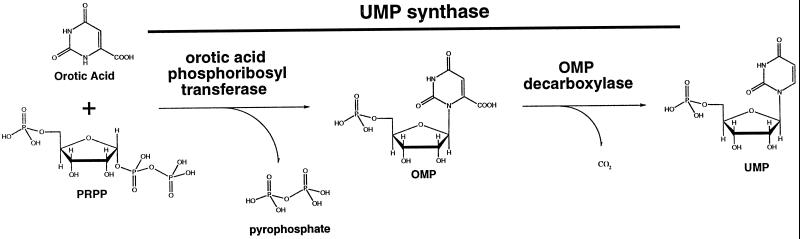
An additional polyprotein, UMPSase, is present in both plants and animals and includes the last two steps of the de novo biosynthetic pathway (Jones, 1980; Nasr et al., 1994; Maier et al., 1995). These last two enzymatic steps are orotate phosphoribosyltransferase and orotidine decarboxylase. During evolution the mRNAs encoding these two enzymes have become fused into a single transcript that encodes one protein having both enzymatic activities (Jones, 1980). UMPSase is one of the key enzymes of the de novo biosynthesis of pyrimidines (Fig. 1). It is the rate-limiting step of the pathway in both mammals (Traut and Jones, 1977) and plants (Santoso and Thornburg, 1992).
Figure 1.
Action of UMPSase. UMPSase has both orotic acid phosphoribosyl transferase activity as well as OMP decarboxylase activity.
We have examined the role of UMPSase in plants in a variety of studies. First, we restructured a UMPSase gene from Dictyostelium discoideum (Shi and Thornburg, 1993) and expressed this protein with a novel form of regulation in transgenic plants. Plants transformed with this gene show altered nucleotide pool sizes (L. Zhou and R. Thornburg, unpublished data).
We have also used a negative selection scheme based on pyrimidine metabolism to produce plant cell lines that have stable alterations in gene expression (Santoso and Thornburg, 1992). This selection uses FOA to generate a toxic metabolite, 5-FdUMP, which provides the basis of selection. Because UMPSase is the rate-limiting step in pyrimidine biosynthesis, its expression is frequently affected by FOA selection. By far, the majority of the selected cell lines show reduced levels of UMPSase. This was expected from previous studies of selection in Saccharomyces cerevisiae (Boeke et al., 1984) and D. discoideum (Kalpaxis et al., 1991). However, in recent work with Nicotiana plumbaginifolia, we have isolated some cell lines that show elevated rather than reduced levels of UMPSase activity. Of the 143 cell lines isolated in 8 separate replicates of FOA selection, 14% of the surviving cell lines had 3-fold or higher levels of UMPSase enzyme activity (D. Santoso and R. Thornburg, unpublished data). In an effort to understand the regulation of UMPSase levels in these selected cell lines, we have investigated the expression of UMPSase in wild-type N. plumbaginifolia cells in response to growth on fluoroorotic acid.
MATERIALS AND METHODS
Nicotiana plumbaginifolia plants in sterile culture were kindly provided by Dr. Laszlo Martón (University of South Carolina, Columbia). The Nicotiana tabacum UMPSase cDNA, pRT327, was used routinely as the template to prepare either 32P-labeled DNA or RNA probes (Maier et al., 1995). A polyclonal antibody raised against UMPSase protein isolated from squash fruit was produced in a female New Zealand white rabbit (Santoso and Thornburg, 1995).
Phosphoribosyl pyrophosphate, orotic acid, FOA, uracil, nitrosomethyl urea, nucleotide metabolites, MTX, and AMT were purchased from Sigma. Media for plant tissue culture and plant hormones were purchased from either Sigma or Gibco Laboratories. Restriction enzymes were from Promega. RNA transcription kits were obtained from Stratagene. The radiochemicals, 7-[14C]orotic acid with specific activity of 48.5 mCi/mmol, [125I]rProteinA (9.0 mCi/mg), α-[32P]UTP (800 Ci/mmol), and α-[32P]dCTP (3000 Ci/mmol) were purchased from New England Nuclear. Other materials were of the highest purity available and were obtained either locally or from Fisher Scientific.
Tissue Culture and Induction by FOA
Cell lines were maintained on Murashige-Skoog medium (3% Suc, 100 mg/L myo-inositol, 1 mg/L thiamine, 0.5 mg/L pyridoxine.HCl, 0.5 mg/L nicotinic acid, 2 mg/L IAA, 0.2 mg/L 6-benzyl amino purine, 250 mg/L casein hydrolysate, 20 mL/L sterile coconut water, and 8 g/L agar). To induce UMPSase, small pieces of calli were cultured in Murashige-Skoog induction medium without casein or coconut water. The media were prepared by adding filter-sterilized additives into the Murashige-Skoog medium to the final concentration indicated in each experiment. Small pieces (0.2–0.3 g) of calli were inoculated onto the induction medium with a density of about 1 to 1.5 g of tissue per 10 mL of medium and incubated at 26°C with a 16-h day and 8-h dark period. After the indicated times, the cells were harvested and washed with half-strength Murashige-Skoog liquid medium; the excess liquid was removed with filter paper, and the cells were stored at −70°C until all samples were ready for assay.
UMPSase Assays
UMPSase was determined by a CO2-release assay modified from Walther et al. (1984) as previously described (Santoso and Thornburg, 1992). [14C]Orotic acid labeled at the carboxyl group (7-[14C]orotic acid) was used as primary substrate. The reaction was linear for the first 20 min but, routinely, the reaction was carried out for only 5 to 10 min to ensure that the initial velocity was being measured.
Blots
Western blots were performed as described by Timmons and Dunbar (1990). The incubation of the transferred proteins in the antisera was performed overnight at 4°C.
Total RNA was isolated from cells of N. plumbaginifolia with the method of Wadsworth et al. (1988). The average yield was about 50 μg RNA/g tissue. The RNA samples were denatured and electrophoresed as described in Tirimanne and Colbert (1991). Northern blots were conducted according to the work of Ausubel et al. (1987).
In Vitro Runoff Transcription
Nuclei were isolated from tissue-cultured cells using a method adapted from Watson and Thompson (1986). All operations were performed in RNase-free conditions at 4°C. The conditions for the in vitro transcription assay were according to the method of Kimpel et al. (1990) with the following modification: after the proteinase K digestion step, 50 μg of tRNA was added, and the RNA solution was ethanol precipitated. After centrifugation the pellet was washed with 70% ethanol and air dried. The pellet containing labeled RNA was dissolved in diethyl pyrocarbonate-treated deionized water and counted to determine the amount of radioactivity.
UMPSase-antisense mRNA was transcribed in vitro using a transcription kit (Stratagene) from the plasmid pRT327. Binding of the in vitro-transcribed antisense UMPSase RNA onto the GeneScreen membrane (DuPont) was performed according to the manufacturer's instructions. After prehybridization for 4 h at 58°C in 150 μL of the hybridization solution per cm2 of membrane, the membrane was hybridized overnight at 58°C with 2 to 8 × 106 cpm of 32P-labeled RNA from the runoff transcription per milliliter of hybridization solution (10% dextran sulfate, 1% SDS, 200 μg/mL denatured-salmon sperm DNA, and 1 m NaCl). After washing, the damp membrane was wrapped with plastic and autoradiographed for 1 to 3 d.
RESULTS
Effect of FOA on UMPSase Activity
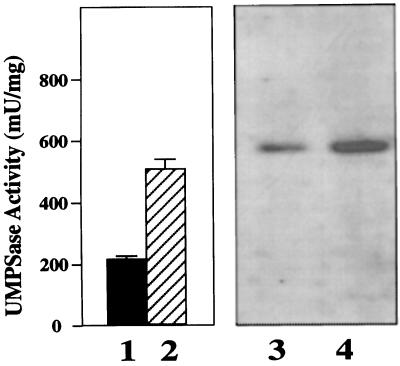
In yeast (Boeke et al., 1984) and Dictyostelium discoideum (Kalpaxis et al., 1991) selection of mutants on FOA plus uracil usually produces mutations in the final two enzymatic steps of pyrimidine biosynthesis (UMPSase). We therefore examined the effect of FOA on UMPSase enzymatic activity. Normal N. plumbaginifolia cells growing on Murashige-Skoog solid medium expressed a relatively low UMPSase activity of about 200 milliunits/mg total protein (Fig. 2, lane 1). After growth on medium containing FOA, however, UMPSase activity increased 2- to 4-fold (lane 2). If orotic acid is added to the medium, UMPSase activity is not increased (data not shown).
Figure 2.
Western-blot analysis of UMPSase protein. Lanes 1 and 2 represent the amount of UMPSase activity from six pooled hNp28 calli grown on the absence (lane 1) and presence (lane 2) of 120 μm FOA for 13 d. Lanes 3 and 4 are western-blot analyses of UMPSase from the same cells as in lanes 1 and 2. Lane 3 is from the same pooled, uninduced cells as lane 1. Lane 4 is from the same pooled FOA-induced cells as lane 2. mU, Milliunit.
To determine whether the increase of UMPSase enzyme activity correlated with an increase in the UMPSase protein levels, we used western blots to examine the expression of UMPSase protein in the presence and absence of FOA. The antiserum was raised against purified UMPSase from summer squash (Curcurbias pepo), and cross-reacted with tobacco UMPSase (Santoso and Thornburg, 1995). The western-blot analysis (Fig. 2, lanes 3 and 4) demonstrates that the level of UMPSase protein accumulated to a higher level in the presence of FOA than in its absence. Because the amount of UMPSase protein changes in the N. plumbaginifolia cells in response to FOA, and because these changes correlated with enzyme activity, we conclude that this increase in UMPSase activity in response to FOA is not regulated posttranslationally, but rather is regulated at some step prior to translation.
Is the UMPSase mRNA Regulated?
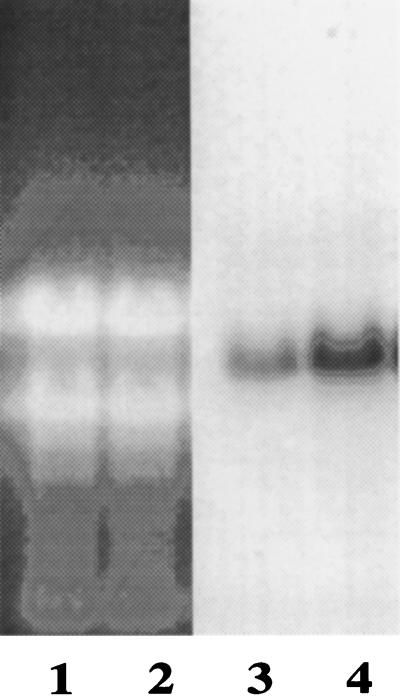
To determine whether the UMPSase mRNA is also regulated, we isolated mRNA from the hNp28 cell lines grown in the absence and presence of FOA for northern-blot analysis. The probe in these studies was 32P-labeled RNA transcribed from the N. tabacum UMPSase cDNA (Maier et al., 1995). These studies, presented in Figure 3, again show that FOA caused an increase in the level of UMPSase mRNA present in the wild-type cells. Quantification of these bands by cutting them from the blots and counting by liquid scintillation revealed that at least 3 times the amount of UMPSase mRNA was present in the cells incubated in the presence of FOA than in the cells incubated on induction medium without FOA.
Figure 3.
Northern-blot analysis of transcripts isolated from the hNp28 cells. Lanes 1 and 2 are the ethidium bromide-stained gel before transfer. Lanes 3 and 4 are the same lanes as lanes 1 and 2 after transfer to Genescreen Plus and hybridization with radiolabeled UMPSase probe. Lanes 1 and 3 are from the hNp28 cells without FOA preculture. Lanes 2 and 4 are from the hNp28 cells with preculture on 120 μm FOA.
UMPSase Transcription
Northern-blot analyses alone cannot determine how the UMPSase gene is regulated. To determine whether the UMPSase gene was transcriptionally regulated, runoff transcription experiments were conducted on nuclei isolated from FOA-induced and uninduced hNp28 cells. UMPSase enzyme activities were determined before the isolation of nuclei so that we could correlate this information with the run-on transcription.
Following nuclear isolation and mRNA radiolabeling, the labeled mRNAs were hybridized to UMPSase antisense transcripts bound to nylon membranes. After washing away unbound counts, the membranes were exposed to x-ray film to localize the bands, and subsequently the bands were cut from the membranes and counted in a liquid scintillation counter. These data are presented in Table I. In a total of nine hybridizations from three independent nuclear isolations, we consistently observed a 2- to 3-fold higher level of labeled transcripts produced in the nuclei from the induced cells than in the nuclei from the uninduced calli. In addition, we observed good agreement between the induction of UMPSase activity by FOA (2.38 ± 0.19-fold) and the induced UMPSase transcripts in cells grown on FOA (2.37 ± 0.59-fold). Thus, analysis of the RNAs transcribed from isolated nuclei of FOA-induced and uninduced hNp28 tissues indicate that the observed increase in expression of UMPSase is transcriptionally regulated.
Table I.
The levels of UMPSase mRNA transcribed from uninduced and FOA-induced hNp28 nuclei
| Experiment | UMPSase
Activitya
|
Hybridization | Radioactivity
Bound
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Uninduced | FOA-Induced | Uninduced | FOA-Induced | ||||||
| milliunit | % | milliunit | %b | cpm | % | cmp | %c | ||
| 1 | 330 | 100 | 714 | 216 | 1 | 43 | 100 | 101 | 234 |
| 2 | 94 | 100 | 206 | 220 | |||||
| 2 | 208 | 100 | 625 | 251 | 3 | 371 | 100 | 960 | 259 |
| 4 | 736 | 100 | 928 | 126 | |||||
| 5 | 1148 | 100 | 2658 | 232 | |||||
| 6 | 573 | 100 | 2035 | 355 | |||||
| 3 | 270 | 100 | 672 | 248 | 7 | 47 | 100 | 93 | 198 |
| 8 | 90 | 100 | 196 | 218 | |||||
| 9 | 85 | 100 | 246 | 290 | |||||
Activity is expressed in milliunits per milligram of protein. One unit is defined as the amount of enzyme activity that liberates 1 nmol of CO2 per min at 25°C.
Average (±sd) was 238 ± 19 (n =3).
Average (±sd) was 237 ± 59 (n = 9).
FOA Toxicity Is Regulated by Thymine Starvation
Thymine starvation is the mechanism responsible for FOA-mediated cell death. FOA is converted into 5- fluorouridine-5′-monophosphate by the action of UMPSase. Further cellular metabolism converts 5-fluorouridine-5′-monophosphate into FdUMP, which is a suicide inhibitor of the enzyme TS (Chouini-Lalanne et al., 1989; Rathod et al., 1992). Thus, during selection on FOA, inhibition of TS by FdUMP causes a severe reduction of thymine biosynthesis. This thymine starvation results in cell death.
As a direct test of this thymine starvation hypothesis, we sought alternative methods unrelated to FOA toxicity that would also modify thymine levels in vivo. DHFR inhibitors are known to block the synthesis of thymine nucleotides by limiting the regeneration of tetrahydrofolate. Tetrahydrofolate is the precursor of N5,N10- methylenetetrahydrofolate, which is the one-carbon donor required for synthesis of dTMP from dUMP. We therefore tested two inhibitors of DHFR to determine whether inhibition of thymine synthesis by blocking DHFR would also up-regulate the levels of UMPSase.
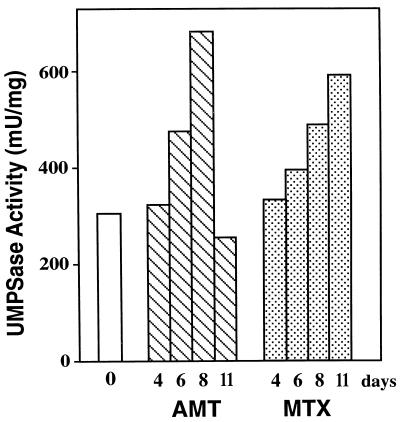
As shown in Figure 4, when we grew the wild-type hNp28 cells on medium containing 0.2 mm AMT or 0.2 mm MTX, UMPSase was induced by both metabolic inhibitors. The kinetics of induction of UMPSase by these metabolic inhibitors were similar to the induction by FOA. Thus, methods that alter thymine levels in vivo via processes unrelated to the mechanism of toxicity of FOA also resulted in the up-regulation of UMPSase.
Figure 4.
The effect of DHFR inhibitors on the UMPSase activity of the N. plumbaginifolia cell. Numbers under the bars represent the culture time in days. The open bar represents UMPSase activity in the absence of inhibitors (0 d). Hatched bars represent UMPSase activity in the presence of 200 μm AMT. Stippled bars represent UMPSase activity in the presence of 200 μm MTX at the indicated times. mU, Milliunit.
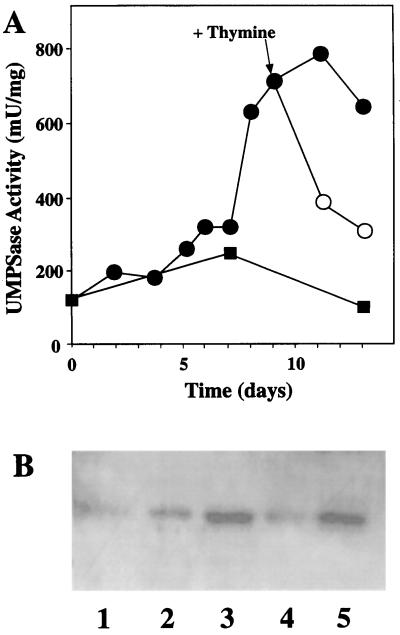
To further demonstrate the role of thymine in the regulation of UMPSase, we examined the effects of added thymine on FOA-induced UMPSase levels in wild-type cells. Figure 5 shows the kinetics of UMPSase up-regulation following the addition of FOA. UMPSase increased slowly for about 1 week prior to a rapid induction of UMPSase activity after about 7 d. This lag phase of induction is due to the time required for the cell to use up its thymine reserves.
Figure 5.
Regulation of N. plumbaginifolia UMPSase by FOA and thymine. A, UMPSase activity of the wild-type cells after being cultured for several days in the Murashige-Skoog medium with no additional supplement (▪) or with 0.12 mm FOA (•). After 9 d, some cells were placed on medium containing 120 μm FOA and 200 μm thymine (○). B, Western-blot analysis of the callus extracts using anti-C. pepo UMPSase. Lane 1 is from cells at 0 d. Lane 2 is from cells after 6 d of inoculation. Lane 3 is from cells after 9 d of inoculation. Lane 4 is from cells after 13 d of inoculation as follows: on FOA for 9 d then 4 d on FOA plus thymine. Lane 5 is from cells after 13 d of inoculation on FOA.
After UMPSase had been induced on d 9, some of the calli were moved onto media in the presence and absence of thymine. As can be seen in Figure 3, the addition of thymine caused UMPSase activity to decline immediately to near wild-type levels. In the absence of added thymine, the UMPSase activity remained high. As shown in Figure 5B, western-blot analysis demonstrated that the expression of UMPSase protein correlated with UMPSase enzymatic activity, both during the up-regulation of UMPSase and the down-regulation phase following addition of thymine. Lanes 1, 2, and 3 show the expression of UMPSase protein on d 0, 6, and 9 prior to transfer onto thymine-containing medium. Lanes 4 and 5 show the expression of UMPSase protein on d 13 in the presence and absence of thymine, respectively. Thus, the addition of thymine results in a rapid down-regulation of the UMPSase protein and corresponding UMPSase enzymatic activity to near wild-type levels.
To determine whether this effect is specific for thymine, we grew cells on Murashige-Skoog medium containing the FOA metabolite 5-fluorouracil, plus other pyrimidines. As can be seen from Table II, fluorouracil caused a 2-fold increase in UMPSase activity. The addition of thymine abrogated the fluorouracil induction of UMPSase. The addition of cytosine resulted in an intermediate expression of UMPSase activity, lower than fluorouracil alone, but twice the level of fluorouracil plus thymine.
Table II.
Effect of pyrimidines on 5-fluorouracil induction of UMPSase
| Treatment | UMPSase Activity | Control |
|---|---|---|
| milliunits/mg protein | % | |
| None | 234 ± 7 | 100 |
| 5-FU | 497 ± 9 | 212 |
| 5-FU + thymidine | 144 ± 1 | 62 |
| 5-FU + cytidine | 339 ± 10 | 145 |
Cells were grown on Murashige-Skoog solid media supplemented with 400 μm 5-fluorouracil (5-FU) alone or 400 μm 5-fluorouracil plus 400 μm nucleotide for 12 d. Then all cells were frozen at −70°, thawed, and assayed for UMPSase activity as described in Methods (n = 4).
DISCUSSION
We have produced a large number of cell lines selected to grow in the presence of FOA. To better comprehend the mechanisms of toxicity of FOA and the types of lesions that selection on FOA provides, we have examined the effect of FOA on UMPSase expression. UMPSase is the rate-limiting step of pyrimidine biosynthesis in plants. When we tested wild-type cells grown in the presence and absence of FOA, both the UMPSase enzymatic activity and the protein itself were induced 2- to 3-fold by the addition of FOA. Northern-blot analysis of the UMPSase mRNA from uninduced and induced tissues demonstrated that the regulation of UMPSase occurred at the mRNA level.
This observation led us to investigate whether the induction of UMPSase mRNA is transcriptionally regulated. We used nuclear runoff transcription experiments with either uninduced or FOA-treated nuclei to examine transcriptional expression. These studies demonstrated that the regulation of UMPSase gene following treatment by FOA occurs transcriptionally.
When FOA is metabolized in cells, the normal pyrimidine metabolic machinery converts this compound into FdUMP. Inhibition of TS enzyme leads to thymine starvation. We therefore directly tested the effect of thymine starvation using MTX and AMT, which are metabolic inhibitors of DHFR. The mechanism of action of MTX and AMT is dissimilar to the mechanism of action of FOA. Inhibition of DHFR also resulted in an up-regulation of UMPSase. These experiments confirm that the normal cellular response to thymine starvation is a transcriptional up-regulation of the rate-limiting step of the de novo pyrimidine biosynthetic pathway.
It is possible that other enzymes are also up-regulated in plants in response to FOA. TS is the enzyme that is blocked by FdUMP. We have preliminary evidence to show that TS is also up-regulated after growth of cells on FOA. Whether the kinetics of this up-regulation is similar to those of UMPSase or whether thymine also alleviates the up-regulation of TS are not known at this time.
When high levels of thymine are provided to the cells following the induction of UMPSase, the induced UMPSase rapidly declines. Thus, either thymine or a thymine metabolite is responsible for the transcriptional regulation of UMPSase in tobacco cells.
In other examples of metabolite-mediated transcriptional regulation in eukaryotes, transcriptional activation proteins are involved (Giniger and Ptashne, 1988; Gronemeyer, 1991; White et al., 1997). Although we have no direct biochemical evidence that such a regulatory cascade is functioning in these tobacco cells, it is clear that they do have a mechanism to detect the level of thymine and to transcriptionally regulate UMPSase and possibly other genes in this pathway.
To account for this regulation, we propose the existence of one or more proteins within the cell that must be able to detect the level of thymine and to transmit that information to the promoters of UMPSase, and perhaps to other members of this pathway. Intracellular measurement of thymine could occur via one of several mechanisms. At least two different thymidine triphosphatases are known from mammalian sources. One is a dimer of identical subunits with specificity toward TTP (Dahlmann, 1982). The other is a multifunctional enzyme with specificity toward both TTP and dCTP (Schultes et al., 1992). Whether the enzymatic hydrolysis of these pyrimidine triphosphates is linked to other regulatory functions has not been explored. It is conceivable that binding or hydrolysis of TTP could trigger a regulatory cascade similar to the kinase cascades so well characterized in yeast (Nishida and Gotoh, 1993).
Second, TTP is known to bind to and regulate the activity of several enzymes in the pyrimidine biosynthetic pathway, including thymidine kinase (Jansson et al., 1992) and ribonucleotide reductase (Hruby, 1985; Ji et al., 1991). The binding of thymine or a thymine metabolite by a nuclear protein could measure the thymine levels within the cell. Numerous examples of metabolite-binding proteins that regulate gene expression are known. Some of these, such as the steroid receptor superfamily, directly bind to and affect promoter activity (Kumar et al., 1987; Gronemeyer, 1991). Others, such as the GAL genes of Saccharomyces cerevisiae, require multiple factors to achieve similar gene regulation.
ACKNOWLEDGMENT
The authors would like to thank Mr. Chris Kafer for his reading of the manuscript.
Abbreviations:
- AMT
aminopterin
- DHFR
dihydrofolate reductase
- FdUMP
5-fluorodeoxy UMP
- FOA
5-fluoroorotic acid
- MTX
methotrexate
- TS
thymidylate synthase
- UMPSase
UMP synthase
Footnotes
This work was sponsored by grant no. 91-37301-6208 from the U.S. Department of Agriculture. This is journal paper no. J-16512 of the Iowa Agriculture and Home Economics Experiment Station, Ames, IA.
LITERATURE CITED
- Ausubel FM, Brent R, Kingston RE, Moore DD, Smith JA, Seidman JG, Struhl K. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1987. [Google Scholar]
- Boeke JD, Lacroute F, Fink GR. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoroorotic acid resistance. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Chouini-Lalanne N, Malet-Martino C, Martino R, Michel G. Study of the metabolism of flucytosine in Aspergillus by 19K nuclear magnetic resonance spectroscopy. Antimicrob Agents Chemother. 1989;33:1939–1945. doi: 10.1128/aac.33.11.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlmann N. Human serum thymidine triphosphate nucleotidohydrolase: purification and properties of a new enzyme. Biochemistry. 1982;21:6634–6639. doi: 10.1021/bi00269a004. [DOI] [PubMed] [Google Scholar]
- Giniger E, Ptashne M. Cooperative DNA binding of the yeast transcriptional activator GAL4. Proc Natl Acad Sci USA. 1988;85:382–386. doi: 10.1073/pnas.85.2.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronemeyer H. Transcriptional activation by estrogen and progesterone receptors. Annu Rev Genet. 1991;25:89–123. doi: 10.1146/annurev.ge.25.120191.000513. [DOI] [PubMed] [Google Scholar]
- Hruby DE. Inhibition of vaccinia virus thymidine kinase by the distal products of its own metabolic pathway. Virus Res. 1985;2:151–156. doi: 10.1016/0168-1702(85)90245-x. [DOI] [PubMed] [Google Scholar]
- Jansson O, Bohman C, Munch-Petersen B, Eriksson S. Mammalian thymidine kinase 2. Direct photoaffinity labeling with [32P]-dTTP of the enzyme from spleen, liver, heart and brain. Eur J Biochem. 1992;206:485–490. doi: 10.1111/j.1432-1033.1992.tb16951.x. [DOI] [PubMed] [Google Scholar]
- Ji JP, Sargent RG, Mathews CK. T4 phage ribonucleotide reductase: allosteric regulation in vivo by thymidine triphosphate. J Biol Chem. 1991;266:16289–16292. [PubMed] [Google Scholar]
- Jones GE, Hann J. Haplopappus graciliscell strains resistant to pyrimidine analogs. Theor Appl Genet. 1979;54:81–87. doi: 10.1007/BF00265474. [DOI] [PubMed] [Google Scholar]
- Jones ME. Pyrimidine nucleotide biosynthesis in animals: genes, enzymes and regulation of UMP biosynthesis. Annu Rev Biochem. 1980;49:253–279. doi: 10.1146/annurev.bi.49.070180.001345. [DOI] [PubMed] [Google Scholar]
- Kalpaxis D, Zundorf I, Werner H, Reindl N, Boy-Marcotte E, Jacquet M, Dingermann T. Positive selection for Dictyostelium discoideummutants lacking UMP synthase activity based on resistance to 5-fluoroorotic acid. Mol Gen Genet. 1991;225:492–500. doi: 10.1007/BF00261692. [DOI] [PubMed] [Google Scholar]
- Kim H, Kelly RE, Evans DR. The structural organization of the hamster multifunctional protein CAD. J Biol Chem. 1992;267:7717–7784. [PubMed] [Google Scholar]
- Kimpel JA, Nagao RT, Goekjian U, Key JL. Regulation of the heat shock response in soybean seedlings. Plant Physiol. 1990;94:988–995. doi: 10.1104/pp.94.3.988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Green S, Stack G, Berry M, Jia-Rui J, Chambon P. Functional domains of the human estrogen receptor. Cell. 1987;51:941–951. doi: 10.1016/0092-8674(87)90581-2. [DOI] [PubMed] [Google Scholar]
- Maier T, Zhou L, Thornburg R. Nucleotide sequence of a cDNA encoding UMP synthase from Nicotiana tabacum (GenBank U22260) (PGR 95-025) Plant Physiol. 1995;108:1747. [Google Scholar]
- Nasr F, Berthauche N, Dufour M-E, Minet M, Lacroute F. Heterospecific cloning of Arabidopsis thaliana cDNAs by direct complementation of pyrimidine auxotrophic mutants of Saccharomyces cerevisiae. I. Cloning and sequence analysis of two cDNAs catalyzing the second, fifth and sixth steps of the de novo pyrimidine biosynthesis pathway. Mol Gen Genet. 1994;244:23–32. doi: 10.1007/BF00280183. [DOI] [PubMed] [Google Scholar]
- Neuhard J, Nygaard P (1987) Purines and Pyrimidines. In FC Neidhardt, JL Ingraham, BK Low, B Magasanik, M Schaechter, HE Umbarger, eds, Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. American Society for Microbiology, Washington, DC, pp 445–473
- Nishida E, Gotoh Y. The MSP kinase cascade is essential for diverse signal transduction pathways. Trends Biochem Sci. 1993;18:128–131. doi: 10.1016/0968-0004(93)90019-j. [DOI] [PubMed] [Google Scholar]
- Rathod PK, Leffers NP, Young RD. Molecular target of 5-fluoroorotate in human malaria parasite Plasmodium falciparum. Antimicrob Agents Chemother. 1992;36:704–711. doi: 10.1128/aac.36.4.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoso D, Thornburg RW. Isolation and characterization of UMPSase mutants from haploid cell suspensions of Nicotiana tabacum. Plant Physiol. 1992;99:1216–1225. doi: 10.1104/pp.99.3.1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoso D, Thornburg RW. The use of a heterologous antibody to characterize UMP synthase mutant of tobacco cells. Menara Perkebunan. 1995;63:71–79. [Google Scholar]
- Schultes BC, Fischbach E, Dahlmann N. Purification and characterization of two different thymidine-5′-triphosphate-hydrolyzing enzymes in human serum. Biol Chem Hoppe Seyler. 1992;373:237–247. doi: 10.1515/bchm3.1992.373.1.237. [DOI] [PubMed] [Google Scholar]
- Shi N-Q, Thornburg RW. Construction of a UMP synthase expression cassette from Dictyostelium discoideum. Gene. 1993;127:199–202. doi: 10.1016/0378-1119(93)90719-j. [DOI] [PubMed] [Google Scholar]
- Timmons ED, Dunbar BS. Protein blotting and immunodetection. In: Deutscher MP, editor. Guide to Protein Purification. San Diego, CA: Academic Press; 1990. [Google Scholar]
- Tirimanne TS, Colbert JT. Transient down-regulation of phytochrome mRNA abundance in etiolated cucumber cotyledons in response to continuous white light. Plant Physiol. 1991;97:1581–1584. doi: 10.1104/pp.97.4.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traut TW, Jones ME. Inhibitors of orotate phosphoribosyl-transferase and orotidine-5′-phosphate decarboxylase from mouse Ehrlich ascites cells: a procedure for analyzing the inhibition of a multi-enzyme complex. Biochem Pharmacol. 1977;26:2281–2291. doi: 10.1016/0006-2952(77)90293-3. [DOI] [PubMed] [Google Scholar]
- van den Hoff MJ, Jonker A, Beintema JJ, Lamers WH. Evolutionary relationships of the carbamoylphosphate synthase genes. J Mol Evol. 1995;41:813–832. doi: 10.1007/BF00173161. [DOI] [PubMed] [Google Scholar]
- Wadsworth GJ, Redinbaugh MG, Scandalios JG. A procedure for the small-scale isolation of plant RNA suitable for RNA blot analysis. Anal Biochem. 1988;172:279–283. doi: 10.1016/0003-2697(88)90443-5. [DOI] [PubMed] [Google Scholar]
- Walther R, Wald K, Glund K, Tewes A. Evidence that a single polypeptide catalyzes the two step conversion of orotate to UMP in cells from a tomato suspension culture. J Plant Physiol. 1984;116:301–311. doi: 10.1016/S0176-1617(84)80109-1. [DOI] [PubMed] [Google Scholar]
- Watson JC, Thompson WF. Purification and restriction endonuclease analysis of plant nuclear DNA. Methods Enzymol. 1986;118:57–75. [Google Scholar]
- White KP, Hurban P, Watanabe T, Hogness DS. Coordination of Drosophilametamorphosis by two ecdysone-induced nuclear receptors. Science. 1997;276:114–117. doi: 10.1126/science.276.5309.114. [DOI] [PubMed] [Google Scholar]
- Williamson CL, Lake MR, Slocum RD. A cDNA encoding carbamoyl phosphate synthetase large subunit (carB) from Arabidopsis (accession no. U40341) (PGR 96-055) Plant Physiol. 1996;111:1354. [Google Scholar]
- Williamson CL, Slocum RD. Molecular cloning and characterization of the pyrB1 and pyrB2 genes encoding aspartate transcarbamoylase in pea (Pisum sativumL.) Plant Physiol. 1994;105:377–384. doi: 10.1104/pp.105.1.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Lacroute F, Thornburg RW. Characterization of the Arabidopsis thaliana cDNA encoding dihydroorotase (accession no. AF000146) (PGR 97-115) Plant Physiol. 1997;114:1569. [Google Scholar]