Abstract
The Nkrp1 (Klrb1)-Clr (Clec2) genes encode a receptor-ligand system utilized by NK cells as an MHC-independent immunosurveillance strategy for innate immune responses. The related Ly49 family of MHC-I receptors displays extreme allelic polymorphism and haplotype plasticity. In contrast, previous BAC-mapping and aCGH studies in the mouse suggest the neighboring and related Nkrp1-Clr cluster is evolutionarily stable. To definitively compare the relative evolutionary rate of Nkrp1-Clr vs. Ly49 gene clusters, the Nkrp1-Clr gene clusters from two Ly49 haplotype-disparate inbred mouse strains, BALB/c and 129S6, were sequenced. Both Nkrp1-Clr gene cluster sequences are highly similar to the C57BL/6 reference sequence, displaying the same gene numbers and order, complete pseudogenes, and gene fragments. The Nkrp1-Clr clusters contain a strikingly dissimilar proportion of repetitive elements compared to the Ly49 clusters, suggesting that certain elements may be partly responsible for the highly disparate Ly49 vs. Nkrp1 evolutionary rate. Focused allelic polymorphisms were found within the Nkrp1b/d (Klrb1b), Nkrp1c (Klrb1c), and Clr-c (Clec2f) genes, suggestive of possible immune selection. Cell-type specific transcription of Nkrp1-Clr genes in a large panel of tissues/organs was determined. Clr-b (Clec2d) and Clr-g (Clec2i) showed wide expression, while other Clr genes showed more tissue-specific expression patterns. In situ hybridization revealed specific expression of various members of the Clr family in leukocytes/hematopoietic cells of immune organs, various tissue-restricted epithelial cells (including intestinal, kidney tubular, lung, and corneal progenitor epithelial cells), as well as myocytes. In summary, the Nkrp1-Clr gene cluster appears to evolve more slowly relative to the related Ly49 cluster, and likely regulates innate immunosurveillance in a tissue-specific manner.
Introduction
Natural killer cells are lymphocytes that are intimately involved in innate host resistance to pathogens and cancer. They patrol the host and survey for the presence of cell surface markers that distinguish healthy versus diseased cells through a variety of receptors [1]. The best characterized examples of receptors include members of the Ly49 family, which bind to class I MHC (MHC-I) and inhibit NK cell responses, and the NKG2D receptor, which binds to MHC-related ligands that are induced following genotoxic or infectious stress, resulting in the activation of NK cells, in turn leading to target cell death and/or induction of cytokine production [2]. The NKR-P1 family of receptors is evolutionarily related to the Ly49 family, but does not survey MHC-I ligands, instead binding to various members of the C-type lectin-related (Clr, also called Ocil) family [3]. Interestingly, the Nkrp1 (Klrb1) and Clr (Clec2) gene families are closely linked, and are in fact intermingled in the Natural Killer gene Complex (NKC) [4].
The known expressed NKR-P1 receptors in the mouse include NKR-P1A (encoded by Klrb1a), NKR-P1B (Klrb1b), NKR-P1C (Klrb1c), NKR-P1F (Klrb1f), and NKR-P1G (Klrb1g) [5], [6]. NKR-P1D (Klrb1b) likely represents an allelic (B6) form of NKR-P1B [7], and Nkrp1e (Klrb1-ps1) is a pseudogene in all mouse strains studied to date (see Table 1). The known Clr ligands include Clr-a (encoded by Clec2e), Clr-b (Clec2d), Clr-c (Clec2f), Clr-d (Clec2g), Clr-f (Clec2h), and Clr-g (Clec2i). Clr-e (a.k.a., Clec2d5) is a pseudogene [8], while the functional status of Clr-h (Clec2j) and Clr-i (no official gene name) are unknown, although Clr-h potentially encodes for a full-length protein in the B6 mouse strain [6]. The available mAb for the mouse NKR-P1 receptors (B6 strain) have revealed that NKR-P1A, NKR-P1B(D), NKR-P1C, NKR-P1F are all expressed on NK cells, but with interesting variation [9]–[11]. Specifically, the putative activating, ITIM-lacking NKR-P1A and NKR-P1F surface proteins are expressed at low levels on all NK cells, but not NKT cells [9]. However, it should be noted that neither IFN-γ nor redirected lysis could be induced with mAb to NKR-P1A or NKR-P1F [9], unlike NKR-P1C [12]. In contrast to NKR-P1A and NKR-P1F, the activating NKR-P1CB6 (NK1.1B6) is expressed at high levels on all NK and NKT cells, while NKR-P1B is stochastically expressed (similar to the Ly49) on approximately 60% of NK cells [9], [10]. At present, mAb for the Clr family are not commercially available, but mAb recognizing Clr-b show that it is broadly expressed on most leukocytes [13], and is down-regulated in response to genotoxic stress and viral infection [14], [15], suggesting it acts as a marker of ‘health’, similar to MHC-I, to inhibit NK cell responses. The known receptor-ligand pairs have been identified through colorimetric reporter cell assays, receptor down-regulation assays, and in some cases through NK cell cytotoxicity assays. These binding pairs include: NKR-P1B/D:Clr-b, NKR-P1F:Clr-c,d,g and NKR-P1G:Clr-f,d,g [6], [10], [13], [16]. In the rat, among 4 Nkrp1 and 11 Clr genes, the known interactions include NKR-P1A:Clr11, NKR-P1B:Clr11, NKR-P1F:Clr2,-3,-4,-6,-7, and NKR-P1G:Clr2,-6,-7 and weak binding to Clr-4 [16]. In humans, there is a single gene encoding NKR-P1A (KLRB1/CD161), which shows inhibitory function upon binding the Clr-related ligand LLT1 (CLEC2D) [17], [18]. However, KLRF1 (NKp80) and KLRF2 (NKp65) have been suggested recently to represent possible stimulatory NKR-P1/KLRB1 homologs, and they bind to the genetically linked ligands, CLEC2B (AICL) and CLEC2A (KACL), respectively [19].
Table 1. Nkrp1/Clr gene nomenclature.
| Common Name(s) | Official Gene Symbol (or Suggested Name) |
| Nkrp1a | Klrb1a |
| Nkrp1b | Klrb1b |
| Nkrp1c | Klrb1c |
| Nkrp1d | Klrb1b |
| Nkrp1e | Klrb1-ps1 |
| Nkrp1f | Klrb1f |
| Nkrp1g | Klrb1 (Klrb1g) |
| Clr-a (Clec2d7) | Clec2e |
| Clr-b (Ocil/Clec2d8) | Clec2d |
| Clr-c (Clec2d6) | Clec2f |
| Clr-d (Ocilrp1/Ddv10//Clec2d4) | Clec2g |
| Clr-e (Clec2d5) | – (Clec2-ps1) |
| Clr-f (Clec2d2) | Clec2h |
| Clr-g (Ocilrp2/Dcl1/LCL-1/Clec2d3) | Clec2i |
| Clr-h (Clec2d1) | Clec2j |
| Clr-i | – (Clec2-ps2) |
| Clr-j | – (Clec2-ps3) |
Although related, there are additional notable differences between the Ly49 and NKR-P1 receptor families, in addition to their unrelated ligands. Firstly, while the majority of Ly49 contain ITIM motifs, this is not the case for the NKR-P1 family of receptors. NKR-P1B(D) contain an ITIM and inhibit NK cell activation upon binding to the ligand, Clr-b [20]; and a second ITIM-containing NKR-P1, NKR-P1G, has been identified at the gene and cDNA level [6], but its cell-type expression and function remain undocumented to date. The remaining NKR-P1 receptors (NKR-P1A/C/F) do not contain ITIM motifs, but instead possess a positively charged residue in the transmembrane domain, presumably for adaptor molecule association (as has been shown for NKR-P1C and FcRγ [21]), and variably contain other features (such as single tyrosine residues, CxCP Lck-recruitment motifs, and proline-rich regions [22], [23]). Secondly, the Ly49 gene clusters of various inbred mouse strains are highly polymorphic at the level of allelic diversity and in terms of gene numbers, with known haplotypes ranging from 8–22 genes [3]. In contrast, the Nkrp1-Clr haplotypes appear to be highly conserved with little or no variation in gene organization or content, and only focused allelic diversity [3]. However, only a single Nkrp1-Clr cluster sequence, from B6 mice, has so far been available; nonetheless, BAC-based gene maps for the 129S6 and BALB/c mouse strains have been constructed [6], [7]. In addition, recent array-based comparative genomic hybridization (aCGH) studies support the contention that the Nkrp1-Clr region shows less inbred mouse-strain variability than the Ly49 region [24]. However, definitive multi-strain genomic sequence data is lacking for this gene cluster.
To further characterize the genetic organization and function of the NKR-P1-Clr self-recognition system utilized by NK cells, we report here the high-resolution sequencing of two Nkrp1-Clr haplotypes from 129S6 and BALB/c mice, and confirm the relative stability of this region of the NKC, including the characterization of novel genes. In order to provide insight into the functional roles of different NKR-P1-Clr receptor-ligand pairs, RT-PCR was performed for all Nkrp1-Clr genes in a large panel of tissues and organs. Additionally, the lack of specific commercially available mAb for the Clr family prompted us to use in situ hybridization of various organs to identify specific cell-types expressing Clr transcripts. We report here the expression of various Clr members in the epithelial cells lining several organs and other non-hematopoietic cell types, supporting a novel function for NK cells in tissue immunosurveillance.
Materials and Methods
Mice
C57BL6/J, BALB/c, and 129S1 mice were obtained from The Jackson Laboratory (Bar Harbor, ME), and maintained in a specific-pathogen-free environment. All breeding and manipulations performed on animals were in accordance with university guidelines and approved by the University of Ottawa Animal Ethics Committee.
BAC Sequencing and Analyses
BACs were sequenced to 20–30x coverage using a Roche 454 GS FLX Titanium 2 instrument at Genome Quebec Innovation Centre (Montreal, QC) and The Centre for Applied Genomics, SickKids Hospital (Toronto, ON). The final cluster sequence was assembled using GS data analysis software (Roche). Gaps in the assembled sequence are primarily due to the presence of low complexity regions of lengths that could not be precisely defined, therefore gaps were introduced to indicate this uncertainty. Ordering of contigs was based on the order of known coding regions that were confirmed from cDNA clones obtained from the identical mouse strain. Final assemblies of the BALB/c and 129S6 strains were compared to each other and the complete B6 sequence to confirm that there were no assembly errors. The following BACs from the 129S6 genomic library RPCI22 were sequenced: 570e7, 525h12, 381m24, 519p18, 410a1, 475e14, and 439c16. The accession number for the BAC sequences covering the 129S6 Nkrp1-Clr cluster is JQ948150. For the BALB/c Nkrp1-Clr region, the following BACs from library CHORI-28 were sequenced: 387p19, 358o7, 451l24, 371k12, and 445a21. The accession number for the BAC sequences covering the BALB/c Nkrp1-Clr cluster is JX026279. For gene sequence comparison, the ‘align two or more sequences’ feature of Blastn was used to align the region of interest from the first to last exon as annotated for each gene. The assembled cluster sequences were analyzed for unknown Nkrp1- and Clr-related genes by taking 10–20 nucleotide segments of the known exons and searching the genomic sequence using Lasergene sequence analysis software (DNASTAR Inc., Madison WI). Repetitive elements were identified with RepeatMasker version 3.1.8 (A.F.A. Smit and P. Green unpublished; available at http://www.repeatmasker.org). A PIP of the repeat-masked Nkrp1-Clr cluster sequences from the various inbred mouse strains (versus the B6 sequence) was constructed using Advanced Pipmaker with a setting of single coverage (available at http://pipmaker.bx.psu.edu/pipmaker).
Tissue RT-PCR
Adult B6, 129S1 or BALB/c mice were anesthetized with avertin, perfused transcardially with PBS and different tissues were collected. Tissues were rinsed with PBS, frozen with liquid nitrogen and transferred to −80° before RNA isolation. RNA was isolated from tissues using RiboZol RNA extraction reagent (AMRESCO). RNA (1 µg) was reverse-transcribed into cDNA using Verso cDNA kit (Thermo Scientific). PCR amplification was performed on 1 µl of the above cDNA product using primers specific for Nkrp1-Clr and Gapdh genes (Table 2). PCR products were visualized on a 1% agarose gel stained with ethidium bromide. Primer specificity was checked using serial dilution of plasmids (1 ng, 10−1 ng, 10−2 ng, 10−3 ng) containing different Nkrp1 and Clr cDNAs as template for PCR amplification. Previous microarray expression studies done for the mouse are available online (http://biogps.org) [25], [26], and were used to confirm RT-PCR results.
Table 2. Primers used to amplify Nkrp1- Clr genes from tissue cDNA and plasmids.
| Primer specificity | Sense primer | Antisense primer |
| Clr-a | CAAAGGTTGAAGAGGCTTCC | TCACGCATGCTTTGGCACAT |
| Clr-b | ACTCAGCTCCTCAGCTCTGA | GGCTAAAAAGCGTCTCTTGG |
| Clr-c | GTTATGACAGCCTCACAGGA | GCTAGCACTGTAACATATAG |
| Clr-d | GTTATGAATGCACAGTGCCT | GCTAGACAGGAACAGGAGTT |
| Clr-f | TTGAAACGAGTTCCATGGGC | GGTCATAGAGCATCTGATTG |
| Clr-g | AGATTGCTTGGAGACAGGAG | GAAGAGTCTCTTGGTAAGTG |
| Clr-h | GTTATGCAGATGAAGGCGAA | GCTAGCACAGTAAGGTGTAG |
| Nkrp1a | TGATGCATCTCCTATGCACA | CTCTTGGACAGGATCTGAGT |
| Nkrp1dB6 | GGTGTCAAGTCCCTCCATCT | CATCAGAATTGAAAGCTGTG |
| Nkrp1b129&BALB | ATGGATTCAACAACACTGGT | TCAGGAGTCATTACACGGGG |
| Nkrp1c | CAGTGGATCCCCATCAAGAGAAAAATGCTG | ACTGGAATTCTCAGGAGTCATTACTTGGGG |
| Nkrp1f | GTTCCCATCTCCTGTCTACA | AGGAATCAGGACACAGGCTT |
| Nkrp1g | ATGGATGCACCAGTGCTCTA | TCAGACGTGTTTCAGTGTCT |
| Gapdh | ACTCACGGCAAATTCAACGGC | ATCACAAACATGGGGGCATCG |
In situ Hybridization
In situ hybridization analysis for Clr genes in different tissues was performed as described previously [27]. Digoxigenin-labeled antisense and sense RNA probes were transcribed from linearized plasmids containing Clr genes with T7 polymerase using DIG RNA labeling mix (Roche). Tissues were collected from adult B6 mice and were fixed either in 10% formalin at RT or in 4% paraformaldehyde at 4° with gentle shaking for up to 24 h. Tissues were then embedded in paraffin and 4 µm sections were prepared on glass slides. Tissue slides were deparaffinated, treated with 0.2 N HCl for 15 min and 30 µg/ml proteinase K at 37° for 20 min, fixed in 4% PFA for 10 min and incubated with 0.25% acetic anhydride in 0.1 M triethanolamine twice for 5 min each. Slides were then pre-hybridized with hybridization solution (50% (v/v) formamide/5× SSC, pH 4.5; 2% (w/v) blocking powder (Roche); 0.05% (w/v) CHAPS; 5 mM EDTA; 50 µg/ml heparin; 1 µg/ml yeast RNA) in a 58° oven for at least 1 h, and then incubated with hybridization solution containing 500 ng/ml digoxigenin-labeled probe overnight at 58°. Post-hybridization washes were performed with 50% formamide/2X SSC, pH 4.5 at 55° 3x for 20 min each and slides were stained with sheep anti-digoxigenin alkaline phosphatase-conjugated antibody at 1/1000 dilution in blocking solution (Roche). Color development was performed by adding nitro blue tetrazolium/5-bromo, 4-chloro, 3-indoylphosphate (NBT/BCIP, Roche) substrates to the slides and slides were kept in the dark at RT until optimal signals appeared on slides. Slides were then counterstained with lightgreen or methylgreen and visualized under a light microscope.
Results
Organization and Content of the B6 Nkrp1-Clr Gene Cluster
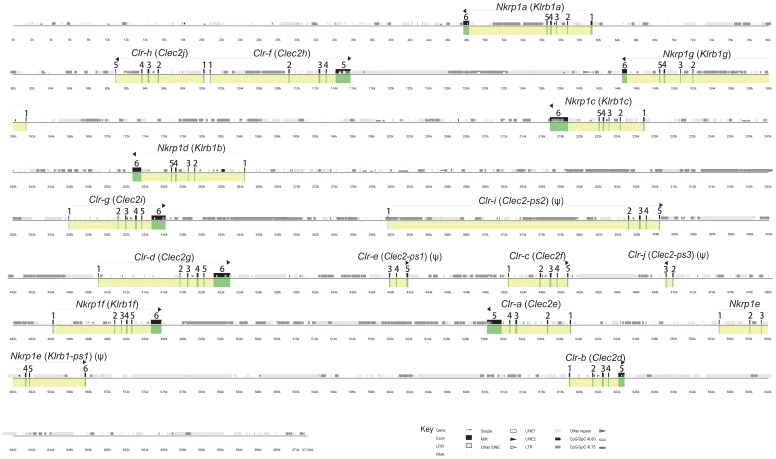
The Nkrp1-Clr (Klrb1-Clec2) cluster of B6 mice has been characterized previously in moderate detail [4], [5], [24]. To compare the Nkrp1-Clr gene cluster sequences in the B6, 129S6, and BALB/c strains, the reference B6 Nkrp1-Clr region was fully assembled using publicly available genomic data ( Figure 1 ). The relative order of genes in the B6 genome is: Nkrp1a (Klrb1a), Clr-h (Clec2j), Clr-f (Clec2h), Nkrp1g (Klrb1g), Nkrp1c (Klrb1c), Nkrp1b/d (Klrb1b), Clr-g (Clec2i), Clr-i (Clec2-ps2), Clr-d (Clec2g), Clr-e (Clec2-ps1), Clr-c (Clec2f), Clr-j (Clec2-ps3), Nkrp1f (Klrb1f), Clr-a (Clec2e), Nkrp1e (Klrb1-ps1), and Clr-b (Clec2d) [6]–[8]. The cluster is approximately 580 kb in length and has a gene order and content consistent with the previously published 129S6 and BALB/c BAC-based maps, including a new gene fragment we term Clr-j (Clec2-ps3) [6], [7]. Similar to the Ly49 (Klra) genes, the protein coding regions of the Nkrp1 genes are divided over 6 exons. In contrast, the Clr genes usually contain 5 coding exons, except for Clr-d (Clec2g) and Clr-g (Clec2i), which have 6 exons due to the presence of an additional 5′ coding exon.
Figure 1. A graphical representation of the organization of the Nkrp1-Clr gene cluster in the C57BL/6 (B6) inbred mouse strain.
The UCSC Genome Browser was used to capture the genomic sequence between and including the Nkrp1a and Clr-b genes. Genes were identified by Lasergene, using short homology searches of B6 Nkrp1/Clr cDNAs. The locations of various kinds of repetitive elements were revealed with RepeatMasker and are shown (see key). Advanced Pipmaker was used to construct a scale diagram of the exons and a PIP vs. itself (single coverage). Genes and exons are marked in yellow and green, respectively. The sequence is demarcated below the plot in kilobases. Arrows show gene orientation, and the common gene name followed by the official gene name (in parentheses) or suggested official name are indicated above the arrow. Obvious pseudogenes are denoted with a ψ.
Sequence database searches for Nkrp1-Clr-related exons in the B6 genome using 10–20 nucleotide segments of all known B6 Nkrp1- and Clr-related cDNAs facilitated the discovery of a novel gene fragment consisting of two exons (exons-2 and -3, relative to other complete 6-exon Clr genes), which we have termed Clr-j (tentatively Clec2-ps3). Another obvious Clr pseudogene is Clr-i (Clec2-ps2), which we identified previously from two exon fragments in 129S6 mice [6]. We have found that the Clr-i gene in B6 mice contains all necessary exons but has multiple in-frame stop codons in all three reading frames (data not shown). Clr-e (Clec2-ps1) is also a pseudogene, as only three exons (exons-3, -4 and -5) have been identified. In contrast, only a single Nkrp1 gene, Nkrp1e (Klrb1-ps1), is a pseudogene in B6 mice, as it contains multiple early stop codons (data not shown). It is noteworthy that the Clr-h (Clec2j) and Clr-f (Clec2h) genes are arranged in a head-to-head orientation and their respective first exons are less than 1 kb apart. When the Clr-f cDNA was used to query the EST database, multiple Clr-f clones from different tissues were discovered: AF350410.1, NM_053165.5, AK090364.1, BC021766.1, AK017207.1, HQ875061.1, HQ713437.1. However, all these cDNAs contained exon-1 (data not shown), suggesting that the promoter for Clr-f is either in the 1 kb region separating the Clr-f and Clr-h genes or may include part of the Clr-h gene. The functional status of the Clr-h gene is unknown; no transcripts have been reported in the EST database or otherwise in the literature, although a complete Clr-h open reading frame is present in B6 mice.
Sequencing of the related Ly49 gene cluster in several inbred strains of mice has revealed an overrepresentation of repetitive elements [28], [29]. Specifically, the LINE1 family of retroelements represents ∼36% of the Ly49 region in all of the four distinct sequenced haplotypes (B6, BALB/c, 129, NOD), which range in size from 8–22 genes. In contrast, the mouse genome contains ∼19% LINE1 elements. We have hypothesized that intra-cluster meiotic recombination due to LINE1 overrepresentation helps to drive Ly49 evolution [3]. The relative conservation of the neighboring and related Nkrp1-Clr gene cluster provides an opportunity to test this hypothesis. The presence of various types of repeats in the B6 Nkrp1 versus Ly49 region is summarized in Table 3 . While the overall percentage of interspersed repeats is similar in the two gene regions (∼54% vs. ∼51%), there are two striking differences: 1) the Nkrp1-Clr cluster consists of only ∼22% (vs. ∼36% in the Ly49 clusters) LINE1 sequences, which is similar to the total genome average, and 2) there are more than twice as many LTR elements in the Nkrp1-Clr region as in the Ly49 region (∼26% vs. ∼12%). The first difference supports the hypothesis that LINE1 elements may drive unequal crossing-over between sister chromatids, but the implications for LTR overrepresentation in the Nkrp1-Clr region are unclear.
Table 3. Comparison of repeats in the Nkrp1-Clr and Ly49 gene clusters from B6 mice.a .
| Type of element | Number of elements | Length occupied (bp) | Percentage of sequence | |||
| Nkrp1 | Ly49 | Nkrp1 | Ly49 | Nkrp1 | Ly49 | |
| SINES | ||||||
| Alu/B1 | 94 | 99 | 11303 | 11148 | 1.68 | 1.95 |
| B2–B4 | 106 | 35 | 15205 | 4936 | 2.27 | 0.86 |
| IDs | 12 | 5 | 832 | 376 | 0.12 | 0.07 |
| MIRs | 9 | 0 | 1207 | 0 | 0.18 | 0.00 |
| Total | 221 | 139 | 28547 | 16460 | 4.25 | 2.87 |
| LINEs | ||||||
| LINE1 | 148 | 264 | 147150 | 203094 | 21.92 | 35.46 |
| LINE2 | 9 | 4 | 1133 | 348 | 0.17 | 0.06 |
| L3/CR1 | 1 | 0 | 46 | 0 | 0.01 | 0.00 |
| Total | 158 | 268 | 148329 | 203442 | 22.10 | 35.52 |
| LTR elements | ||||||
| ERVL | 21 | 2 | 5500 | 791 | 0.82 | 0.14 |
| ERVL-MaLRs | 57 | 26 | 17394 | 12857 | 2.59 | 2.24 |
| ERV_classI | 38 | 18 | 18508 | 14752 | 2.76 | 2.58 |
| ERV_classII | 182 | 43 | 136954 | 40436 | 20.40 | 7.06 |
| Total | 298 | 89 | 178356 | 68836 | 26.57 | 12.02 |
| DNA elements | ||||||
| hAT-Charlie | 6 | 4 | 902 | 305 | 0.13 | 0.05 |
| TcMar-Tigger | 17 | 0 | 3784 | 0 | 0.56 | 0.00 |
| Total | 25 | 4 | 4939 | 305 | 0.74 | 0.05 |
| Unclassified | 2 | 1 | 1866 | 45 | 0.28 | 0.01 |
| Total interspersed repeats | 362037 | 289088 | 53.93 | 50.48 | ||
| Small RNA | 14 | 8 | 1028 | 513 | 0.15 | 0.09 |
| Satellites | 3 | 2 | 313 | 197 | 0.05 | 0.03 |
| Simple repeats | 186 | 199 | 10491 | 12840 | 1.56 | 2.24 |
| Low complexity | 64 | 110 | 3236 | 5281 | 0.48 | 0.92 |
As detected by RepeatMasker.
Sequence Analysis of the 129S6 and BALB/c Nkrp1-Clr Gene Clusters
In agreement with our Ly49 and Nkrp1-Clr haplotype studies, a recent report utilized aCGH analyses of a large panel of inbred mouse strains in support of the hypothesis that the Nkrp1-Clr cluster is less diverse compared to the Ly49 region [24]. As discussed above, the 129S6 and BALB/c Nkrp1-Clr genes have been mapped using BAC clones [6], [7]. The organization, number, and type of genes appear to be identical to the B6 cluster, as shown in Figure 1 ; however, only high resolution sequencing can definitively reveal the degree of relatedness among all three clusters and any potential hidden genes. Therefore, a minimal BAC tiling assembly was chosen for both the 129S6 and BALB/c clusters and sequenced. The exon organization of the BALB/c and 129S6 Nkrp1-Clr regions is shown in Figure 2 . The Nkrp1 and Clr genes in both the 129S6 and BALB/c haplotypes were identical to the B6 haplotype in terms of gene content (Nkrp1a-Nkrp1g and Clr-a-Clr-j), including all functional genes, as well as complete and fragmentary pseudogenes ( Figure 2 ).
Figure 2. A graphical representation of the organization of the Nkrp1-Clr gene cluster in (A) 129S6 and (B) BALB/c inbred mouse strains.
A minimal BAC tiling path was selected from previous BAC mapping reports and next-generation sequencing was performed. Contigs were assembled using GS data analysis software. Genes were identified by Lasergene, using 10–20 nucleotide sequences of Nkrp1/Clr cDNAs from 129S6 or BALB/c mice. Advanced Pipmaker was used to construct a scale diagram of the exons and a PIP vs. the B6 Nkrp1-Clr cluster sequence (single coverage). Genes and exons are marked in yellow and green, respectively. The sequence is demarcated below the plot in kilobases. Arrows show gene orientation, and the common gene name is above the arrow. Obvious pseudogenes are denoted with a ψ. The locations of various kinds of repetitive elements were revealed with RepeatMasker and are shown (see key in Figure 1). Vertical red bars indicate gaps between contigs. Accession numbers for the cluster sequences are given in the Materials and Methods.
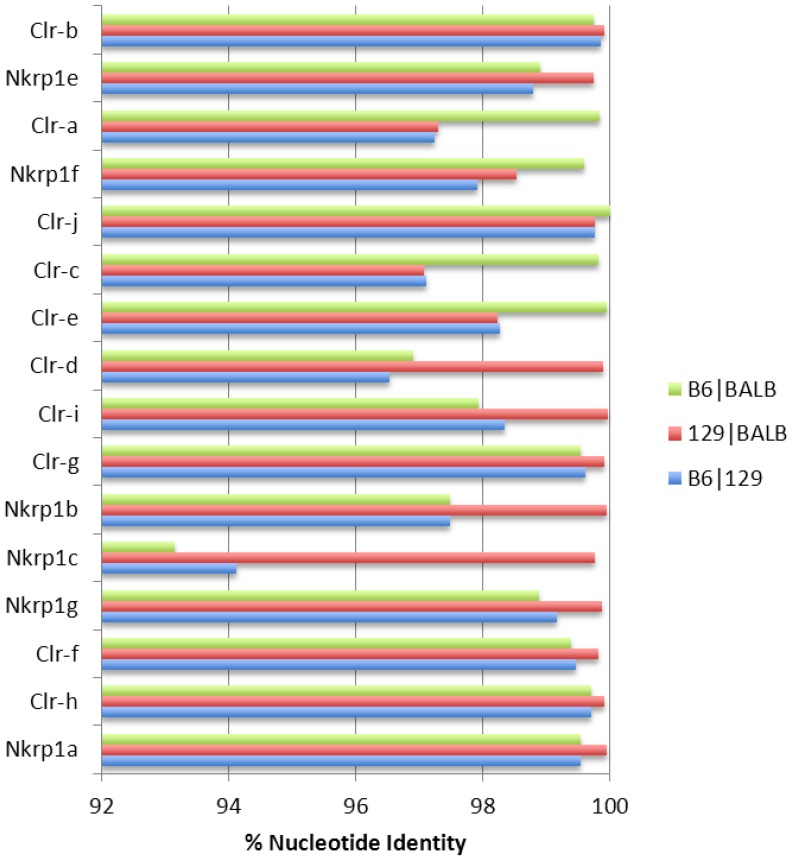
The three haplotypes are highly related and share ≥98% sequence identity overall ( Figure 3 ), but with some gene-specific differences. For example, for the Nkrp1a gene, the 129S6 allele is 99.54% identical to the B6 allele, whereas the BALB/c allele is 99.65% identical to the B6 allele, and the 129S6 and BALB/c alleles are 99.96% identical to each other. In fact, from Nkrp1a to Clr-d, 129S6 and BALB/c are >99.5% identical and B6 identity to either 129S6 or BALB/c is more variable ( Figure 3 ). In particular, BALB/c and B6 Nkrp1c alleles are ∼93% identical, while the BALB/c and B6 Nkrp1b/d alleles are ∼97% identical. In direct contrast, the BALB/c segment encompassing Clr-e, Clr-c, Clr-j, Nkrp1f, and Clr-a is highly homologous (>99.5%) to the corresponding B6 segment, suggesting a common origin, possibly as a result of a double cross-over event resulting in intra-cluster recombination.
Figure 3. Comparisons of Clr and Nkrp1 gene homologies between three inbred mouse strains.
The genomic region of each gene from the translational start codon to the termination codon was subjected to pair-wise comparison (B6/BALB; 129/BALB; B6/129) using Blastn. The percent nucleotide identity of each gene for the three strain pairs is shown.
Previous studies have identified cloned Nkrp1-Clr cDNA sequences from various mouse strains [6]; however, some cDNAs from intact genes remain unreported, possibly due to restricted or unknown tissue-specific expression. Therefore, the putative exonic sequences of all BALB/c and 129S6 Nkrp1 and Clr genes were collected and assembled from the regional sequencing, and were used to ascertain the degree of polymorphism between the cloned and uncloned alleles of all three mouse strains ( Table 4 ). We used these data to confirm, at the genomic level, the sequences of the cloned cDNAs of all transcribed Nkrp1 and Clr genes in both mouse strains, including the Clr-c alleles, which we previously observed were highly divergent (38/206 aa differences) between the 129-strain allele compared to the BALB/c and B6 Clr-c alleles, which are identical [6]. These data are thus far also consistent with the 129S1 and 129S6 substrains having identical Nkrp1, Clr, and Ly49 gene sequences, as the isolated cDNA (129S1 strain) and genomic DNA templates (129S6 strain) were identical to each other. Among the three inbred strains, the degrees of relatedness for most alleles were ∼99% or greater (nucleotide or aa), except for the previously noted divergence of the Clr-c, Nkrp1b/d, and Nkrp1c alleles.
Table 4. Nkrp1 and Clr homology in 129S6, B6, and BALB/c mice.
| cDNAa | amino acid | |||||
| 129 vs B6 | BALB vs B6 | 129 vs BALB | 129 vs B6 | BALB vs B6 | 129 vs BALB | |
| Clr-a | 98% | 99% | 99% | 97% | 99% | 97% |
| Clr-b | 100% | 100% | 100% | 100% | 100% | 100% |
| Clr-c | 92% | 100% | 92% | 82% | 100% | 82% |
| Clr-d | 98% | 98% | 100% | 97% | 97% | 100% |
| Clr-f | 99% | 99% | 100% | 99% | 99% | 100% |
| Clr-g | 100% | 100% | 100% | 100% | 100% | 100% |
| Clr-h | 100% | 100% | 100% | 100% | 100% | 100% |
| Nkrp1a | 99% | 99% | 100% | 99% | 99% | 100% |
| Nkrp1b(d) | 96% | 96% | 100% | 90% | 90% | 100% |
| Nkrp1c | 94% | 94% | 100% | 88% | 88% | 100% |
| Nkrp1f | 99% | 100% | 99% | 99% | 100% | 99% |
| Nkrp1g | 99% | 99% | 100% | 99% | 99% | 100% |
cDNAs were compared from the translational start to stop codons to ascertain sequence identity.
Interestingly, we found that the fragmentary pseudogenes Clr-e (Clec2-ps1) and Clr-j (Clec2-ps3) and the complete pseudogene Clr-i (Clec2-ps2) were present in all three haplotypes with the same number of exons (Clr-e: exons-3, -4, and -5; Clr-i: exons-1-5; Clr-j: exons-2 and -3), which is somewhat surprising, as it is expected that there would be no selective pressure to retain these non-coding genes. As previously noted, the Clr-h gene appears to be intact, as there are no premature stop codons in the open reading frame, but it might represent a pseudogene, as no transcript was detectable by RT-PCR in NK cells. The closest EST database match for Clr-h is Clr-b (∼95% identity). However, Clr-h is 100% conserved in exonic nucleotide sequence among all three strains ( Table 4 ), which would be remarkable for a non-functional gene, and strongly suggests that a candidate cell-type that expresses Clr-h has not yet been identified.
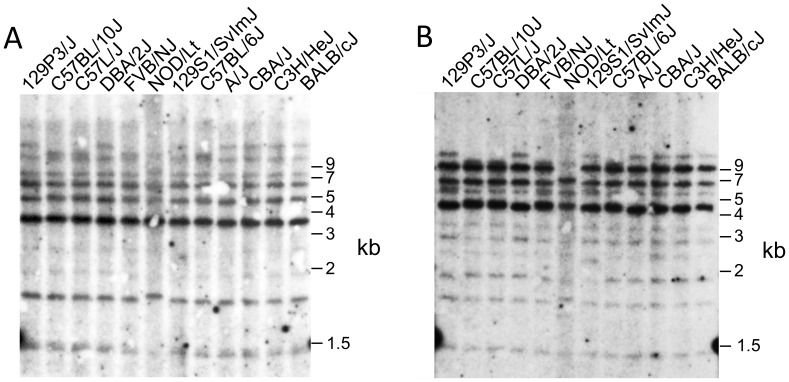
In line with our BAC sequencing results and previous studies [7], Southern blot analyses of genomic DNA from a panel of inbred mouse strains, using the Clr-b and Clr-f cDNA sequences as probes, indicated that these Clr gene RFLP patterns are highly similar. In turn, these results suggest that not only are the Clr-b and Clr-f genes well conserved, but in addition, due to cross-hybridization, that most if not all Clr genes in general are quite conserved, although some polymorphisms are present ( Figure 4 ). Finally, the location and frequency of repetitive elements in the three Nkrp1-Clr haplotypes was found to be similar ( Figures 1 , 2 and Table 3 ). In summary, in agreement with our previous BAC mapping and cDNA cloning studies [6], [7], as well as aCGH studies from another group [24], the Nkrp1-Clr gene cluster appears to be quite conserved and evolutionarily more stable relative to the Ly49 cluster; nonetheless, specific genes appear to be evolving more rapidly than others, and like the Ly49 region [3], [30], there is evidence of recombination between distinct Nkrp1-Clr haplotypes.
Figure 4. Evolutionary conservation of Clr genes in the mouse.
Thymic genomic DNA from the indicated inbred mouse strains was digested with EcoRI and subjected to southern blot analyses using 32P-labelled cDNA probes for (A) Clr-b, and (B) Clr-f. The membrane was stripped between hybridizations.
Characterization of Nkrp1-Clr Transcript Expression in Tissues and Organs
The genomic sequencing of the Nkrp1-Clr region has facilitated the identification and mapping of conserved genes and allelic/regional polymorphisms, including exonic and intronic sequences for the three inbred strains. Applying these findings, the specific tissues and cell types expressing Nkrp1-Clr genes were analyzed next. Tissue-specific, and, in some cases, cell-type-specific micro-array analyses have been performed previously to characterize global transcript expression of mouse genes, including most Nkrp1 and Clr genes, and these findings are publicly available on the BioGPS website [26]. To independently confirm and extend previous tissue expression studies [4], the transcript expression of all functional Nkrp1 and Clr genes in a panel of tissues and organs from the three inbred strains was analyzed by RT-PCR.
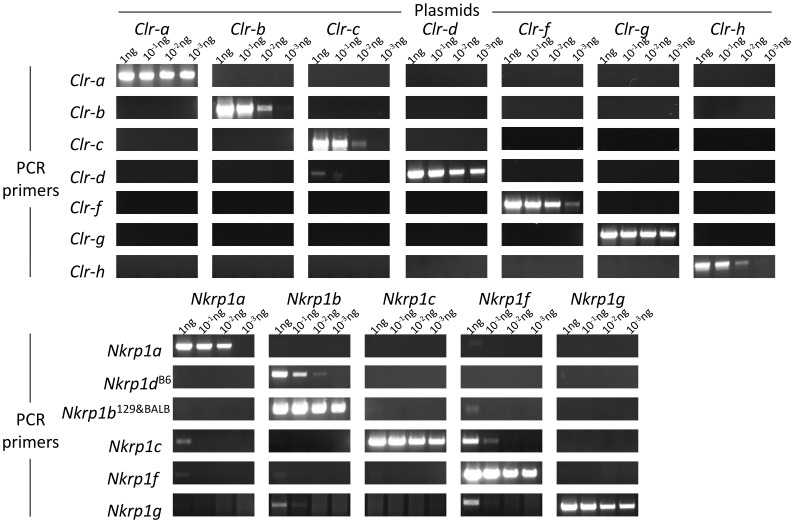
First, each set of gene-specific primers was tested for specificity against cloned cDNAs by serial dilution of template plasmid DNA. Overall, it was determined that the primers utilized were quite specific for the particular genes they were originally designed to amplify ( Figure 5 ). Minor cross-amplification was observed for select primer sets, including faint detection of Clr-c cDNA (by Clr-d primers), Nkrp1a cDNA (by Nkrp1c primers), Nkrp1b/d cDNA (by Nkrp1g primers), and Nkrp1f cDNA (by Nkrp1a,b,c,g primers); however, this was typically observed only at the highest amounts of template plasmid DNA (1 ng), and is thus unlikely to be a confounding factor for ex vivo tissue-specific RT-PCR analysis, where typically ∼50 ng or less of total cDNA is used per reaction.
Figure 5. Testing of Nkrp1-Clr primers for PCR specificity.
Cloned cDNAs for each of the known Nkrp1 and Clr genes were used as a template at the indicated amounts in PCR amplification experiments with primers for each of the genes. The resultant DNA amplification product is shown after agarose gel electrophoresis. Primer specificity is indicated on the left and plasmid template across the top.
Next, RT-PCR for each Nkrp1 and Clr gene product was performed on RNA isolated from 15 tissues and organs: tongue, muscle (hind leg), liver, bladder, spleen, thymus, ovaries, testes, eye, kidney, heart, lung, lymph node, brain, and intestine. The analyses were performed for the 129S1, B6, and BALB/c strains, and the expression results are shown in Table 5 and Table 6 . The corresponding micro-array results, as previously reported [25], [26], are shown underneath the RT-PCR results for each gene/tissue, but micro-array data was not available for Clr-c, Clr-h, or Nkrp1g. Overall, the RT-PCR results agreed with the micro-array data, as well as previous expression studies for the Nkrp1-Clr genes [4].
Table 5. Tissue and organ expression of Clr genes as determined by RT-PCR.
| tongue | muscle | liver | bladder | spleen | thymus | ovaries/testes | eye | kidney | heart | lung | lymph node | brain | intestine | |
| Clr-a | ||||||||||||||
| B6 | -a | – | – | – | – | – | – | – | – | – | – | – | – | + |
| BALB | - | – | – | – | – | – | – | – | – | – | – | – | – | + |
| microarrayb | - | – | – | – | – | – | – | – | – | – | – | – | – | + |
| Clr-b | ||||||||||||||
| B6 | + | +++ | ++ | + | ++ | ++ | + | + | + | + | + | ++ | – | + |
| 129 | + | +++ | +++ | + | + | + | + | −/+ | ++ | ++ | ++ | ++ | – | + |
| BALB | + | +++ | +++ | + | ++ | ++ | + | −/+ | ++ | +++ | ++ | ++ | – | + |
| microarray | + | + | + | + | + | + | + | - | + | + | + | + | – | + |
| Clr-c | ||||||||||||||
| B6 | −/+ | – | – | −/+ | + | + | + | – | – | – | – | −/+ | – | + |
| 129 | −/+ | – | – | −/+ | + | + | −/+ | – | −/+ | – | – | + | – | + |
| BALB | + | – | – | – | + | + | + | – | −/+ | – | + | −/+ | - | + |
| microarray | NDc | |||||||||||||
| Clr-d | ||||||||||||||
| B6 | + | – | – | – | −/+ | −/+ | – | + | – | – | – | – | – | – |
| 129 | – | – | – | – | −/+ | – | – | + | – | – | – | – | – | – |
| BALB | −/+ | – | −/+ | −/+ | −/+ | −/+ | – | + | – | – | – | – | – | – |
| microarray | – | – | – | – | – | – | – | + | – | – | – | – | – | – |
| Clr-f | ||||||||||||||
| B6 | −/+ | −/+ | ++ | – | −/+ | −/+ | – | – | +++ | – | −/+ | −/+ | – | +++ |
| 129 | – | – | ++ | −/+ | + | – | – | – | +++ | + | – | −/+ | −/+ | +++ |
| BALB | – | −/+ | – | – | – | – | – | – | +++ | – | – | −/+ | – | +++ |
| microarray | – | + | + | – | – | – | – | – | + | – | – | – | – | + |
| Clr-g | ||||||||||||||
| B6 | – | – | – | – | ++ | ++ | – | – | −/+ | – | + | ++ | – | + |
| 129 | – | – | −/+ | – | ++ | +++ | −/+ | – | −/+ | – | + | +++ | – | + |
| BALB | – | −/+ | −/+ | – | ++ | ++ | – | – | – | – | + | ++ | −/+ | ++ |
| microarray | – | – | – | – | + | + | – | – | – | – | – | + | – | – |
| Clr-h | ||||||||||||||
| B6 | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| 129 | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| BALB | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| microarray | ND | |||||||||||||
-, no expression; −/+, very weak expression; +, weak to moderate expression; ++, strong expression; +++, very strong expression.
Microarray data were obtained from the Mouse MOE430 Gene Atlas database at BioGPS portal server.
ND, not determined.
Table 6. Tissue and organ expression of Nkrp1 genes as determined by RT-PCR.
| tongue | muscle | liver | bladder | spleen | thymus | ovaries/testes | eye | kidney | heart | lung | lymph node | brain | intestine | ||||||||||||||||||
| Nkrp1a | |||||||||||||||||||||||||||||||
| B6 | -a | – | – | – | + | + | + | – | – | –+ | + | ++ | – | – | |||||||||||||||||
| 129 | – | – | – | – | + | + | ++ | – | – | – | + | ++ | – | – | |||||||||||||||||
| BALB | – | – | −/+ | – | −/+ | −/+ | + | – | – | – | ++ | + | – | – | |||||||||||||||||
| microarrayb | – | – | – | – | + | – | – | – | – | – | – | + | – | – | |||||||||||||||||
| Nkrp1c | |||||||||||||||||||||||||||||||
| B6 | – | – | + | −/+ | +++ | ++ | −/+ | − | −/+ | - | ++ | ++ | – | + | |||||||||||||||||
| 129 | – | – | + | −/+ | ++ | ++ | −/+ | −/+ | −/+ | −/+ | + | ++ | – | + | |||||||||||||||||
| BALB | – | – | ++ | −/+ | + | + | + | − | −/+ | + | ++ | + | – | + | |||||||||||||||||
| microarray | + | – | + | – | + | + | – | – | – | – | + | + | – | – | |||||||||||||||||
| Nkrp1d/b | |||||||||||||||||||||||||||||||
| B6 | + | −/+ | + | + | ++ | ++ | + | −/+ | + | + | ++ | ++ | - | ++ | |||||||||||||||||
| 129 | + | – | – | −/+ | + | + | + | −/+ | – | + | ++ | ++ | – | + | |||||||||||||||||
| BALB | + | – | + | + | + | + | −/+ | −/+ | – | + | ++ | + | – | + | |||||||||||||||||
| microarray | + | + | + | + | + | + | – | + | + | + | + | + | – | + | |||||||||||||||||
| Nkrp1f | |||||||||||||||||||||||||||||||
| B6 | – | – | + | – | ++ | + | – | – | – | – | ++ | ++ | – | + | |||||||||||||||||
| 129 | – | – | + | – | + | + | – | – | – | – | + | +++ | - | + | |||||||||||||||||
| BALB | – | – | ++ | – | + | + | – | – | – | – | ++ | ++ | – | + | |||||||||||||||||
| microarray | – | – | – | – | + | – | – | – | – | – | – | + | – | + | |||||||||||||||||
| Nkrp1g | |||||||||||||||||||||||||||||||
| B6 | – | – | – | – | + | + | – | – | – | – | + | −/+ | – | + | |||||||||||||||||
| 129 | – | – | – | – | + | + | + | – | – | – | – | −/+ | −/+ | + | |||||||||||||||||
| BALB | – | – | – | – | + | + | + | – | – | – | – | – | – | + | |||||||||||||||||
| microarray | NDc | ||||||||||||||||||||||||||||||
-, no expression; −/+, very weak expression; +, weak to moderate expression; ++, strong expression; +++, very strong expression.
Microarray data were obtained from the Mouse MOE430 Gene Atlas database at BioGPS portal server in order to compare with results of RT-PCR.
ND, not determined.
Clr-a expression was found only in the intestine, and only at low levels. Clr-b, which is known to be expressed by most nucleated hematopoietic cells [10], [13], was expressed broadly and strongly in almost all tissues except the brain ( Table 5 ), and is in agreement with northern blot analysis showing Clr-b (mOcil) expression in the liver, kidney, gut, spleen, heart, and smooth muscle [31]. The brain was the only organ negative for all Nkrp1 and Clr transcripts, as assessed by both RT-PCR and micro-array ( Table 5 and Table 6 ). Tissues used for our RT-PCR experiments and the micro-array data were isolated from PBS-perfused animals to avoid misleading interpretations from resident blood cells. However, it is possible that the wide expression of Clr-b may have been due to tissue-embedded resident leukocytes, which have been previously reported to express Clr-b [10]. Clr-c was expressed at low levels in the tongue, spleen, thymus, ovaries, testes, and lymph node tissues. Clr-d transcripts were present specifically in the eye. Clr-f was expressed in the liver and very highly in the kidney and intestine. Clr-g was consistently expressed in the spleen, thymus, and lymph node. Our RT-PCR results clearly indicated that Clr-g is also expressed in the lung and intestine, but these findings disagreed with micro-array results, which reported no Clr-g expression in these tissues. This could be due, in part, to alternate splicing of Clr-g gene products [32], in turn detected differentially by distinct primer and probe sets. Finally, while the Clr-h gene appears to be complete with an intact coding region, as well as being 100% conserved across all three mouse strains, no expression was detected in any of the tissues analyzed. Thus, most Clr genes appear to be expressed in a tissue-specific manner, with some overlap between isoforms and some discrepancies revealed by distinct detection methods. Whether the overall Clr expression patterns are dependent upon non-hematopoietic cell types or tissue-resident leukocytes, which remains possible for Clr-b [10], [13], requires further inquiry.
In contrast, the Nkrp1 family of genes was more consistent in terms of expression patterns. In general, wherever NK cells (or related lymphocytes) have been reported in abundance, both the RT-PCR and micro-array results were positive ( Table 6 ). Transcripts for all Nkrp1 genes were detected in the spleen, thymus, lung, and lymph node tissues. Additionally, based on our RT-PCR results, the intestine showed expression of almost all Nkrp1 transcripts, with the exception of Nkrp1a. Furthermore, we found the liver to be positive for Nkrp1b/d, Nkrp1c, and Nkrp1f transcripts, although the micro-array results were not always consistent with these RT-PCR results. However, given the well-known abundance of liver-resident NK (and NKT) cells, it is likely that the micro-array results may not have sufficient sensitivity to detect expression in subsets of these cells; alternatively, PBS perfusion may have inadvertently removed these immune populations from the liver tissue. Finally, Nkrp1b/d was somewhat unique in showing expression within the tongue and bladder. Like the Clr transcripts, no Nkrp1 transcripts were detected in the brain, in contrast to the related Ly49 transcripts, which have been reported to be abundantly expressed in neurons [33].
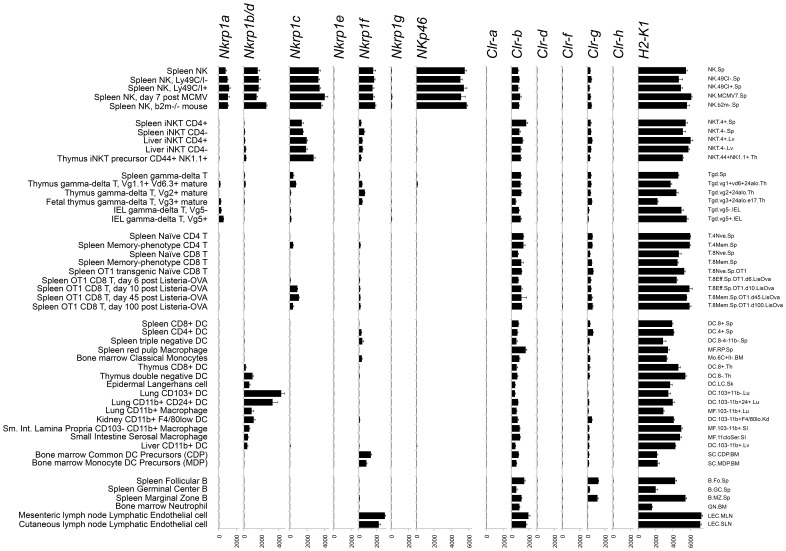
Micro-array data comparing many immune cell types is also publicly available as part of the Immunological Genome Project [34]. Expression of Nkrp1 and Clr genes in selected populations is depicted in Figure 6 . As previously observed, Nkrp1a, Nkrp1b/d, Nkrp1c, and Nkrp1f are expressed by NK cells, while Nkrp1c is additionally expressed in NKT cells and some activated/memory T cells. γδ T cells show some signal for several Nkrp1 receptors, but subsets differ in the predominant receptor expressed. Interestingly, several populations isolated to be tissue dendritic cells or macrophages show substantial expression of Nkrp1b/d. The markers used to sort these cells (including MHC-II+, CD11b+, CD11c+) can also be found on NK cells under certain conditions [35], [36]. However, these Nkrp1b/d + populations lacked expression of the classical NK cell markers Nkrp1c (NK1.1 in B6 mice) and Ncr1 (NKp46). This implies that lung, kidney, skin, thymus, and small intestine may contain either: a) Nkrp1b/d + DC and macrophages or b) Nkrp1b/d + Nkrp1c - Ncr1 - non-conventional NK or lymphoid cells that co-purify with DC and/or macrophages in these isolations. Additionally, Nkrp1f transcripts were detected in bone marrow DC/monocyte precursors and lymph node endothelial cells ( Figure 6 ). Within the Clr family, widespread Clr-b expression was confirmed in many hematopoietic cell types with levels correlated with MHC-I. Clr-g was also detected in many populations, but no substantial signal was detected for Clr-a, Clr-d, Clr-f, and Clr-h in any hematopoietic cell types tested.
Figure 6. Expression of Nkrp1 and Clr genes in immune cell types revealed by transcriptome micro-array.
Normalized hybridization is shown ± SEM. Text at the left indicates brief subset descriptions, while text at the right indicates Immunological Genome Project cell population names. Data was obtained from the Immunological Genome Project (NCBI GEO Series GSE15907) [34]. iNKT, invariant NKT cell; IEL, intra-epithelial lymphocyte; DC, dendritic cell; MCMV, murine cytomegalovirus.
Identification of Specific Cell Types Expressing Clr Genes
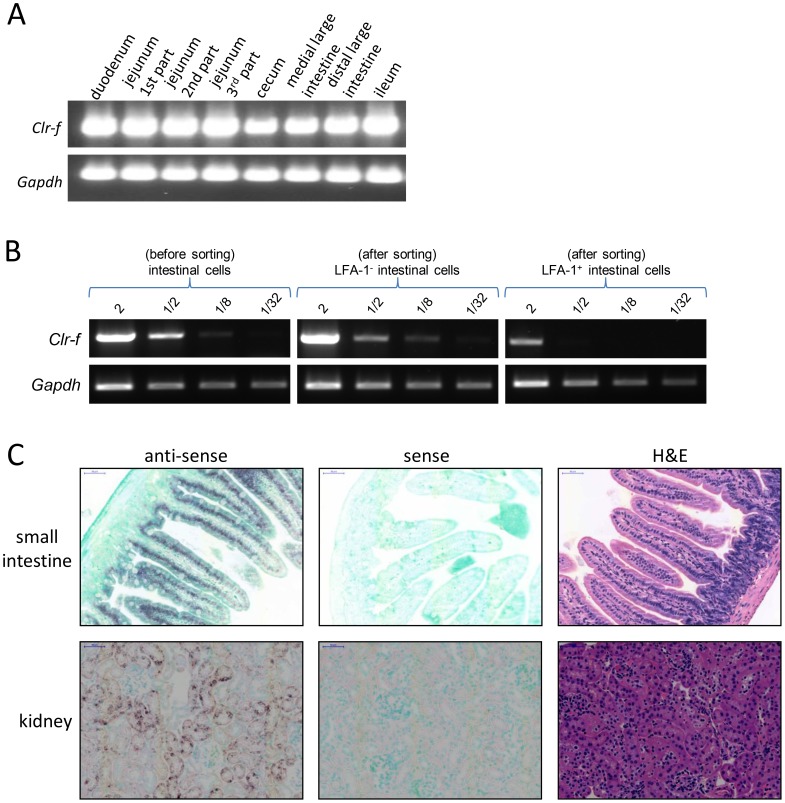
The high-level expression of Clr-f mRNA in the intestine is in agreement with previous studies [4], and prompted us to attempt to identify the specific cell types expressing this gene. Interestingly, Clr-f was expressed in all tissue sections of the small and large intestine ( Figure 7A ), as detected by RT-PCR. Therefore, enzymatic separation of intestinal cells followed by sorting into epithelial (non-hematopoietic) and leukocyte (hematopoietic) subsets based on the expression of the LFA-1 integrin marker (CD11a/CD18) was performed, and documented that Clr-f transcripts were mainly expressed within the LFA-1– epithelial (non-hematopoietic) cells ( Figure 7B ). To confirm the specific cell type in the intestine expressing Clr-f transcripts, in situ RNA hybridization was employed using a Clr-f anti-sense RNA probe. This method was selected because mAb are not generally or commercially available for most members of the Clr family. The anti-sense Clr-f probe clearly and specifically hybridized to the nuclei of the epithelial cells lining the villi of the small intestine ( Figure 7C ). Importantly, no staining was detected in the lamina propria, which contains resident lymphoid cells. In situ RNA hybridization results also revealed that the previous detection of Clr-f transcripts by RT-PCR in the kidney was likely due to expression by kidney tubular epithelial cells ( Figure 7C ).
Figure 7. Clr-f is expressed in intestinal epithelial cells.
(A) Different sections of the small and large intestine from a PBS-perfused B6 mouse were separated and processed for RNA isolation. RT-PCR was performed on the resulting cDNAs with Clr-f-specific and Gapdh-specific primers. (B) Enriched Clr-f expression in non-lymphocytes of the intestine. A section of the small intestine was collagenase-digested and a single-cell suspension was prepared and stained with anti-LFA-1 mAb followed by sorting into lymphocyte and non-lymphocyte (epithelial) populations from which RNA was isolated and used for semi-quantitative Clr-f RT-PCR. The amount of cDNA in micrograms used in each reaction is indicated. (C) In situ hybridization reveals that Clr-f is abundantly expressed in intestinal epithelial cells and kidney tubular epithelium. DIG-labeled sense (control) and anti-sense RNA probes were used for hybridization to paraffin-embedded intestinal (jejunum) and kidney sections. Hybridization location of the probes was revealed by an alkaline-phosphatase-conjugated anti-DIG secondary followed by (brown) color substrate development. H&E staining of different section of the same organ reveals cellular organization of the tissue. The scale bars in the intestine and kidney sections indicate a length of 50 µm.
In situ RNA hybridizations were subsequently conducted to identify the specific cell types expressing the remaining Clr transcripts in tissues that were documented to be positive by RT-PCR and microarray. The results are collectively as follows:
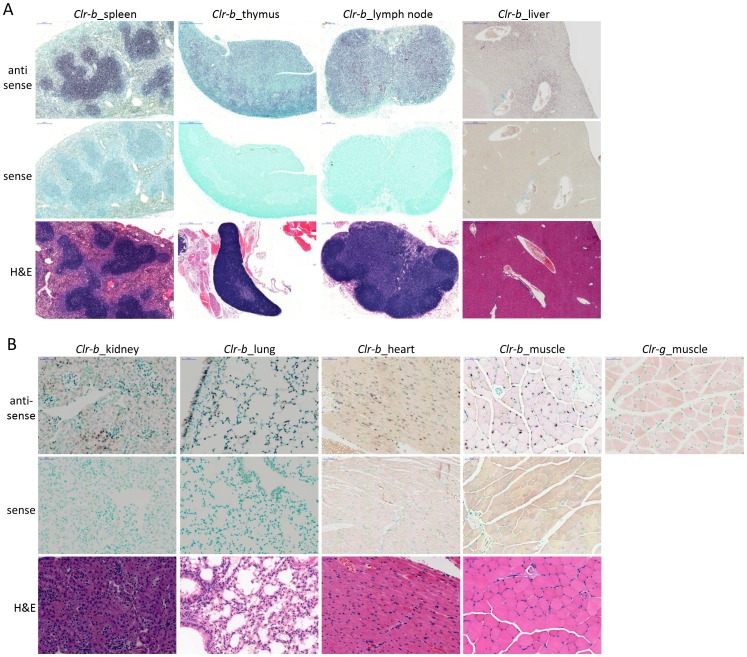
Clr-b. As shown in Figure 8A , Clr-b transcripts are detected at high levels in splenic lymphoid follicle (B cell zone) cells and less obviously in the periarteriolar lymphoid sheath (T cell zone). Clr-b transcripts are also abundantly present in the thymus and in lymph node tissues, in agreement with previous cell-type specific flow cytometric analyses [13]. In the liver, Clr-b is expressed highly and specifically around the central vein area, portal triad area, and hepatocytes close to the epithelial stroma, with weaker expression in other areas as well. Non-lymphoid organ Clr-b hybridizations are shown in Figure 8B . Like Clr-f, Clr-b transcripts were detected in the kidney. However, unlike Clr-f, kidney Clr-b staining was restricted to the glomerular and interstitial areas in a pattern indicative of capillary endothelial cells. Clr-b was detected in bronchial epithelial and endothelial cells, as well as endothelial cells of the lung parenchyma. Surprisingly, myocyte nuclei of skeletal muscle and heart tissue are also clearly stained with the Clr-b probe. As a specificity control, muscle section hybridization was also carried out using a Clr-g probe and found to be negative, in agreement with RT-PCR and microarray results. Our results are consistent with previous reports documenting Clr-b expression by in situ RNA hybridization in lung, heart, small intestine, skeletal muscle, and spleen [31], although the specific cell types involved were not identified in this report.
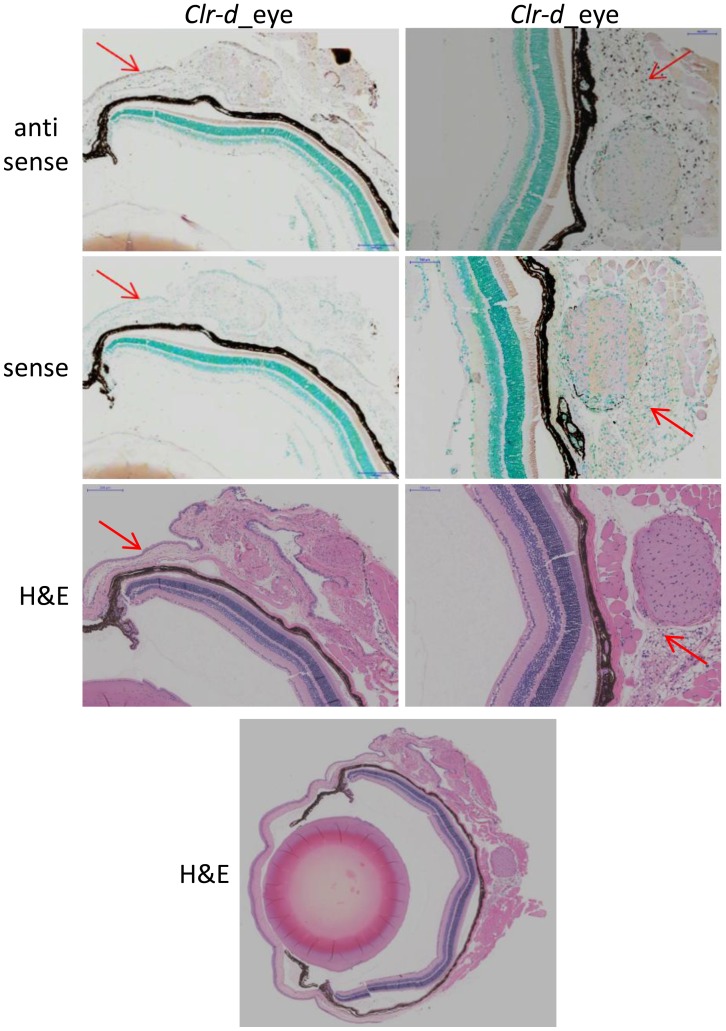
Clr-d . The antisense Clr-d probe detected expression by different cell types in the eye, specifically the epithelial layer of the sclera (the outermost cells; Figure 9, left panel) and the choroid body, which is the large mass of cells surrounding the optic nerve ( Figure 9, right panel). The latter are possibly glial cells (astrocytes or oligodendrocytes), but could also be residual blood cells (after PBS perfusion) circulating in the tissue or endothelial cells from blood vessels. Low levels of Clr-d transcripts could also be detected in the corneal epithelium (data not shown).
Clr-f . As described above, Clr-f was detected specifically in intestinal epithelial cells and kidney tubular epithelial cells ( Figure 7C ).
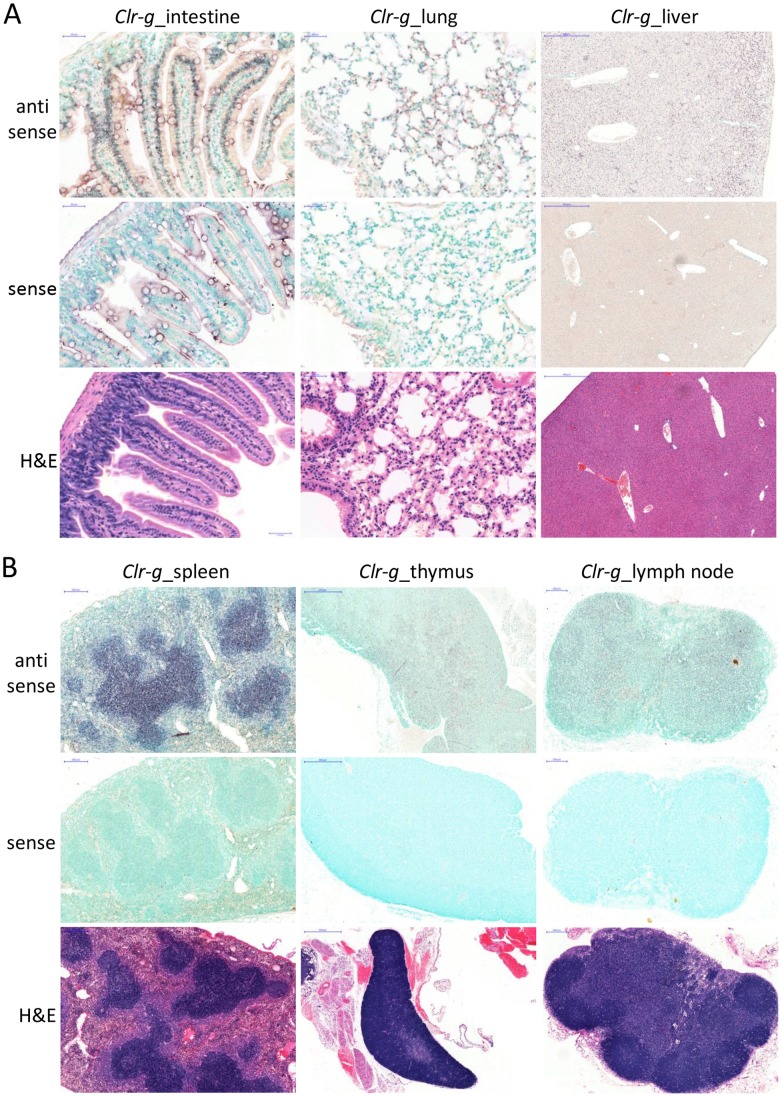
Clr-g. Clr-g transcripts were found to be present in intestinal epithelial cells, although at apparently lower levels than Clr-f, as well as an unidentified cell subset within the lamina propria ( Figure 10A ). Interestingly, Clr-g appears to be transcribed in bronchial epithelial cells, endothelial cells, and alveolar epithelial cells of the lung ( Figure 10A ), but to a lesser extent than Clr-b. Although the lung epithelial cells are clearly stained in the bronchi, the expression is sporadic. The lung endothelial cells also show more diffuse expression, as well as alveolar epithelial cells (pneumocytes), specifically type-2 alveolar cells, which are large surfactant-producing cells. Similar to Clr-b, Clr-g transcripts were also detectable in the B cell areas of the spleen, and the majority of the thymus and lymph node tissues, but the latter two organs appear to express lower levels of Clr-g compared to Clr-b ( Figure 10B ). Like Clr-b, Clr-g transcripts were also detected in the liver ( Figure 10A ), despite RT-PCR and microarray both being negative. However Clr-g was expressed more evenly throughout the liver than Clr-b. The reason for the in situ hybridization versus RT-PCR/microarray discrepancy may be due to blood cells that were removed for the latter analyses.
Figure 8. Wide-spread expression of Clr-b in lymphoid and non-lymphoid tissues and cell types.
In situ hybridization was performed on tissue sections from a PBS-perfused B6 mouse. Hybridization of the indicated (A) primary and secondary lymphoid organs and (B) non-lymphoid organs was performed with a DIG-labeled anti-sense Clr-b RNA probe and revealed with an alkaline phosphatase-conjugated anti-DIG secondary mAb. Control in situ hybridization with a DIG-labeled Clr-b sense RNA probe is shown. Anti-sense Clr-g staining of hind leg muscle tissue is provided as a hybridization specificity control. H&E staining is provided to identify cell types and reveal organ structure. The scale bars indicate 200 µm in the lymph node and spleen, 500 µm in the liver and thymus, and 50 µm in the kidney, lung, heart, and muscle.
Figure 9. Clr-d is expressed in distinct areas of the mouse eye.
A DIG-labeled RNA probe for Clr-d was used to identify regions of the eye expressing this gene. Expression was detected in the corneal epithelial progenitor cells (left column) and the cell mass surrounding the optic nerve (right column). Red arrows indicate areas of positive staining. Sense (control) RNA hybridizations are shown along with H&E stains to elucidate eye cell types and architecture. For organ orientation an H&E stain of the whole eye is provided at the bottom. The scale bar in the sclera epithelial cell section indicates 200 µm, and 100 µm in the optic nerve section.
Figure 10. Lymphoid and non-lymphoid cell expression of Clr-g.
Paraffin-embedded tissue sections of the (A) small intestine (jejunum), lung, and liver, and (B) spleen, thymus, and inguinal lymph node were prepared from a PBS-perfused B6 mouse followed by hybridization with DIG-labeled sense (control) or anti-sense RNA probes. Hybridization was visualized with an alkaline phosphatase-conjugated anti-DIG mAb followed by a (brown) color substrate. An H&E stain of a subsequent tissue section is provided to visualize organ architecture. The scale bars of each tissue section are as indicated for the previous figures.
Discussion
The observation that the Nkrp1 and Ly49 genes are closely linked within mouse chromosome 6 led to the designation of this region as the NK gene complex, or NKC [37]. Although related, the two receptor families encoded by these genes have undergone different evolutionary strategies for their contribution to the innate immune system and expression by NK cells. Firstly, the ligands for the two receptor families are unrelated, with the Ly49 receptors recognizing type-I-transmembrane, Ig-superfamily-related MHC-I molecules, while the NKR-P1 receptors recognize type-II-transmembrane, C-type lectin-related Clr molecules. Secondly, the genes encoding the Clr (Clec2) family are intermingled among the Nkrp1 (Klrb1) genes, while the genes encoding the MHC and Ly49 receptors are found on separate chromosomes, with concomitant ramifications for the strength and number of possible self-specific receptors that are responsible, in part, for NK cell responsiveness [38], [39]. Thirdly, the Ly49 genes are highly polymorphic at the allelic level, as well as highly elastic at the haplotype level, with a known structural gene content variation of between 8–22 genes documented to date per cluster [30], [40]. In contrast, as shown in the present and previous studies [6], [7], the Nkrp1/Klrb1 (and Clr/Clec2) gene number appears to be stable in at least three different inbred mouse strains, with minimal but focused allelic variation. It is possible that while the B6, 129, and BALB/c strains all represent a single categorical Nkrp1 haplotype, there may be other mouse strains possessing alternate Nkrp1 clusters with different gene numbers. In limited support of this, aCGH and Southern analyses of a large panel of mouse strains suggest that there may be at least four distinct Nkrp1 haplotypes (compared to five Ly49 haplotypes), and that the B6 and 129/BALB haplotypes may broadly characterize two different groups [24], although this study concluded that Nkrp1 group haplotype diversity is significantly less than that displayed by Ly49. The Nkrp1 and Clr genes are highly conserved, as suggested by comparison of the Southern blot analyses of the Clr/Clec2 family genomic RFLP patterns reported here ( Figure 4 ), and by Nkrp1/Klrb1 Southern blot analyses reported previously [24]. Finally, whereas the ligands for the Ly49 receptors are expressed on almost all nucleated cell types, thus excluding mature erythrocytes and thrombocytes, we find that the ligands for the NKR-P1 receptors appear to be expressed in a tissue- and organ-restricted manner, perhaps with the exclusion of Clr-b (and possibly Clr-g). Specifically, we observed isoform/locus-specific Clr expression not only among hematopoietic cells (leukocytes), but also among non-hematopoietic epithelial cells (intestine, kidney, eye), endothelial cells (lung, kidney), myocytes (hind leg muscle, heart), and possibly neuronal cell types (eye). Thus, while Ly49/MHC-I immunosurveillance is global, the NKR-P1/Clr system appears to be more tissue-specific.
Role of Nucleotide Repeat Elements in Nkrp1/Ly49 Evolution
We had previously hypothesized that the high percentage of LINE-1 elements in the Ly49 cluster, calculated to be double the genomic average, contributes to the rapid contraction and expansion of this region by acting as ‘sliding’ regions of homology during meiotic sister chromatid cross-overs [3]. LINE-1 elements are endogenous retro-elements that contain promoters and frequently code for functional reverse transcriptase and/or endonuclease activities, but don’t contain additional gag-pro-env gene products like standard retroviruses. Since the Nkrp1/Clr cluster appears to be relatively stable, as concluded by the present sequencing and previous BAC contig and aCGH analyses of multiple inbred strains [6], [7], [24], it can serve as a natural test of this hypothesis. Indeed, it was observed that the LINE-1 make-up of the Nkrp1/Clr region was close to the genomic average. However, an unexpected finding was the high percentage of LTR retro-elements in the Nkrp1/Clr cluster, which was approximately twice the genomic average, as well as twice the percentage observed within the sequenced Ly49 clusters ( Table 3 ) [29]. LTR retro-elements (or endogenous retroviruses) are the result of germ-line infection and transmission of bona fide retroviruses (with LTR regions and coding regions for gag, pro, pol, env). There are few to no LTR retro-elements still capable of replication in humans (called HERVs), as they appear to be heavily mutated or contain deletions [41]. In contrast, mice have hundreds of functional LTR elements (e.g., MusD and IAP) that appear to be still actively translocating within the mouse genome [41]. Despite being largely non-infectious, human cells can still express gene products derived from these LTR retro-elements that can assemble in virus-like particles upon activation. Indeed, some autoimmune diseases have been suspected to be linked to these retro-element activities [42]. The reason for the over-representation of LTR sequences in the Nkrp1/Clr region is unclear at present.
Clr Expression in Non-hematopoietic Cells
The expression of several Clr family members in non-hematopoietic cells, specifically within various kinds of tissue-specific epithelial cells, myocytes, endothelial cells, and possibly neurons, suggests that NKR-P1 receptor regulation of NK cell function may in fact be tailored for immunosurveillance of many if not most organs and tissues. This may represent a paradigm shift for NK cell biology, as KIR/Ly49 regulation via MHC-I expression appears to be more global, at least for most nucleated cells. Various Clr transcripts were detected by RT-PCR and in situ RNA hybridization in non-hematopoietic cells, including epithelial cells of the kidney, intestine, lung, and eye. Clr transcript expression was also detected in myocytes of the heart and hind leg muscle. Contaminating leukocytes may account for some of the signal by RT-PCR analysis, but in situ RNA hybridization of tissue sections may be used to visually exclude blood cells. It is possible that the antisense RNA probe hybridization detected in the various organs/tissues is not due to annealing with a functional transcript; for example, hybridization could result from binding to non-functional alternatively spliced transcripts or RNA molecules with coincidental complementarity. However, full-length sequence-verified cDNAs were amplified and cloned from multiple tissues (which were PBS perfused to eliminate blood cells), thus supporting the finding that the tissue-specific expression of the genes appears to coincide with detectable in situ RNA hybridization within specific cell types. Additionally, while intact transcripts may be detectable, it is also possible that the corresponding protein may not be properly translated or expressed at the cell surface until the appropriate cellular stimuli are provided. This possibility awaits the generation of mAb specific for the remaining Clr family members. However, it remains likely that the cellular transcripts detected in the present study are translated at least in part.
Species Conservation of Clr Expression and Function
The Clr/Clec2 loci have also been documented to be part of a multi-gene family in the rat. The Clr/Clec2 gene closest to the Cd69 locus and the Ly49 cluster is Clec2d11/Clr11 in the rat and Clec2d/Clr-b in the mouse. These two genes are clearly homologous and likely orthologous, but also most likely function similarly since rat Clec2d11/Clr11 expression has been broadly documented in the spleen, thymus, liver, heart, muscle, and kidney, similar to mouse Clec2d/Clr-b [43]. The recognition of rClr11 by the inhibitory rNKR-P1B receptor also highlights conservation of function with mClr-b, which is recognized by mNKR-P1B [44], [45]. Rat Clr10 is thought to be orthologous to mClr-a, based on sequence and location, and was previously reported to be highly expressed in the thymus and moderately expressed in the spleen and liver [43]. On the other hand, while we detected mClr-a to be only weakly expressed in the intestine, our RT-PCR results were otherwise in agreement with previous microarray analyses of mClr-a expression. The expression of rClr10 in the intestine remains unknown. The only other somewhat likely orthologous relationship between the mouse and rat systems is mClr-f and rClr9; however, no expression data is currently available on the rat counterpart for functional comparison.
While human LLT1 is thought to be the closest homologue and potential orthologue of mClr-b due to its binding of the inhibitory hNKR-P1A (CD161) receptor [17], [18], the LLT1 expression pattern clearly contrasts with that of mClr-b in that it is more highly restricted in resting leukocytes, with significant inducible expression documented in TLR-activated pDC and B cells, as well as TCR-activated T cells and IL-2-activated NK cells [46], [47]. Mouse Clr-b, on the other hand, appears to be constitutively expressed on leukocytes [13], but may also have evolved additional functions on non-hematopoietic cells. As Clr-b expression is down-regulated upon cell stress due to transformation, DNA damage, or infection, it seems to function as a marker of ‘health’ for NK cells [14], [15], [44]; however, its inducible expression on the above cell populations in response to activation still remains undocumented. It thus remains possible that LLT1 may more closely represent a ligand for the other inhibitory NKR-P1 receptor in the mouse, mNKR-P1G, which in turn may more closely represent a hNKR-P1A homologue. These possibilities await further detailed bioinformatic and functional characterization of the NKR-P1/Clr system.
Likewise, the constitutive expression of Clr-f ligand for the inhibitory NKR-P1G receptor in the intestine and kidney suggests that it may also act as a marker of health in these organs, which may contain NK cells or other lymphocytes expressing NKR-P1G. Clr-f joins ICOS-L and B7-H1 as a probable immune modulator expressed by renal tubular epithelial cells [48]. The tubular epithelium of the kidney and villus epithelium of the small intestine are susceptible to injury from not only bacterial and viral pathogens, but also ischemia-reperfusion injury, oxidative stress, nephrotoxins, inflammation, and immune disorders. We further found constitutive expression of Clr-g in various organs and tissues, suggesting that it also may act as a regulator of cell health for NK cells or other lymphocytes via NKR-P1G. However, Clr-g is also a common ligand for the putative stimulatory NKR-P1F receptor [6], [16]; moreover, it is not yet clear whether NKR-P1F is functionally active in the same way that the NKG2D or Ly49D receptors signal, since NKR-P1F does not appear to induce cytokine production or cytotoxicity by NK cells [9]. Similarly, Clr-d, which we found to be uniquely expressed in the eye, is a common ligand for the NKR-P1F and NKR-P1G receptors [6], but the functional role of receptor-ligand interactions in this organ is unclear.
In summary, sequencing analyses in three different inbred mouse strains confirms previous gene-mapping and aCGH studies, suggesting that the Nkrp1/Clr gene cluster is highly conserved compared to the nearby Ly49 region, especially with respect to gene number stability, which the Ly49 region lacks. Genome sequencing further indicates a correlation between conservation and specific repetitive element prevalence, suggesting that these elements may also be involved in immune gene evolution, but as a facilitating mechanism rather than immune selection pressure. Complementary expression analysis of transcripts from this cluster suggest that NKR-P1-mediated immunosurveillance may be tissue- and ligand-specific, rather than global as per the Ly49/KIR system in recognition of MHC-I molecules.
Acknowledgments
We thank Dr. Stephanie Bennett (University of Ottawa) for assistance with in situ hybridization, Drs. Michel Cayouette (IRCM, Montreal, QC), Wael Maharsy, Marc-Andre Langlois, Alain Stinzi, and Earl Brown (University of Ottawa) for helpful discussions, and Louise Pelletier (University of Ottawa) for assistance with histology slide preparation.
Funding Statement
This work was supported by an operating grant from the Canadian Institutes of Health Research (CIHR 86630; to A.P.M. & J.R.C), and an Early Researcher Award from the Ontario Ministry of Research and Innovation (to J.R.C). A.P.M. holds a Canada Research Chair in Innate Pathogen Resistance. J.R.C. holds a CIHR New Investigator Award and an Investigator in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund, United States of America. This research was supported in part by the Intramural Research Program of National Institutes of Health (NIH), Frederick National Lab, Center for Cancer Research. This project has been funded in part with federal funds from the Frederick National Laboratory for Cancer Research, NIH, under contract HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the United States government. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Lanier LL (2005) NK cell recognition. Annu Rev Immunol 23: 225–274. [DOI] [PubMed] [Google Scholar]
- 2. Gasser S, Orsulic S, Brown EJ, Raulet DH (2005) The DNA damage pathway regulates innate immune system ligands of the NKG2D receptor. Nature 436: 1186–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Carlyle JR, Mesci A, Fine JH, Chen P, Belanger S, et al. (2008) Evolution of the Ly49 and Nkrp1 recognition systems. Semin Immunol 20: 321–330. [DOI] [PubMed] [Google Scholar]
- 4. Plougastel B, Dubbelde C, Yokoyama WM (2001) Cloning of Clr, a new family of lectin-like genes localized between mouse Nkrp1a and Cd69. Immunogenetics 53: 209–214. [DOI] [PubMed] [Google Scholar]
- 5. Plougastel B, Matsumoto K, Dubbelde C, Yokoyama WM (2001) Analysis of a 1-Mb BAC contig overlapping the mouse Nkrp1 cluster of genes: cloning of three new Nkrp1 members, Nkrp1d, Nkrp1e, and Nkrp1f. Immunogenetics 53: 592–598. [DOI] [PubMed] [Google Scholar]
- 6. Chen P, Belanger S, Aguilar OA, Zhang Q, St-Laurent A, et al. (2011) Analysis of the mouse 129-strain Nkrp1-Clr gene cluster reveals conservation of genomic organization and functional receptor-ligand interactions despite significant allelic polymorphism. Immunogenetics 63: 627–640. [DOI] [PubMed] [Google Scholar]
- 7. Carlyle JR, Mesci A, Ljutic B, Belanger S, Tai LH, et al. (2006) Molecular and genetic basis for strain-dependent NK1.1 alloreactivity of mouse NK cells. J Immunol 176: 7511–7524. [DOI] [PubMed] [Google Scholar]
- 8. Hao L, Klein J, Nei M (2006) Heterogeneous but conserved natural killer receptor gene complexes in four major orders of mammals. Proc Natl Acad Sci U S A 103: 3192–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Aust JG, Gays F, Mickiewicz KM, Buchanan E, Brooks CG (2009) The expression and function of the NKRP1 receptor family in C57BL/6 mice. J Immunol 183: 106–116. [DOI] [PubMed] [Google Scholar]
- 10. Iizuka K, Naidenko OV, Plougastel BF, Fremont DH, Yokoyama WM (2003) Genetically linked C-type lectin-related ligands for the NKRP1 family of natural killer cell receptors. Nat Immunol 4: 801–807. [DOI] [PubMed] [Google Scholar]
- 11. Ryan JC, Turck J, Niemi EC, Yokoyama WM, Seaman WE (1992) Molecular cloning of the NK1.1 antigen, a member of the NKR-P1 family of natural killer cell activation molecules. J Immunol 149: 1631–1635. [PubMed] [Google Scholar]
- 12. Arase H, Arase N, Saito T (1996) Interferon gamma production by natural killer (NK) cells and NK1.1+ T cells upon NKR-P1 cross-linking. J Exp Med 183: 2391–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Carlyle JR, Jamieson AM, Gasser S, Clingan CS, Arase H, et al. (2004) Missing self-recognition of Ocil/Clr-b by inhibitory NKR-P1 natural killer cell receptors. Proc Natl Acad Sci U S A 101: 3527–3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fine JH, Chen P, Mesci A, Allan DS, Gasser S, et al. (2010) Chemotherapy-induced genotoxic stress promotes sensitivity to natural killer cell cytotoxicity by enabling missing-self recognition. Cancer Res 70: 7102–7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Williams KJ, Wilson E, Davidson CL, Aguilar OA, Fu L, et al. (2012) Poxvirus Infection-Associated Downregulation of C-Type Lectin-Related-b Prevents NK Cell Inhibition by NK Receptor Protein-1B. J Immunol 188: 4980–4991. [DOI] [PubMed] [Google Scholar]
- 16. Kveberg L, Dai KZ, Inngjerdingen M, Brooks CG, Fossum S, et al. (2011) Phylogenetic and functional conservation of the NKR-P1F and NKR-P1G receptors in rat and mouse. Immunogenetics 63: 429–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rosen DB, Bettadapura J, Alsharifi M, Mathew PA, Warren HS, et al. (2005) Cutting edge: lectin-like transcript-1 is a ligand for the inhibitory human NKR-P1A receptor. J Immunol 175: 7796–7799. [DOI] [PubMed] [Google Scholar]
- 18. Aldemir H, Prod’homme V, Dumaurier MJ, Retiere C, Poupon G, et al. (2005) Cutting edge: lectin-like transcript 1 is a ligand for the CD161 receptor. J Immunol 175: 7791–7795. [DOI] [PubMed] [Google Scholar]
- 19. Vogler I, Steinle A (2011) Vis-a-vis in the NKC: genetically linked natural killer cell receptor/ligand pairs in the natural killer gene complex (NKC). J Innate Immun 3: 227–235. [DOI] [PubMed] [Google Scholar]
- 20. Carlyle JR, Martin A, Mehra A, Attisano L, Tsui FW, et al. (1999) Mouse NKR-P1B, a novel NK1.1 antigen with inhibitory function. J Immunol 162: 5917–5923. [PubMed] [Google Scholar]
- 21. Arase N, Arase H, Park SY, Ohno H, Ra C, et al. (1997) Association with FcRgamma is essential for activation signal through NKR-P1 (CD161) in natural killer (NK) cells and NK1.1+ T cells. J Exp Med 186: 1957–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ljutic B, Carlyle JR, Filipp D, Nakagawa R, Julius M, et al. (2005) Functional requirements for signaling through the stimulatory and inhibitory mouse NKR-P1 (CD161) NK cell receptors. J Immunol 174: 4789–4796. [DOI] [PubMed] [Google Scholar]
- 23. Ryan JC, Seaman WE (1997) Divergent functions of lectin-like receptors on NK cells. Immunol Rev 155: 79–89. [DOI] [PubMed] [Google Scholar]
- 24. Higuchi DA, Cahan P, Gao J, Ferris ST, Poursine-Laurent J, et al. (2010) Structural variation of the mouse natural killer gene complex. Genes Immun 11: 637–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lattin JE, Schroder K, Su AI, Walker JR, Zhang J, et al. (2008) Expression analysis of G Protein-Coupled Receptors in mouse macrophages. Immunome Res 4: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wu C, Orozco C, Boyer J, Leglise M, Goodale J, et al. (2009) BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol 10: R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gregorieff A, Clevers H (2010) In situ hybridization to identify gut stem cells. Curr Protoc Stem Cell Biol Chapter 2: Unit. [DOI] [PubMed]
- 28. Wilhelm BT, Gagnier L, Mager DL (2002) Sequence analysis of the Ly49 cluster in C57BL/6 mice: a rapidly evolving multigene family in the immune system. Genomics 80: 646–661. [DOI] [PubMed] [Google Scholar]
- 29. Makrigiannis AP, Patel D, Goulet ML, Dewar K, Anderson SK (2005) Direct sequence comparison of two divergent class I MHC natural killer cell receptor haplotypes. Genes Immun 6: 71–83. [DOI] [PubMed] [Google Scholar]
- 30. Belanger S, Tai LH, Anderson SK, Makrigiannis AP (2008) Ly49 cluster sequence analysis in a mouse model of diabetes: an expanded repertoire of activating receptors in the NOD genome. Genes Immun 9: 509–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhou H, Kartsogiannis V, Hu YS, Elliott J, Quinn JM, et al. (2001) A novel osteoblast-derived C-type lectin that inhibits osteoclast formation. J Biol Chem 276: 14916–14923. [DOI] [PubMed] [Google Scholar]
- 32. Tian W, Nunez R, Cheng S, Ding Y, Tumang J, et al. (2005) C-type lectin OCILRP2/Clr-g and its ligand NKRP1f costimulate T cell proliferation and IL-2 production. Cell Immunol 234: 39–53. [DOI] [PubMed] [Google Scholar]
- 33. Zohar O, Reiter Y, Bennink JR, Lev A, Cavallaro S, et al. (2008) Cutting edge: MHC class I-Ly49 interaction regulates neuronal function. J Immunol 180: 6447–6451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Heng TS, Painter MW (2008) The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol 9: 1091–1094. [DOI] [PubMed] [Google Scholar]
- 35. Blasius AL, Barchet W, Cella M, Colonna M (2007) Development and function of murine B220+CD11c+NK1.1+ cells identify them as a subset of NK cells. J Exp Med 204: 2561–2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vosshenrich CA, Lesjean-Pottier S, Hasan M, Richard-Le GO, Corcuff E, et al. (2007) CD11cloB220+ interferon-producing killer dendritic cells are activated natural killer cells. J Exp Med 204: 2569–2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yokoyama WM, Ryan JC, Hunter JJ, Smith HR, Stark M, et al. (1991) cDNA cloning of mouse NKR-P1 and genetic linkage with LY-49. Identification of a natural killer cell gene complex on mouse chromosome 6. J Immunol 147: 3229–3236. [PubMed] [Google Scholar]
- 38. Brodin P, Lakshmikanth T, Johansson S, Karre K, Hoglund P (2009) The strength of inhibitory input during education quantitatively tunes the functional responsiveness of individual natural killer cells. Blood 113: 2434–2441. [DOI] [PubMed] [Google Scholar]
- 39. Joncker NT, Fernandez NC, Treiner E, Vivier E, Raulet DH (2009) NK cell responsiveness is tuned commensurate with the number of inhibitory receptors for self-MHC class I: the rheostat model. J Immunol 182: 4572–4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Proteau M-F, Rousselle E, Makrigiannis AP (2004) Mapping of the BALB/c Ly49 cluster defines a minimal natural killer cell receptor gene repertoire. Genomics 84: 669–677. [DOI] [PubMed] [Google Scholar]
- 41. Stoye JP (2012) Studies of endogenous retroviruses reveal a continuing evolutionary saga. Nat Rev Microbiol 10: 395–406. [DOI] [PubMed] [Google Scholar]
- 42. Balada E, Vilardell-Tarres M, Ordi-Ros J (2010) Implication of human endogenous retroviruses in the development of autoimmune diseases. Int Rev Immunol 29: 351–370. [DOI] [PubMed] [Google Scholar]
- 43. Flornes LM, Nylenna O, Saether PC, Daws MR, Dissen E, et al. (2010) The complete inventory of receptors encoded by the rat natural killer cell gene complex. Immunogenetics 62: 521–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Voigt S, Mesci A, Ettinger J, Fine JH, Chen P, et al. (2007) Cytomegalovirus evasion of innate immunity by subversion of the NKR-P1B:Clr-b missing-self axis. Immunity 26: 617–627. [DOI] [PubMed] [Google Scholar]
- 45. Kveberg L, Dai KZ, Westgaard IH, Daws MR, Fossum S, et al. (2009) Two major groups of rat NKR-P1 receptors can be distinguished based on chromosomal localization, phylogenetic analysis and Clr ligand binding. Eur J Immunol 39: 541–551. [DOI] [PubMed] [Google Scholar]
- 46. Rosen DB, Cao W, Avery DT, Tangye SG, Liu YJ, et al. (2008) Functional consequences of interactions between human NKR-P1A and its ligand LLT1 expressed on activated dendritic cells and B cells. J Immunol 180: 6508–6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Germain C, Meier A, Jensen T, Knapnougel P, Poupon G, et al. (2011) Induction of lectin-like transcript 1 (LLT1) protein cell surface expression by pathogens and interferon-gamma contributes to modulate immune responses. J Biol Chem 286: 37964–37975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. de HS, Woltman AM, Trouw LA, Bakker AC, Kamerling SW, et al. (2005) Renal tubular epithelial cells modulate T-cell responses via ICOS-L and B7-H1. Kidney Int 68: 2091–2102. [DOI] [PubMed] [Google Scholar]