Abstract
Circadian rhythm disorders constitute a group of phenotypes that usually present as altered sleep-wake schedules. Until a human genetics approach was applied to investigate these traits, the genetic components regulating human circadian rhythm and sleep behaviors remained mysterious. Steady advances in the last decade have dramatically improved our understanding of the genes involved in circadian rhythmicity and sleep regulation. Finding these genes presents new opportunities to use a wide range of approaches, including in vitro molecular studies and in vivo animal modeling, to elevate our understanding of how sleep and circadian rhythms are regulated and maintained. Ultimately, this knowledge will reveal how circadian and sleep disruption contribute to various ailments and shed light on how best to maintain and recover good health.
Keywords: Circadian rhythm, Circadian rhythm sleep disorders, Advanced sleep phase, Delayed sleep phase, Advanced sleep phase disorder
Introduction
Behavioral studies using model organisms are stimulating and relatively approachable since the researchers can, for the most part, control both external and internal variables. These studies have provided us novel insights into the genetic nature of many interesting behaviors. Predictably, gaps remain when trying to translate what scientists learn in model organisms to something applicable or useful for human health conditions. In contrast to model organisms, studying behavior in humans such as sleep requirement and preferred sleep-wake times is a daunting task due to the complexity of confounding co-morbidities, environmental factors, and the polygenic nature of behavioral phenotypes.
Like most organisms, humans exhibit daily behaviors that are regulated in a circadian (24-hour) manner. Early pioneers of circadian biology research, such as Jurgen Aschoff, observed that human behaviors under constant conditions exhibited rhythms with an approximately 24-hour periodicity (Aschoff, et al., 1971). Aschoff’s studies in humans revealed the existence of an endogenous biological time-keeping mechanism. Decades later, studies from model organisms uncovered a molecular clock comprised of many genetic players that governs the circadian oscillation of physiology and behaviors through complex methods of regulation (Dunlap, 2004, Hastings, et al., 2008). Not surprisingly, there are shared molecular mechanisms for the molecular clock amongst diverse organisms, including Neurospora, Drosophila, rodents and humans. However, there are also salient differences. For instance, core clock mechanisms are presumably more widely integrated with other forms of physiological regulation in metazoans compared with unicellular organisms due to the inherent complexity of intercellular interactions. In addition, while molecular differences between vertebrates and invertebrates cannot be overlooked (Hardin, 2011, Lowrey and Takahashi, 2011), differences between closely related species, such as rodents and humans, are also expected. For instance, mice are nocturnal and with circadian period average approximately 23.5 hours under constant darkness (Lowrey and Takahashi, 2011), whereas humans are diurnal with an average slightly longer than 24-hours (Czeisler, et al., 1999). More importantly, the highly polyphasic nature of sleep where dozens to hundreds of sleep-wake transitions occur every hour in mice directly contrasts with the single 5–9 hour block of consolidated sleep in most modern adult humans with only a few sleep to wake transitions.
Identification of the underlying genetic basis of human circadian rhythm behaviors was first advanced with the characterization of the first Mendelian circadian trait, Familial Advanced Sleep Phase (FASP), in 1999 (Jones, et al., 1999). This study began with meticulous phenotypic characterization of a 69 year old woman who had life-long early sleep-wake onset, which led to the identification of a large family segregating this behavior. This story pioneered the field of human sleep genetics at the molecular level, including the search for rare Mendelian single gene/mutation forms and genome-wide association studies aimed at discovering novel variants in larger populations. Since the initial FASP findings, other human circadian/sleep phenotypes have been attributed to underlying genetic components, such as Familial Natural Short Sleep (FNSS) (He, et al., 2009, Zhang, et al., 2011). Therefore, studies of rare and extreme Mendelian behavioral traits have established a foundation for identifying human genetic components for circadian rhythms and sleep behaviors, which then provide further opportunities for understanding the molecular mechanisms of these behaviors. This review will outline the current understanding of the field of circadian rhythm disorders.
Human Circadian Rhythm Sleep Disorders
Human alertness demonstrates a circadian rhythmicity with a seemingly paradoxical nadir of sleepiness at the end of the day (the “Maintenance of Wakefulness Zone”) (Strogatz, et al., 1987), followed by a peak in difficulty sustaining wakefulness in the second third of the sleep period (approximately 3–5 A.M.) and then a gradual increase in alertness until the next evening. Pineal release of melatonin is stimulated by the suprachiasmatic nucleus of the hypothalamus (SCN) starting about 1–2 hours before habitual sleep onset time and continuing through the night, unless such stimulation is masked by light of more than 50–100 lux intensity (Lewy, et al., 1980).
The International Classification of Sleep Disorders lists approximately 60 disorders of human sleep (ASDA, 1997) including Circadian Rhythm Sleep Disorders (CRSD). CRSD usually present as a social problem in a person’s sleep/wake timing. The most common complaints for CRSD are difficulty initiating or ending sleep at appropriate social times.
Circadian Rhythm Sleep Disorders are classified into the following types according to the American Academy of Sleep Medicine (AASM, 2012):
1. Advanced Sleep Phase Disorder (ASPD)
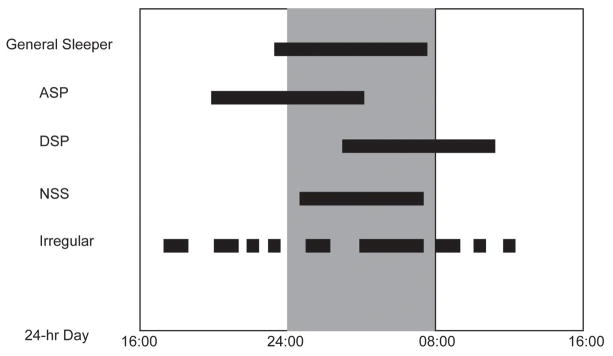
Individuals with ASPD have earlier sleep/wake times than a majority of the population, whereby the entire sleep-wake cycle of ASPD is shifted up to several hours earlier with respect to solar time (Fig. 1) (AASP, 2005). These people are considered “morning larks”. People with ASPD fall asleep during the “Maintenance of Wakefulness Zone” for conventional sleepers and tend to wake up alert and energetic in the early morning hours when most people are the sleepiest. ASPD patients usually present with both difficulty in staying awake to satisfy domestic responsibilities in the evening and an obligate early morning awakening before others are active. This can result in significant sleep deprivation if social responsibilities keep the patient awake late and their biological clock wakes them up early. ASPD is more common in the elderly. It is notable that many people have extremely early sleep and wake times and in whom it is not considered a disorder. Such individuals may in fact feel virtuous for being the ‘early bird’ that gets the worm. Thus, perception of waking early may be positive or negative and is really in the eye of the beholder, underscoring the fact that these clinical classifications are truly behavioral phenotypes.
Fig. 1.
Schematic sleep schedules for ASP. DSP, NSS, ISW in comparison to general sleepers during each 24 hour period. Black-filled bars represent time spent in sleep.
The International Classification of Sleep Disorders criteria for the clinical diagnosis of ASPD are based on the report of a patient who seeks medical attention specifically because of a complaint of a sleep time that is earlier than his/her “desired” sleep schedule (ASDA, 1997). No (solar) clock times of the habitual morning wake up are specified, and no measurements of melatonin, temperature, or other rhythms are required.
2. Delayed Sleep Phase Disorder (DSPD)
DSPD individuals are “night owls” and routinely have later sleep/wake onset compared to the rest of the population (Fig. 1) (AASP, 2005). Their entire sleep-wake cycle is shifted later with respect to solar time, and DSPD patients feel wide awake, energetic and motivated until late in the night. Depending on the severity, sleep onset may be delayed until 01:00 to 06:00 A.M., and the circadian “morning” increase in alertness does not occur until approximately 10:00 A.M. to 2:00 P.M. DSPD individuals are often sleep deprived because sleep onset is delayed by the biological clock and morning waking time is enforced by the alarm clock and social responsibilities. DSPD is common in adolescents and young adults with an estimated prevalence of 7–16%. Again, it is important to point out that there are many night owls who do not consider it a disorder.
3. Free-running sleep disorder (FRSD)
People with free-running disorder have a variable but predictable sleep-wake cycle that shifts approximately one hour later every day (AASP, 2005). FRSD is usually associated with a history of either total retinal blindness or sleep phase delay.
4. Irregular Sleep-wake disorder (ISWD)
These individuals have undefined sleep-wake cycles and their sleep is fragmented into a series of naps that occur throughout the 24-hour day (Fig. 1) (AASP, 2005). A mild symptom of irregular sleep-wake rhythm is sometimes seen in many neurological disorder patients (e.g. dementia) and in mentally retarded children. Individuals with this disorder suffer from excessive sleepiness, chronic insomnia, or both.
5. Jet lag disorder
This happens when the sleep/wake times of the new location for a traveler are misaligned with the traveler’s internal body clock derived from his/her original place of abode. The severity increases with the number of time zones crossed; symptoms are temporary.
6. Shift work disorder
When an individual’s work hours are during their optimal sleep time, this causes sleepiness during the work shift and difficulty remaining asleep during the new required sleep time. Depending on the timing of the shift, a person’s inherent sleep preference can influence his/her ability to adjust to the shift work. For example, morning larks will be able to adjust to early shift work easier than the rest of population.
Phenotyping CRSD
The mammalian circadian rhythm system is complicated (Saper, et al., 2005) and whole- organism circadian behavior is further confounded by strong interactions between the organism and its environment, especially the perceived photic regime (Takahashi, et al., 2001). That the core circadian “clock” mechanism is relatively inaccessible inside the central nervous system creates a further investigative challenge to measuring intrinsic or “biological” time relative to the timing of the perceived light-dark cycle. Under laboratory confinement, it is possible to measure the timing of objective markers of human biological circadian time (e.g. core body temperature or plasma melatonin levels) relative to the timing of the most recently-imposed light-dark cycle (the so-called “phase angle of circadian entrainment”) in a “constant routine” protocol (Mills, et al., 1978). The more prolonged “forced desynchrony” protocol has been successfully employed in many human studies to measure the intrinsic period length, or speed, of the hypothalamic circadian clock (Czeisler, et al., 1986). These laboratory procedures are expensive for the investigator and time-consuming for the participant. Therefore, many human circadian studies are ambulatory and report measurements of various phase angles of entrainment for either self-reported phase markers (e.g. times of first falling asleep and final awakening) or objective phase markers (e.g. the time of saliva melatonin secretion onset) (Burgess and Fogg, 2008) or the onset time of the daily rhythm of wrist movement (Littner, et al., 2003). Questionnaires such as the Morningness-Eveningness Questionnaire (Horne and Ostberg, 1976) and the Munich ChronoType Questionnaire (Roenneberg, et al., 2003) also have moderately strong correlations with objective assessments (Duffy, et al., 2001, Horne and Ostberg, 1976) of phase-advanced or delayed sleep-wake rhythms (i.e. morning or evening “chronotype”) (Roenneberg, 2012). However, inferring intrinsic circadian physiology from observed rhythms in daily behavior outside the laboratory is confounded in humans by self-imposed artificial (i.e. non-solar) photic regimes and by the circadian influence of idiosyncratic developmental, psycho-social, physical/medical, and non-circadian sleep characteristics. Therefore, careful individual human chronotyping includes consideration of these multiple categories of information.
Familial Advanced Sleep Phase Disorder
Molecular genetic studies of CRSD began from Familial Advanced Sleep Phase Disorder (FASPD, formerly familial advanced sleep phase syndrome or FASPS) since it is the first CRSD with identified mutations. FASPD is an autosomal dominant human sleep trait characterized by stable entrainment of sleep and wakefulness to early solar times. In the mid-1990s, FASPD was first genetically characterized in a Utah family, due to its very dramatic advancement of sleep phase in the proband (Jones, et al., 1999). The proband’s sleep phase was advanced ~4–6 hours, in comparison to controls; however, FASPD subjects’ sleep quality and quantity were measurably normal. We have since then identified a large number of families that exhibit the FASPD phenotype segregating as an autosomal dominant trait. Identification of these families afforded the opportunity to search for underlying genes causative for the advanced sleep phase phenotype.
PER2
The first FASPD family was large enough to allow for linkage analysis, which localized the mutant allele to the chromosome 2q telomeric region (Toh, et al., 2001). Positional cloning, cDNA identification, and mutation screening of genes within the critical region identified a single base change in the human Period 2 gene (hPER2). PER2 is one of three homologs of the Drosophila per gene, and the mutation substitutes a serine with a glycine residue at amino acid position 662 of PER2.
The proband was willing to participate in a time-isolation (under an environment without any time cues) experiment for three weeks and showed an ~1 hour shortening in her circadian period (“tau”). Following the identification of the mutation, mouse models carrying the hPER2S662G variant were engineered and period length for these mice were measured under laboratory conditions (Xu, et al., 2007). Under standard “free-running” constant darkness (DD), S662G mutant transgenic mice exhibited a near-2-hour shorter free-running period (tau) and a ~4–6 hour phase advancement of the behavioral phenotype, which recapitulates the behavioral phenotype seen in human subjects.
Mechanistic insights regarding the human clock regulation via hPER2 were partially revealed using molecular and biochemical approaches (Xu, et al., 2007). PER2 serine 662 is followed by four additional serines, each separated by two amino acids (a recognition motif for CKIδ/ε). The S662G mutation renders PER2 hypo-phosphorylated by blocking phosphorylation by CKIδ/ε (Toh, et al., 2001, Xu, et al., 2007). The mutation also affected transcript levels for both endogenous mouse Per2 and the human PER2 transgene in the transgenic mice. Both showed a reduced level with phase advancement compared to wild-type control mice, indicating that PER2S662G is a stronger transcriptional repressor than wild-type PER2. To further enhance our understanding of the role of serine 662, parallel investigations with S662D in addition to S662G were performed. Importantly, S662D transgenic mice demonstrated a longer tau and exhibited increased mPer2 and hPER2 transcript levels with delayed phase in relation to wild-type controls, indicating that PER2S662D is a weaker transcriptional repressor than wild-type PER2. Together, these results demonstrate the direct impact of S662 phosphorylation on PER2 transcriptional activity, which then plays a critical role in setting the speed of the molecular clock.
PER2 S662G and S662D transgenic mice were also crossed with CKIδ wild-type transgenic (with 4–5 copies of CKIδ) and CKIδ heterozygous knock-out mice (one copy of CKIδ) to investigate the genetic interactions between Per2 and CKIδ. The copy number of wild-type CKIδ does not have any effect on the tau of wild type mouse behavior. However, a lower copy number of CKIδ led to longer tau for mutant PER2 transgenic mice, and a higher copy number of CKIδ caused the tau of PER2 mutant transgenic mice to be shortened. These results implied that phosphorylation by CKIδ on another region(s) of PER2 can modulate tau independently from the S662 region. The phosphorylation status of S662 also influenced PER2 stability; PER2S662G is less stable and PER2S662D is more stable than wild type PER2 (Vanselow, et al., 2006). Hence, the stronger repressor is less stable and the weaker repressor is more stable. This finding raised the possibility that the suicide model of transcriptional factors is a likely mechanism for regulating PER2 (and possibly for other circadian repressors), and this mode of regulation can also provide a highly sensitive system to track the timing of the clock (Fu, 2008).
Casein Kinase I delta (CKIδ)
In the early 2000s, with a growing number of human genome sequences in the public databases, it became reasonable to search for causative mutations using a candidate gene approach since the number of candidate genes had dramatically increased. A second causative mutation for FASPD was identified in human CKIδ; the mutation substitutes threonine at amino acid position 44 by alanine (T44A) (Xu, et al., 2005). The conservation of threonine at amino acid 44 and the mutation-induced decrease in phosphorylation of alpha-casein and PER protein substrates by in vitro kinase assays strongly suggested the biological relevance of this genetic variant. A BAC transgenic mouse model carrying CKIδT44A was generated and the “free-running” period (tau) was shorter than control mice by ~20 minutes. The mutant transgene on a CKIδ knock-out background demonstrated a much shorter tau (22.7hrs) in addition to rescuing the lethal phenotype of the CKIδ knock-out. Intriguingly, expressing the human CKIδT44A mutation in Drosophila generated flies with a significantly longer period than control flies (Xu, et al., 2005), which highlighted the underlying differences between mammals and invertebrates.
CKIδ is a critical core component of the molecular clock. To further explore the clock composition through CKIδ, proteomic studies were conducted using CKIδ and CKIε. These studies were carried out at different circadian time points to reveal the dynamics of CKI proteomes, and distinctive potential interacting partners and/or substrates were found for these enzymes at different circadian times. Importantly, different pools of proteins were identified for CKIδ and CKIε, though with some overlap, supporting the partially redundant roles for these enzymes. Prohibitin2 (PHB2) was chosen after the initial in vitro screening process for further characterization. Interestingly, PHB2 protein levels were regulated by CKIε, yet its RNA levels were modulated by CKIδ. Characterization revealed that PHB2 is a novel clock component and acts as a transcriptional repressor for clock controlled genes in a CLOCK/BMAL1 independent manner (Kategaya, et al., 2012).
Familial Natural Short Sleepers
While screening genes for the “extreme early bird” phenotype, a mutation was identified in human DEC2 (hDEC2). Variant carriers in this small family were further evaluated and found to possess a Natural Short Sleeper (NSS) phenotype with a lifelong daily sleep time requirement of approximately ~6 hours (He, et al., 2009). Similar to the FASP human subjects, the FNSS individuals woke up at an extremely early hour in the morning; however, their sleep onset time was comparable to conventional sleepers, thus shortening their total sleep duration by ~2 hours each day compared to conventional sleepers (Fig. 1).
DEC2
Using a candidate gene sequencing approach, a point mutation that changed the amino acid at position 384 from a proline to arginine in hDEC2 was found to co-segregate with the FNSS phenotype. In vitro luciferase reporter experiments showed that the mutation mitigated transcriptional inhibitory function of hDEC2. Transgenic mice carrying this P384R mutation displayed a similarly shorter total sleep time, including an activity period (alpha) ~1.2-hours longer than wild-type mice. When the mutant transgene was bred onto a mouse Dec2 knock-out background, the mice exhibited an even more dramatic activity phenotype with alpha lengthened by ~2.5 hours.
Electroencephalography (EEG) and electromyography (EMG) were also performed on DEC2 P384R mutant transgenic mice and their littermate controls to further characterize the sleep phenotype. Mutant transgenic mice were awake for significantly longer periods of time during the light phase (when mice are supposed to be sleeping). Consistently, they showed a significant reduction of both non-rapid eye movement (NREM) and REM sleep in the light phase (He, et al., 2009). NREM sleep in the light phase was more fragmented in the mutant transgenic mice than control mice. In addition, the EEG study also demonstrated that mutant transgenic mice accumulated less recovery sleep after sleep deprivation.
The function of DEC2 in sleep regulation was also tested in Drosophila to see whether it gives a conserved phenomenon across species. This was carried out by generating transgenic flies with either wild-type or P384R mDec2 expressed in either whole brain or the mushroom bodies. In agreement with the findings from mammalian systems, flies with mutant mDec2 expressed in their mushroom bodies demonstrated lengthened activity duration (i.e. a shortened sleep length) compared to equivalent control flies. All this evidence supported a role for hDEC2 in regulating sleep homeostasis.
Other clock genes
Interestingly, some mouse models of clock genes have been shown to have effects on sleep architecture. In mice, the Clock mutation was shown to alter sleep homeostasis (Naylor, et al., 2000) and mice that are deficient for both Cry1 and Cry2 sleep longer than wild-type mice by almost 100 minutes (Wisor, et al., 2002). Similarly, Bmal1 (Laposky, et al., 2005) and Npas2 (Franken, et al., 2006) knock-out mice were reported to have altered sleep time and architecture. These findings raise the possibility that the molecular circuitry used to set internal time-of-day might also be utilized, at least in part, to track and anticipate sleep need (Franken and Dijk, 2009).
Genetic Association Studies
In addition to searching for genes with Mendelian forms of CRSD, the rapid advance in human genome sequencing and SNP genotyping during the last two decades allowed for association studies of human CRSD (or morningness/eveningness preference) to become possible.
I. Sleep phase
CLOCK
Association between a polymorphism in the 3′-untranslated region of CLOCK (3111C to T) and evening preference was first reported in 1998 using human volunteers (Katzenberg, et al., 1998). Individuals with C/C or C/T genotypes showed an increased evening preference compared with individuals with the T/T genotype. This CLOCK 3111C/T polymorphism was later found not to be associated with eveningness by Robilliard et al. (Robilliard, et al., 2002). In addition, Robilliard et al. reported that CLOCK 3111C/T is neither associated with tau length, nor with delayed sleep phase. Their molecular study further demonstrated that this polymorphism does not affect CLOCK mRNA translation, hence weakening the value of CLOCK 3111C/T as a marker for diurnal preference, tau, or delayed sleep phase disorder in humans. However, the Clock mutation mouse model was shown to demonstrate phase delays of the rhythm for body temperature, activity and wake duration in one study (Sei, et al., 2001) and delayed locomotor activity under constant light and during lactation in a separate study (Wakatsuki, et al., 2007). These results do support the possibility that CLOCK mutations/polymorphisms may be associated with evening preference.
PER3
A length polymorphism in PER3 was reported by Archer, von Schantz, and colleagues to be linked to delayed sleep phase disorder and extreme diurnal preference (Archer, et al., 2003). This length polymorphism is due to a variable-number tandem repeat (VNTR, 4 or 5 repeats) in PER3. The shorter 4 allele was found to have a strong association with delayed sleep phase disorder. They followed up this study and reported that polymorphisms in the PER3 promoter are also associated with diurnal preference and delayed sleep phase disorder (Archer, et al., 2010). However, a recent Norwegian study with 432 subjects was not able to reproduce these findings (Osland, et al., 2011).
II. Sleep duration
Several genetic association studies for sleep duration have been conducted. The first report was for more than 700 participants from the Framingham Heart Study Offspring Cohort using the Affymetrix 100K SNP GeneChip. This family-based study was able to confirm significant heritability of sleepiness, usual bedtime, and usual sleep duration (Gottlieb, et al., 2007). Allebrandt et al. applied a more focused approach using high-density markers from 19 clock genes to perform a two-stage design association study on two European populations, South Tyrol and Estonia. In this study, variants in the CLOCK gene were found to have an association with sleep duration for these two independent populations (Allebrandt, et al., 2010). Another genome-wide association study was carried out for seven European populations, totaling 4251 subjects. The meta-analysis for this study revealed a genome-wide significant signal in the ABCC9 locus that encodes a pore-forming subunit of an ATP-sensitive potassium channel (KATP) involved in energy metabolism and in the etiology of cardiomyopathies (Allebrandt, et al., 2011). Interestingly, the authors also reported that knocking down Drosophila ABCC9 caused shortened nighttime but not daytime duration of sleep-like behavior in the fly.
Concluding Remarks
Identification of mutations that are responsible for various human diseases has revolutionized the field of biomedical research during the last twenty years. Many of these are rare diseases where large families are used to map and clone the causative genes/mutations. Knowledge of genes and pathways contributing to the pathophysiology of rare diseases often opens new areas for studying more common and non-Mendelian forms of the disease. Genes causing rare familial Alzheimer’s and Parkinson’s disease encode proteins that are central to the pathophysiology of the common sporadic forms of these diseases (Bertram, et al., 2010, Hardy, 2010). A logical extension of this kind of work in disease genetics is to apply this approach to even more complex issues such as human behavioral phenotypes which are subject to large influences of environmental and social factors. Using human genetics to identify specific genes/proteins that are critical for specific behavioral phenotypes (especially those that are associated with human health) are windows into novel biological pathways relevant to regulation of human behaviors.
As with genetic studies in all organisms, the ultimate proof of functional mutations vs. polymorphisms lies in establishing causality of the phenotype by the mutation. Many Mendelian human genetic disease mutations were shown to be causative with multiple unrelated families co-segregating the DNA variant and disease phenotype. However, with human behavioral phenotypes, it is inherently difficult to obtain multiple families that have the same mutations. One way to circumvent this is by engineering the DNA variant into model organisms and to observe whether it produces a similar phenotype in the model organism. It is also difficult to demonstrate the causality of a mutation for a behavioral phenotype if the mutation is found in multiple random subjects with no family relation and does not produce a similar phenotype in model organisms. Conversely, cloning a variant found in only a few individuals but that replicates the phenotype in every single animal of the model organism (mouse, fly, etc) proves causality with no reservations.
Although genetic association studies have provided new insights into human phenotypes, the inconsistency of the results remains a major difficulty. This complication may stem from the obstacle in finding large enough cohorts of subjects with proper phenotyping data which renders the replication for association study arduous. Also, the effect sizes are often quite small which calls to question the cost of such studies vs. the information learned. Rare heritable CRSD families have so far proven to be useful and offer new genetic and mechanistic information. However, one persistent challenge in studying these families is the amount of effort and difficulty in finding large families to define the potential mutation region with relative ease. This was one of the major factors limiting researchers to use the candidate gene approach in the last decade. With the advancement of sequencing technology and the decreasing cost, whole exome sequencing and whole genome sequencing are becoming more realistic approaches today. However, new challenges present themselves with these new technologies such as the enormous effort required to sort through the large number of SNP variants in order to identify true functional mutations. Regardless, these new methods do offer hope of finding novel human circadian/sleep genes that were previously impossible.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.AASM. 2012 http://www.aasmnet.org/
- 2.AASP; Medicine., A. A. o. S. Diagnostic & Coding Manual. 2. Westchester, Illinois: 2005. International Classification of Sleep Disorders. [Google Scholar]
- 3.Allebrandt KV, Amin N, Muller-Myhsok B, Esko T, Teder-Laving M, Azevedo RV, Hayward C, van Mill J, Vogelzangs N, Green EW, Melville SA, Lichtner P, Wichmann HE, Oostra BA, Janssens AC, Campbell H, Wilson JF, Hicks AA, Pramstaller PP, Dogas Z, Rudan I, Merrow M, Penninx B, Kyriacou CP, Metspalu A, van Duijn CM, Meitinger T, Roenneberg T. A K(ATP) channel gene effect on sleep duration: from genome-wide association studies to function in Drosophila. Mol Psychiatry. 2011 doi: 10.1038/mp.2011.142. [DOI] [PubMed] [Google Scholar]
- 4.Allebrandt KV, Teder-Laving M, Akyol M, Pichler I, Muller-Myhsok B, Pramstaller P, Merrow M, Meitinger T, Metspalu A, Roenneberg T. CLOCK gene variants associate with sleep duration in two independent populations. Biol Psychiatry. 2010;67:1040–1047. doi: 10.1016/j.biopsych.2009.12.026. [DOI] [PubMed] [Google Scholar]
- 5.Archer SN, Carpen JD, Gibson M, Lim GH, Johnston JD, Skene DJ, von Schantz M. Polymorphism in the PER3 promoter associates with diurnal preference and delayed sleep phase disorder. Sleep. 2010;33:695–701. doi: 10.1093/sleep/33.5.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Archer SN, Robilliard DL, Skene DJ, Smits M, Williams A, Arendt J, von Schantz M. A length polymorphism in the circadian clock gene Per3 is linked to delayed sleep phase syndrome and extreme diurnal preference. Sleep. 2003;26:413–415. doi: 10.1093/sleep/26.4.413. [DOI] [PubMed] [Google Scholar]
- 7.Aschoff J, Fatranska M, Giedke H, Doerr P, Stamm D, Wisser H. Human circadian rhythms in continuous darkness: entrainment by social cues. Science. 1971;171:213–215. doi: 10.1126/science.171.3967.213. [DOI] [PubMed] [Google Scholar]
- 8.ASDA, A. S. D. A . International classification of sleep disorders: Diagnostic and coding manual. American Sleep Disorders Association; Rochester: 1997. [Google Scholar]
- 9.Bertram L, Lill CM, Tanzi RE. The genetics of Alzheimer disease: back to the future. Neuron. 2010;68:270–281. doi: 10.1016/j.neuron.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 10.Burgess HJ, Fogg LF. Individual differences in the amount and timing of salivary melatonin secretion. PLoS One. 2008;3:e3055. doi: 10.1371/journal.pone.0003055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Czeisler CA, Allan JS, Kronauer RE. A method to assess the intrinsic period of the endogenous circadian oscillator. Sleep Research. 1986;15:266. [Google Scholar]
- 12.Czeisler CA, Duffy JF, Shanahan TL, Brown EN, Mitchell JF, Rimmer DW, Ronda JM, Silva EJ, Allan JS, Emens JS, Dijk DJ, Kronauer RE. Stability, precision, and near-24-hour period of the human circadian pacemaker [see comments] Science. 1999;284:2177–2181. doi: 10.1126/science.284.5423.2177. [DOI] [PubMed] [Google Scholar]
- 13.Duffy JF, Rimmer DW, Czeisler CA. Association of intrinsic circadian period with morningness-eveningness, usual wake time, and circadian phase. Behav Neurosci. 2001;115:895–899. doi: 10.1037//0735-7044.115.4.895. [DOI] [PubMed] [Google Scholar]
- 14.Dunlap JCL, JJ, DeCoursey PJ. Chronobiology: Biological Timekeeping. Sinauer Associate; Sunderland: 2004. p. 406. [Google Scholar]
- 15.Franken P, Dijk DJ. Circadian clock genes and sleep homeostasis. Eur J Neurosci. 2009;29:1820–1829. doi: 10.1111/j.1460-9568.2009.06723.x. [DOI] [PubMed] [Google Scholar]
- 16.Franken P, Dudley CA, Estill SJ, Barakat M, Thomason R, O’Hara BF, McKnight SL. NPAS2 as a transcriptional regulator of non-rapid eye movement sleep: genotype and sex interactions. Proc Natl Acad Sci U S A. 2006;103:7118–7123. doi: 10.1073/pnas.0602006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu YH. Oscillating per-cision. PLoS Biol. 2008;6:e192. doi: 10.1371/journal.pbio.0060192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gottlieb DJ, O’Connor GT, Wilk JB. Genome-wide association of sleep and circadian phenotypes. BMC Med Genet. 2007;8(Suppl 1):S9. doi: 10.1186/1471-2350-8-S1-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hardin PE. Molecular genetic analysis of circadian timekeeping in Drosophila. Adv Genet. 2011;74:141–173. doi: 10.1016/B978-0-12-387690-4.00005-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hardy J. Genetic analysis of pathways to Parkinson disease. Neuron. 2010;68:201–206. doi: 10.1016/j.neuron.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hastings MH, Maywood ES, Reddy AB. Two decades of circadian time. J Neuroendocrinol. 2008;20:812–819. doi: 10.1111/j.1365-2826.2008.01715.x. [DOI] [PubMed] [Google Scholar]
- 22.He Y, Jones CR, Fujiki N, Xu Y, Guo B, Holder JL, Jr, Rossner MJ, Nishino S, Fu YH. The transcriptional repressor DEC2 regulates sleep length in mammals. Science. 2009;325:866–870. doi: 10.1126/science.1174443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horne JA, Ostberg O. A self-assessment questionnaire to determine morningness-Eveningness in human circadian rhythms. International Journal of Chronobiology. 1976;4:97–110. [PubMed] [Google Scholar]
- 24.Jones CR, Campbell SS, Zone SE, Cooper F, DeSano A, Murphy PJ, Jones B, Czajkowski L, Ptacek LJ. Familial advanced sleep-phase syndrome: A short-period circadian rhythm variant in humans [see comments] Nat Med. 1999;5:1062–1065. doi: 10.1038/12502. [DOI] [PubMed] [Google Scholar]
- 25.Kategaya LS, Hilliard A, Zhang L, Asara JM, Ptacek LJ, Fu YH. Casein kinase 1 proteomics reveal prohibitin 2 function in molecular clock. PLoS One. 2012;7:e31987. doi: 10.1371/journal.pone.0031987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katzenberg D, Young T, Finn L, Lin L, King DP, Takahashi JS, Mignot E. A CLOCK polymorphism associated with human diurnal preference. Sleep. 1998;21:569–576. doi: 10.1093/sleep/21.6.569. [DOI] [PubMed] [Google Scholar]
- 27.Laposky A, Easton A, Dugovic C, Walisser J, Bradfield C, Turek F. Deletion of the mammalian circadian clock gene BMAL1/Mop3 alters baseline sleep architecture and the response to sleep deprivation. Sleep. 2005;28:395–409. doi: 10.1093/sleep/28.4.395. [DOI] [PubMed] [Google Scholar]
- 28.Lewy AJ, Wehr TA, Goodwin FK, Newsome DA, Markey SP. Light suppresses melatonin secretion in humans. Science. 1980;210:1267–1269. doi: 10.1126/science.7434030. [DOI] [PubMed] [Google Scholar]
- 29.Littner M, Kushida CA, Anderson WM, Bailey D, Berry RB, Davila DG, Hirshkowitz M, Kapen S, Kramer M, Loube D, Wise M, Johnson SF. Practice parameters for the role of actigraphy in the study of sleep and circadian rhythms: an update for 2002. Sleep. 2003;26:337–341. doi: 10.1093/sleep/26.3.337. [DOI] [PubMed] [Google Scholar]
- 30.Lowrey PL, Takahashi JS. Genetics of circadian rhythms in Mammalian model organisms. Adv Genet. 2011;74:175–230. doi: 10.1016/B978-0-12-387690-4.00006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mills JN, Minors DS, Waterhouse JM. Adaptation to abrupt time shifts of the oscillator(s) controlling human circadian rhythms. J Physiol. 1978;285:455–470. doi: 10.1113/jphysiol.1978.sp012582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naylor E, Bergmann BM, Krauski K, Zee PC, Takahashi JS, Vitaterna MH, Turek FW. The circadian clock mutation alters sleep homeostasis in the mouse. J Neurosci. 2000;20:8138–8143. doi: 10.1523/JNEUROSCI.20-21-08138.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Osland TM, Bjorvatn BR, Steen VM, Pallesen S. Association study of a variable-number tandem repeat polymorphism in the clock gene PERIOD3 and chronotype in Norwegian university students. Chronobiol Int. 2011;28:764–770. doi: 10.3109/07420528.2011.607375. [DOI] [PubMed] [Google Scholar]
- 34.Robilliard DL, Archer SN, Arendt J, Lockley SW, Hack LM, English J, Leger D, Smits MG, Williams A, Skene DJ, Von Schantz M. The 3111 Clock gene polymorphism is not associated with sleep and circadian rhythmicity in phenotypically characterized human subjects. J Sleep Res. 2002;11:305–312. doi: 10.1046/j.1365-2869.2002.00320.x. [DOI] [PubMed] [Google Scholar]
- 35.Roenneberg T. What is chronotype? Sleep and Biological Rhythms. 2012;10:75–76. [Google Scholar]
- 36.Roenneberg T, Wirz-Justice A, Merrow M. Life between clocks: daily temporal patterns of human chronotypes. J Biol Rhythms. 2003;18:80–90. doi: 10.1177/0748730402239679. [DOI] [PubMed] [Google Scholar]
- 37.Saper CB, Lu J, Chou TC, Gooley J. The hypothalamic integrator for circadian rhythms. Trends Neurosci. 2005;28:152–157. doi: 10.1016/j.tins.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 38.Sei H, Oishi K, Morita Y, Ishida N. Mouse model for morningness/eveningness. Neuroreport. 2001;12:1461–1464. doi: 10.1097/00001756-200105250-00033. [DOI] [PubMed] [Google Scholar]
- 39.Strogatz SH, Kronauer RE, Czeisler CA. Circadian pacemaker interferes with sleep onset at specific times each day: role in insomnia. Am J Physiol. 1987;253:R172–178. doi: 10.1152/ajpregu.1987.253.1.R172. [DOI] [PubMed] [Google Scholar]
- 40.Takahashi JS, Turek FW, Moore RY. Handbook of Behavioral Neurobiology:Circadian Clocks. Kluwer Academic/Plenum Publishers; New York: 2001. p. 770. [Google Scholar]
- 41.Toh KL, Jones CR, He Y, Eide EJ, Hinz WA, Virshup DM, Ptacek LJ, Fu YH. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science. 2001;291:1040–1043. doi: 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- 42.Vanselow K, Vanselow JT, Westermark PO, Reischl S, Maier B, Korte T, Herrmann A, Herzel H, Schlosser A, Kramer A. Differential effects of PER2 phosphorylation: molecular basis for the human familial advanced sleep phase syndrome (FASPD) Genes Dev. 2006;20:2660–2672. doi: 10.1101/gad.397006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wakatsuki Y, Kudo T, Shibata S. Constant light housing during nursing causes human DSPS (delayed sleep phase syndrome) behaviour in Clock-mutant mice. Eur J Neurosci. 2007;25:2413–2424. doi: 10.1111/j.1460-9568.2007.05490.x. [DOI] [PubMed] [Google Scholar]
- 44.Wisor JP, O’Hara BF, Terao A, Selby CP, Kilduff TS, Sancar A, Edgar DM, Franken P. A role for cryptochromes in sleep regulation. BMC Neurosci. 2002;3:20. doi: 10.1186/1471-2202-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu Y, Pdaiath Q, Shapiro R, Jones CR, Wu SM, Saigoh N, Saitgoh K, Ptacek L, Fu Y. Functional consequences of a CKId mutation causing familial advanced sleep phase syndrome. Nature. 2005 doi: 10.1038/nature03453. [DOI] [PubMed] [Google Scholar]
- 46.Xu Y, Toh KL, Jones CR, Shin JY, Fu YH, Ptacek LJ. Modeling of a human circadian mutation yields insights into clock regulation by PER2. Cell. 2007;128:59–70. doi: 10.1016/j.cell.2006.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang L, Jones CR, Ptacek LJ, Fu YH. The genetics of the human circadian clock. Adv Genet. 2011;74:231–247. doi: 10.1016/B978-0-12-387690-4.00007-6. [DOI] [PubMed] [Google Scholar]