Abstract
FtsZ is a guanosine triphosphatase (GTPase) that mediates cytokinesis in bacteria. FtsZ is homologous in structure to eukaryotic tubulin and polymerizes in a similar head-to-tail fashion. The study of tubulin’s function in eukaryotic cells has benefited greatly from specific and potent small molecule inhibitors, including colchicine and taxol. Although many small molecule inhibitors of FtsZ have been reported, none has emerged as a generally useful probe for modulating bacterial cell division. With the goal of establishing a useful and reliable small molecule inhibitor of FtsZ, a broad biochemical cross-comparison of reported FtsZ inhibitors was undertaken. Several of these molecules, including phenolic natural products, are unselective inhibitors that seem to derive their activity from the formation of microscopic colloids or aggregates. Other compounds, including the natural product viriditoxin and the drug development candidate PC190723, exhibit no inhibition of GTPase activity using protocols in this work or under published conditions. Of the compounds studied, only zantrin Z3 exhibits good levels of inhibition, maintains activity under conditions that disrupt small molecule aggregates, and provides a platform for exploration of structure-activity relationships (SAR). Preliminary SAR studies have identified slight modifications to the two sidechains of this structure that modulate the inhibitory activity of zantrin Z3. Collectively these studies will help focus future investigations toward the establishment of probes for FtsZ that fill the roles of colchicine and taxol in studies of tubulin.
Introduction
Cell division in bacteria is controlled by several proteins that make up the “divisome” in which FtsZ, the bacterial homolog of eukaryotic tubulin, plays a central role (Figure 1) 1–4. The possibility of modulating the activity of this protein in order to better understand the cell division process in prokaryotes and possibly advance FtsZ as a target of new antibiotics has led to many reports of small molecule inhibitors. Given the close structural homology between FtsZ and tubulin, the prospects for finding selective small molecule inhibitors has always seemed good. Among the many potent inhibitors of tubulin’s function, colchicine, taxol and vinblastine all exhibit selective effects that can be correlated to their molecular interactions with the protein. In addition, the crystallographic structures of tubulin bound to all three of these inhibitors5–7 enables new inhibitors to be classified by their similarity to colchicine, which destabilizes tubulin polymers, and taxol, which stabilizes polymers of tubulin. Both molecules are used as drugs and as chemical probes for cell biology experiments. Analogous information for FtsZ in bacteria is still lacking. None of the known inhibitors exhibits potency that approaches that of taxol’s and colchicine’s inhibition of tubulin’s function. Aside from GTP and close analogs, for which the structures of complexes with FtsZ have been solved with X-ray crystallography8, there is little direct structural information for the basis of perturbing FtsZ’s function with small molecules. The lone example lies in the recent co-crystal structure of PC190723 with FtsZ from Staphylococcus aureus (SaFtsZ), which confirms the structural inferences made by resistant mutants of Bacillus subtilis in the original disclosure of this inhibitor9. The fact that this compound preferentially affects SaFtsZ limits the extent to which this result impacts the majority of studies that employ FtsZ from Escherichia coli (EcFtsZ) and B. subtilis (BsFtsZ). Importantly, there is little information for how well a small molecule inhibitor of FtsZ from one species of bacteria inhibits that from other species. This gap prevents both the development of small molecule inhibitors of FtsZ as drugs and the use of small molecules to better understand bacterial cell division. Here we report a critical assessment of small molecule inhibitors of FtsZ to date and document that only a small number currently represent good starting points for finding small molecules that will be as useful to the study of FtsZ as colchicine, taxol, and others have proven to the study of tubulin.
Figure 1.
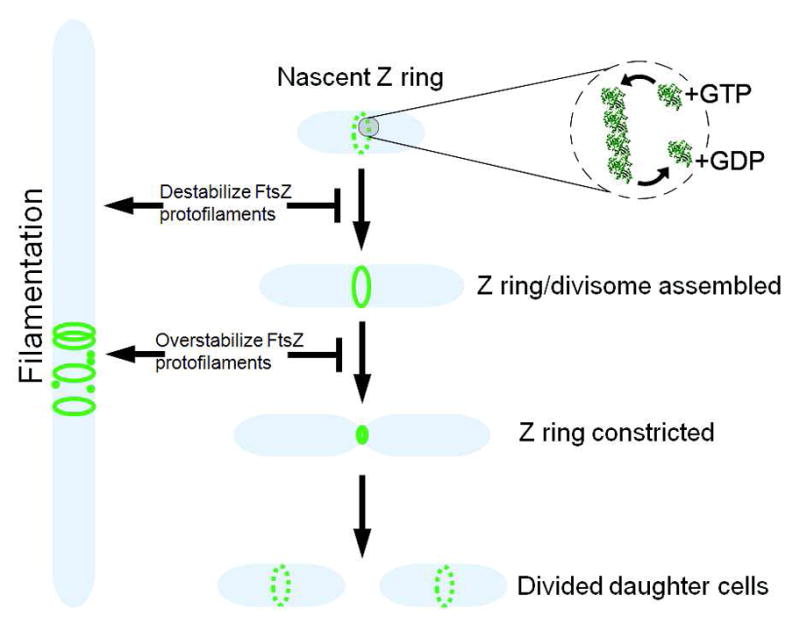
FtsZ in bacterial cell division and possible pathways for inhibition.
The lack of generally useful FtsZ inhibitors might seem strange, given that the literature would suggest that investigators are surrounded by useful small molecule probes of bacterial cell division that vary widely in their chemical structures (Figure 2)10, 11. Careful scrutiny of these molecules suggests that they can be divided into seven groups based on structure and origin. For instance, 4′,6-diamidino-2-phenylindole (3, DAPI), a widely used DNA dye, resembles zantrins Z5 (1) and Z2 (2) in that they all have extended heterocyclic structures that are cationic at biological pH (Figure 2A)12, 13. Compounds 48, 14 and 515 were designed as GTP analogs (Figure 2B). Although several different inhibitors (6–10)13, 16–23 resemble “drug-like” heterocyclic compounds, they bear little structural resemblance to each other. It is notable that PC190723 (8) and (1a-G7, 10) represent optimizations of previously reported bacterial cell division inhibitors 3-methoxybenzamide (3MBA)24 and thiabendazole, respectively (not shown).25 Many FtsZ inhibitors have been isolated from natural sources. The majority identified to date are non-alkaloid structures that are almost all phenolic (Figure 2D)26–32. Similarly, screening of libraries of small molecules has provided a wealth of structures united only by the ease of their chemical synthesis or commercial availability (19–24, Figure 2E) 13, 19, 33–36. Finally, the anionic dye bis-ANS (25) and two quaternary alkaloids (26, 27) do not structurally resemble most of the other compounds and are relegated to their own “classes” (Figure 2F,G)37–39. The consistent traits of all of the molecules in Figure 2 is that most have not been the subject of additional studies since their discovery and almost none have been compared directly in biochemical or cell-based assays.
Figure 2.
Small molecule inhibitors of FtsZ divided by structural class. A) cationic dyes, B) nucleoside and nucleotide analogs, C) “drug like” heterocycles, D) phenolic natural products, E) miscellaneous high-throughput screening hits, F) anionic dye, G) quaternary alkaloid natural products. [sized for two-column formatting]
Central to the problem of studying FtsZ with small molecule inhibitors is cross-species reactivity, i.e. the extent to which an inhibitor reported for protein from one species of bacteria might affect the activity of protein from another species. Most inhibitors reported to date have documented activity for protein from one species of bacterium, usually E. coli or B. subtilis. A small number of experiments are reported for S. aureus and M. tuberculosis FtsZ, the latter of which is known to be a much slower enzyme and, as a result, generally requires higher concentrations of inhibitor and protein. Confounding the issue further is the multitude of assays for the function of FtsZ. The most consistent assay is for GTP hydrolysis, which is usually measured either by an enzyme-coupled assay that consumes NADH or by the detection of free phosphate with malachite green. The latter assay has largely replaced the use of radiolabeled GTP in studies of FtsZ. Displacement of GTP analogs, including the fluorescent mant-GTP40 and GTP-γ-S41, has also been reported. Finally, the interpretation of biochemical studies is complicated by differences in buffers and additives that prevent comparison between results from different labs. Another problem is the complex coupling between GTPase activity and filament formation. The direct impact of small molecules on polymerization can be examined with electron microscopy, light scattering or centrifugation to separate free FtsZ from polymers, or so-called “protofilaments.” A highly selective inhibitor would be expected to phenocopy the temperature sensitive mutants of FtsZ from which many of the genes controlling cell division derive their name (fts, filamentous temperature sensitive). That said, assessing the chemically induced phenotype is complicated by the many other mechanisms by which filamentation might be induced, including: 1) induction of the SOS response by genotoxic compounds, 2) inhibition of one or more of the other cell division proteins and 3) decoupling of the Z-ring from the cell wall, which can be induced by several different classes of small molecules42. Furthermore, the inability of bacteria to function without FtsZ prohibits the examination of mutants that lack this protein, which is a standard control experiment in chemical biology by which one can assign the target of a small molecule. Collectively, these caveats greatly limit the extent to which induction of filamentation can be used as evidence for selective chemical inhibition of FtsZ or any other cell division protein.
We set out to discover new and better inhibitors of FtsZ with the long-term goal of ascertaining the molecular interactions by which these compounds affect FtsZ using NMR or X-ray crystallography. As a starting point, we needed to be able to compare and contrast any new compounds with known inhibitors that we assumed would help establish structure-activity relationships (SAR) and enable the rational design of compounds to synthesize. Among the compounds in Figure 2, we opted not to study compounds with known liabilities associated with low activity or promiscuity and instead focused on a subset of nine representative compounds with the potential for further refinement. Close examination of the activity of these compounds has revealed some important conclusions that will impact future studies of small molecule inhibitors of FtsZ. First, there is compelling evidence that many inhibitors derive their activity from the formation of aggregates, as described by Shoichet.43 Second, several reported inhibitors do not exhibit the reported levels of inhibition when examined under standardized conditions. Third, zantrin Z3 (9) was found to be a robust and reliable inhibitor of FtsZ from two different species (E. coli and B. subtilis) and it does not appear to form aggregates that result in unselective inhibition. Synthetic analogs of 9 exhibit informative SAR, with one in particular exhibiting enhanced biochemical inhibition of FtsZ. These data cement this compound as an effective reagent to use as a positive control and as a starting point for our search for better inhibitors. Herein we describe all of the experiments that led to these conclusions and, hopefully, establish consistent protocols for assaying future inhibitors so that useful comparisons across FtsZ from various organisms are straightforward.
RESULTS AND DISCUSSION
Identification of FtsZ inhibitors that likely form aggregates
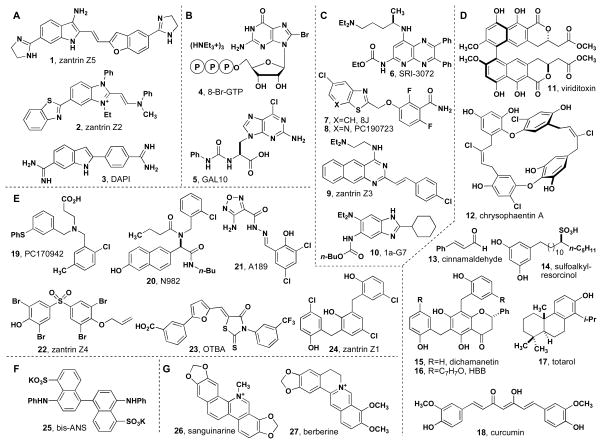
Small molecules discovered in high-throughput screens sometimes exhibit enzyme inhibitory activity that results from the formation of aggregates or colloids.44, 45 Shoichet has documented many cases of this effect46–48 and established experiments for assessing the presence of aggregates.43, 49 We have been interested in the activity of totarol, which has a long history of antimicrobial activity50, 51 and was recently shown to inhibit FtsZ.28 Using the enzyme-coupled GTPase assay, we were able to reproduce the IC50 of totarol (Figure 3). Oddly, several close analogs of totarol showed absolutely no inhibition of FtsZ in the same assay (data not shown). We suspected that this dichotomy could be due to the formation of aggregates and repeated the assay in the presence of 0.01% Triton X-100, a neutral detergent shown to disrupt aggregate formation in enzymatic assays. In the presence of Triton X-100, totarol’s activity vanished. Similar behavior was observed when a structurally unrelated eukaryotic GTPase was examined. Dnm1, which mediates mitochondrial division,52, 53 showed significant reduction in GTPase activity in the presence of micromolar concentrations of totarol and the activity was completely restored by the addition of 0.01% Triton X-100 (see supporting information). Similar reduction in FtsZ’s activity was observed for 4′-hydroxydichamanetin (4HD) and zantrin Z1 each of which exhibited the published activity in the absence of Triton X-100. In our control experiments, i.e. those in which no inhibitor is added, GTPase activity remained constant in the presence or absence of Triton X-100, indicating that the protein largely unaffected by this low concentration of detergent. Dichamanetin was substituted with 4HD, which is prepared in a single synthetic step from commercially available materials and behaves identically to dichamanetin (See supporting information).
Figure 3.
Inhibition of GTPase activity of FtsZ by totarol (17), zantrin Z1 (24), and 4′-hydroxydichamanetin (15b) in the absence and presence of 0.01% Triton X-100. A) BsFtsZ, B) EcFtsZ.
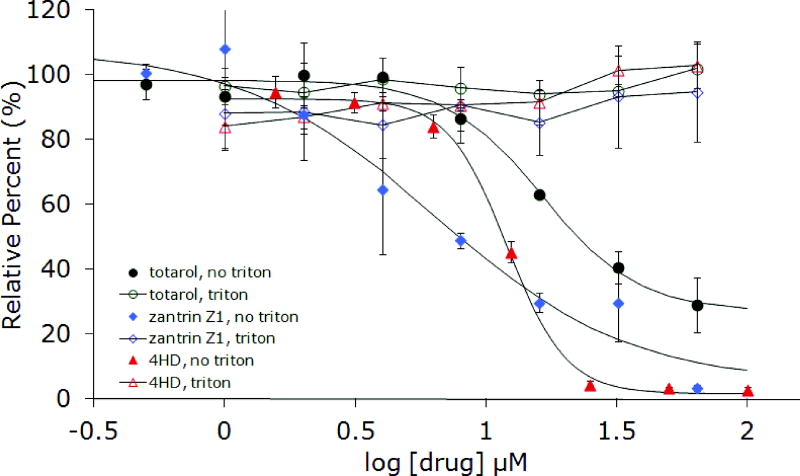
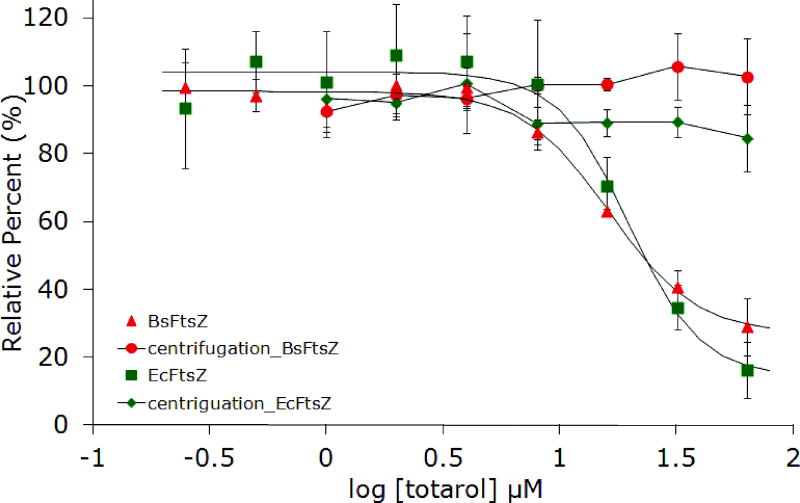
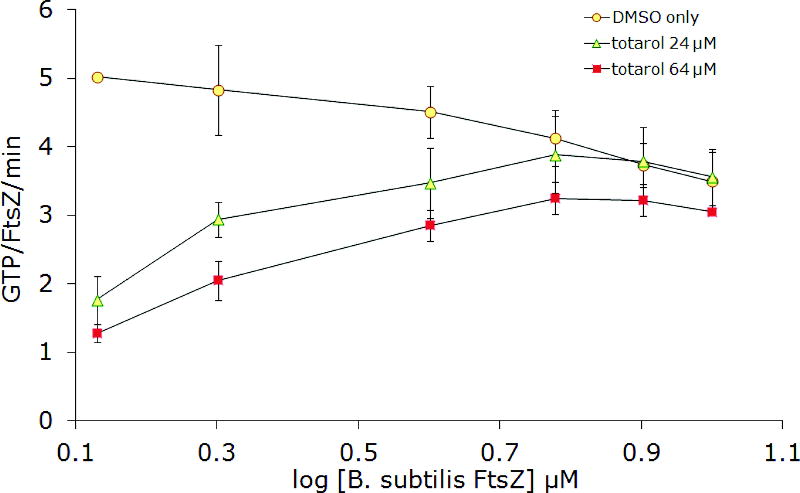
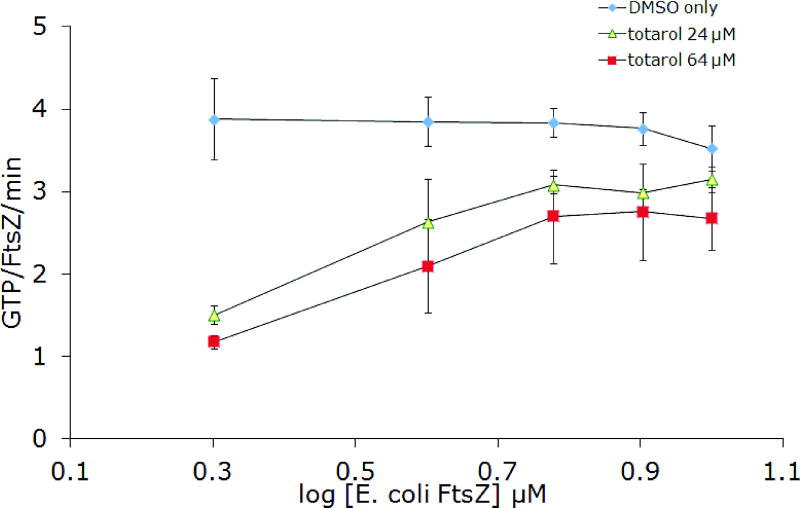
In order to confirm aggregate formation as the underlying mechanism for inhibition of FtsZ by totarol, several additional experiments were conducted. Stock solutions of totarol in buffer were centrifuged before adding aliquots to the assay wells (Figure 4). Again, no activity was observed, consistent with the ability of small molecule colloids to be removed by centrifugation. Next, we varied the concentration of protein and noticed that as the concentration increased, the inhibitory activity of totarol decreased nearly to the point of having no effect well above the established IC50 (Figure 5). This result is consistent with the formation of particles that are becoming saturated with protein as opposed to soluble small molecules that are still present in gross molar excess relative to the protein. Collectively, these results support the conclusion that totarol’s inhibition of FtsZ is an artifact of aggregate formation. Although the additional experiments were not performed on dichamanetin and zantrin Z1, the Triton experiment provides strong evidence in favor of aggregate formation and we conclude that all future reports of FtsZ inhibition by small molecules should include this important control experiment. Based on the structural appearance and similar bulk properties, zantrin Z4 (22) and sulfoalkylresorcinol 14 should be viewed as aggregation suspects until proven otherwise.
Figure 4.
Inhibition of EcFtsZ and BsFtsZ GTPase activity by totarol before and after centrifugation of the drug in assay buffer.
Figure 5.
Inhibition of FtsZ GTPase activity by 24 μM and 64 μM totarol at increasing FtsZ concentrations. A) BsFtsZ, B) EcFtsZ. The slight downward trend for DMSO only can be explained by increased bundling of FtsZ protofilaments at the higher concentrations, which can lead to slower nucleotide exchange73.
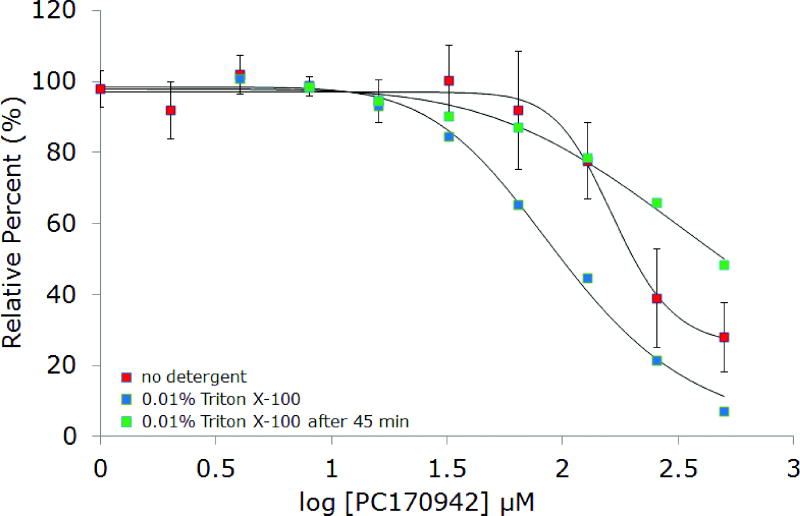
Screening compound PC170942, which is structurally unrelated to PC190723 (see Figure 2), exhibits unusual behavior consistent with some level of aggregation. Under standard conditions, an IC50 of approximately 160 μM is observed, which is 3-fold higher than originally reported (Figure 6). In the presence of 0.01% Triton X-100, inhibition is similar and, oddly, the IC50 is slightly lower. The enzyme-coupled assay was used for this experiment and it was noted that inhibition seemed to consistently decrease over time when examining the level of enzymatic activity at longer time points. We hypothesized that this effect was due to slow de-aggregation of PC170942 by Triton X-100. Aliquots of the compound were treated with the detergent and stored for 45 minutes before repeating the dose response. The IC50 was shifted significantly higher, suggesting that the primary mode of inhibition was by the formation of small molecule aggregates. This result suggests that in some cases, longer incubation times for the Triton X-100 control experiment might be necessary to completely rule out unselective inhibition by aggregates.
Figure 6.
Inhibition of BsFtsZ activity by PC170942 in the absence of Triton X-100, with 0.01% Triton X-100, and after 45 minutes incubation with 0.01% Triton X-100.
Assessment of benzamides (8J, PC190723), viriditoxin, SRI-3072, and zantrin Z3
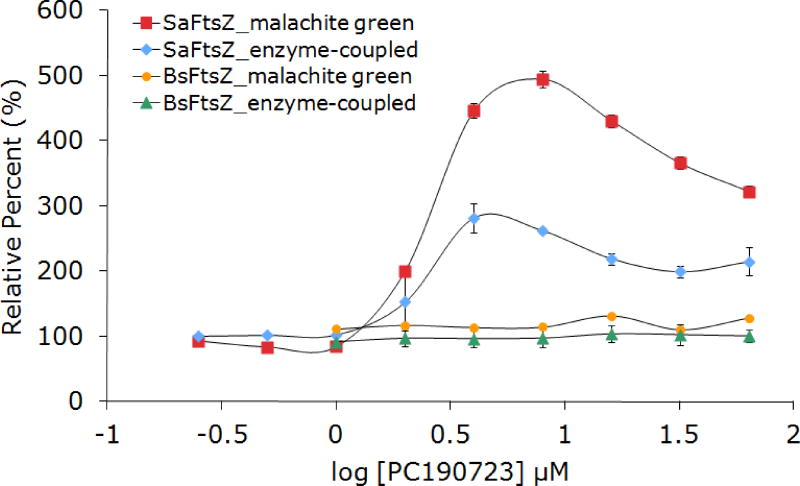
The search for new small molecule inhibitors relies on the ability to make comparisons to positive controls, i.e. compounds that are known to inhibit FtsZ to a certain degree under an established set of conditions. We initially looked to PC190723, given that this is the compound with the lowest reported IC50 and allegedly the best known inhibitor of FtsZ GTPase activity. We reported a practical synthesis of this compound in order to ensure a steady supply for our laboratory and others with whom we have shared this reagent54. Although the reported activity is for S. aureus FtsZ, the close analog 8J (7) was shown to also inhibit BsFtsZ.55 We observed no inhibition of B. subtilis FtsZ by PC190723 at concentrations up to 100 μM using either the enzyme-coupled or malachite green assays (Figure 7). The latter result conflicts with the observations of Andreu and co-workers who observed widely variant activity, including dose-dependent increase or decrease of enzymatic activity, while studying the biochemical effects of PC190723 on BsFtsZ and EcFtsZ56. As a point of direct comparison to the original work of Haydon et al, the activity of PC190723 was examined for S. aureus FtsZ (SaFtsZ) and still no inhibition was observed. In fact, a dose-dependent and reproducible increase in activity for SaFtsZ was observed (Figure 7). In this case, the basal activity of the enzyme is significantly lower (~0.5 GTP/FtsZ/min) than we observed for EcFtsZ or BsFtsZ. The original report includes only the relative change in activity (as a percentage of control) without noting the actual enzymatic turnover in the absence of inhibitor. The activity of BsFtsZ and EcFtsZ can also vary greatly depending on conditions (vide infra), making these data important when assessing inhibition of the enzymatic function. The lack of reproducibility of PC190723’s inhibition of SaFtsZ has been confirmed in at least one other laboratory to whom we have provided this compound (data not shown). Very recently (i.e. during the review of this manuscript), a detailed study of the mechanism of action of PC190723 by scientists at Merck documents this compound as an activator of the GTPase activity of SaFtsZ, consistent with our results.57 We did observe antimicrobial activity against B. subtilis strain 168 that closely matched the original report (MIC 1 μM).20 Although we currently have no explanation for these results, it can be safely concluded that PC190723 cannot serve as a positive control for in vitro experiments involving FtsZ from S. aureus or any other organism tested to date.
Figure 7.
Effect of PC190723 on SaFtsZ and BsFtsZ as observed by enzyme-coupled and malachite green assays for GTPase activity. Basal FtsZ GTPase activity (no inhibitor) was 0.5 FtsZ/GTP/min for SaFtsZ and 4.4 FtsZ/GTP/min for BsFtsZ.
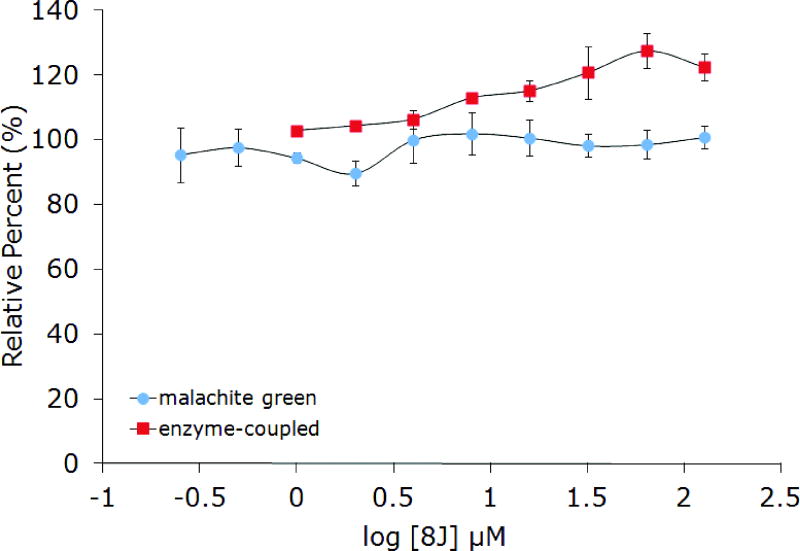
Compound 8J was also examined as a point of direct comparison. This compound differs from PC190723 by a single atom substitution (carbon for nitrogen on the heterocyclic appendage) involving replacement of a pyridine ring in the side chain with a benzene ring. As a followup to the studies of PC190723, this compound was examined in B. subtilis and inhibition of FtsZ from that organism was reported55, albeit without documenting the specific activity of the enzyme. We observed no inhibitory activity using the enzyme-coupled assay with up to 128 μM of 8J (Figure 8). Direct comparison was possible by switching to the malachite green assay. During the optimization of this assay for this protein, a variety of different responses were observed. On one occasion there was a dose-dependent reduction in activity, albeit with similarly large error bars when compared to the published data, shallow sloping of the dose response curve, and only 50% reduction in enzymatic activity (data not shown). In our case, this was apparently due to incomplete quenching of the reactions and unequal times on ice before malachite green reagent addition. The most consistently observed behavior for 8J was a slight dose-dependent increase in enzymatic activity. This trend was observed reproducibly with either the malachite green or enzyme-coupled assays, with slight variation between the two for the magnitude of the increase. Compound 8J exhibited better antimicrobial activity than PC190723 against B. subtilis strain 168 (MIC 0.25 μM), consistent with the reported value for S. aureus.22
Figure 8.
Effect of 8J (7) on the GTPase activity of BsFtsZ using the enzyme-coupled and malachite green assays.
Viriditoxin is one of the most potent inhibitors from a natural source reported to date with an IC50 of 10 μM. Our lab had previously completed a synthesis of this compound and confirmed the identity by isolating this metabolite from a culture of Aspergillus viridinutans58. Under standard conditions, no inhibition of FtsZ was observed up to a concentration of 200 μM for either B. subtilis or E. coli FtsZ. Careful inspection of the report from Merck revealed unique buffer conditions that included only 40 μM GTP, 0.75 mM Mg(OAc)2 and 1 μM FtsZ at a pH of 7.4. In addition, the authors included 2.5 mM CaCl2 and 50 μg/mL DEAE-Dextran, which favor extensive FtsZ polymerization in order to facilitate rapid separation of polymeric FtsZ from monomers and short oligomers in a filter binding assay59. Based on the premise that the activity of viriditoxin could only be observed under these unusual conditions, we attempted to replicate the assay that is documented in the original report and still observed no inhibition at concentrations up to 128 μM. In this second experiment, the baseline enzymatic activity (i.e. in the absence of inhibitor) was reduced by two orders of magnitude (from 5 FtsZ/GTP/min down to 0.05, see supporting information), which greatly decreases the signal-to-noise of any biochemical experiment under these conditions. We cannot rule out the possibility that viriditoxin may inhibit the GTPase activity of FtsZ or otherwise interfere with the function of this protein under a very specific and not completely documented set of conditions. Furthermore, we have noted that viriditoxin is chemically unstable and degrades if it is not stored free of solvent and at low temperature. The toxicity of this compound prevents further development of this compound for drug discovery and our data indicates that it is also not useful as a biochemical probe for SAR studies.
SRI-3072 was reported in a study that focused specifically on inhibitors of M. tuberculosis FtsZ (MtbFtsZ). The GTPase activity of this protein is significantly lower than that of E. coli or B. subitilis, which is probably related, on some level, to the slow division of this organism. Recent SAR studies on this compound identified a few analogs that have lower IC50 values for FtsZ inhibition, but in all cases the MICs were higher for two strains of M. tuberculosis and in one case a modest level of tubulin inhibition was observed. The effects of these compounds on FtsZ were mostly studied by light scattering and in one case, the GTPase activity was shown to decrease by 20% at 100 μM. Given the 5-10-fold higher concentrations at which the zantrins inhibit MtbFtsZ relative to EcFtsZ, it was possible that significant inhibition of the latter protein might be possible at a much lower concentration. This compound exhibited no inhibition of E. coli or B. subtilis FtsZ up to 128 μM (See supporting information).
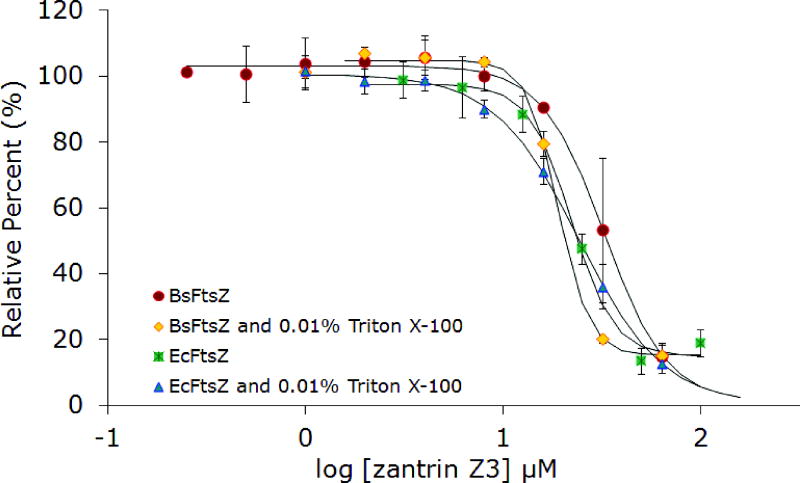
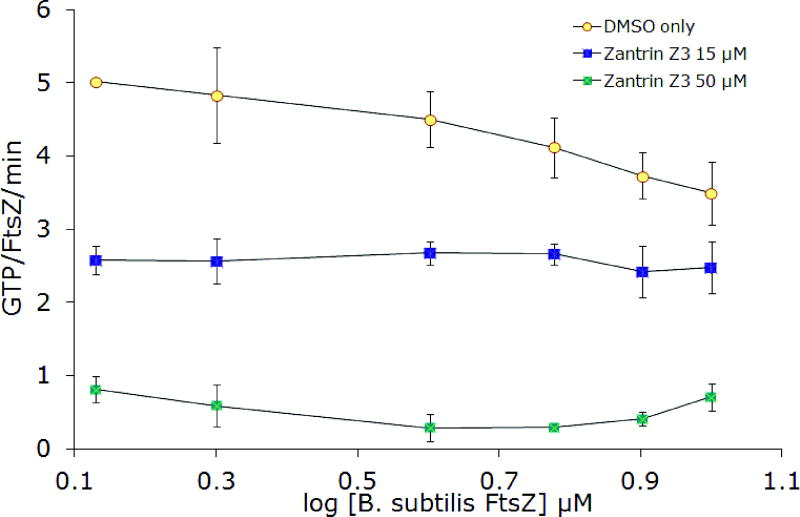
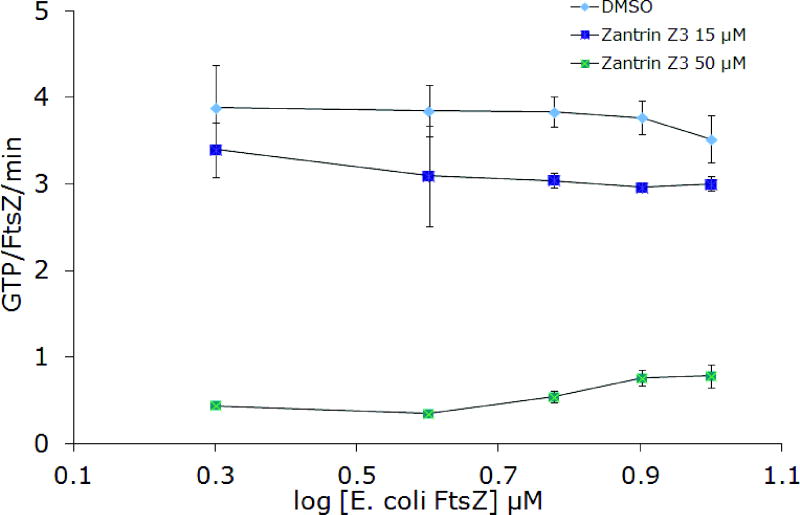
Zantrin Z3, along with the other zantrins, was discovered in a high-throughput screen for inhibitors of the GTPase activity of FtsZ. In spite of the shared name for these compounds, their chemical structures are largely unrelated except in the cases of zantrins Z3 and Z5, which are both structurally similar to DAPI (Figure 2). Zantrin Z3 stands out for having a structure that is drug-like, i.e. it has no offending electrophilic or phenolic functionality and has a passing resemblance to the antimalarial drug chloroquine. Inhibition of EcFtsZ was observed and the IC50 value (20 μM) compares favorably with the reported value (Figure 9). Zantrin Z3 also effectively inhibited the function of BsFtsZ with an IC50 of 32 μM. Inhibition was maintained in the presence of 0.01% Triton X-100 (Figure 9) and at increasing concentrations of protein (Figure 10). Collectively, these experiments suggest that the activity is probably not an artifact of aggregate formation. Of all the compounds examined in this study, zantrin Z3 is the best in terms of overall performance against protein from multiple species of bacteria under a variety of conditions.
Figure 9.
Inhibition of GTPase activity of BsFtsZ and EcFtsZ by zantrin Z3 in the absence and presence of 0.01% Triton X-100.
Figure 10.
Inhibition of FtsZ GTPase activity by 15 μM and 50 μM zantrin Z3 at increasing FtsZ concentrations. A) BsFtsZ, B) EcFtsZ.
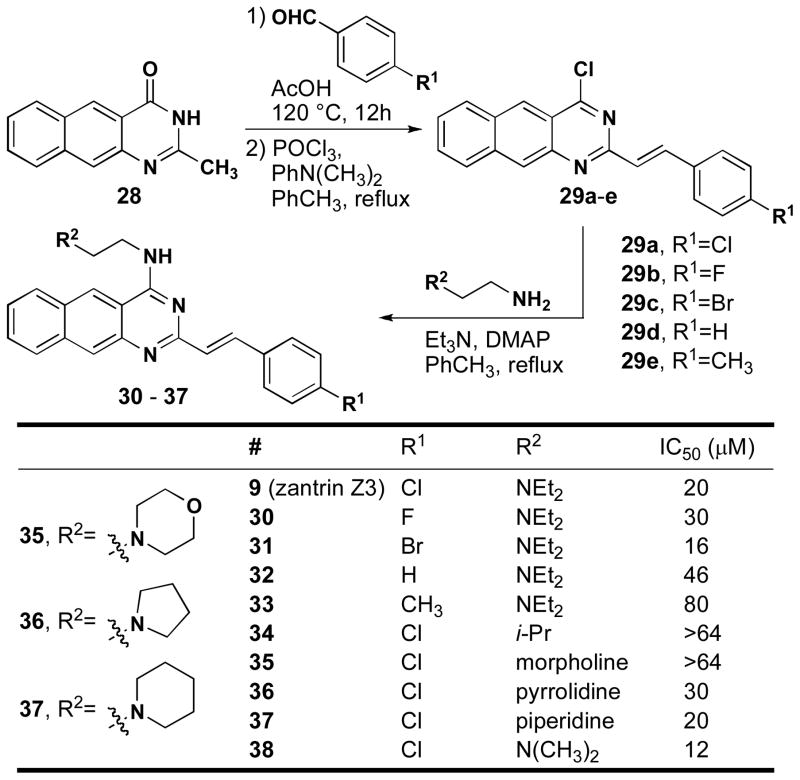
We have developed a modular synthesis of zantrin Z3 and conducted preliminary structure-activity relationship (SAR) studies (Figure 11). Although an exact synthesis for zantrin Z3 has not been published, the route employed was adapted from Okano and co-workers60. Naphthopyrimidinone 28 is made in a single step from commercially available 3-amino-2-naphthoic acid. This compound can be carried through an aldol-like condensation with a variety of benzaldehydes to provide compounds 29a–e in modest to good yields. These intermediates were converted to their corresponding chlorides and then treated with various N,N-dialkylaminoethylamines. Variation of the styryl sidechain revealed that the para-substituent significantly influenced the activity and that halogens (F, Cl & Br) were all superior to H or methyl. Replacement of the aminoethyl sidechain with a less basic (35) or non-basic (34) abolished the inhibition observed with the other side chains. Finally, replacement of the diethylamino group with a dimethyl amino group resulted in the best inhibitor of the series (38, 12 μM). This compound is currently being used for co-crystallization with BsFtsZ and EcFtsZ as well as isolation of resistant strains of E. coli and B. subtilis. Current efforts are directed at replacing the styryl side chain with a different linker that will enhance the inhibitory activity.
Figure 11.
Synthesis of zantrin Z3 analogs and IC50s for inhibition of EcFtsZ.
Implications for Future Studies of FtsZ Inhibition
Zantrin Z3 and the related compounds with similar activity (31 and 37) are uniquely active and selective in their inhibition of FtsZ when compared to the other molecules examined in this study or described previously. Many of the compounds in Figure 2 were not examined based on established liabilities. One of the first inhibitors was bis-ANS, a sulfonate dye molecule37 (25, Figure 2), is known to occupy nucleotide binding sites on several proteins, including myosin61 and tubulin62. DAPI (3), a dye used for staining double-stranded DNA, has been reported as an inhibitor of FtsZ12 and the structural similarity to two other zantrins (Z2/5 and Z5/1)13 suggested these two could be similarly promiscuous. Two nucleotide analogs have been documented as inhibitors of FtsZ. The first, 8-methoxy GTP (4), inhibits the GTPase activity of FtsZ at 15 μM and a crystal structure of a related compound in the series (8-morpholino-GTP, not shown) documents binding of this compound in the active site of FtsZ8. Although all of the GTP analogs discussed in this study showed some level of inhibition of FtsZ GTP hydrolysis, the compounds were roughly split between analogs that promote or inhibit tubulin assembly. Finally, although they were not examined in this study, the extremely high IC50 values for GAL10 (5, 450 μM)15 and A189 (21, 260 μM)33 test the limits of solubility in aqueous media.
Some FtsZ inhibitors are not specific and modulate the function of other proteins. OTBA (23)34 and a recently disclosed analog (not shown)35 share a common core (rhodanine) that is included in a list of pan-assay interference compounds (PAINS) due to the promiscuity with which certain recurrent compounds appear as hits in high-throughput screening experiments63. The fact that OTBA has an IC50 for FtsZ of approximately 20 μM and is cytotoxic to mammalian cells at 8 μM is consistent with PAIN behavior. Although zantrin Z3 may have additional targets, the substructure of this molecule is not among the PAINS.
Natural product inhibitors of FtsZ all carry significant liabilities to further development as selective probes, which is surprising given the natural origin of the best tubulin inhibitors (e.g. taxol, colchicine, vinblastine). Curcumin (18)32 and cinnamaldehyde (13)29 have both been described as FtsZ inhibitors, but also as inhibitors of many other proteins and pathways, including tubulin in the case of curcumin64. Dichamanetin (15)65 and HBB (16)66 are antimicrobial natural products whose close structural similarity to zantrin Z1 (24) suggested that they also inhibited FtsZ and their activities were reported to be 12.5 and 8.3 μM, respectively31. Our studies demonstrate that these compounds, along with totarol, are all probably inhibiting FtsZ by forming aggregates and call into question the origin of the inhibitory activity of sulfoalkylresorcinol derivative 14 and the recently-isolated chrysophaentin A27. Although viriditoxin (11) was found in a high-throughput screen for compounds that disrupted polymerization of FtsZ tagged with a fluorescent reporter26, this activity is not easily replicated using more standard conditions. Two cationic alkaloid natural products, berberine (27)38 and sanguinarine (26)39, also inhibit the function of FtsZ. The latter compound is also known to inhibit tubulin assembly whereas the former does not do so, even at high concentration (100 μM)67, 68. Finally, a series of taxol derivatives (not shown) was designed and synthesized on the premise that the similar structures between FtsZ and tubulin would result in similar structures of inhibitors69. These compounds are described as FtsZ inhibitors solely on the filamented phenotype that is observed by bacteria treated with the compounds. Given the large number of mechanisms that can result in filamentation, conclusions regarding a single protein target based solely on this (or any) phenotype should be made cautiously. All told, Mother Nature has been far less generous with selective and potent inhibitors of FtsZ when compared to tubulin.
The activity of PC190723, the only sub-micromolar inhibitor if FtsZ GTPase activity reported to date, is the most difficult to interpret in light of the subsequent studies of this molecule and its close analog 8J. The original publication does not include a detailed protocol for the assay. Although one was eventually provided by one of the authors (N. Stokes) the specific activity of the SaFtsZ was not recorded, thus making it difficult to directly compare our results with theirs. Subsequent reports from Andreu56 and Errington55 on PC190723 and 8J, respectively, using EcFtsZ and/or BsFtsZ do not contain any obvious explanation for the discrepancy in reported and observed inhibition of SaFtsZ. In our studies, the activity of SaFtsZ is quite low, making any interpretation of its biochemical behavior more error-prone than analogous studies with EcFtsZ or BsFtsZ. However, an interaction between PC190723 with SaFtsZ is clearly indicated by the recent publication of the co-crystal structure55, which is the only structure of FtsZ published to date in a complex with a non-nucleotide small molecule. Moreover, this structure confirms the binding region predicted by the original resistant mutant studies. We have indeed observed antimicrobial activity of PC190723 against B. subtilis that was very similar to that originally reported. The conclusion from all of these data is that the behavior of FtsZ in vitro can be highly erratic and assay-dependent, even in cases where all other indicators point to strong interaction that disrupts cell division.
Zantrin Z3’s biochemical inhibition of FtsZ makes it an important probe for future investigations of the bacterial cell division machinery. Demonstration that this molecule does not inhibit FtsZ through an unselective aggregation mechanism is an important first step in understanding how inhibition of the GTPase activity of FtsZ impacts bacterial cell division. Ideally, a compound with significantly better inhibition will be identified in future SAR studies for both cell-based studies and crystallography. The activity against EcFtsZ, BsFtsZ and MtFtsZ13 make this a useful compound for studying all of these organisms, on which the vast majority of studies to date are based. Although FtsZ still lacks the potent and selective small molecule tools available for studying tubulin, these studies demonstrate a clear path forward for discovery of new FtsZ inhibitors.
METHODS
Protein Expression and Purification
E. coli FtsZ in pET11b was the kind gift of Harold Erickson (Duke University). B. subtilis FtsZ was cloned from B. subtilis strain 168 by colony PCR using primers 5′-AATTAACATATGTTGGAGTTCGAAACAAACATAG-3′ and 5′-ATAGGATCCTTAGCCGCGTTTATTACGGTTTC-3′. The FtsZ PCR fragment was purified, digested with NdeI and BamHI, then ligated into NdeI/BamHI-digested pET11b plasmid. The final product was verified by sequencing. S. aureus FtsZ in pET11a was the kind gift of Carole Bewley (NIH/NIDDK).
C41(DE3) cells (Lucigen) were used as the host strain for protein expression. Cells were grown at 37 °C in LB with 100 μg mL-1 ampicillin to an OD(600 nm) of approximately 0.6, then induced with 0.5 mM IPTG for 3 hours at 37 °C, with the exception of S. aureus FtsZ, which was induced at 30 °C for 5 hours. Cells were pelleted at 6000 rpm in a FIBERLite F9-4x1000y rotor (Thermo Scientific) and either frozen overnight before resuspension or directly resuspended in 50 mM Tris, 1 mM EDTA, 50 mM KCl, 10%(v/v) glycerol, pH 7.9 (Buffer A). Resuspended cells were treated with lysozyme (Sigma-Aldrich) for 30 min–1 h on ice, then lysed by probe sonication for 1.5 minutes on ice (5 minutes with 30% duty cycle). The lysate was clarified by centrifugation at 35,000 rpm in a Sorvall T-1250 rotor for 1 h at 4 °C and the resulting supernatant brought to 35%-saturated (Ec, Bs) or 50%-saturated (Sa) ammonium sulfate. After incubation at 0 °C for at least 30 min with periodic mixing, precipitated protein was collected by centrifugation at 20,000 rpm in a Sorvall SS-34 rotor for 20 min at 4 °C. Ammonium sulfate pellets were resuspended in Buffer A lacking KCl and dialyzed against the same buffer before loading onto a HiPrep Q Sepharose FastFlow anion exchange column (GE Healthcare Life Sciences). Fractions were eluted with a 0–800 mM KCl gradient and analyzed by SDS-PAGE. Peak FtsZ fractions were aliquotted and stored at −80 °C. Prior to enzymatic assays, FtsZ protein was dialyzed into the appropriate assay buffer and the concentration determined using the BCA protein assay (Thermo Fisher). The calculated concentration of FtsZ was divided by 0.75 (~1.33-fold increase in estimated concentration) due to the lower color development of FtsZ relative to BSA in the assay70. Specific activity ranged from 4–6 GTP/FtsZ/min for BsFtsZ and from 3–5 GTP/FtsZ/min for EcFtsZ. The specific activity for SaFtsZ was more variable and generally lower, ranging from 0.1 to 1 GTP/FtsZ/min.
Small molecule inhibitors
Small molecule inhibitors were either purchased or synthesized as described in the supporting information. Zantrin Z1 (24) was purchased from Ryan Scientific. Although zantrin Z3 (9) was initially purchased from Ryan Scientific, it was later discontinued and was synthesized by the route developed for compounds 30–38 (see supporting information). 4HD (15b) was prepared in analogy to dichamanetin31 using naringenin as a starting material. Compound 8j (7) was prepared in analogy to PC190723 (see supporting information). SRI-3072 was kindly provided by Dr. Robert Reynolds (Southern Research Institute). The synthesis of PC170942 (19) was not reported in the literature and the route developed for this work is described in supporting information. Viriditoxin (12) and PC190723 (8) were prepared by previously described routes54, 58, 71.
Enzyme-Coupled GTPase Assay
This assay was calibrated and performed generally as described72. Specifically, a 50x concentrate of the inhibitor in DMSO (4 μL) was mixed into the appropriate volume of assay buffer (50 mM MES buffer, pH 6.5, 5 mM Mg(OAc)2, and 100 mM KOAc), then FtsZ from E. coli or B. subtilis (2 μM) or S. aureus (10 μM) was added and incubated at 37 °C for 5 min. Next, 40 μL of a 5x “master mix” of coupling enzymes and substrates (containing 100 U mL-1 each of lactate dehydrogenase/pyruvate kinase, 2.5 mM phosphoenolpyruvate and 2.0 mM NADH, all from Sigma-Aldrich) was added. GTP hydrolysis was initiated with 1 mM GTP (2 μL) to give a final volume of 200 μL. The solution was thoroughly mixed and 150 μL of each reaction was pipetted into a microplate well. Absorbance readings were taken over time at room temperature (~25 °C) on a Biotek EL808 microplate reader at 340 nm.
Malachite Green GTPase Assay
This assay was performed approximately as described13 without the addition of sodium citrate due to the high amounts of phosphates known to contaminate many sources of citrate. FtsZ from E. coli or B. subtilis (2 μM) or S. aureus (10 μM) was mixed with a 50x concentrate of the inhibitor (4 μL) in 50 mM MES buffer, pH 6.5, 5 mM Mg(OAc)2, and 100 mM KOAc to a final volume of 198 μL and incubated for 5 min. GTP hydrolysis was initiated with 1 mM GTP (2 μL) to give a final volume of 200 μL. At appropriate time points after incubation at 30 °C, 20 μL of reaction mixture was transferred to a tube containing 5 μL of 100 mM EDTA on ice. 20 μL of a series of phosphate standards were also transferred into EDTA on ice. 600 μL of freshly prepared malachite green working reagent [2 volumes malachite green (0.8 mg mL-1)/1 volume polyvinyl alcohol (23.2 mg mL-1)/1 volume ammonium molybdate (57.2 mg mL-1 in 2H2O:3HCl)/2 volumes H2O, allowed to sit for 30 mins] was then added to each tube, mixed and incubated for 30 mins at room temperature. Finally, 200 μL of each sample was pipetted into a microplate well and absorbance readings were taken on a Biotek EL808 microplate reader at 630 nm.
Approximate recapitulation of Merck viriditoxin conditions
To repeat as closely as possible the reported GTPase inhibition by viriditoxin26, without using a radioactive assay as reported, we used the malachite green assay described above under the following reaction conditions: 50 mM Tris, 34 mM KCl, 0.75 mM Mg(OAc)2, 2.5 mM CaCl2, 50 μg mL-1 DEAE-Dextran (Sigma D9885) at 37 °C.
Curve fitting of IC50 data
Inhibition data was fit to a logistic equation, y = A2 + (A1-A2)/(1+(x/x0)p), by the method of least squares, using the Solver function of Microsoft Excel and allowing all parameters (A1, A2, x0, p) to vary. Occasionally, the lower assymtote/maximally inhibited value was fixed at 0 if the first solution gave a negative number. Several iterations were performed to confirm convergence.
Aggregation tests
When testing for compound aggregation43 in the GTPase assay, the following conditions were used: for detergent addition, 0.01%(v/v) Triton X-100 (from a freshly prepared 1%(v/v) stock) was added at the beginning along with buffer and compound stock; for centrifugation, the buffer/compound mixture was centrifuged at 14,100 × g for 20 minutes before addition of protein; for increasing enzyme concentration, the amount of FtsZ was varied as noted in the figure.
Supplementary Material
Acknowledgments
This work was supported by the NIH/NIAID (R01AI08093), National Institutes of Health/ National Eye Institute (EY012347 to JBA), and by NIH/NIGMS (T32-GM008799), which provided JTM with predoctoral fellowship support. MBK and TEO acknowledge support in the form of GAANN fellowships. TEO and JTM acknowledge support from Bradford Borge fellowships from UC Davis. NAS acknowledges fellowship support from Bristol-Meyers Squibb and from Proctor and Gamble through the National GEM consortium. NAS, TEO and CIG were also supported by fellowships from the Alfred P. Sloan Foundation. C. Bewley (NIH/NIDDK) is acknowledged for helpful discussions, conducting confirmatory experiments with PC190723, and for providing the plasmid for SaFtsZ. H. Erickson (Duke University) is acknowledged for providing the plasmid for EcFtsZ. R. Reynolds (Southern Research Institute) is acknowledged for providing a sample of SRI-3072 (6).
Footnotes
Supporting Information Available: Additional details for biochemical experiments, syntheses of 8J, PC190723, PC170942, zantrin Z3 and compounds 30-38, XXX This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.RayChaudhuri D, Park JT. Escherichia coli cell-division gene ftsZ encodes a novel GTP-binding protein. Nature (London, UK) 1992;359:251–254. doi: 10.1038/359251a0. [DOI] [PubMed] [Google Scholar]
- 2.De Boer P, Crossley R, Rothfield L. The essential bacterial cell-division protein FtsZ is a GTPase. Nature (London, UK) 1992;359:254–256. doi: 10.1038/359254a0. [DOI] [PubMed] [Google Scholar]
- 3.Mukherjee A, Dai K, Lutkenhaus J. Escherichia coli cell division protein FtsZ is a guanine nucleotide binding protein. Proc Natl Acad Sci USA. 1993;90:1053–1057. doi: 10.1073/pnas.90.3.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Erickson HP, Anderson DE, Osawa M. FtsZ in bacterial cytokinesis: cytoskeleton and force generator all in one. Microbiology and molecular biology reviews: MMBR. 2010;74:504–528. doi: 10.1128/MMBR.00021-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Löwe J, Li H, Downing KH, Nogales E. Refined structure of alpha beta-tubulin at 3.5 Å resolution. J Mol Biol. 2001;313:1045–1057. doi: 10.1006/jmbi.2001.5077. [DOI] [PubMed] [Google Scholar]
- 6.Ravelli RB, Gigant B, Curmi PA, Jourdain I, Lachkar S, Sobel A, Knossow M. Insight into tubulin regulation from a complex with colchicine and a stathmin-like domain. Nature. 2004;428:198–202. doi: 10.1038/nature02393. [DOI] [PubMed] [Google Scholar]
- 7.Gigant B, Wang C, Ravelli RBG, Roussi F, Steinmetz MO, Curmi PA, Sobel A, Knossow M. Structural basis for the regulation of tubulin by vinblastine. Nature. 2005;435:519–522. doi: 10.1038/nature03566. [DOI] [PubMed] [Google Scholar]
- 8.Läppchen T, Pinas VA, Hartog AF, Koomen GJ, Schaffner-Barbero C, Andreu JM, Trambaiolo D, Löwe J, Juhem A, Popov AV, den Blaauwen T. Probing FtsZ and tubulin with C8-substituted GTP analogs reveals differences in their nucleotide binding sites. Chem Biol. 2008;15:189–199. doi: 10.1016/j.chembiol.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 9.Tan CM, Therien AG, Lu J, Lee SH, Caron A, Gill CJ, Lebeau-Jacob C, Benton-Perdomo L, Monteiro JM, Pereira PM, Elsen NL, Wu J, Deschamps K, Petcu M, Wong S, Daigneault E, Kramer S, Liang L, Maxwell E, Claveau D, Vaillancourt J, Skorey K, Tam J, Wang H, Meredith TC, Sillaots S, Wang-Jarantow L, Ramtohul Y, Langlois E, Landry F, Reid JC, Parthasarathy G, Sharma S, Baryshnikova A, Lumb KJ, Pinho MG, Soisson SM, Roemer T. Restoring Methicillin-Resistant Staphylococcus aureus Susceptibility to β-Lactam Antibiotics. Sci Transl Med. 2012;4:126ra135. doi: 10.1126/scitranslmed.3003592. [DOI] [PubMed] [Google Scholar]
- 10.Foss MH, Eun Y-J, Weibel DB. Chemical-biological studies of subcellular organization in bacteria. Biochemistry. 2011;50:7719–7734. doi: 10.1021/bi200940d. [DOI] [PubMed] [Google Scholar]
- 11.Ma S, Ma S. The development of FtsZ inhibitors as potential antibacterial agents. ChemMedChem. 2012;7:1161–1172. doi: 10.1002/cmdc.201200156. [DOI] [PubMed] [Google Scholar]
- 12.Nova E, Montecinos F, Brunet JE, Lagos R, Monasterio O. 4′,6-Diamidino-2-phenylindole (DAPI) induces bundling of Escherichia coli FtsZ polymers inhibiting the GTPase activity. Arch Biochem Biophys. 2007;465:315–319. doi: 10.1016/j.abb.2007.06.032. [DOI] [PubMed] [Google Scholar]
- 13.Margalit DN, Romberg L, Mets RB, Hebert AM, Mitchison TJ, Kirschner MW, RayChaudhuri D. Targeting cell division: Small-molecule inhibitors of FtsZ GTPase perturb cytokinetic ring assembly and induce bacterial lethality. Proc Natl Acad Sci USA. 2004;101:11821–11826. doi: 10.1073/pnas.0404439101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laeppchen T, Hartog AF, Pinas VA, Koomen G-J, Den Blaauwen T. GTP Analogue Inhibits Polymerization and GTPase Activity of the Bacterial Protein FtsZ without Affecting Its Eukaryotic Homologue Tubulin. Biochemistry. 2005;44:7879–7884. doi: 10.1021/bi047297o. [DOI] [PubMed] [Google Scholar]
- 15.Paradis-Bleau C, Beaumont M, Sanschagrin F, Voyer N, Levesque RC. Parallel solid synthesis of inhibitors of the essential cell division FtsZ enzyme as a new potential class of antibacterials. Bioorg Med Chem. 2007;15:1330–1340. doi: 10.1016/j.bmc.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 16.White EL, Suling WJ, Ross LJ, Seitz LE, Reynolds RC. 2-Alkoxycarbonylaminopyridines: inhibitors of Mycobacterium tuberculosis FtsZ. J Antimicrob Chemother. 2002;50:111–114. doi: 10.1093/jac/dkf075. [DOI] [PubMed] [Google Scholar]
- 17.Reynolds RC, Srivastava S, Ross LJ, Suling WJ, White EL. A new 2-carbamoyl pteridine that inhibits mycobacterial FtsZ. Bioorg Med Chem Lett. 2004;14:3161–3164. doi: 10.1016/j.bmcl.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 18.Mathew B, Srivastava S, Ross LJ, Suling WJ, White EL, Woolhiser LK, Lenaerts AJ, Reynolds RC. Novel pyridopyrazine and pyrimidothiazine derivatives as FtsZ inhibitors. Bioorg Med Chem. 2011;19:7120–7128. doi: 10.1016/j.bmc.2011.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stokes NR, Sievers J, Barker S, Bennett JM, Brown DR, Collins I, Errington VM, Foulger D, Hall M, Halsey R, Johnson H, Rose V, Thomaides HB, Haydon DJ, Czaplewski LG, Errington J. Novel inhibitors of bacterial cytokinesis identified by a cell-based antibiotic screening assay. J Biol Chem. 2005;280:39709–39715. doi: 10.1074/jbc.M506741200. [DOI] [PubMed] [Google Scholar]
- 20.Haydon DJ, Stokes NR, Ure R, Galbraith G, Bennett JM, Brown DR, Baker PJ, Barynin VV, Rice DW, Sedelnikova SE, Heal JR, Sheridan JM, Aiwale ST, Chauhan PK, Srivastava A, Taneja A, Collins I, Errington J, Czaplewski LG. An inhibitor of FtsZ with potent and selective anti-staphylococcal activity. Science (Washington, DC, U S) 2008;321:1673–1675. doi: 10.1126/science.1159961. [DOI] [PubMed] [Google Scholar]
- 21.Czaplewski LG, Collins I, Boyd EA, Brown D, East SP, Gardiner M, Fletcher R, Haydon DJ, Henstock V, Ingram P, Jones C, Noula C, Kennison L, Rockley C, Rose V, Thomaides-Brears HB, Ure R, Whittaker M, Stokes NR. Antibacterial alkoxybenzamide inhibitors of the essential bacterial cell division protein FtsZ. Bioorg Med Chem Lett. 2009;19:524–527. doi: 10.1016/j.bmcl.2008.11.021. [DOI] [PubMed] [Google Scholar]
- 22.Haydon DJ, Bennett JM, Brown D, Collins I, Galbraith G, Lancett P, Macdonald R, Stokes NR, Chauhan PK, Sutariya JK, Nayal N, Srivastava A, Beanland J, Hall R, Henstock V, Noula C, Rockley C, Czaplewski L. Creating an antibacterial with in vivo efficacy: synthesis and characterization of potent inhibitors of the bacterial cell division protein FtsZ with improved pharmaceutical properties. J Med Chem. 2010;53:3927–3936. doi: 10.1021/jm9016366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar K, Awasthi D, Lee SY, Zanardi I, Ruzsicska B, Knudson S, Tonge PJ, Slayden RA, Ojima I. Novel Trisubstituted Benzimidazoles, Targeting Mtb FtsZ, as a New Class of Antitubercular Agents. J Med Chem. 2011;54:374–381. doi: 10.1021/jm1012006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohashi Y, Chijiiwa Y, Suzuki K, Takahashi K, Nanamiya H, Sato T, Hosoya Y, Ochi K, Kawamura F. The lethal effect of a benzamide derivative, 3-methoxybenzamide, can be suppressed by mutations within a cell division gene, ftsZ, in Bacillus subtilis. J Bacteriol. 1999;181:1348–1351. doi: 10.1128/jb.181.4.1348-1351.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sarcina M, Mullineaux CW. Effects of tubulin assembly inhibitors on cell division in prokaryotes in vivo. FEMS Microbiol Lett. 2000;191:25–29. doi: 10.1111/j.1574-6968.2000.tb09314.x. [DOI] [PubMed] [Google Scholar]
- 26.Wang J, Galgoci A, Kodali S, Herath KB, Jayasuriya H, Dorso K, Vicente F, Gonzalez A, Cully D, Bramhill D, Singh S. Discovery of a small molecule that Inhibits cell division by blocking FtsZ, a novel therapeutic target of antibiotics. J Biol Chem. 2003;278:44424–44428. doi: 10.1074/jbc.M307625200. [DOI] [PubMed] [Google Scholar]
- 27.Plaza A, Keffer JL, Bifulco G, Lloyd JR, Bewley CA. Chrysophaentins A-H, antibacterial bisdiarylbutene macrocycles that inhibit the bacterial cell division protein FtsZ. J Am Chem Soc. 2010;132:9069–9077. doi: 10.1021/ja102100h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jaiswal R, Beuria TK, Mohan R, Mahajan SK, Panda D. Totarol inhibits bacterial cytokinesis by perturbing the assembly dynamics of FtsZ. Biochemistry. 2007;46:4211–4220. doi: 10.1021/bi602573e. [DOI] [PubMed] [Google Scholar]
- 29.Domadia P, Swarup S, Bhunia A, Sivaraman J, Dasgupta D. Inhibition of bacterial cell division protein FtsZ by cinnamaldehyde. Biochem Pharmacol. 2007;74:831–840. doi: 10.1016/j.bcp.2007.06.029. [DOI] [PubMed] [Google Scholar]
- 30.Kanoh K, Adachi K, Matsuda S, Shizuri Y, Yasumoto K, Kusumi T, Okumura K, Kirikae T. New sulfoalkylresorcinol from marine-derived fungus, Zygosporium sp. KNC52. J Antibiot. 2008;61:192–194. doi: 10.1038/ja.2008.29. [DOI] [PubMed] [Google Scholar]
- 31.Urgaonkar S, La Pierre HS, Meir I, Lund H, RayChaudhuri D, Shaw JT. Synthesis of Antimicrobial Natural Products Targeting FtsZ: (±)-Dichamanetin and (±)-2‴-Hydroxy-5″-benzylisouvarinol-B. Org Lett. 2005;7:5609–5612. doi: 10.1021/ol052269z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rai D, Singh JK, Roy N, Panda D. Curcumin inhibits FtsZ assembly: an attractive mechanism for its antibacterial activity. Biochem J. 2008;410:147–155. doi: 10.1042/BJ20070891. [DOI] [PubMed] [Google Scholar]
- 33.Ito H, Ura A, Oyamada Y, Tanitame A, Yoshida H, Yamada S, Wachi M, Yamagishi J-i. A 4-aminofurazan derivative-A189-inhibits assembly of bacterial cell division protein FtsZ in vitro and in vivo. Microbiol Immunol. 2006;50:759–764. doi: 10.1111/j.1348-0421.2006.tb03851.x. [DOI] [PubMed] [Google Scholar]
- 34.Beuria TK, Singh P, Surolia A, Panda D. Promoting assembly and bundling of FtsZ as a strategy to inhibit bacterial cell division: a new approach for developing novel antibacterial drugs. Biochem J. 2009;423:61–69. doi: 10.1042/BJ20090817. [DOI] [PubMed] [Google Scholar]
- 35.Singh P, Jindal B, Surolia A, Panda D. A rhodanine derivative CCR-11 inhibits bacterial proliferation by inhibiting the assembly and GTPase activity of FtsZ. Biochemistry. 2012;51:5434–5442. doi: 10.1021/bi201813u. [DOI] [PubMed] [Google Scholar]
- 36.Li Z, Garner AL, Gloeckner C, Janda KD, Carlow CK. Targeting the Wolbachia cell division protein FtsZ as a new approach for antifilarial therapy. PLoS Neglected Trop Dis. 2011;5:e1411. doi: 10.1371/journal.pntd.0001411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu XC, Margolin W. Inhibition of assembly of bacterial cell division protein FtsZ by the hydrophobic dye 5,5′-bis-(8-anilino-1-naphthalenesulfonate) J Biol Chem. 1998;273:10216–10222. doi: 10.1074/jbc.273.17.10216. [DOI] [PubMed] [Google Scholar]
- 38.Domadia PN, Bhunia A, Sivaraman J, Swarup S, Dasgupta D. Berberine targets assembly of escherichia coli cell division protein FtsZ. Biochemistry. 2008;47:3225–3234. doi: 10.1021/bi7018546. [DOI] [PubMed] [Google Scholar]
- 39.Beuria TK, Santra MK, Panda D. Sanguinarine blocks cytokinesis in bacteria by inhibiting FtsZ assembly and bundling. Biochemistry. 2005;44:16584–16593. doi: 10.1021/bi050767+. [DOI] [PubMed] [Google Scholar]
- 40.Schaffner-Barbero C, Gil-Redondo R, Ruiz-Avila LB, Huecas S, Lappchen T, den Blaauwen T, Diaz JF, Morreale A, Andreu JM. Insights into nucleotide recognition by cell division protein FtsZ from a mant-GTP competition assay and molecular dynamics. Biochemistry. 2010;49:10458–10472. doi: 10.1021/bi101577p. [DOI] [PubMed] [Google Scholar]
- 41.Scheffers DJ, den Blaauwen T, Driessen AJ. Non-hydrolysable GTP-gamma-S stabilizes the FtsZ polymer in a GDP-bound state. Mol Microbiol. 2000;35:1211–1219. doi: 10.1046/j.1365-2958.2000.01791.x. [DOI] [PubMed] [Google Scholar]
- 42.Foss MH, Eun Y-J, Grove CI, Pauw DA, Sorto NA, Rensvold JW, Pagliarini DJ, Shaw JT, Weibel DB. Inhibitors of bacterial tubulin target bacterial membranes in vivo. MedChemComm. 2012 doi: 10.1039/C2MD20127E. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shoichet BK. Screening in a spirit haunted world. Drug Disc Today. 2006;11:607–615. doi: 10.1016/j.drudis.2006.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McGovern SL, Caselli E, Grigorieff N, Shoichet BK. A common mechanism underlying promiscuous inhibitors from virtual and high-throughput screening. J Med Chem. 2002;45:1712–1722. doi: 10.1021/jm010533y. [DOI] [PubMed] [Google Scholar]
- 45.Coan KED, Shoichet BK. Stoichiometry and physical chemistry of promiscuous aggregate-based inhibitors. J Am Chem Soc. 2008;130:9606–9612. doi: 10.1021/ja802977h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seidler J, McGovern SL, Doman TN, Shoichet BK. Identification and prediction of promiscuous aggregating inhibitors among known drugs. J Med Chem. 2003;46:4477–4486. doi: 10.1021/jm030191r. [DOI] [PubMed] [Google Scholar]
- 47.Feng BY, Shelat A, Doman TN, Guy RK, Shoichet BK. High-throughput assays for promiscuous inhibitors. Nat Chem Biol. 2005;1:146–148. doi: 10.1038/nchembio718. [DOI] [PubMed] [Google Scholar]
- 48.Feng BY, Simeonov A, Jadhav A, Babaoglu K, Inglese J, Shoichet BK, Austin CP. A high-throughput screen for aggregation-based inhibition in a large compound library. J Med Chem. 2007;50:2385–2390. doi: 10.1021/jm061317y. [DOI] [PubMed] [Google Scholar]
- 49.Feng BY, Shoichet BK. A detergent-based assay for the detection of promiscuous inhibitors. Nat Protoc. 2006;1:550–553. doi: 10.1038/nprot.2006.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kubo I, Muroi H, Himejima M. Antibacterial activity of totarol and its potentiation. J Nat Prod. 1992;55:1436–1440. doi: 10.1021/np50088a008. [DOI] [PubMed] [Google Scholar]
- 51.Muroi H, Kubo I. Bactericidal effects of anacardic acid and totarol on methicillin-resistant Staphylococcus-aureus (MRSA) Biosci Biotechnol, Biochem. 1994;58:1925–1926. [Google Scholar]
- 52.Ingerman E, Perkins EM, Marino M, Mears JA, McCaffery JM, Hinshaw JE, Nunnari J. Dnm1 forms spirals that are structurally tailored to fit mitochondria. J Cell Biol. 2005;170:1021–1027. doi: 10.1083/jcb.200506078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cassidy-Stone A, Chipuk JE, Ingerman E, Song C, Yoo C, Kuwana T, Kurth MJ, Shaw JT, Hinshaw JE, Green DR, Nunnari J. Chemical inhibition of the mitochondrial division dynamin reveals its role in bax/bak-dependent mitochondrial outer membrane permeabilization. Dev Cell. 2008;14:193–204. doi: 10.1016/j.devcel.2007.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sorto NA, Olmstead MM, Shaw JT. Practical synthesis of PC190723, an inhibitor of the bacterial cell division protein FtsZ. J Org Chem. 2010;75:7946–7949. doi: 10.1021/jo101720y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Adams DW, Wu LJ, Czaplewski LG, Errington J. Multiple effects of benzamide antibiotics on FtsZ function. Mol Microbiol. 2010;80:68–84. doi: 10.1111/j.1365-2958.2011.07559.x. [DOI] [PubMed] [Google Scholar]
- 56.Andreu JM, Schaffner-Barbero C, Huecas S, Alonso D, Lopez-Rodriguez ML, Ruiz-Avila LB, Nunez-Ramirez R, Llorca O, Martin-Galiano AJ. The antibacterial cell division inhibitor PC190723 is an FtsZ polymer-stabilizing agent that induces filament assembly and condensation. J Biol Chem. 2010;285:14239–14246. doi: 10.1074/jbc.M109.094722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Elsen NL, Lu J, Parthasarathy G, Reid JC, Sharma S, Soisson SM, Lumb KJ. Mechanism of Action of the Cell-Division Inhibitor PC190723: Modulation of FtsZ Assembly Cooperativity. J Am Chem Soc. 2012;134:12342–12345. doi: 10.1021/ja303564a. [DOI] [PubMed] [Google Scholar]
- 58.Park YS, Grove CI, González-López M, Urgaonkar S, Fettinger JC, Shaw JT. Synthesis of (−)-viriditoxin: A 6,6′-binaphthopyran-2-one that targets the bacterial cell division protein FtsZ. Angew Chem, Int Ed. 2011;50:3730–3733. doi: 10.1002/anie.201007298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Trusca D, Bramhill D. Fluorescent assay for polymerization of purified bacterial FtsZ cell-division protein. Anal Biochem. 2002;307:322–329. doi: 10.1016/s0003-2697(02)00036-2. [DOI] [PubMed] [Google Scholar]
- 60.Okano M, Mito J, Maruyama Y, Masuda H, Niwa T, Nakagawa S-i, Nakamura Y, Matsuura A. Discovery and structure-activity relationships of 4-aminoquinazoline derivatives, a novel class of opioid receptor like-1 (ORL1) antagonists. Bioorg Med Chem. 2009;17:119–132. doi: 10.1016/j.bmc.2008.11.012. [DOI] [PubMed] [Google Scholar]
- 61.Takashi R, Tonomura Y, Morales MF. 4,4′-Bis(1-anilinonaphthalene 8-sulfonate) (bis-ANS): A new probe of the active site of myosin. Proc Natl Acad Sci USA. 1977;74:2334–2338. doi: 10.1073/pnas.74.6.2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Horowitz P, Prasad V, Luduena RF. Bis(1,8-anilinonaphthalenesulfonate). A novel and potent inhibitor of microtubule assembly. J Biol Chem. 1984;259:14647–14650. [PubMed] [Google Scholar]
- 63.Baell JB, Holloway GA. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J Med Chem. 2010;53:2719–2740. doi: 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]
- 64.Gupta KK, Bharne SS, Rathinasamy K, Naik NR, Panda D. Dietary antioxidant curcumin inhibits microtubule assembly through tubulin binding. FEBS J. 2006;273:5320–5332. doi: 10.1111/j.1742-4658.2006.05525.x. [DOI] [PubMed] [Google Scholar]
- 65.Hufford CD, Lasswell WL., Jr Antimicrobial activities of constituents of Uvaria chamae. Lloydia. 1978;41:156–160. [PubMed] [Google Scholar]
- 66.Anam EM, Ekpa OD, Gariboldi PV, Morah FNI, Dosunmu MI. 6″ ″-Hydroxybenzyldiuvaretins and related compounds from Xylopia africana (Benth) Oliver. Indian J Chem, Sect B: Org Chem Incl Med Chem. 1993;32B:1051–1054. [Google Scholar]
- 67.Wolff J, Knipling L. Antimicrotubule properties of benzophenanthridine alkaloids. Biochemistry. 1993;32:13334–13339. doi: 10.1021/bi00211a047. [DOI] [PubMed] [Google Scholar]
- 68.Lopus M, Panda D. The benzophenanthridine alkaloid sanguinarine perturbs microtubule assembly dynamics through tubulin binding. A possible mechanism for its antiproliferative activity. FEBS J. 2006;273:2139–2150. doi: 10.1111/j.1742-4658.2006.05227.x. [DOI] [PubMed] [Google Scholar]
- 69.Huang Q, Kirikae F, Kirikae T, Pepe A, Amin A, Respicio L, Slayden RA, Tonge PJ, Ojima I. Targeting FtsZ for antituberculosis drug discovery: noncytotoxic taxanes as novel antituberculosis agents. J Med Chem. 2006;49:463–466. doi: 10.1021/jm050920y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lu C, Stricker J, Erickson HP. FtsZ from Escherichia coli, Azotobacter vinelandii, and Thermotoga maritima--quantitation, GTP hydrolysis, and assembly. Cell Motil Cytoskeleton. 1998;40:71–86. doi: 10.1002/(SICI)1097-0169(1998)40:1<71::AID-CM7>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 71.Grove CI, Fettinger JC, Shaw JT. Second generation synthesis of viriditoxin. Synthesis. 2012;44:362–371. doi: 10.1055/s-0031-1289651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ingerman E, Nunnari J. A continuous, regenerative coupled GTPase assay for dynamin-related proteins. Methods Enzymol. 2005;404:611–619. doi: 10.1016/S0076-6879(05)04053-X. [DOI] [PubMed] [Google Scholar]
- 73.Marrington R, Small E, Rodger A, Dafforn TR, Addinall SG. FtsZ fiber bundling is triggered by a conformational change in bound GTP. J Biol Chem. 2004;279:48821–48829. doi: 10.1074/jbc.M404944200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.