Abstract
Rationale
Hyperhomocysteinemia is a risk factor of atherogenesis. Soluble epoxide hydrolase (sEH) is a major enzyme hydrolyzing epoxyeicosatrienoic acids and attenuates their cardiovascular protective effects. Whether homocysteine (Hcy) regulates sEH and the underlying mechanism remains elusive.
Objective
To elucidate the mechanism by which Hcy regulates sEH expression and endothelial activation in vitro and in vivo.
Methods and Results
Hcy treatment in cultured human endothelial cells dose- and time-dependently upregulated sEH mRNA and protein. Hcy increased the expression of adhesion molecules, which was markedly reversed by inhibiting sEH activity. Hcy-induced sEH upregulation is associated with activation of activating transcription factor 6 (ATF6). Bioinformatics analysis revealed a putative ATF6-binding motif in the promoter region of the sEH gene, which was found being a methylation site. Site-directed mutagenesis and chromatin immunoprecipitation assays demonstrated that Hcy treatment or ATF6 overexpression promoted ATF6 binding to the promoter of sEH and increased its activity. Result of methylation-specific PCR revealed that the ATF6 binding site on the sEH promoter was partially methylated and was demethylated with Hcy. SiRNA knockdown of ATF6α and/or SP1 blocked, and ATF6 overexpression and DNA methyltransferase inhibitor mimicked, the effect of homocysteine on sEH upregulation. In vivo, immunofluorescence assay revealed elevated expression of sEH and adhesion molecules in the aortic intima of mice with mild hyperhomocysteinemia, which was attenuated by sEH deletion or inhibition.
Conclusion
ATF6 activation and DNA demethylation may coordinately contribute to Hcy-induced sEH expression and endothelial activation. Inhibition of sEH may be a therapeutic approach for treating Hcy-induced cardiovascular diseases.
Keywords: sEH, homocysteine, ATF6, demethylation, endothelial cells
Introduction
Hyperhomocysteinemia (HHcy) is a well-known independent risk factor for cardiovascular disease1, but the underlying mechanisms remain unclear. Endothelial dysfunction plays a major role in the vascular pathology associated with homocysteine (Hcy). Oxidative stress2, endoplasmic reticulum (ER) stress3, inflammation4,5, telomerase inactivation6, cell apoptosis7 and epigenetics regulation8 are involved in this process. Accumulating data suggests that ER stress is the major process linking the level of Hcy with apoptosis and inflammation. ER stress is associated with development of Hcy-induced atherosclerotic lesions in apoE−/− mice9. Increased intracellular Hcy content could increase the expression of several ER stress-response genes, including GRP78, GRP94, Herp, RTP and GADD153, which are involved in ER stress-induced cell death, for further evidence of a mechanism involved in Hcy-induced cell dysfunction and programmed death10. The specific conversion of elevated Hcy to S-adenosyl homomcysteine (SAH) further inducing DNA hypomethylation represents another major mechanism8. Clinical and animal model studies revealed that DNA hypomethylation can modulate atherosclerosis-related gene transcription and their protein function. Sharma et al. reviewed the literature for 135 genes that modulated the blood level of Hcy or were regulated by elevated level of Hcy11. The dysregulation of these genes and their respective pathways involved in the development of atherosclerosis could affect cell function by oxidative and ER stress.
Recently, soluble epoxide hydrolase (sEH) functioning to enzymatically hydrolyze epoxyeicosatrienoic acids (EETs) and other fatty acid epoxides to their respective diols has attracted great interest as a potential therapeutic target for renal, cardiovascular and inflammatory disease12. Under physiological conditions, EETs are produced by the cytochrome P450 (CYP) epoxygenase pathway of arachidonic acid metabolism and tend to be anti-hypertensive, anti-inflammatory and protective against insulin resistance and ischemic injury13–15, which suggests the importance of EETs-sEH regulation in human health and diseases. We previously demonstrated that angiotensin II transcriptionally upregulated sEH by activating AP-1 in vascular endothelial cells (ECs) and cardiomyocytes in vitro and in vivo, which contributed to hypertension and cardiac hypertrophy16,17. Moreover, the transcriptional factor SP1 was also involved in the regulation of sEH expression as a result of sEH promoter demethylation18.
Given the evidence of endothelial activation by Hcy and the protective effect of exogenous EETs on ECs19,20, we investigated the potential role of sEH in Hcy-induced endothelial activation and the underlying mechanism. sEH mRNA was significantly upregulated by Hcy through the coordinated effects of ER stress and DNA demethylation by activating the transcriptional factor activating transcription factor 6 (ATF6). Further, Hcy-induced EC activation, evidenced by upregulated vascular cell adhesion molecule 1 (VCAM-1) and intercellular adhesion molecule 1 (ICAM-1), could be markedly attenuated by sEH inhibition and gene deletion. Transcriptional downregulation of sEH, as well as its inhibition, may be a therapeutic tool in the treatment of Hcy-induced cardiovascular diseases.
Materials and Methods
The chemicals and reagents used in this study and all experimental techniques, including cell cultures and treatment protocols, quantitative real-time RT-PCR (qRT-PCR), western blot analysis, inhibitors of signal transduction pathways, adenovirus infection, chromatin immunoprecipitation (ChIP) assay, immunohistochemical and immunofluorescence staining, transfection of siRNA, reporter gene assays, and methylation-specific PCR (MSP) are described in the online supplement methods and all PCR primers used are in the Online Table I.
Statistical Analysis
Results are expressed as means±SD from at least 3 replicates for each experiment. Statistical analysis involved two-tailed Student t test, one-way ANOVA, Student-Newman-Keuls test and Dunnett multiple comparison test as appropriate. P < 0.05 was considered statistically significant. All statistical analysis involved use of SPSS 16.0 (SPSS Inc., Chicago, IL, USA).
Results
Hcy upregulated sEH expression and endothelial activation in human ECs
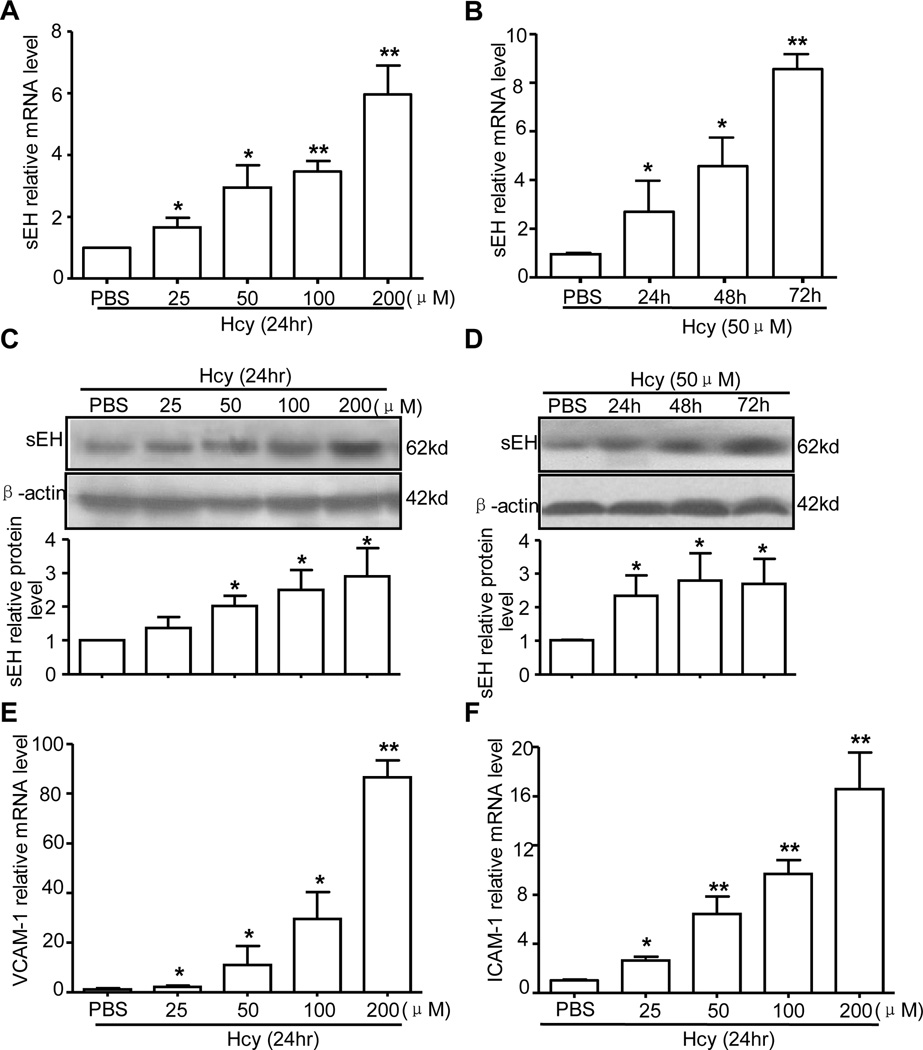
Elevated Hcy content causes vasculature injury and atherogenesis by inducing endothelial activation and dysfunction3. sEH hydrolyzes EETs, which decreases the protective role of EETs21,22. We first examined the effect of Hcy on sEH expression in ECs. Human umbilical vein ECs (HUVECs) were treated with Hcy (25~200 µmol/L) for 24 hr or with 50 µmol/L Hcy for various times. Real-time qRT-PCR and western blot analysis revealed that Hcy dose- and time-dependently upregulated the mRNA and protein levels of sEH: Hcy at 50 µmol/L significantly upregulated sEH expression at both mRNA and protein levels, with peak expression at 200 µmol/L (Fig. 1A, C); and Hcy at 50 µmol/L upregulated sEH beginning at 24 hr and lasting for at least 72 hr (Fig. 1B, D). In parallel, Hcy increased the expression of VCAM-1 and ICAM-1, markers of endothelial activation, in a dose-dependent manner (Fig. 1E, F). Hcy-induced sEH upregulation was confirmed in human aortic endothelial cells (Online Figure Ia, Ib).
Figure 1. Effect of homocysteine (Hcy) on soluble epoxide hydrolase (sEH) expression.
Human umbilical vein endothelial cells (HUVECs) were treated with various concentrations of Hcy for different times to examine the mRNA level of sEH (A, B) and protein level of sEH (C, D), vascular cell adhesion molecule 1 (VCAM-1) (E) and intercellular adhesion molecule 1 (ICAM-1) (F). β-actin cDNA and protein were the internal controls, respectively. Quantification of protein levels was by densitometry. Data are means ± SD of the relative mRNA normalized to that of β-actin from 3 independent experiments. *P<0.05, **P<0.01, vs. phosphate buffered saline (PBS) control.
EETs and sEH inhibitor (TUPS) prevented Hcy-induced endothelial activation
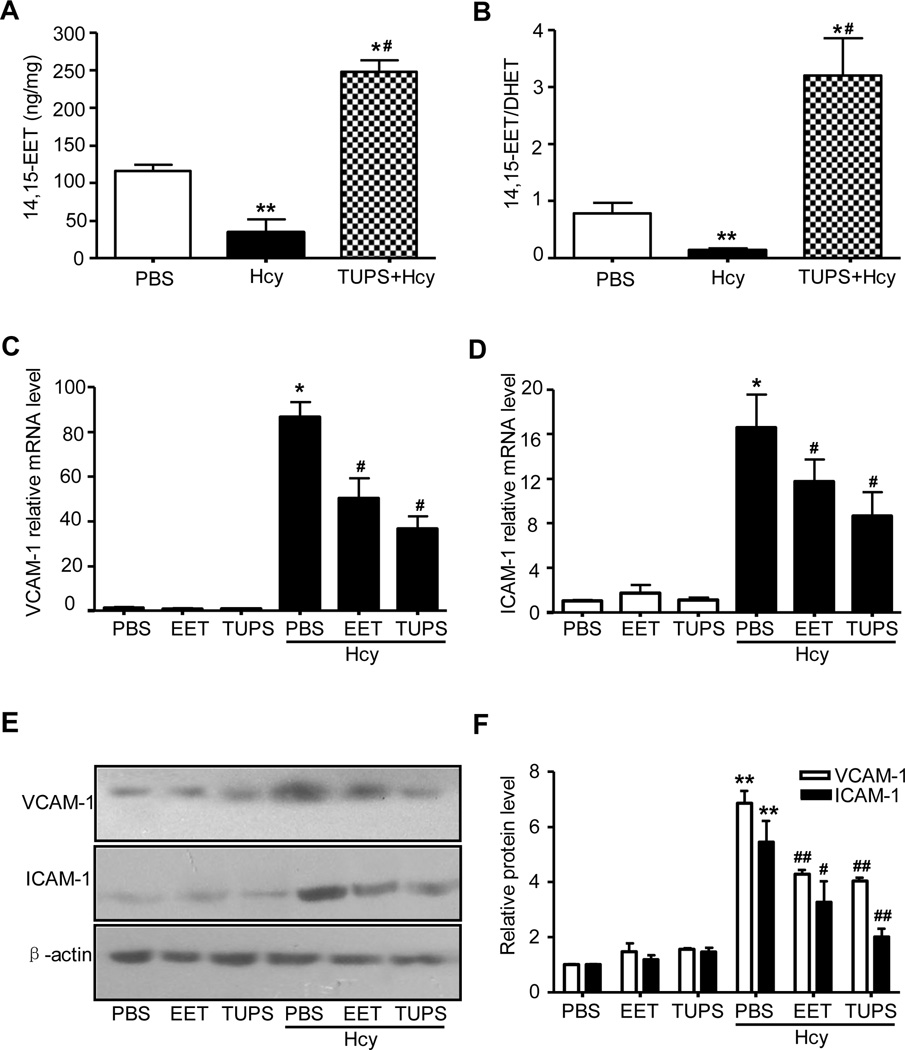
Given that Hcy-induced increase in sEH expression could reduce the amount of EETs in cells, we measured the levels of EETs and the ratio of EETs to DHETs in HUVECs. Indeed, Hcy decreased the levels of 14,15-EET and the ratio of 14,15-EET to 14,15-DHET, which could be reversed by treatment with the sEH inhibitor 1-(1-methanesulfonyl-piperidin-4-yl)-3-(4-trifluoro methoxy-phenyl)-urea (TUPS, 1 µmol/L)23 (Fig. 2A and B). Further, Hcy-induced VCAM-1 and ICAM-1 upregulation was reversed by pre-treatment with TUPS or 14,15-EET (100 nmol/L) 1 hr before Hcy stimulation (Fig. 2 C–F and Online Figure Ic–e). Therefore sEH induction may contribute to Hcy-induced endothelial activation, and inhibition of sEH activity can prevent the effect of Hcy, at least in part, through the increased protective effect of EETs and possibly other epoxylipides in HUVECs.
Figure 2. TUPS prevents Hcy-induced reduction in 14,15-EET and endothelial activation.
HUVECs (5×105) were incubated with PBS, Hcy (200µmol/L), or plus TUPS (1µmol/L) or 14,15-EET (100 nmol/L) for 24 hr. Intracellular levels of 14,15-EET and 14,15-EET/DHET were detected by ELISA (A, B). Quantitative RT-PCR of mRNA expression (C, D) or western blot analysis (E) of protein expression of VCAM-1 or ICAM-1. (F) Quantification of protein levels was by densitometry. Data are means±SD from 3 dependent experiment. *P<0.05, **P<0.01 vs. PBS controls, # P<0.05 vs. Hcy.
ATF6 pathway involved in Hcy-induced sEH expression in HUVECs
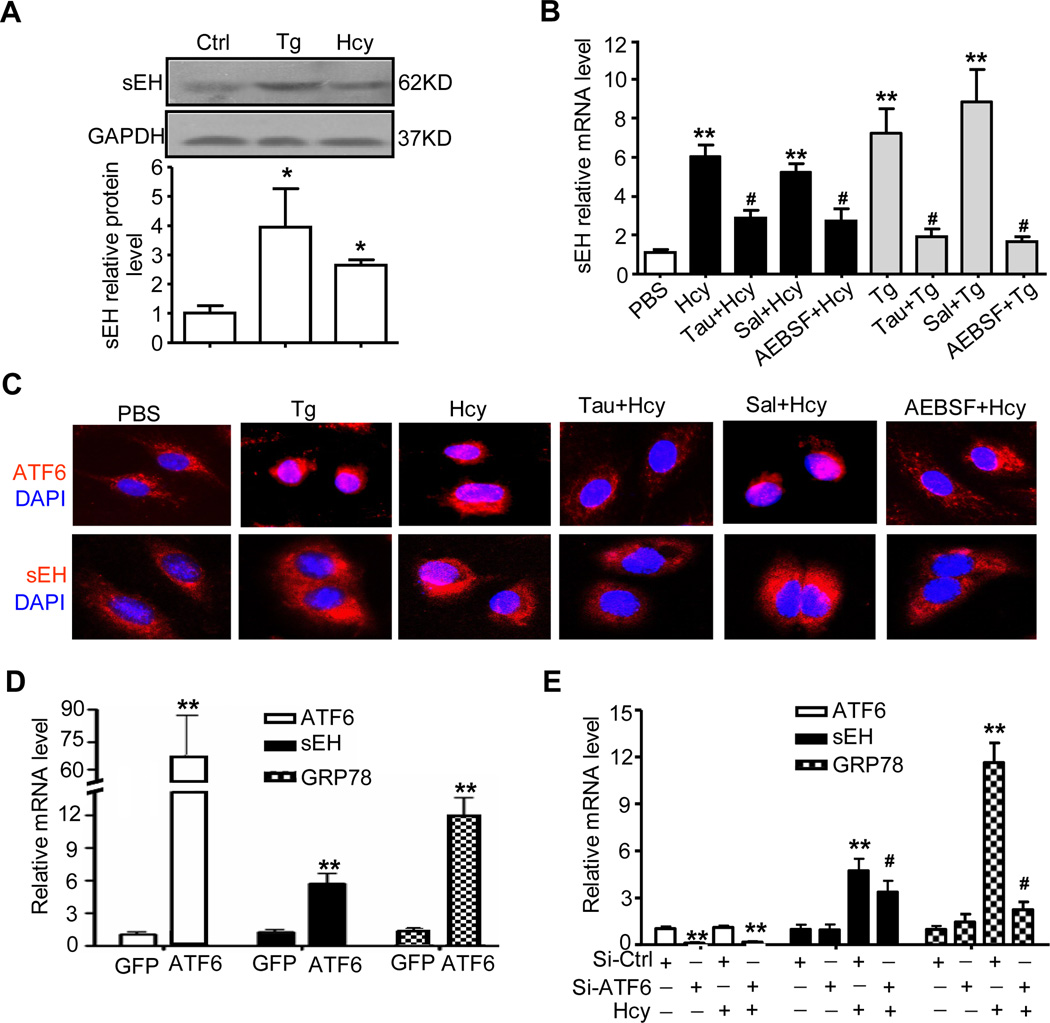
Hcy can alter the cellular redox state and induce ER stress24. To determine whether ER stress plays a role in Hcy-upregulated sEH expression, we detected markers of ER stress with an ER inducer, thapsigargin (Tg), used as a control. A high concentration of Hcy (200 µmol/L) increased the protein expression of GRP78, JNK and caspase-12, which was associated with increased sEH expression and activity (Fig. 3A and Online Figure IIa,b). Three ER stress inhibitors; taurine, serine protease inhibitor 4-(2-aminoethyl) benzenesulfonyl fluoride (AEBSF), and salubrinal, were reported to have a protective effect against Hcy-induced ER stress25, ATF6 spliced by S1P26 or eIF2α dephosphorylation during ER stress27, respectively. We found the Hcy-increased mRNA level of GRP78 and sEH attenuated by taurine and AEBSF but not salubrinal (Fig. 3B and Online Figure IIc). Immunofluorescence staining revealed that taurine and AEBSF blocked both Tg- and Hcy-induced nuclear translocation of ATF6 and upregulation of sEH (Fig. 3C), so activation of ATF6 is involved in Hcy-induced sEH expression.
Figure 3. Activating transcription factor 6 (ATF6) pathway is involved in Hcy-induced sEH expression in ECs.
(A) HUVECs were lysed after stimulation with 200 µmol/L Hcy for 24 hr, or 4 µmol/L thapsigargin (Tg) for 8 hr. Western blot analysis of protein level of sEH with GAPDH as an internal control. Quantification of protein levels was by densitometry from 3 independent experiments. Data are means±SD. *P<0.05 vs. PBS controls. (B) HUVECs were pre-treated with Tau (10 mmol/L), AEBSF (100 µmol/L) or Sal (50 µmol/L) for 1 hr before treatment with Hcy (200 µmol/L) and Tg (4 µmol/L). Real-time RT-PCR analysis of mRNA expression of sEH. β-actin was an internal control. *P<0.05, **P<0.01 vs. PBS control; #P<0.05 vs. Hcy. (C) Immunostaining of ATF6 and sEH (red) in treated HUVECs. Nuclei were visualized by DAPI staining (blue). (D, E) qRT-PCR analysis of mRNA expression of ATF6, sEH and GRP78. HUVECs were infected with adenovirus ATF6(N) [Ad-ATF6(N)] or Ad-GFP for 24 hr (D), or HUV-EC-C cells were transfected with siRNA of ATF6α (si-ATF6α) or scramble siRNA control (si-Ctrl) for 48 hr (E). *P<0.05, **P<0.01 vs. Ad-GFP infection or si-Ctrl transfection; #P<0.05 vs. Hcy. Data are mean±SD from at least 3 independent experiments. β-actin was an internal control.
To further study the effects of ATF6 on sEH expression, we examined the overexpression of ATF6 and siRNA knockdown of endogenous ATF6α, a potent transcriptional factor of ER stress-response genes. The mRNA levels of GRP78 and sEH in HUVECs were significantly higher with adenovirus-mediated overexpression and nuclear translocation of N-terminal ATF6 [Ad-ATF6(N)] for 24 hr than with Ad-GFP-infected control (Fig. 3D and Online Figure IIIa). Also, western blot analysis revealed increased protein levels of sEH in Ad-ATF6(N)-infected HUVECs (Online Figure IIIb). Further, in HUV-EC-C cells (ATCC #CRL-1730), a human umbilical vein vascular endothelium cell line, siRNA knockdown of ATF6α (to 11.35%) attenuated Hcy-induced sEH expression at protein level (Online Figure IIId) and mRNA level, with no change in basal levels of GRP78 and sEH (Fig. 3E).
ATF6 binding site on the sEH promoter contributed to the induction of sEH in ECs
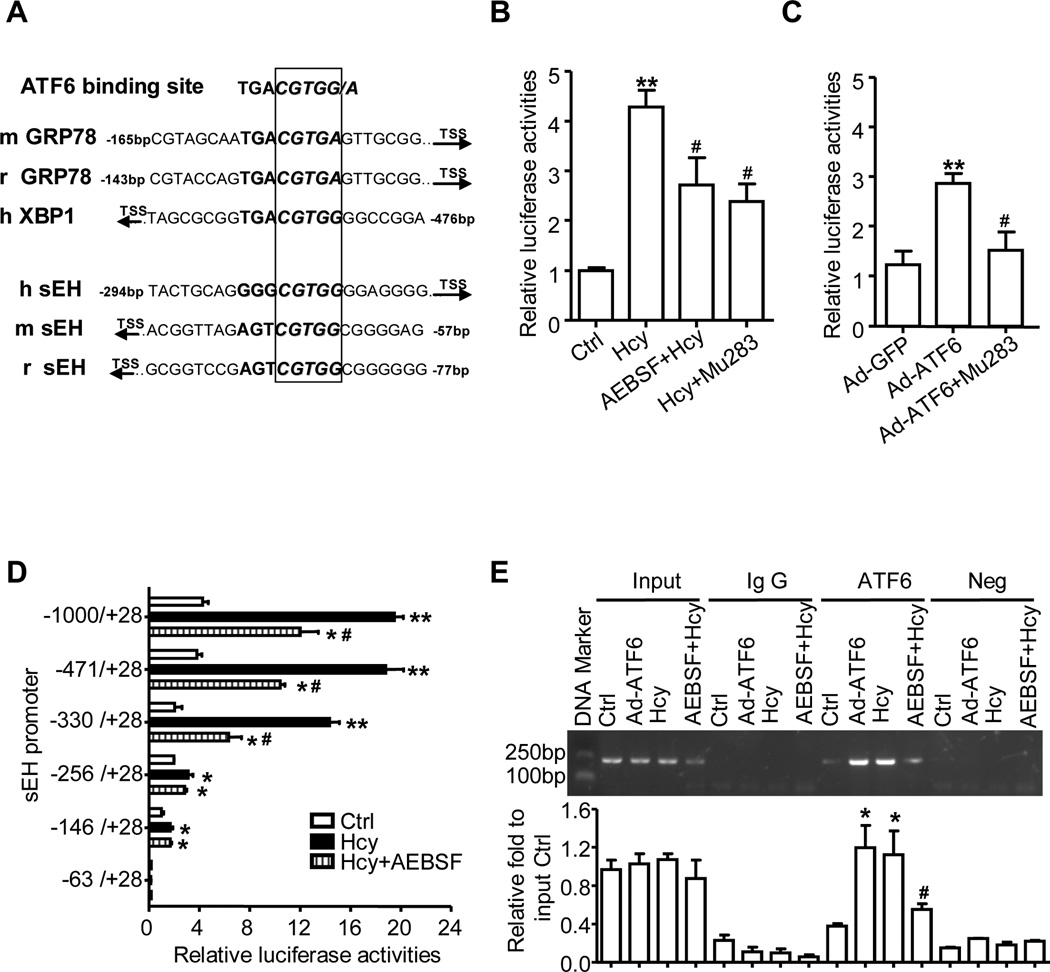
ATF6 was reported to bind directly to the unfolded protein response element (UPRE) (TGACGTGG/A) of target genes and activate the transcription of UPR genes such as GRP78 in rodents and X-box binding protein 1 (XBP1) in humans, in which the G flanking the “TGACGT” is critical for ATF6-specific binding28. The human sEH promoter region has multiple transcription factor binding sites, including sites for AP-1 and SP1, as we reported16,18. To determine whether ATF6 can directly regulate the promoter activity of sEH, we used bioinformatics analysis. Sequence analysis revealed one UPRE-like region “GGGCGTGG” within −279 to −286 bp upstream of the transcription initiation site of the sEH promoter region in humans, −43 to −50 bp in mouse and −63 to −70 bp in rat (Fig. 4A), which suggests a common regulatory mechanism of ATF6 on sEH in rodents and humans. Interestingly, nucleotide “GGGCG” in the UPRE-like region on the human sEH promoter is a methylated site by a bioinformatics analysis (http://www.urogene.org/methprimer/index1.html), so a multi-mechanism including ER stress and DNA methylation could be involved in sEH gene regulation.
Figure 4. ATF6 binds to sEH promoter and activates promoter activity.
(A) Homology analysis of ATF6 binding site on the promoter of sEH among of human, rat and mouse by UCSC Genome Browser. Plasmids of sEH-1000-Luc or ATF6 site mutation (Mu283) were transfected into EA.hy926 cells, and then treated with Hcy, with or without AEBSF in (B), or infected with Ad-GFP or Ad-ATF6(N) for 24 hr in (C). Serial deletion constructs of sEH-Luc were transfected into EA.hy926 cells and treated with Hcy, with or without AEBSF for 24 hr (D). The β-gal plasmid was co-transfected as a transfection control. Promoter activities were measured by luciferase activity, which was normalized to that of β-galactosidase. Data are mean±SD of relative luciferase activities from 4 independent experiments. (E) ChIP assay involved use of anti-ATF6 antibody for immunoprecipitation in treated HUVECs; normal rabbit IgG was a control. Semi-quantitative PCR involved sEH promoter-specific primers to detect the binding of ATF6 to the sEH promoter region. Data are mean±SD of the percentage of input control from 3 independent experiments. *P<0.05, **P<0.01 vs. Ctrl; #P<0.05 vs. Hcy or Ad-ATF6.
To elucidate the role of this motif in response to ATF6, we used plasmids of the human sEH promoter sEH-1000-Luc, as well as deletion29 and ATF6 binding-site point mutation constructs, for transient transfection. The promoter activities of these constructs were evaluated in EA.Hy926 cells, a human endothelial cell line for better transfection efficiency, with or without Hcy or Ad-ATF6(N). As shown in Figure 4B, the sEH promoter activity of sEH-1000-Luc was increased approximately 4.3-fold with Hcy treatment for 24 hr, which was reversed in part by pretreatment with AEBSF or mutated ATF6 site (Mu283) in the transfection. Ad-ATF6(N) infection significantly increased sEH promoter activity on the wild-type plasmid but lost the responsiveness on transfection with the mutation plasmid Mu283 (Fig. 4C). Further, AEBSF decreased Hcy-induced sEH promoter activity (from 7.1- to 3.1-fold) for sEH-330-Luc constructs containing ATF6 binding sites (−279~−286 bp) but had no effect on sEH-256-Luc (Fig. 4D), which also suggested that the UPRE-like motif plays critical roles in the regulation of sEH by ATF6 and Hcy. Chromatin immunoprecipitation (ChIP) assay further confirmed that Ad-ATF6(N) and Hcy increased ATF6 binding to the sEH promoter (−343~−144bp) as compared with controls; AEBSF could partially block the Hcy-induced binding of ATF6 to the UPRE-like region (Fig. 4E). The negative control was reported previously18.
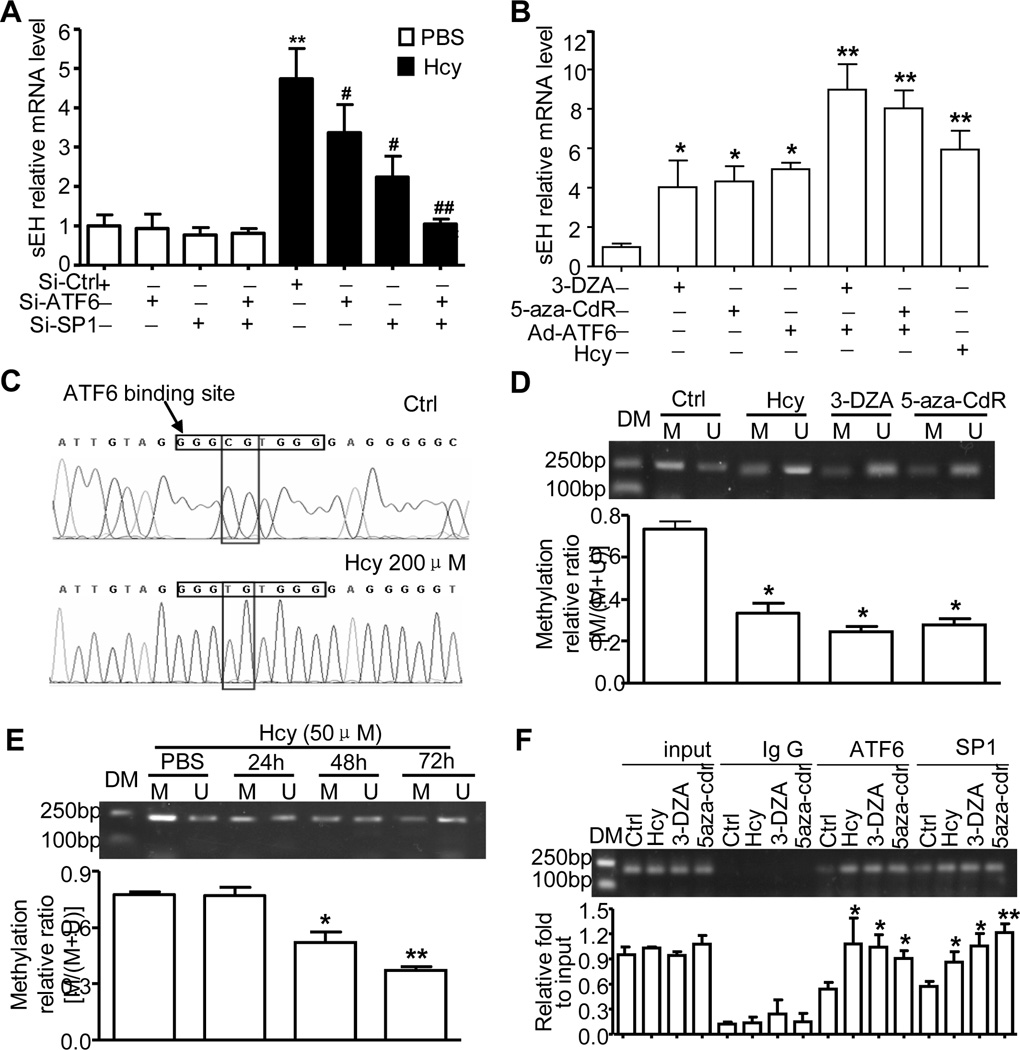
Hcy-induced DNA demethylation of ATF6 binding site facilitated the ATF6-induced sEH upregulation
We previously reported that DNA demethylation could increase the binding of SP1 to its motifs in the sEH promoter and induce sEH expression18. Further bioinformatics analysis revealed DNA methylation sites in the UPRE-like region on the sEH promoter. To study the role of DNA methylation in Hcy-induced sEH expression, we used siRNA knockdown of ATF6α and/or SP1 level in HUV-EC-C cells, which was decreased to 11.4% and/or 38.7%, respectively. Compared with their corresponding siRNA controls, siRNA knockout of ATF6α or SP1 in HUV-EC-C cells could partially attenuate Hcy-induced sEH expression at both mRNA and protein levels, whereas knockdown of both ATF6α and SP1 completely inhibited the effect of Hcy (Fig. 5A and Online Figure IVa). When HUVECs were treated with DNA methyl transferase inhibitors 5-aza-2' deoxycytidine (5-aza-CdR) or 3-deazadenosine (3-DZA), or Ad-ATF6(N) infection, the mRNA level of sEH was upregulated by 4.1±1.9, 4.3±1.1, or 4.9±0.5 times, respectively. Ad-ATF6(N) and 3-DZA or 5-aza-CdR treatment combined increased sEH mRNA level by 9.0±1.8 or 8.0±1.3 times (Fig. 5B), which was consistent with sEH protein expression (Online Figure IVb). Thus, both ATF6 and SP1 contributed to Hcy-induced sEH expression via DNA demethylation.
Figure 5. Hcy-induced DNA demethylation of ATF6 binding site contributed to sEH upregulation.
(A) si-ATF6 and/or si-SP1 and their corresponding si-Ctrl were transfected into cultured HUV-EC-C for 48 hr and treated with Hcy (200 µmol/L) for 24 hr. (B) HUVECs were exposed to Hcy (200 µmol/L), 5-aza-CdR (8 µmol/L), 3-DZA (100 µmol/L), or Ad-ATF6(N) for 24 hr. RT-PCR analysis of sEH mRNA expression. (C-E) Methylation-specific PCR (MSP) analysis of the methylation status of ATF6 binding site on the sEH promoter by treatment with Hcy, 5-aza-CdR and 3-DZA, or for different times with 50µmol/L Hcy. (C) Representative sequencing after MSP for the ATF6 binding site on the sEH promoter from Hcy-treated HUVECs. Densitometry quantified the relative DNA methylation abundance; the histogram represents the ratio of DNA methylation to total of methylation and unmethylation. DM: DNA marker, M: methylated, U: unmethylated. (F) ChIP assay to detect the binding of ATF6 or SP1 to the sEH promoter in HUVECs with different treatment as indicated. Data are mean±SD of the percentage of input control from 3 repeated experiments. *P<0.05, **P<0.01 vs. Ctrl; *P<0.05, vs. Hcy treatment.
To determine the changes in DNA methylation status with Hcy treatment, HUVECs underwent MSP after treatment with Hcy (200 µmol/L) for 24 hr. As shown in Figure 5C, the methylation of ATF6 binding site were increased in Hcy-treated ECs, compared with control. The DNA methylation rate for the identified methylated site within the UPRE-like region on the sEH promoter was decreased from 73.4% to 33.2% with Hcy treatment. Similar effects were also found with 3-DZA or 5-aza-CdR, for a decrease to 24.4% or 27.7%, respectively (Fig. 5D). As well, a lower concentration of Hcy (50 µmol/L) could time dependently (from 24 to 72 hr) promote changes in DNA methylation of the site on the sEH promoter (Fig. 5E). ChIP assay further confirmed that DNA demethylation by Hcy, 3-DZA, and 5-aza-CdR increased ATF6 or SP1 binding to the sEH promoter (−343~−144 bp) as compared with the PBS control (Fig. 5F). Therefore, the ATF6 binding site (UPRE-like region) on the sEH promoter was partially methylated with Hcy, and Hcy switched the binding site from methylated to unmethylated to facilitate ATF6 binding and promoter activation.
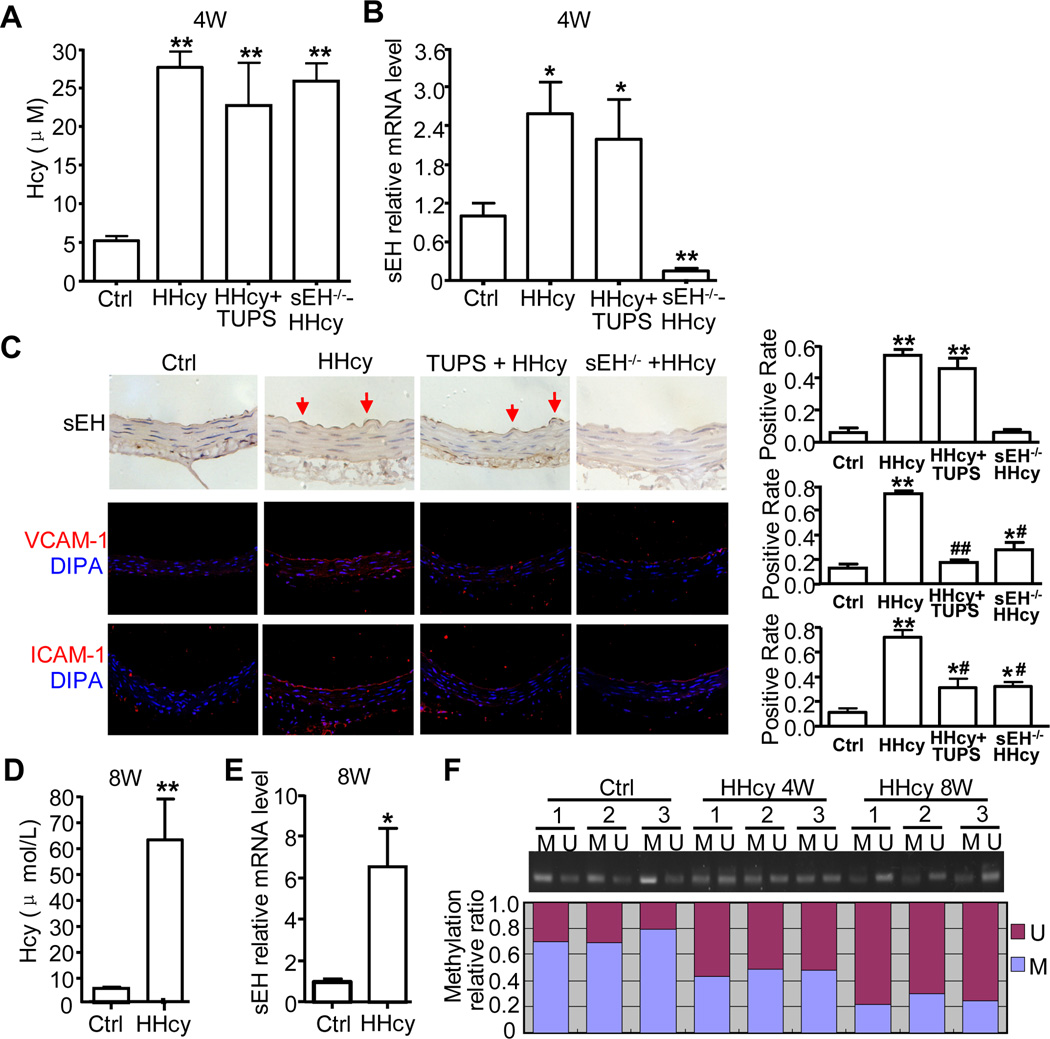
Hyperhomocysteinemia (HHcy) upregulated sEH expression in aortic intima in vivo
We next determined the pathophysiological relevance of Hcy-induced sEH expression and endothelial activation in vivo by establishing an HHcy model in C57BL/6J and sEH−/− mice with 2% (wt/wt) L-methionine in a chow diet for 4 weeks. We also used the sEH inhibitor TUPS (20 mg/L in drinking water, HHcy+TUPS) for treatment. The mean plasma level of total Hcy was significantly elevated in L-methionine–fed mice (HHcy group) as compared with controls (27.6±4.5 vs. 5.2±1.3 µmol/L, P<0.001) (Fig. 6A). TUPS treatment and sEH gene deletion did not change the elevated plasma Hcy levels in HHcy mice. Real-time RT-PCR revealed a 2.6-fold increase in sEH mRNA level in the aortic arteries of HHcy mice as compared with controls (Fig. 6B). Immunostaining also revealed increased expression of sEH in the aortic endothelium of HHcy mice (Fig. 6C). Although TUPS did not alter the expression of sEH, the increased protein levels of VCAM-1 and ICAM-1 were decreased with TUPS in aortic intima of HHcy mice and in sEH−/− mice, as detected by immunofluorescence staining (Fig. 6C). Therefore, inhibition of sEH activity or sEH gene deletion could protect against Hcy-induced aortic endothelial activation and injury in vivo. With longer treatment of L-methionine in C57BL/6J mice for 8 weeks, the mean plasma level of total Hcy was increased up to 61.5 ±31.4 µmol/L, and sEH mRNA increased 6.5-fold as compared with the control group (Fig. 6D, E). Furthermore, mouse aortas showed partial methylation of the ATF6 binding site on the sEH promoter. MSP analyses revealed the methylation level up to 73% in the control group but decreased to 47% and 23% in HHcy mice for 4 and 8 weeks, respectively (Fig. 6F). Thus, our in vitro and in vivo data suggested that sEH upregulation by both ER stress and DNA demethylation plays an important role in Hcy-induced endothelial activation.
Figure 6. The expression of sEH, VCAM-1 and ICAM-1 and methylation status of sEH in the aortic intima of HHcy mice.
Male C57BL/6J or sEH−/− mice (8 weeks old) were fed standard chow diet with 2% (wt/wt) L-methionine with or without TUPS in drinking water (20 mg/L/day) for 4–8 weeks. (A) Mean total plasma Hcy levels and (B) sEH mRNA expression (mouse β-actin was an internal control) in aorta intima in 4 groups of mice (n=8) for 4 weeks. (C) Representative immunohistochemical staining for sEH or confocal images of immunofluorescence staining for VCAM-I and ICAM-I (red) and nuclei (blue) in cross sections of pectoral aortas from 4 groups of mice. The arrows indicate the endothelium. Positive immunostaining of sEH, ICAM-1 and VCAM-1 in aorta intima was evaluated semiquantitatively. (D) Mean total plasma Hcy levels and (E) sEH mRNA expression in aorta intima detected in control and HHcy mice for 8 weeks. (F) MSP analysis of methylation status of sEH promoter with a focus on ATF6 binding site in the aortic intima of the mice. Histogram represents the ratio of DNA methylation to total methylation and unmethylation on the sEH promoter. *P<0.05, **P<0.01 vs. Ctrl; #P<0.05, ##P<0.01 vs. Hcy treatment. Data are mean ± SD. M: methylated, U: unmethylated.
Discussion
An optimal level of EETs has several beneficial effects on cardiovascular homeostasis, including hyperpolarizing vascular smooth muscle cells, dilating coronary arteries, and suppressing adhesion molecules. An imbalance in the metabolism of EETs by increased sEH expression or activity may lead to impaired vascular protection. Our previous studies showed that AP-1 or SP1 activation participated in sEH transcription upregulation, which contributes to hypertension and cardiac hypertrophy16–18. Here, we defined the role of sEH in Hcy-induced endothelial dysfunction and the underlying mechanism. Our novel findings include the following: 1) Hcy transcription upregulates sEH expression in human ECs through ER stress and the ATF6 pathway; 2) Hcy-induced DNA demethylation enhances ATF6 binding to the sEH promoter and increases sEH expression; 3) ATF6 mediates Hcy-induced sEH expression via the synergistic effect between DNA demethylation and ER stress; and 4) increased sEH expression plays an important role in Hcy-induced endothelial activation, which can be prevented by inhibition of sEH enzyme activity.
Previous studies reported that exogenous CYP epoxygenases increased human EC survival and protected bovine aortic ECs against cytokine/lipopolysaccharide-induced apoptosis by mitogen-activated protein kinase and PI3K/Akt signaling pathways19,20. However, the levels of sEH expression and endogenous EETs with insults such as HHcy on endothelial injury were unclear. Our results reveal the mechanism of Hcy-regulated sEH expression in ECs and show that sEH inhibition could reverse in part the Hcy-induced increase in expression of the endothelial activators VCAM-1 and ICAM-1 in vitro and in vivo. Because 14,15-EET could also reduce the Hcy-induced increase in expression of adhesion molecules, increased sEH expression may accelerate the degradation of EETs, thereby losing the protective effect of EETs on ECs. Moshal and colleagues reported that Hcy treatment downregulated CYP2J2 protein expression and reduced the generation of EETs30. We also measured the mRNA levels of the major isoforms of CYPs generating EETs (e.g., CYP2C8, 2C9 and 2J2) and did not detect a significant change in mRNA levels with Hcy treatment in human ECs (data not shown). However, the possibility that Hcy may also decrease the generation of EETs cannot be excluded.
We and others10,31,32 have demonstrated that high levels of Hcy and thapsigargin, an ER stress inducer, cause ER stress and increase the expression of UPR genes, including GRP78, JNK, Caspase12, and p-eIF2α, in human ECs. ATF6 is a member of the basic-leucine zipper family of transcription factors and is activated by cleavage from the ER membrane and translocation to the nucleus under ER stress33. We found that ATF6 nuclear translocation contributed to Hcy-induced sEH induction: N-terminal ATF6 overexpression could mimic and the inhibitor of ATF6 activation (AEBSF), but not p-eIF2α inhibitor, could inhibit the effect of Hcy. That Hcy-induced GRP78 and sEH upregulation was reduced with siRNA knockdown of ATF6 also supported the ATF6 pathway involved in Hcy-induced sEH transcription upregulation. A consensus DNA binding sequence “TGACGTGG/A” for ATF6 in the rat or mouse GRP78 promoter and human XBP1 promoter has been identified28. In this study, we found a core sequence, “CGTGG,” on the sEH promoter in human, rat and mouse (Fig. 4A). The DNA binding of ATF6 and activity of sEH promoter was increased by ATF6(N) overexpression and Hcy treatment in the presence of the UPRE-like motif (e.g., sequence of sEH-330~−256 in human). This effect was significantly attenuated by mutation of ATF6 binding site and treatment with the ATF6 inhibitor AEBSF, which suggests that this UPRE-like motif contributes to the regulation of sEH by ATF6, as well as Hcy.
Notably, Hcy-induced sEH expression was only partially inhibited by ATF6 inhibition, which indicates the involvement of other mechanisms such as DNA methylation or reactive oxygen species. We previously reported that sEH-325~−316 was an SP1 binding site, which was completely methylated in a hepatocellular carcinoma cell line18. Our results reveal a mechanism of DNA methylation-dependent perturbation of SP1 binding to the sEH promoter as a cell type-specific epigenetic regulation of the sEH gene. Similarly, this phenomenon exists in human ECs. This site could be demethylated by Hcy and facilitate SP1 binding and sEH promoter activation. However, many other transcription factors also have CG-rich binding sites in their DNA recognition element. DNA methylation may interfere with the binding of SP1 to DNA and repress gene transcription and expression. We found DNA methylation on the ATF6 binding site of the sEH promoter, as detected by MSP sequencing. The synergistic effects of ATF6 activation and DNA demethylation, as well as the combined effect of knockdown of both SP1 and ATF6, support that Hcy treatment demethylates the SP1 binding site and the ATF6 binding site, which largely increases SP1 and ATF6 binding to the sEH promoter and is an argument for its transcriptional regulation.
Patients with vascular disease, especially atherosclerosis, show disturbed global DNA methylation associated with elevated plasma level of Hcy (>75 µmol/L) and SAH concentrations34. Only 16 genes related to atherosclerotic diseases have been reported to be regulated at least in part by DNA methylation8. Given the important role of sEH in EETs metabolism and HHcy in the development of atherosclerosis, we established a mild or moderate HHcy mouse model with plasma Hcy level 27.6 or 62.5 µmol/L, respectively. In this model, the expression of ER stress markers ATF6, GRP78 or caspase-12 (Online Figure V) and sEH was elevated in the aortic intima, and that of ICAM-1 and VCAM-1 was elevated in the intima, which suggests injury or activation of ECs in mice fed L-methionine. Because humans and rodents share high homology for the sEH promoter sequence, we found partial methylation of the ATF6 binding site on the sEH promoter also in mouse, and the methylation status switch by HHcy may reflect a permission effect for full induction of sEH by Hcy. However, we established the role of sEH in Hcy-induced EC activation because the induction of VCAM-1 and ICAM-1 in the mouse aortic intima was largely attenuated by sEH inhibition and sEH gene deletion. Although we did not detect the plasma concentration of SAH and global methylation status in HHcy mice, Liu et al found the global DNA methylation status of the aortic tissue lower in HHcy mice than in controls, which supports our results. However, plasma SAH concentrations were negatively associated with global DNA hypomethylation and DNA methyltransferase activity in the aortic tissue of the authors’ apoE-deficient mice, which suggested that plasma SAH is a better biomarker of atherosclerosis than Hcy35. The authors indicated that variations in the plasma SAH levels might reflect changes in the intracellular concentrations of SAH, which led to global DNA hypomethylation in the aortic tissue of their atherosclerotic mouse model35. Our results suggest that both ER stress and DNA hypomethylation contribute to the atheroprone effect of Hcy, in which sEH transcription upregulation might play an important role and lead to endothelial dysfunction.
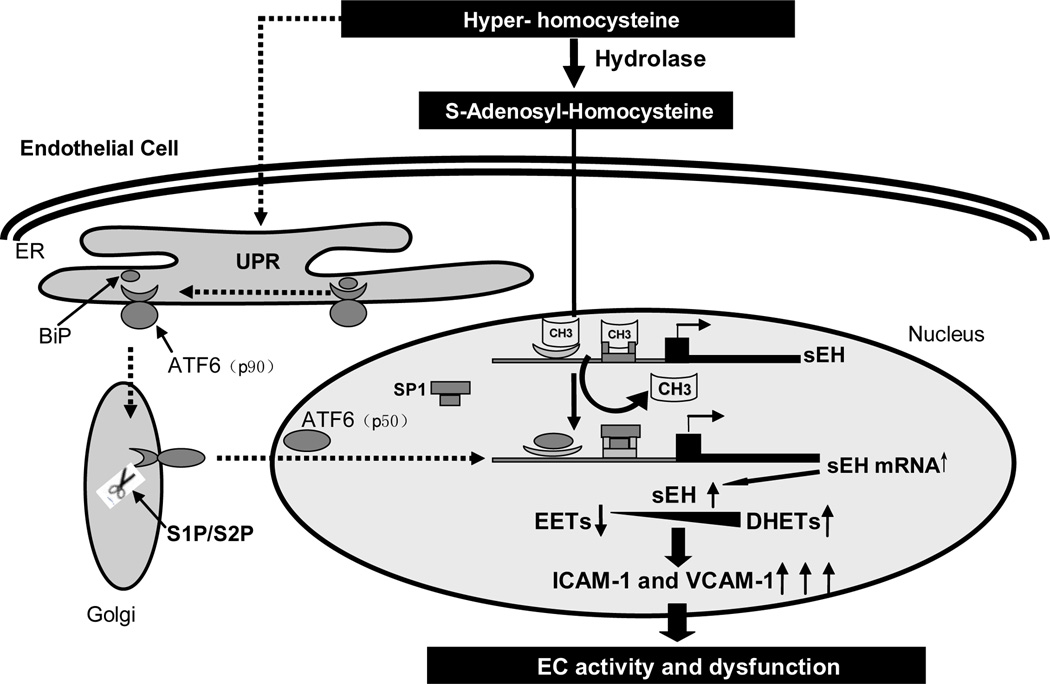
In conclusion, our study revealed that sEH could be transcriptionally upregulated by Hcy by both ER stress or ATF6 activation and DNA methylation in vitro and in vivo. As shown in Figure 7, in ECs, high levels of Hcy induce ER stress to activate ATF6; Hcy also hydrolyzes to SAH, which decreases the methylation status of SP1 and ATF6 binding motifs on the sEH promoter, facilitates both ATF6 and SP1 binding to the sEH promoter and augments sEH expression. In turn, the increased sEH expression hydrolyzes EETs and reduces the protective effect of EETs, which increases the expression of VCAM-1 and ICAM-1 and activates ECs. Our results may reveal a novel mechanism of Hcy-induced endothelial injury that may contribute to the development of atherosclerosis. Our findings warrant further studies to uncover the molecular mechanism underlying the protective effect of sEH inhibition in clinical applications.
Figure 7. Schematic model of signaling pathway of sEH induction by Hcy in ECs.
High levels of Hcy induce ER stress to activate ATF6. Hcy also hydrolyzes to SAH, which decreases the methylation status of SP1 and ATF6 binding motifs on the sEH promoter, which facilitates both ATF6 and SP1 binding to the sEH promoter, and augments sEH expression. In turn, sEH upregulation hydrolyzes EETs and reduces the protective effect of EETs, which increases the expression of VCAM-1 and ICAM-1 and activates ECs.
Supplementary Material
Novelty and Significance.
What is known?
High level of homocysteine in blood is a risk factor foratherosclerosis in humans. Homocysteine can cause DNA demethylation and endoplasmic reticulum stress, including activation of activating transcription factor 6 (ATF6).
Epoxyeicosatrienoic acids (EETs) have broad cardiovascular protective effects and are hydrolyzed by an enzyme called soluble epoxide hydrolase (sEH).
Endothelial dysfunction caused by athero-prone stimuli is one of the earliest vascular events leading to atherogenesis.
What new information does this article contribute?
In vitro, homocysteine upregulates sEH in endothelial cells and increases the expression of adhesion molecules, a marker of endothelial dysfunction, which isreversed by inhibiting sEH activity.
Homocysteine-induced activation of activating transcription factor 6 (ATF6) and demethylation of sEH promoter coordinately contribute to homocysteine-induced sEH expression and endothelial activation.
In vivo, elevated expression of sEH and adhesion molecules in the aortic intima of mice with mild hyperhomocysteinemia could be attenuated by sEH gene deletion or inhibition.
High level of homocysteine in blood is a risk factor for atherosclerosis. EETs exert a broad cardiovascular protective effect. The EETs are hydrolyzed by she, which attenuates their effects. Whether homocysteine regulates sEH expression and the underlying mechanism remains elusive. We found that homocysteine treatment in cultured human endothelial cells upregulated sEH and adhesion molecules, which was reversed by inhibiting sEH activity. Homocysteine-induced sEH upregulation was associated with the activation of ATF6, which binds a site in the sEH promoter that is partially is methylatedunder normal conditions. This ATF6-binding site was shown to be demethylated upon elevation of homocysteine in vitro and in vivo. Knockdown ATF6 blocked, and overexpression of ATF6 and DNA methyltransferase inhibitor mimicked, the effect of homocysteine on sEH upregulation. In a mouse model with elevated blood level of homocysteine, expression of sEH and adhesion molecules in the aortic intima was increased, which was attenuated by sEH deletion or inhibition. This study provides the first evidence that ATF6 activation and DNA demethylation coordinately contribute to homocysteine-induced sEH upregulation and endothelial activation. Our findings suggest that inhibition of sEH may be a therapeutic approach for treating homocysteine-induced cardiovascular diseases.
Acknowledgments
Sources of Funding: This work was supported in part by grants from the Major National Basic Research Grant of China [No. 2010BC912500], the National Natural Science Foundation of China [30971063, 81070113, 81028002, 81121061], the “111” plan of China and the National Institute of Environmental Health Sciences, NIH Grants [RO1 ES013933; RO1 ES002710]. B.D.H. is a George and Judy Marcus Fellow of the American Asthma Society.
Non-standard Abbreviations and Acronyms
- ECs
Endothelial cells
- sEH
Soluble epoxide hydrolase
- EETs
Epoxyeicosatrienoic acids
- Hcy
Homocysteine
- HHcy
Hyperhomocysteinemia
- HUVECs
Human umbilical vein endothelial cells
- ER
Endoplasmic reticulum
- ATF6
Activating transcription factor 6
- SAH
S-adenosyl homomcysteine
- VCAM-1
Vascular cell adhesion molecule 1
- ICAM-1
Intercelluar cell adhesion molecule 1
- MSP
Methylation-specific PCR
- TUPS
1-(1-methanesulfonyl-piperidin-4-yl)-3-(4-trifluoro methoxy-phenyl)-urea
- AEBSF
4-(2-aminoethyl) benzenesulfonyl fluoride
- UPRE
Unfolded protein response element
- 5-aza-CdR
5-aza-2' deoxycytidine
- 3-DZA
3-deazadenosine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: None.
References
- 1.Splaver A, Lamas GA, Hennekens CH. Homocysteine and cardiovascular disease: biological mechanisms, observational epidemiology, and the need for randomized trials. Am Heart J. 2004;148:34–40. doi: 10.1016/j.ahj.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 2.Perna AF, Ingrosso D, De Santo NG. Homocysteine and oxidative stress. Amino Acids. 2003;25:409–417. doi: 10.1007/s00726-003-0026-8. [DOI] [PubMed] [Google Scholar]
- 3.Austin RC, Lentz SR, Werstuck GH. Role of hyperhomocysteinemia in endothelial dysfunction and atherothrombotic disease. Cell Death Differ. 2004;11:S56–S64. doi: 10.1038/sj.cdd.4401451. [DOI] [PubMed] [Google Scholar]
- 4.Lu ZY, Jensen LE, Huang Y, Kealey C, Blair IA, Whitehead AS. The up-regulation of monocyte chemoattractant protein-1 (MCP-1) in Ea.hy 926 endothelial cells under long-term low folate stress is mediated by the p38 MAPK pathway. Atherosclerosis. 2009;205:48–54. doi: 10.1016/j.atherosclerosis.2008.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ross R. Atherosclerosis--an inflammatory disease. N Engl J Med. 1999;340:115–126. doi: 10.1056/NEJM199901143400207. [DOI] [PubMed] [Google Scholar]
- 6.Zhu JH, Chen JZ, Wang XX, Xie XD, Sun J, Zhang FR. Homocysteine accelerates senescence and reduces proliferation of endothelial progenitor cells. J Mol Cell Cardiol. 2006;40:648–652. doi: 10.1016/j.yjmcc.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 7.Suhara T, Fukuo K, Yasuda O, Tsubakimoto M, Takemura Y, Kawamoto H, Yokoi T, Mogi M, Kaimoto T, Ogihara T. Homocysteine enhances endothelial apoptosis via upregulation of Fas-mediated pathways. Hypertension. 2004;43:1208–1213. doi: 10.1161/01.HYP.0000127914.94292.76. [DOI] [PubMed] [Google Scholar]
- 8.Turunen MP, Aavik E, Yla-Herttuala S. Epigenetics and atherosclerosis. Biochim Biophys Acta. 2009;1790:886–891. doi: 10.1016/j.bbagen.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 9.Zhou J, Werstuck GH, Lhotak S, de Koning AB, Sood SK, Hossain GS, Moller J, Ritskes-Hoitinga M, Falk E, Dayal S, Lentz SR, Austin RC. Association of multiple cellular stress pathways with accelerated atherosclerosis in hyperhomocysteinemic apolipoprotein E-deficient mice. Circulation. 2004;110:207–213. doi: 10.1161/01.CIR.0000134487.51510.97. [DOI] [PubMed] [Google Scholar]
- 10.Ji C, Kaplowitz N. Hyperhomocysteinemia, endoplasmic reticulum stress, and alcoholic liver injury. World J Gastroenterol. 2004;10:1699–1708. doi: 10.3748/wjg.v10.i12.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharma P, Senthilkumar RD, Brahmachari V, Sundaramoorthy E, Mahajan A, Sharma A, Sengupta S. Mining literature for a comprehensive pathway analysis: a case study for retrieval of homocysteine related genes for genetic and epigenetic studies. Lipids Health Dis. 2006;5:1. doi: 10.1186/1476-511X-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen HC. Soluble epoxide hydrolase inhibitors: a patent review. Expert Opin Ther Pat. 2010;20:941–956. doi: 10.1517/13543776.2010.484804. [DOI] [PubMed] [Google Scholar]
- 13.Luria A, Bettaieb A, Xi Y, Shieh GJ, Liu HC, Inoue H, Tsai HJ, Imig JD, Haj FG, Hammock BD. Soluble epoxide hydrolase deficiency alters pancreatic islet size and improves glucose homeostasis in a model of insulin resistance. Proc Natl Acad Sci U S A. 2011;108:9038–9043. doi: 10.1073/pnas.1103482108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fleming I. Cytochrome P450 epoxygenases as EDHF synthase(s) Pharmacol Res. 2004;49:525–533. doi: 10.1016/j.phrs.2003.11.016. [DOI] [PubMed] [Google Scholar]
- 15.Imig JD, Hammock BD. Soluble epoxide hydrolase as a therapeutic target for cardiovascular diseases. Nat Rev Drug Discov. 2009;8:794–805. doi: 10.1038/nrd2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ai D, Fu Y, Guo D, Tanaka H, Wang N, Tang C, Hammock BD, Shyy JY, Zhu Y. Angiotensin II up-regulates soluble epoxide hydrolase in vascular endothelium in vitro and in vivo. Proc Natl Acad Sci U S A. 2007;104:9018–9023. doi: 10.1073/pnas.0703229104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ai D, Pang W, Li N, Xu M, Jones PD, Yang J, Zhang Y, Chiamvimonvat N, Shyy JY, Hammock BD, Zhu Y. Soluble epoxide hydrolase plays an essential role in angiotensin II-induced cardiac hypertrophy. Proc Natl Acad Sci U S A. 2009;106:564–569. doi: 10.1073/pnas.0811022106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang D, Ai D, Tanaka H, Hammock BD, Zhu Y. DNA methylation of the promoter of soluble epoxide hydrolase silences its expression by an SP-1-dependent mechanism. Biochim Biophys Acta. 2010;1799:659–667. doi: 10.1016/j.bbagrm.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 19.Yang S, Lin L, Chen JX, Lee CR, Seubert JM, Wang Y, Wang H, Chao ZR, Tao DD, Gong JP, Lu ZY, Wang DW, Zeldin DC. Cytochrome P-450 epoxygenases protect endothelial cells from apoptosis induced by tumor necrosis factor-alpha via MAPK and PI3K/Akt signaling pathways. Am J Physiol Heart Circ Physiol. 2007;293:H142–H151. doi: 10.1152/ajpheart.00783.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dhanasekaran A, Al-Saghir R, Lopez B, Zhu D, Gutterman DD, Jacobs ER, Medhora M. Protective effects of epoxyeicosatrienoic acids on human endothelial cells from the pulmonary and coronary vasculature. Am J Physiol Heart Circ Physiol. 2006;291:H517–H531. doi: 10.1152/ajpheart.00953.2005. [DOI] [PubMed] [Google Scholar]
- 21.Spiecker M, Liao JK. Vascular protective effects of cytochrome p450 epoxygenase-derived eicosanoids. Arch Biochem Biophys. 2005;433:413–420. doi: 10.1016/j.abb.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 22.Pokreisz P, Fleming I, Kiss L, Barbosa-Sicard E, Fisslthaler B, Falck JR, Hammock BD, Kim IH, Szelid Z, Vermeersch P, Gillijns H, Pellens M, Grimminger F, van Zonneveld AJ, Collen D, Busse R, Janssens S. Cytochrome P450 epoxygenase gene function in hypoxic pulmonary vasoconstriction and pulmonary vascular remodeling. Hypertension. 2006;47:762–770. doi: 10.1161/01.HYP.0000208299.62535.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones PD, Tsai HJ, Do ZN, Morisseau C, Hammock BD. Synthesis and SAR of conformationally restricted inhibitors of soluble epoxide hydrolase. Bioorg Med Chem Lett. 2006;16:5212–5216. doi: 10.1016/j.bmcl.2006.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Outinen PA, Sood SK, Liaw PC, Sarge KD, Maeda N, Hirsh J, Ribau J, Podor TJ, Weitz JI, Austin RC. Characterization of the stress-inducing effects of homocysteine. Biochem J. 1998;332:213–221. doi: 10.1042/bj3320213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nonaka H, Tsujino T, Watari Y, Emoto N, Yokoyama M. Taurine prevents the decrease in expression and secretion of extracellular superoxide dismutase induced by homocysteine: amelioration of homocysteine-induced endoplasmic reticulum stress by taurine. Circulation. 2001;104:1165–1170. doi: 10.1161/hc3601.093976. [DOI] [PubMed] [Google Scholar]
- 26.Okada T, Haze K, Nadanaka S, Yoshida H, Seidah NG, Hirano Y, Sato R, Negishi M, Mori K. A serine protease inhibitor prevents endoplasmic reticulum stress-induced cleavage but not transport of the membrane-bound transcription factor ATF6. J Biol Chem. 2003;278:31024–31032. doi: 10.1074/jbc.M300923200. [DOI] [PubMed] [Google Scholar]
- 27.Boyce M, Bryant KF, Jousse C, Long K, Harding HP, Scheuner D, Kaufman RJ, Ma D, Coen DM, Ron D, Yuan J. A selective inhibitor of eIF2alpha dephosphorylation protects cells from ER stress. Science. 2005;307:935–939. doi: 10.1126/science.1101902. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y, Shen J, Arenzana N, Tirasophon W, Kaufman RJ, Prywes R. Activation of ATF6 and an ATF6 DNA binding site by the endoplasmic reticulum stress response. J Biol Chem. 2000;275:27013–27020. doi: 10.1074/jbc.M003322200. [DOI] [PubMed] [Google Scholar]
- 29.Tanaka H, Kamita SG, Wolf NM, Harris TR, Wu Z, Morisseau C, Hammock BD. Transcriptional regulation of the human soluble epoxide hydrolase gene EPHX2. Biochim Biophys Acta. 2008;1779:17–27. doi: 10.1016/j.bbagrm.2007.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moshal KS, Zeldin DC, Sithu SD, Sen U, Tyagi N, Kumar M, Hughes WM, Jr, Metreveli N, Rosenberger DS, Singh M, Vacek TP, Rodriguez WE, Ayotunde A, Tyagi SC. Cytochrome P450 (CYP) 2J2 gene transfection attenuates MMP-9 via inhibition of NF-kappabeta in hyperhomocysteinemia. J Cell Physiol. 2008;215:771–781. doi: 10.1002/jcp.21356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kokame K, Agarwala KL, Kato H, Miyata T. Herp, a new ubiquitin-like membrane protein induced by endoplasmic reticulum stress. J Biol Chem. 2000;275:32846–32853. doi: 10.1074/jbc.M002063200. [DOI] [PubMed] [Google Scholar]
- 32.Outinen PA, Sood SK, Pfeifer SI, Pamidi S, Podor TJ, Li J, Weitz JI, Austin RC. Homocysteine-induced endoplasmic reticulum stress and growth arrest leads to specific changes in gene expression in human vascular endothelial cells. Blood. 1999;94:959–967. [PubMed] [Google Scholar]
- 33.Haze K, Yoshida H, Yanagi H, Yura T, Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell. 1999;10:3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Castro R, Rivera I, Struys EA, Jansen EE, Ravasco P, Camilo ME, Blom HJ, Jakobs C, Tavares de Almeida I. Increased homocysteine and S-adenosylhomocysteine concentrations and DNA hypomethylation in vascular disease. Clin Chem. 2003;49:1292–1296. doi: 10.1373/49.8.1292. [DOI] [PubMed] [Google Scholar]
- 35.Liu C, Wang Q, Guo H, Xia M, Yuan Q, Hu Y, Zhu H, Hou M, Ma J, Tang Z, Ling W. Plasma S-adenosylhomocysteine is a better biomarker of atherosclerosis than homocysteine in apolipoprotein E-deficient mice fed high dietary methionine. J Nutr. 2008;138:311–315. doi: 10.1093/jn/138.2.311. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.