Abstract
Chromosome ends are complex structures, consisting of repetitive DNA sequence terminating in an ssDNA overhang with many associated proteins. Because alteration of these ends is a hallmark of cancer, telomeres and telomere maintenance have been prime drug targets. The universally conserved ssDNA overhang is sequence-specifically bound and regulated by Pot1 (protection of telomeres), and perturbation of Pot1 function has deleterious effects for proliferating cells. The specificity of the Pot1/ssDNA interaction and the key involvement of this protein in telomere maintenance have suggested directed inhibition of Pot1/ssDNA binding as an efficient means of disrupting telomere function. To explore this idea, we developed a high-throughput time-resolved fluorescence resonance energy transfer (TR-FRET) screen for inhibitors of Pot1/ssDNA interaction. We conducted this screen with the DNA-binding subdomain of S. pombe Pot1 (Pot1pN), which confers the vast majority of Pot1 sequence-specificity and is highly similar to the first domain of human Pot1 (hPOT1). Screening a library of ~20,000 compounds yielded a single inhibitor, which we found interacted tightly with submicromolar affinity. Furthermore, this compound, subsequently identified as the bis-azo dye Congo red, was able to competitively inhibit hPOT1 binding to telomeric DNA. ITC and NMR chemical shift analysis suggest that CR interacts specifically with the ssDNA-binding cleft of Pot1, and that alteration of this surface disrupts CR binding. The identification of a specific inhibitor of ssDNA interaction establishes a new pathway for targeted telomere disruption.
Keywords: high-throughput screen, telomeres, Pot1, OB-fold, ssDNA binding, Congo red, small molecule recognition
Telomeres are the nucleoprotein structures at the ends of eukaryotic linear chromosomes that function in genome maintenance and cellular survival by distinguishing the chromosome ends from sites of DNA damage and ensuring complete DNA replication (1-5). These unique structures minimally consist of repetitive G-rich DNA sequence terminating in a 3' ssDNA overhang and an associated six-protein complex called shelterin (6). Telomeric ssDNA is particularly vulnerable to misrecognition by the DNA damage machinery, and its protection is necessary for proper cellular function (7, 8). In a broad range of species spanning humans to the model organism Schizosaccharomyces pombe, this overhang is tightly and specifically bound by the shelterin component Pot1, which safeguards against inappropriate ssDNA processing (7, 9). Deletion of S. pombe Pot1 (SpPot1) results in nearly complete loss of telomeric DNA and cell viability, with a small population of cells surviving via chromosome circularization (10). The protective role of Pot1 in chromosomal maintenance is highly conserved. Alteration of human Pot1 (hPOT1) function can lead to G-strand overhang loss, chromosomal-end fusions, chromosomal rearrangements, and rapid cell cycle arrest, ultimately leading to senescence and apoptosis (7, 9, 11-13).
In addition to providing a protective cap for the ssDNA overhang, Pot1 is an essential regulatory protein, allowing controlled access to the 3' end in order to facilitate complete chromosome replication (14-17). As a result of the end-replication problem, the terminal nucleotides cannot be duplicated, leading to progressive sequence loss with each round of DNA replication (4, 18, 19). As telomeres shorten in somatic cells, a critical length is reached, at which point genomic integrity can no longer be assured and cells undergo cell cycle arrest and senescence (20, 21). In stem cells and unicellular organisms, this problem is averted through the action of the reverse transcriptase enzyme telomerase, which recognizes and extends the telomeric DNA from the 3'-ssDNA overhang, allowing for continued replication (22-24). While crucial for stem cell function, this mechanism is often hijacked by cancer cells, providing an avenue for the uncontrolled replication required for cancer progression (25, 26).
Telomerase is activated in > 85% of human cancer cells (26, 27), and, as a result, has been a major focus of cancer therapeutic research (28-30). A common approach to therapeutic intervention has been the use of small molecule inhibitors to decrease or block telomerase activity. Many small molecules have been discovered that function through a variety of mechanisms, including decreasing telomerase expression (31-33), inhibition of telomerase catalytic activity (34-37) and disruption of telomerase/ssDNA interaction (38, 39). Because cell proliferation halts only when telomeres become critically short, cellular response to telomerase inhibition is dependent upon initial telomere length, with an average response time of ~50 days (40-46).
A second widely studied class of small molecule therapeutics targets the telomerase substrate ssDNA. These compounds have been proposed to alter chromosome end accessibility by inducing G-quadruplex formation at the telomere ends thus restricting access of telomerase to its substrate (29, 47-51). This class of compounds proved quite effective in tumor cell lines, resulting in a surprisingly rapid loss of telomeric DNA and apoptosis in only a few cell cycles, while having no effect on normal cell lines (52-57). While striking, the rapid time-action of these compounds was inconsistent with direct inhibition of telomerase and instead suggested an alternative mechanism of action. Subsequent studies into this rapid mechanism of action suggested that the induced G-quadruplex formation negatively impacted the ability of hPOT1 to bind to the single-stranded telomere ends, resulting in telomere deprotection (58-64). Confirmation for this proposed mechanism came with the discovery that overexpression of hPOT1 provided resistance to these compounds (58, 59, 61). Additionally, hPOT1 expression is altered in many tumors and cancer cell lines (65-67) and is specifically upregulated in therapeutic and radiation-resistant cell lines (68, 69), suggesting a role for hPOT1 in cancer progression.
As a result of caveats associated with RNAi/shRNA knockdown and overexpression experiments, the exact role of the DNA-binding activity of hPOT1 in telomere maintenance has not been defined (9, 11-14, 70, 71). If the G-quadruplex promoting ligands act by displacing hPOT1, direct inhibition of hPOT1 activity may prove to be a more effective strategy to impede telomere protection and provide important insight into hPOT1 function. The Pot1 proteins utilize OB fold motifs to bind telomeric ssDNA with high affinity and specificity (72). While the minimal ssDNA substrates of the Pot1 proteins are large compared to most small molecules (10-12 nucleotides), mutagenesis of the protein and modification of the DNA suggest that disruption of a small region of the interface is sufficient to drastically reduce binding (73-77). To explore this idea, we designed a small molecule screen for inhibitors of Pot1/ssDNA interaction. We identified an inhibitor, which binds specifically to the Pot1 ssDNA-binding cleft, demonstrating that Pot1/ssDNA interactions are amenable to specific inhibition by small molecules.
Experimental Procedures
Chemicals, Reagents, and Proteins
All chemicals and reagents were obtained from Fisher Scientific unless otherwise indicated. Oligonucleotides were commercially synthesized by Integrated DNA Technologies and [γ-32P]ATP was purchased from PerkinElmer. Pot1pN, Pot1pN_F88A, Pot1-DBD, and Cdc13-DBD were obtained from as previously described (76-78). hPOT1 was a generous gift from Dr. Derek Taylor, Elaine Podell, and Prof. Thomas Cech.
High-throughput Screen
The Europium-chelate donor (LANCE Eu-W1024-labeled anti-6xHis antibody) and ULight acceptor dye (ULight-streptavidin) FRET reagents were purchased from PerkinElmer. Our combined screening library consisted of 20,480 compounds from the maximum chemical diversity TimTec Diversity set (10,240 compounds) and the “universal” hand-synthesized ChemBridge DIVERSet™ (10,240 compounds) libraries. Screening was conducted at room temperature in low-volume 384-well plate format using a Biomek FX liquid handling workstation equipped with a 96-well pipeting head and a plate stacker unit (Beckman Coulter). Total sample volumes were 20 μL and each well contained 200 nM Pot1pN, 200 nM biotin-6mer, 1.25 nM Eu-W1024-labeled anti-6xHis antibody (EC), 25 nM ULight-streptavidin (UL), and 250 μM test compound. Compound concentrations were approximate and based on a library molecular weight average. Assay conditions were 50 mM Tris, pH 7.5, 50 mM NaCl, 0.02% Pluronic F-68 detergent (Sigma-Aldrich), and 5% DMSO. Pluronic F-68 is known to limit promiscuous aggregation, and we found that inclusion eliminated DMSO concentration-dependent effects on the signal-to-background ratio.
A total of 10,240 compounds were screened per run, with 320 compounds per 384-well plate (OptiPlate, PerkinElmer), and 32 wells each of positive control samples (addition of excess unlabeled 6mer DNA) and negative controls (addition of DMSO). Pot1pN, EC and compounds were sequentially pipetted into plates and incubated for 5 min prior to addition of biotin-6mer and UL. The TR-FRET signal was read by a PerkinElmer EnVision plate reader with an excitation wavelength of 320 nm and detection at both 615 nm (EC emission) and 665 nm (EC-UL FRET emission). A delay time of 100 μsec before detection allowed non-specific fluorescence to decay, while preserving the EC-UL FRET signal. Data quantification was performed using ActivityBase software (IDBS Ltd.) to obtain percent activity from the ratio of EC to EC-UL FRET emission signals.
Data from the controls were used to normalize compound activity, and calculate signal-to-background ratios (at 665 nm) and Z′- and Z-factors (79). The Z′-factor, which is a statistical measure of the quality of the assay itself using control data only, is calculated using the equation:
| (1) |
where Z′ is the Z′-factor, σp is the standard deviation of the positive controls, σn is the standard deviation of the negative controls, μp is the mean of the positive controls, and μn is the mean of the negative controls. The Z-factor parameter reports on the quality and suitability of an assay for HTS format using the data variability and signal dynamic range of the assay once the compounds are added and is calculated using the following equation:
| (2) |
where Z is the Z-factor, σs is the standard deviation of the samples, σc is the standard deviation of the controls, μs is the mean of the sample, and μc is the mean of the controls. TimTec compounds were screened a single time, while the ChemBridge library underwent two rounds of screening. Primary hits from the HTS assay were defined as those that exhibited ≥ 50% reduction in FRET signal for the TimTec screening runs, and ≥ 55% reduction for both Chembridge screens.
Validation of TimTec Screen Hits and FRET Measurements of the Dose-dependence of Compound Inhibition
The TimTec primary hits were independently validated in a 96-well format. These assays were conducted as described for initial screening, but manually set up and scaled up in volume for the 96-well plate format. Following validation, dose-dependence of small molecule activity was determined similarly, but using 10 serial 2-fold dilutions from 250 μM to 470 nM of each compound, such that eight compounds could be tested per plate. Each plate included positive and negative controls in the outer two columns. Compounds that exhibited dose-dependent loss of FRET signal with ≥ 50% reduction at 250 μM were selected for secondary assay screening.
Secondary Screening of Compounds by Double-filter Binding Assay
All screen hits that displayed dose-dependent inhibition of Pot1pN/6mer FRET were independently analyzed for inhibition by double-filter binding (80). The 6mer oligonucleotide was 5’-end labeled with [γ-32P]ATP (PerkinElmer) using T4 polynucleotide kinase according to manufacturer's protocol (New England Biolabs). Labeled 6mer was separated from unincorporated ATP using G25 spin columns (GE Healthcare) and stored at -20°C. Binding was performed in 20 mM potassium phosphate, pH 8.0, 15 mM NaCl, 3 mM βME, 10% DMSO. We determined that DMSO concentrations up to 20% had no effect on Pot1pN binding activity (data not shown). Compounds were titrated in 40 μL reactions consisted of 10 μM Pot1pN, 200 pM cognate ssDNA (6mer), and approximate compound concentrations of 0, 1, 5, 10, 50, 100, 250, and 500 μM. Pot1pN was incubated for 30 m with compounds prior to addition of DNA. Final reactions were incubated for another 30 m. 30 μL of each reaction was loaded onto a dot-blot filter-binding apparatus (Whatman) assembled with a sandwich of nitrocellulose (protein and protein/ssDNA complex binding; GE Healthcare), HyBond-XL (GE Healthcare), and Whatman paper (filters presoaked in binding buffer for 1 h). Vacuum was applied to the assembled apparatus and a 12-channel multi-channel pipettor was used to wash each well two times with 90 μL of binding buffer. 30 μL of each reaction mixture was loaded, followed by 2 × 90 μL washes. After all liquid was pulled from each well, the vacuum was turned off and the nitrocellulose and HyBond-XL filters were placed on plastic wrap, dried with a hot air dryer, exposed to phosphorimaging screens (GE Healthcare) and visualized on a Typhoon Imager (GE Healthcare). SpPot1-DBD and hPOT1 competition assays were conducted as described above, with the exception that protein and DNA concentrations were 500 nM and 400 nM, respectively. Data were quantified, plotted as a function of fraction bound vs. compound concentration, and fit to the following equation using KaleidaGraph (version 4.0; Synergy Software) to determine relative inhibitory values:
| (3) |
where FB is fraction bound, S is a scaling factor, P is the protein concentration, IC50 is the relative apparent half maximal inhibitory concentration, h is the Hill coefficient for cooperative binding, and O is the background offset. Because the technical aspects of measuring IC50 values (large excess of DNA ligand) preclude the use of the proper assay for protein/nucleic acid binding, the values reported here do not represent true IC50 values and only report the relative abilities of the compounds to competitively inhibit binding of the ssDNA to the protein (IC50rel).
Determination of CR, Thioflavin T, and TB Concentrations
CR was purchased from MP Biomedicals, TB from Fisher Scientific, and Thioflavin T from from Sigma. CR was resuspended in water and aliquots were lyophilized. The extinction coefficient of CR in phosphate buffer differs from that in water. Therefore, equivalent dry aliquots were resuspended in water or binding buffer (50 mM KPhos, pH 8.0, 50 mM NaCl, 3 mM BME). The concentration of CR in solution was determined by absorbance spectroscopy in water using a molar extinction coefficient of 45000 M-1 cm-1 at 495 nm (81). Thioflavin T concentration was determined using a molar extinction coefficient of 36000 M-1 cm-1 at 412 nm, which is the same in water and phosphate buffer (82, 83). TB was dialyzed in a 100 Da molecular weight cutoff Float-A-Lyzer (Spectrum Laboratories, Inc.) at high concentration in water to remove excess salt and concentration was determined using a molar extinction coefficient of 51550 M-1 cm-1 at 600 nm (84, 85).
ITC of Protein/Ligand Complexes
Binding of CR, Thioflavin T, and TB to Pot1pN and of CR to Pot1pN_F88A and Cdc13-DBD were measured by ITC on an ITC200 MicroCalorimeter (MicroCal), with an active cell volume of 203 μL and a 40 μL syringe. Proteins were dialyzed at 4 °C for >12 h against binding buffer using a 2000 Da molecular weight cutoff Slide-A-Lyzer dialysis cassette (Thermo Scientific) and dry CR, Thioflavin T, and TB were resuspended in the same binding buffer. Reference power was set at 11 μcal/s, the stirring rate was 1000 RPM, and all experiments were conducted at 20 °C. Each experiment consisted of a single 0.2 μL injection followed by 20 injections of 1.95 μL compound with injection intervals of 180 s. Pot1pN/CR and Pot1pN_F88A experiments were performed in triplicate with 1.2 mM CR titrated into 100 μM protein. 2 mM Thioflavin T was titrated into 150 μM Pot1pN as a single replicate; triplicate Cdc13-DBD experiments were performed with 2.1 mM CR titrated into 88 μM protein. Duplicate Pot1pN/TB experiments were performed with 1 mM TB titrated into 103 μM protein. Data analysis was performed using Origin 7 SR4 software (OriginLab Corporation). Thermograms were analyzed using a non-linear least squares one-site of binding model obtain binding stoichiometry (n), binding enthalpy (ΔH), binding entropy (ΔS), and dissociation constant (KD). ITC isotherms were obtained by subtracting reference data (compound titrated into buffer alone) from experimental data. Thermograms and isotherms were plotted using Origin 7.
Inhibition of Pot1/ssDNA Interaction by CR
Experiments were conducted as outlined for secondary screening with the following modifications. Binding was performed in 50 mM Tris, pH 8.0, 50 mM NaCl, 1 mM DTT. CR titrated in 40 μL reactions containing different Pot1 proteins and cognate ssDNA: 500 nM Pot1-DBD + 400 nM d(GGTTACGGTTAC), 1 μM Pot1pN + 800 nM d(GGTTAC), or 500 nM hPOT1 + 400 nM d(GGTTAGGGTTAG). For these assays CR was titrated at concentrations of 0.01, 0.1, 1, 5, 10, 50, 100, 250, 500, and 750 μM. Proteins were incubated on ice with compounds for 15 minutes prior to addition of DNA and then incubated on ice for 1 h further. Filter binding was performed as above, but with cold binding buffer washes containing no salt (50 mM Tris, pH 8.0, 1 mM DTT).
Dynamic Light Scattering
DLS was performed on a DynaPro system (Wyatt Technology Corporation) operating at a wavelength of 633 nm with scattered light detected at 90°. 12 μL samples were taken directly from free and CR-bound 2H-15N-labeled Pot1pN NMR samples (300 μM). The 6mer competition sample consisted of 300 μM Pot1pN, 300 μM CR, and 1.5 mM 6mer. Samples were spun down at maximum speed in a microcentrifuge prior to measurement in quartz cuvettes. Data were analyzed using DynaPro Dynamics V 6.3.40 (Wyatt Technology Corporation) to calculate hydrodynamic radius and molecular weight.
Pot1pN/CR 1H-15N HSQC Chemical Shift Perturbation Analysis
15N-labeled and 2H/15N-labeled Pot1pN were expressed as described previously (86) with the following modifications for doubly labeled Pot1pN. A 10 mL LB overnight culture was inoculated directly into M9 minimal media containing 100% D2O and grown at 37 °C to OD600 of 0.6. Cells were then placed on ice for 1 h, protein expression was induced by addition of 500 μM IPTG and incubation for 20 h at 20 °C. Monomeric protein was concentrated to 1 mL and exchanged into NMR buffer (50 mM K2HPO4, pH 8.0, 50 mM NaCl, 1 mM DTT-d10, 6% D2O) with continued concentration to >300 μM. Final yield was 15 mg/L. The free Pot1pN sample was prepared at 300 μM and the Pot1pN/CR complex was formed as a 1:1 complex at 300 μM. Gradient-selected, sensitivity-enhanced TROSY 1H-15N HSQC spectra were acquired at 30°C on a Varian VNMRS 900 or Inova 600 spectrometer equipped with a HCN cold probe and VnmrJ integrated software using Varian BioPack pulse sequences with minor modifications. Spectra were processed in NMRPipe (87) and analyzed using CcpNmr Analysis 2.0.7 (88).
Results
FRET-based HTS Assay Design
We developed a TR-FRET assay to discover small molecule inhibitors of Pot1/ssDNA binding. TR-FRET is a robust method for screening and has been successfully combined with HTS for probing disruption of a diverse array of biomolecular interactions (89, 90). We designed a TR-FRET HTS assay to identify inhibitors of Pot1/ssDNA interaction using an ssDNA-binding subdomain of the S. pombe Pot1 protein (Pot1pN) as a model for Pot1 proteins. Pot1pN is the first OB fold of the DNA-binding domain of SpPot1 and is structurally similar to the first OB fold (OB1) of hPOT1 (10, 74-76), superposing with an r.m.s.d. of 1.26 Å for 122 of 140 backbone positions (Figure 1A). Binding studies of the full-length protein show that Pot1pN provides the bulk of the specificity of the SpPot1/ssDNA interaction, while the second OB-fold subdomain contributes increased affinity for a longer ssDNA sequence (77). Similarly, five of the six specifically recognized nucleotide positions in the hPOT1/ssDNA complex interact with OB1 (75). This experimental design also takes advantage of the fact that Pot1pN in isolation is highly specific for a short 6-nucleotide cognate ssDNA sequence (d(GGTTAC); 6mer) (73, 93), is extremely stable, and can be produced in abundant quantities suitable for screen development.
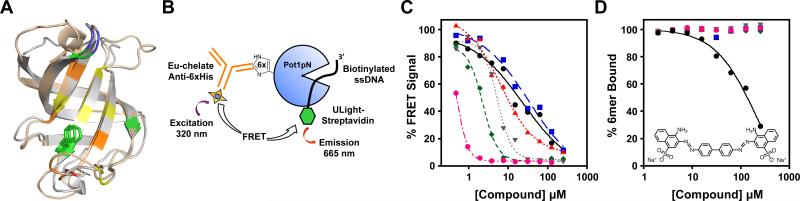
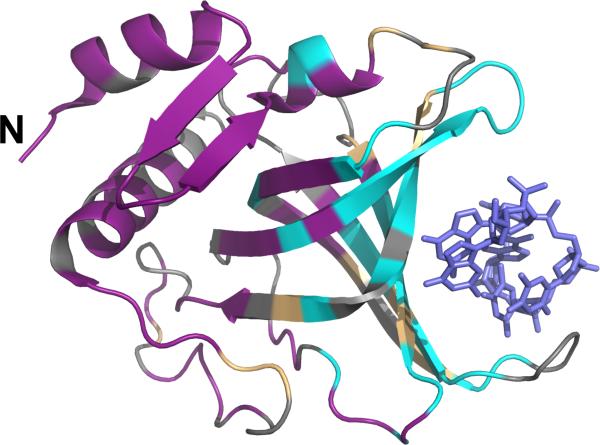
Figure 1.
High-throughput screen and secondary validation of compounds identify as single inhibitor. (A) Pot1pN (wheat) superposed with OB1 of hPOT1 (white) shows structural similarity; the N- and C-terminal portions of each protein have been removed for clarity. Conserved residues on the ssDNA-binding surface are colored according to chemical character: aromatic (green), hydrophobic (yellow), polar (orange), acidic (red), and basic (blue). Pot1pN F88 (and corresponding hPOT1 F62) is shown as green sticks. Superposition was performed using LSQMAN (91) and the image was generated using MacPymol version 1.3 (92). (B) Schematic of the TR-FRET assay used to identify inhibitors of Pot1pN interaction with 6mer ssDNA. 6xHis-tagged Pot1pN is bound by the Eu-chelate anti-6xHis antibody (donor), which transfers energy to ULight-Streptavidin (acceptor) bound to biotinylated ssDNA. Excitation and FRET emission wavelengths are indicated. (C) Plot of the FRET-based dose-dependence of the activity of small molecules identified by the TR-FRET screen. Percent FRET signal is plotted as a function of compound concentration. (D) Plot of the dose-dependent inhibitory activity of the compounds from (C) by secondary filter-binding assay revealing only one compound (Congo red) that directly inhibits the Pot1pN/6mer interaction. Percent 6mer ssDNA bound is plotted as a function of compound concentration.
We first screened several potential FRET pairs for optimal activity and, based on signal-to-background ratios and low sensitivity to changes in assay conditions, we chose the LANCE Europium-chelate/ULight FRET pair (PerkinElmer; data not shown). The FRET donor LANCE Eu-W1024-labeled anti-6xHis antibody (EC) was used to label N-terminally 6xHis-tagged Pot1pN and the streptavidinconjugated FRET acceptor dye ULight (UL) bound the 5′ biotin-tagged 6mer ssDNA ligand (Figure 1B). As the 5′ end of 6mer is critical for binding (73, 74), we inserted a 3-nucleotide linker between the biotin-conjugated threonine and the 6mer sequence, resulting in the oligonucleotide biotind(TAAGGTTAC). Competitive FRET assays confirmed that the biotin-d(TAA) addition and streptavidin binding had little effect on Pot1pN/6mer binding (Supporting Information Figure S1). Both Pot1pN and 6mer were held at 200 nM for screening assays, which is well above the 30 nM KD for this interaction and ensures the Pot1pN/6mer complex is fully formed (76).
We found that the EC-UL FRET pair yielded superb signal-to-background ratios over the course of a 10,240 compound library screen (32 plates) with low concentrations (EC = 1.25 nM; UL = 25 nM) of fresh reagents. We used excess unlabeled 6mer as a positive control for complete inhibition, and, because compound libraries were resuspended in DMSO, DMSO addition served as a negative control. We tested the Pot1pN/6mer interaction for DMSO sensitivity and found that the ssDNA-binding activity of Pot1pN was unaffected by DMSO concentrations up to 20% with 24 h incubation times (Supporting Information Figure S2). Signal-to-background for the first 16 plates averaged > 31, and ratios for plates 17-32 averaged > 20. A 32-plate screening run lasted approximately 6 h, and, while we did observe a drop in signal-to-background over that time, a ratio of 20 is exceptionally high, and indicated excellent assay robustness over time (94). We measured Z′- and Z-factors of 0.86 and 0.57, respectively. As Z′- and Z-factors ≥ 0.5 are considered excellent, these values demonstrate that our assay is of extremely high quality (79, 94).
Screen Implementation and Validation of Dose-dependent Compound Activity
We initially screened the TimTec Diversity set, a 10,240 compound library characterized by a high degree of structural diversity and largely conforming to Lipinski's Rule of 5 (95). Compounds exhibiting ≤ 50% of the normalized control FRET signal were scored as primary hits. This screen positively identified 208 compounds, corresponding to a hit rate of approximately 2%. Because of the large number of primary hits identified in the TimTec library, prior to testing compound activity using an independent secondary assay, we validated these hits manually by replicating the HTS assay in 96-well format. Of the 208 hits, 40 of the compounds had been previously identified in unrelated assays suggesting a non-specific mode of action and were eliminated from the positive hit list (96, 97). Of the remaining set, 25 compounds were confirmed as hits using a repeat assay in 96-well format. Utilizing freshly prepared compound stocks, we measured dose-dependent activity for these compounds with a partially automated FRET-based assay. The majority of these small molecules (19/25) showed no change in signal (data not shown). The remaining six compounds displayed dose-dependent inhibition of signal with apparent IC50rel values < 200 μM (Figure 1C).
We then screened a second library of drug-like structurally diverse compounds (ChemBridge DIVERSet™) in duplicate in order to eliminate spurious errors and minimize false positive hits. In this screen, we scored compounds exhibiting ≤ 55% FRET signal in both replicates as primary hits. Using these criteria, we identified 62 compounds in this library screen, corresponding to a 0.6% hit rate. This significant reduction in the hit rate demonstrates the value of duplicate screening to minimize the list of hits and subsequent analysis of compounds.
Identification of a Compound that Directly Inhibits the Pot1/ssDNA Interaction
While the FRET-based dose-dependent inhibition measurements are convenient and partially automated, they do not alone confirm direct binding inhibition; for example, a compound may interfere with one or both of the fluorophores leading to false indication of activity. Fortunately, protein/nucleic interactions are amenable to relatively high throughput 96-well double filter-binding dot-blot assays (98, 99), providing a convenient means to verify that the compound indeed inhibits ssDNA binding. This assay is a direct readout of protein-ssDNA binding, and the fraction of ssDNA bound by the protein can be measured as a function of compound concentration. We screened the 62 ChemBridge primary hits and the six TimTec compounds displaying a significant dose-dependent change in FRET signal for inhibitory activity by filter binding. Of the 62 ChemBridge primary hits analyzed in this manner, none displayed inhibition of Pot1pN binding to 6mer at any concentration (data not shown). However, analysis of the TimTec compounds revealed that a single molecule (ST012888) functionally inhibited Pot1pN binding to 6mer (Figure 1D).
Congo Red Directly Binds Pot1pN
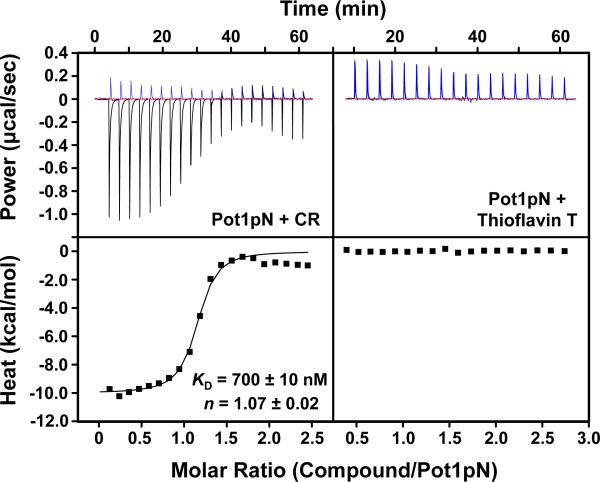
Surprisingly, NMR and mass spectrometry analysis revealed that the active small molecule identified in the screen is the known compound Congo red (Figure 1D; Supporting Information Figure S3). However, the binding assay does not distinguish between CR interaction with Pot1pN and interaction with the ssDNA ligand. To directly assay the ability of CR to interact with Pot1pN, we performed isothermal titration calorimetry (ITC) experiments. Using ITC, we found that Pot1pN/CR binding was comprised of a major exothermic interaction, and continued titration of CR beyond saturation of Pot1pN resulted in a second process that evolved additional heat. The initial exothermic interaction was fit with a one-site binding model to reveal that Pot1pN robustly bound CR with dissociation constant of 700 ± 10 nM and 1:1 stoichiometry (n = 1.07 ± 0.02) (Figure 2). Because CR undergoes micellar-like self-association and is known to cause oligomerization of complexes at high concentration (81, 100-102), we hypothesized that the minor process was a result of oligomerization or aggregation at high CR concentrations toward the end of the titration. To address this, we performed the reverse experiment keeping CR below the aggregation point of 50 μM (103) and titrating Pot1pN. We observed a single exothermic interaction with a KD comparable to that of the forward titration (620 nM; Supporting Information Figure S4). These findings demonstrate that CR directly binds to Pot1pN.
Figure 2.
Pot1pN binds CR but not Thioflavin T. The left panels show baseline corrected raw ITC data (upper) for 1.2 mM CR titrated into buffer (blue) or 100 μM Pot1pN and the reference-subtracted integrated heat of binding (lower). The right panels show baseline corrected raw ITC data (upper) for 2 mM Thioflavin T titrated into buffer (blue) or 100 μM Pot1pN and the reference-subtracted integrated heat of binding (lower). KD and n values for fitting triplicate Pot1pN/CR experiments to a one-site binding model are reported; errors are the standard error of the mean.
CR is also known to bind amyloid fibrils and fibril-forming proteins and peptides (reviewed in (104)). In order to assess the specificity of Pot1pN binding to CR, we tested Pot1pN binding to another amyloid fibril-binding small molecule, Thioflavin T (83, 105). By ITC, we observed no detectable binding of Thioflavin T to Pot1pN (Figure 2). We additionally verified that Thioflavin T has no effect on Pot1pN/ssDNA binding using a double filter-binding assay (data not shown). These data demonstrate that direct binding of Pot1pN by CR inhibits the interaction with ssDNA and that Pot1pN likely does not bind the compound by a mechanism similar to amyloid fibril/CR binding.
CR Promotes Specific Pot1pN Trimerization at High Concentration
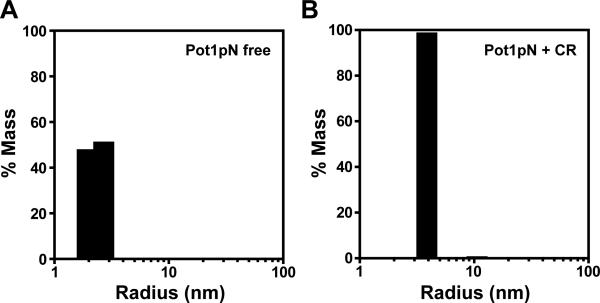
The secondary event observed by ITC was suggestive of CR-mediated higher order complexation. In order to examine this possibility more thoroughly, we used dynamic light scattering (DLS) to probe the oligomerization state of the Pot1pN/CR complex at high concentration. As expected from NMR, EMSA, and gel filtration studies (73, 93), 100% of free Pot1pN existed in solution as a monomer with a calculated radius of 2.3 nm and a calculated MW of 25 kDa (expected MW of 22.6 kDa) (Figure 3). Upon addition of equimolar CR (300 μM), the species fully shifted to a new state with a calculated radius of 3.8 nm and a MW of 77 kDa (Figure 3). This mass is consistent with the MW of a 3:3 Pot1pN/CR trimer complex. This species accounts for 99% of the total sample mass, and thus indicates that the Pot1pN/CR complex exists as a single, discrete species as opposed to a population-weighted average of non-specific aggregates. Addition of a 5-fold excess of 6mer reverted the majority (80%) of the protein to a monomeric state with an average MW of 24 kDa (Supporting Information Figure 5), demonstrating that CR-mediated trimerization is largely reversible.
Figure 3.
Particle size distribution obtained by DLS shows that CR-bound Pot1pN is a trimer. (A) A monomeric species of calculated radius and MW of 1.3 nm and 25 kDa, respectively, accounts for 100% of sample mass for free Pot1pN. (B) The calculated radius and MW for the Pot1pN/CR sample are 3.8 nm and 77 kDa, respectively, and this species accounts for 99% of the total sample mass.
CR Interacts with the ssDNA-binding Cleft of Pot1
Because CR competes with ssDNA to bind Pot1, we hypothesized that CR interacts directly with the ssDNA-binding surface of the Pot1 proteins. We used NMR to probe the direct structural interactions between CR and Pot1pN. 1H-15N HSQC experiments are a powerful strategy to assess structural alterations to a protein by monitoring chemical shift changes that report on differences in the chemical environment of the protein backbone. Backbone residue assignments are available for free Pot1pN at pH 6.15 (93, 106). However, as CR precipitates from solution at this pH, we conducted the analysis at higher pH. Raising the pH to 8.0 resulted in the loss of some resonances from exchange (data not shown), thus we limited our analysis to the 67% of peak assignments we could readily transfer from the low pH assignment (116/174 peaks).
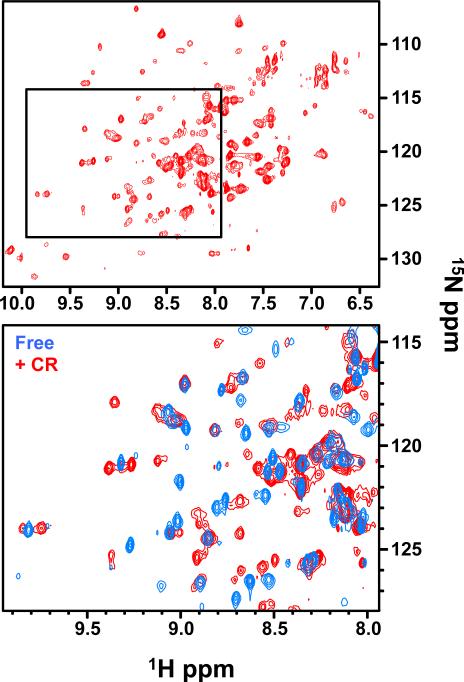
Very few crosspeaks were detected in the initial CR-bound 15N-Pot1pN spectrum, suggesting either line broadening from a large complex size or severe exchange broadening (data not shown). To improve the relaxation properties and increase sensitivity for a larger molecular weight species, we prepared doubly labeled 2H/15N-Pot1pN samples and acquired free and CR-bound TROSY spectra at 900 MHz. For the 2H/15N-Pot1pN/CR complex, we observed well-dispersed peaks in the spectrum, demonstrating that CR addition caused neither protein unfolding nor extreme aggregation (Figure 4). Consistent with DLS data, the differences between deuterated and non-deuterated Pot1pN/CR spectra suggest the formation of a homogeneous stable Pot1pN oligomer. Furthermore, approximately 25 more peaks were observed in the CR-bound spectrum, as compared to the free spectrum, which suggests that several residues undergoing exchange in the free spectrum are visible in the presence of CR or may exist in multiple chemical environments. While we were not able to independently assign the CR-bound spectrum, comparison to the Pot1pN free state allowed us to qualitatively assess the nature of the Pot1pN/CR interface. Superposition of the spectra allowed us to confidently transfer residue assignments for peaks that showed no significant changes between free and CR-bound. This comparison of the free and bound spectra revealed 68 of the 116 assigned residues undergo no chemical shift change upon CR addition (Figure 4). Mapping these residues on the crystal structure of Pot1pN reveals that these residues are all located distal from the ssDNA-binding face (Figure 5), revealing that CR does not interact indiscriminately with the protein surface. Additionally, there were a significant number of peaks in the free spectrum for which there were no obvious CR-bound candidate peaks, suggesting that these are sites of clear chemical shift change in the CR-bound complex. We were able to designate 35 residues as significantly perturbed in the CR spectrum in comparison to the free spectrum. Importantly, these residues cluster within the ssDNA-binding cleft of Pot1pN (Figure 5). These data demonstrate both that CR does not cause global rearrangement of the protein and that it specifically interacts with the ssDNA-binding surface of Pot1pN.
Figure 4.
Comparison of CR-bound (red) and free (blue) Pot1pN 1H-15N HSQC spectra obtained at 900 MHz. reveals differences upon CR binding, but no global changes or aggregation of Pot1pN. Boxed region of CR-bound spectrum in top panel is expanded below and superposed with free Pot1pN.
Figure 5.
Mapping of amide backbone chemical shift changes onto the Pot1pN crystal structure demonstrates that CR specifically binds the Pot1pN ssDNA-binding surface. Pot1pN residues definitively unperturbed by CR binding (magenta), residues that undergo significant chemical shift changes in the presence of CR (teal), and unassigned residues (gray) are shown. Pot1pN N-terminus is indicated (N) and 6mer (blue sticks) is shown for reference in the ssDNA-binding cleft. Images were generated using MacPymol version 1.3 (92).
CR Inhibits SpPot1 and hPOT1 Interaction with ssDNA
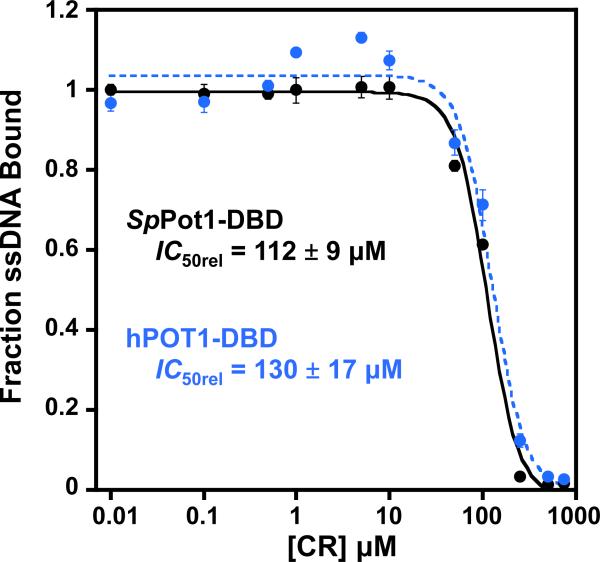
Pot1pN is one of two OB folds that constitute the complete DNA-binding domain of SpPot1 (SpPot1-DBD) (86). Because Pot1pN confers specificity to SpPot1-DBD/ssDNA interactions (76, 77, 86), we hypothesized that the inhibition we observed for Pot1pN applies to the fully active protein. Free SpPot1-DBD is unstable with respect to aggregation at the concentrations required for ITC, and we were unable to obtain direct binding data for SpPot1-DBD/CR. Instead, we performed a competition experiment similar to our secondary screen assay to determine if CR inhibits the interaction of SpPot1-DBD with cognate ssDNA. Small molecule inhibition studies often determine IC50 values to describe the ability of a compound to inhibit protein activity. While accurate values can be determined for enzymatic activities, readout of the absolute inhibition of protein/nucleic acid interactions cannot be determined by traditional means. In order to determine an accurate IC50 for the inhibition of a protein/ssDNA interaction, the ssDNA ligand must be held at a concentration significantly in excess of the dissociation constant (107), such that all of the protein is fully bound. This constraint has the result that the large signal from the free ssDNA masks the much smaller signal from the protein/ssDNA interaction and any changes in binding caused by the inhibitor. However, the assay can be used to determine whether or not a compound inhibits an interaction and to obtain relative compound activities in similar systems (IC50rel), as long as the intrinsic ssDNA-binding affinities are comparable. Using the filter-binding competition assay with SpPot1-DBD and ssDNA at close to 1:1, we found that CR inhibits SpPot1-DBD/ssDNA binding with an IC50rel of 112 ± 9 μM (Figure 6), suggesting that CR directly blocks ssDNA binding by SpPot1-DBD. As mentioned above, assay limitations do not allow for the measurement of absolute IC50 values; however, this result clearly demonstrates the inhibitory activity of CR on SpPot1-DBD interaction with telomeric ssDNA.
Figure 6.
Plotting of the global fits from triplicate filter-binding experiments show that CR equivalently inhibits SpPot1-DBD (black) and hPOT1-DBD (blue) binding cognate telomeric ssDNA. Apparent IC50rel values for triplicate experiments are reported; errors are the standard error of the mean.
SpPot1 and hPOT1 are evolutionarily homologous, structurally similar (Figure 1A), and recognize very similar telomeric DNA-repeat sequences (d(GGTTAC) and d(GGTTAG), respectively) (10, 73, 75, 108). Based on these similarities, we hypothesized that CR would interact with the DNA-binding domain of hPOT1 (hPOT1-DBD). As with SpPot1-DBD, hPOT1-DBD in its free state is unstable at the concentrations required for ITC, thus we performed competition assays to probe binding. Using the same protein and ssDNA concentrations as for SpPot1-DBD, we found that CR inhibits telomeric ssDNA binding by hPOT1-DBD to the same extent as SpPot1-DBD, with an apparent IC50rel of 130 ± 17 μM (Figure 6). As the IC50 is directly related to the KD values of the protein/ssDNA and protein/inhibitor interactions (107) and the the dissociation constants for SpPot1 and hPOT1 interaction with cognate ssDNA are comparable ((77) and data not shown), the apparent IC50rels measured here report on the relative ability of CR to compete for binding to these two proteins. Thus, these data show that CR inhibits SpPot1 and hPOT1 to a similar extent.
CR Binding Shows Specificity for the Pot1 Family of TEP Proteins
S. pombe and human Pot1 are members of the telomere-end protection (TEP) family of proteins. All known TEP proteins utilize structurally homologous OB-fold domains to bind telomeric ssDNA (72). Because we observed inhibition of both SpPot1 and hPOT1 proteins, we examined whether this interaction is specific for Pot1 in comparison to the TEP family in general. We probed the specificity of the Pot1/CR interaction by first testing CR binding to the DNA-binding domain of the evolutionarily distinct TEP protein Cdc13 from Saccharomyces cerevisiae. Even with increased CR concentration, we observed no binding to Cdc13 over the course of the titration (data not shown). This suggests that Pot1pN has specificity for CR, but this specificity does not extend to all TEP proteins.
This led us to further investigate the specificity Pot1/CR binding. Pot1pN and hPOT1 OB1 have highly similar ssDNA-binding surfaces, suggesting that specific residues may be involved in CR binding. Both Pot1pN and hPOT1 OB1 contain a conserved phenylalanine that forms an intermolecular aromatic stack with the ssDNA ligand (F88 and F62, respectively; Figure 1A) (74, 75). Mutation of F88 to alanine in both Pot1pN and SpPot1-DBD results in a significant decrease in binding affinity (76, 77). Similarly, we found that the F88A mutation in Pot1pN (Pot1pN_F88A) had a dramatic effect on Pot1pN binding to CR. This mutation causes a >10-fold reduction in binding affinity (9.6 ± 0.4 μM) and a large unfavorable change in ΔH of ~ 4 kcal/mol (-6.0 ± 0.1 kcal/mol vs. -9.7 ± 0.2 kcal/mol) (Table 1 and Figure 7A). These data suggest that CR interacts specifically with residues in the ssDNA-binding cleft of Pot1 and that perturbations to this surface that impact ssDNA binding also weaken the Pot1/CR interaction.
Table 1.
Thermodynamic parameters of binding for PotlpN/CR, Pot1pN_F88A/CR, and PotlpN/TB interactions measured at 20 °C.
| Protein/ligand interaction | KD (μM) | ΔH (kcal/mol) | -TΔS (kcal/mol) |
|---|---|---|---|
| PotlpN/CR | 0.7 ± 0.01 | -9.7 ± 0.2 | 1.4 ± 0.2 |
| Pot1pN_F88A/CR | 9.6 ± 0.2 | -6.0 ± 0.1 | 0.74 ± 0.09 |
| PotlpN/TB | 1.2 ± 0.05 | -13.6 ± 0.004 | 5.7 ± 0.02 |
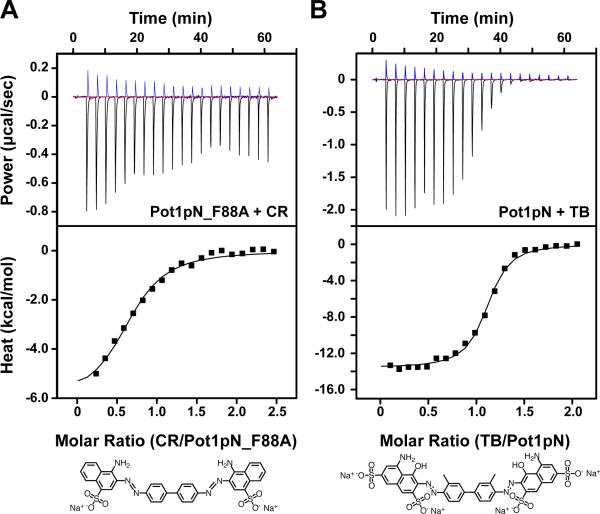
Figure 7.
Mutation of Pot1pN and alteration of CR structure alter the characteristics of binding. (A) Baseline corrected raw ITC data (upper) for 1.2 mM CR titrated into buffer (blue) or 100 μM Pot1pN_F88A and the reference-subtracted integrated heat of binding (lower). (B) Baseline corrected raw ITC data (upper) for 1 mM TB titrated into buffer (blue) or 103 μM Pot1pN and the reference-subtracted integrated heat of binding (lower). The structures of CR and TB are shown below for comparison Titrations were performed at 20 °C with 21 injections. Integrated heat data were fit to a one-site binding model.
Several compounds are known that are structurally similar to CR. For example, the congener Trypan blue (TB) has a structure that is largely similar to that of CR, with the same bis-azo symmetric scaffold, but containing both added and positional alterations of some functional groups (Figure 7B). Specifically, TB contains two additional sulphonate groups and two hydroxyl groups. ITC data show that TB bound to Pot1pN with a KD similar to, but slightly weaker than, CR (1.21 μM ± 0.05 μM) (Table 1 and Figure 7). Additionally, Pot1pN/TB binding was characterized by significantly more favorable enthalpy and compensating unfavorable entropy (Table 1), suggesting that the structural alterations between CR and TB may contribute to differences in binding mode. The ability of related molecules to block ssDNA binding suggests this scaffold may be amenable to further optimization for the specific targeting of Pot1.
Discussion
Human POT1 plays a central role in telomere maintenance; however, understanding this role has been limited by an inability to either fully delete the protein or exclusively disrupt its ssDNA-binding function. The number of hPOT1 binding sites in the genome is quite small, thus, even with 90% knockdown of the protein levels, the remaining hPOT1 may provide enough function to confound results, and, additionally this technique cannot address the separate the functions of hPOT1. Exongenous expression of mutant hPOT1 at endogenous levels is not feasible, and reports of overexpression of DNA-binding deficient hPOT1 have conflicting results likely due to the complications of dominant negative behavior (11, 70, 71). The ability to specifically inhibit endogenous hPOT1 ssDNA-binding activity would provide a means to precisely define the role of this function of hPOT1 in telomere maintenance. Taking advantage of the similarities between the human and yeast Pot1 proteins and the highly stable nature of Pot1pN, we developed and implemented an effective HTS screen utilizing the benefits of time-resolved FRET to quickly and efficiently screen a set of ~20,000 small molecules for inhibition of Pot1pN binding to telomeric ssDNA and identified a single effective inhibitor. This inhibitor, Congo red, functions by specifically and competitively interacting with the ssDNA-binding surface of Pot1. Crucially, CR also inhibits hPOT1 binding to human telomeric DNA. Our data demonstrate the suitability of Pot1pN as a model for small molecule inhibition of hPOT1 and establish that the Pot1/ssDNA interface is an attractive target for small molecule interference.
Successful High-throughput Screening for Inhibitors of Pot1 Interaction with Telomeric ssDNA
The chemically rich surface of the ssDNA-binding cleft of Pot1 provides many potential interaction sites for small molecules capable of specifically inhibiting ssDNA binding. We have developed an HTS assay well optimized for targeting this interaction. The robustness of the selected FRET pair and ease of tagging both the protein and the ssDNA make this screen suitable for application to a wide range of protein/nucleic acid systems. One of the main caveats of this assay design is the potential susceptibility of FRET to direct interference by some small molecules in the screening libraries. Indeed, analysis of the spectral properties of many of the positive hits identified by this screen suggests this to be the case. However, the reasonably high throughput secondary double filter-binding verification step directly assays the ability of a compound to disrupt the protein/nucleic acid interaction, making elimination of false positives rapid and facile. The identification of a single inhibitor from the >20,000 compound library suggests the Pot1 ssDNA interface is highly specific beyond DNA sequence recognition and is not a promiscuous small molecule binding site, making Pot1 an excellent candidate for larger scale screening.
Nature of the Pot1/CR interaction
Our identification of CR as an inhibitor of the ssDNA-binding function of Pot1 is unexpected considering CR is known to bind a number of other proteins and is most recognized for its ability to bind amyloid fibrils. CR and other amyloid-binding small molecules, including Thioflavin T, interact with the β-sheet pockets and channels characteristic of amyloid fibrils (82, 109-111). While CR directly binds the β-barrel surface involved in ssDNA binding, Thioflavin T does not, demonstrating that this surface does not interact indiscriminately with β-sheet-binding small molecules.
Our Pot1 screening assay design was based on the hypothesis that inhibitors of Pot1pN/ssDNA interaction would also inhibit the activity of the human protein. Our data show that indeed CR inhibits the ssDNA-binding activity of hPOT1 to the same degree as SpPot1-DBD, demonstrating that the specific targeting of Pot1pN is sufficient to specify for the human protein. As with Pot1/ssDNA binding, electrostatics do not appear to play a significant role in CR/Pot1 binding. While CR is net negatively charged, the two additional sulfonate groups on Trypan blue did not affect the binding affinity. These findings are consistent with the specific nature of Pot1/ssDNA binding, in which the protein makes numerous base-specific and stacking interactions, but minimal contact with the phosphate backbone (74, 75). Intriguingly, we found that CR does not interact with Cdc13, the TEP from budding yeast. While both Pot1 and Cdc13 contain OB folds with specificity for GT-rich ssDNA, the Cdc13 ssDNA-binding cleft is longer and shallower over much of the surface compared to those of the Pot1 proteins and additionally contains a long flexible loop contributing a large number of contacts (112, 113). These distinctions likely contribute to the difference in binding between the Pot1 proteins and Cdc13. We also observed that a Pot1pN mutant (Pot1pN_F88A) deficient in DNA binding displays reduced binding to CR. Phe88 lies on the surface of the OB fold β-barrel and participates in a key stacking interaction with two nucleotides of the 6mer ligand (74). Considering the aromatic nature of CR, the observed reduction in binding may not be surprising. However, the ssDNA-binding surface of Cdc13 contains seven aromatic residues (in contrast to three for Pot1pN), yet showed no affinity for CR. Taken together these data suggest that while CR binds numerous proteins, the compound is specific for the unique structural characteristics of Pot1.
While CR is known for its amyloid-binding activities, several lines of evidence suggest a distinct mechanism is responsible for its action in inhibiting Pot1/ssDNA activity. The planar nature of the CR structure causes the molecule to readily self-aggregate, and this process is important for interaction with amyloid proteins (101, 114, 115). While CR also triggers trimerization when binding Pot1, we note that the binding constant for CR interaction with Pot1pN is well below the aggregation point of CR (103), and that self-association only occurs at very high concentrations of CR. This suggests that self-assembly of CR is dispensable for interaction with Pot1. The robust interaction of TB with Pot1pN supports this hypothesis. TB does not self-associated as a result of its non-planarity due to the positioning of its additional sulfonate groups (114, 116). As a result, TB poorly binds amyloid-forming proteins (109, 117). In contrast, TB binds Pot1 nearly as well as CR. Furthermore, TB does not cause secondary complexation following saturation of the Pot1pN binding sites, showing that binding is not driven by self-assembly of the compound. Despite the similarities between CR and TB, ITC data revealed large differences in the enthalpic and entropic contributions to binding, demonstrating that these compounds to not interact with Pot1 equivalently.
Potential Biological Impact
The identification of a known amyloid binding protein as a telomere-binding protein inhibitor suggests pleotropic functions for CR. The use of CR and CR derivatives to slow amyloid plaque growth in cell culture and animal models has been studied extensively ((104) and references therein). These studies have shown that low doses of CR (≤ 10 μM) rescue amyloidosis phenotypes and prolong cell survival. However, high doses of CR (~ 100 μM) have been shown to be toxic in cell culture and animal models (118). In addition, prenatal administration of CR and certain analogs to mice during germ cell development resulted in a drastic and permanent reduction in germ cell number (119). Because testicular and ovarian cells are some of the most rapidly proliferating cells, part of this phenotype might stem from specific effects of CR on telomere maintenance. However, CR can interact with many other proteins in the cell that are far more abundant that Pot1, thus any specific effects related to Pot1 cannot be determined from available data. A less promiscuous inhibitor of Pot1 is required to understand the in vivo effects of specific interference with Pot1/ssDNA binding. The differences we observed between TB and CR suggests that probing the structure-function relationship of Pot1pN/CR could lead to higher affinity and more specific interaction with the protein.
Supplementary Material
Acknowledgment
We gratefully acknowledge Drs. Geoffrey Armstrong and Richard Shoemaker for advice and assistance with NMR experiments. We also thank Dr. Garry Dallmann for technical assistance with the Biomek FX workstation and ActivityBase software. We acknowledge Dr. Robert Batey for ITC experimental suggestions and discussion. We thank Dr. Karen Lewis for critical review of this manuscript.
We acknowledge the National Institutes of Health Training Appointment (NIH) GM065103 (to S.E.A.), National Research Service Award Postdoctoral Fellowship GM-071257 (to J.E.C), NIH GM-059414 and NS-059370 (to D.S.W.), and University of Colorado Innovative Seed Grant (to D.S.W) for funding this research, and a shared instrumentation grant from NIH for the ITC (1S10 RR026516).
Abbreviations
- 6mer
oligonucleotide sequence d(GGTTAC)
- βME
β-mercaptoethanol
- BSA
bovine serum albumin
- CR
Congo red
- CTD
C-terminal domain
- DLS
dynamic light scattering
- DMSO
dimethyl sulfoxide
- DTT
dithiothreitol
- EC
Eu-W1024-labeled anti-6xHis antibody
- FRET
fluorescence resonance energy transfer
- HTS
high-throughput screen
- IC50
apparent half-maximal inhibitor concentration
- IC50rel
relative apparent half-maximal inhibitor concentration
- IPTG
isopropyl-beta-D-thiogalactopyranoside
- KD
apparent equilibrium binding constant
- ITC
isothermal titration calorimetry
- OB fold
oligonucleotide/oligosaccharide-binding fold
- Pot1
protection of telomeres 1 protein
- SpPot1-DBD
complete DNA-binding domain of full length S. pombe Pot1
- Pot1pN
first DNA-binding OB fold of full length S. pombe Pot1
- SpPot1
Schizosaccharyomyces pombe protection of telomeres 1
- ssDNA
single-stranded deoxyribonucleic acid
- TB
Trypan blue
- TR-FRET
high-throughput time-resolved fluorescence resonance energy transfer
- UL
ULight-streptavidin
Footnotes
Supporting Information Available
Figures S1-S5 are available free of charge via the Internet at http://pubs.acs.org.
References
- 1.d'Adda di Fagagna F, Teo SH, Jackson S. Functional links between telomeres and proteins of the DNA-damage response. Genes Dev. 2004;18:1781–1799. doi: 10.1101/gad.1214504. [DOI] [PubMed] [Google Scholar]
- 2.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 3.Shampay J, Szostak JW, Blackburn EH. DNA sequences of telomeres maintained in yeast. Nature. 1984;310:154–157. doi: 10.1038/310154a0. [DOI] [PubMed] [Google Scholar]
- 4.Lingner J, Cooper JP, Cech TR. Telomerase and DNA end replication: no longer a lagging strand problem? Science. 1995;269:1533–1534. doi: 10.1126/science.7545310. [DOI] [PubMed] [Google Scholar]
- 5.Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 6.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 7.Denchi EL, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448:1068–1071. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- 8.de Lange T. How telomeres solve the end-protection problem. Science. 2009;326:948–952. doi: 10.1126/science.1170633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hockemeyer D, Sfeir AJ, Shay JW, Wright WE, de Lange T. POT1 protects telomeres from a transient DNA damage response and determines how human chromosomes end. EMBO J. 2005;24:2667–2678. doi: 10.1038/sj.emboj.7600733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baumann P, Cech TR. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science. 2001;292:1171–1175. doi: 10.1126/science.1060036. [DOI] [PubMed] [Google Scholar]
- 11.Loayza D, de Lange T. POT1 as a terminal transducer of TRF1 telomere length control. Nature. 2003;423:1013–1018. doi: 10.1038/nature01688. [DOI] [PubMed] [Google Scholar]
- 12.Veldman T, Etheridge K, Counter C. Loss of hPot1 function leads to telomere instability and a cut-like phenotype. Curr. Biol. 2004;14:2264–2270. doi: 10.1016/j.cub.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 13.Yang Q, Zheng YL, Harris CC. POT1 and TRF2 cooperate to maintain telomeric integrity. Mol. Cell. Biol. 2005;25:1070–1080. doi: 10.1128/MCB.25.3.1070-1080.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Colgin LM, Baran K, Baumann P, Cech TR, Reddel RR. Human POT1 facilitates telomere elongation by telomerase. Curr. Biol. 2003;13:942–946. doi: 10.1016/s0960-9822(03)00339-7. [DOI] [PubMed] [Google Scholar]
- 15.Lei M, Zaug AJ, Podell ER, Cech TR. Switching human telomerase on and off with hPOT1 protein in vitro. J. Biol. Chem. 2005;280:20449–20456. doi: 10.1074/jbc.M502212200. [DOI] [PubMed] [Google Scholar]
- 16.Ye JZ, Hockemeyer D, Krutchinsky AN, Loayza D, Hooper SM, Chait BT, de Lange T. POT1-interacting protein PIP1: a telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes Dev. 2004;18:1649–1654. doi: 10.1101/gad.1215404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shakirov EV, Surovtseva YV, Osbun N, Shippen-Lentz DE. The Arabidopsis Pot1 and Pot2 proteins function in telomere length homeostasis and chromosome end protection. Mol. Cell. Biol. 2005;25:7725–7733. doi: 10.1128/MCB.25.17.7725-7733.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watson JD. Origin of concatemeric T7 DNA. Nature New Biol. 1972;239:197–201. doi: 10.1038/newbio239197a0. [DOI] [PubMed] [Google Scholar]
- 19.Olovnikov AM. A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J. Theor. Biol. 1973;41:181–190. doi: 10.1016/0022-5193(73)90198-7. [DOI] [PubMed] [Google Scholar]
- 20.Hayflick L. The limited in vitro lifetime of human diploid cell strains. Exp. Cell Res. 1965;37:614–636. doi: 10.1016/0014-4827(65)90211-9. [DOI] [PubMed] [Google Scholar]
- 21.Folini M, Venturini L, Cimino-Reale G, Zaffaroni N. Telomeres as targets for anticancer therapies. Expert Opin. Ther. Targets. 2011;15:579–593. doi: 10.1517/14728222.2011.556621. [DOI] [PubMed] [Google Scholar]
- 22.Blackburn EH, Greider CW, Henderson E, Lee MS, Shampay J, Shippen-Lentz DE. Recognition and elongation of telomeres by telomerase. Genome. 1989;31:553–560. doi: 10.1139/g89-104. [DOI] [PubMed] [Google Scholar]
- 23.Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 24.Greider CW, Blackburn EH. The telomere terminal transferase of Tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell. 1987;51:887–898. doi: 10.1016/0092-8674(87)90576-9. [DOI] [PubMed] [Google Scholar]
- 25.Counter CM, Avilion AA, LeFeuvre CE, Stewart NG, Greider CW, Harley CB, Bacchetti S. Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J. 1992;11:1921–1929. doi: 10.1002/j.1460-2075.1992.tb05245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL, Shay JW. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 27.Shay JW, Bacchetti S. A survey of telomerase activity in human cancer. Eur. J. Cancer. 1997;33:787–791. doi: 10.1016/S0959-8049(97)00062-2. [DOI] [PubMed] [Google Scholar]
- 28.Shay JW, Wright WE. Telomerase: a target for cancer therapeutics. Cancer Cell. 2002;2:257–265. doi: 10.1016/s1535-6108(02)00159-9. [DOI] [PubMed] [Google Scholar]
- 29.Tárkányi I, Aradi J. Pharmacological intervention strategies for affecting telomerase activity: future prospects to treat cancer and degenerative disease. Biochimie. 2008;90:156–172. doi: 10.1016/j.biochi.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 30.Buseman CM, Wright WE, Shay JW. Is telomerase a viable target in cancer? Mutat. Res. 2012;730:90–97. doi: 10.1016/j.mrfmmm.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang PR, Yeh YM, Wang TC. Potent inhibition of human telomerase by helenalin. Cancer Lett. 2005;227:169–174. doi: 10.1016/j.canlet.2004.11.045. [DOI] [PubMed] [Google Scholar]
- 32.Eitsuka T, Nakagawa K, Suzuki T, Miyazawa T. Polyunsaturated fatty acids inhibit telomerase activity in DLD-1 human colorectal adenocarcinoma cells: a dual mechanism approach. Biochim. Biophys. Acta. 2005;1737:1–10. doi: 10.1016/j.bbalip.2005.08.017. [DOI] [PubMed] [Google Scholar]
- 33.He H, Xia HH, Wang JD, Gu Q, Lin MC, Zou B, Lam SK, Chan AO, Yuen MF, Kung HF, Wong BC. Inhibition of human telomerase reverse transcriptase by nonsteroidal antiinflammatory drugs in colon carcinoma. Cancer. 2006;106:1243–1249. doi: 10.1002/cncr.21694. [DOI] [PubMed] [Google Scholar]
- 34.Strahl C, Blackburn EH. Effects of reverse transcriptase inhibitors on telomere length and telomerase activity in two immortalized human cell lines. Mol. Cell. Biol. 1996;16:53–65. doi: 10.1128/mcb.16.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hayakawa N, Nozawa K, Ogawa A, Kato N, Yoshida K, Akamatsu K, Tsuchiya M, Nagasaka A, Yoshida S. Isothiazolone derivatives selectively inhibit telomerase from human and rat cancer cells in vitro. Biochemistry. 1999;38:11501–11507. doi: 10.1021/bi982829k. [DOI] [PubMed] [Google Scholar]
- 36.Ueno T, Takahashi H, Oda M, Mizunuma M, Yokoyama A, Goto Y, Mizushina Y, Sakaguchi K, Hayashi H. Inhibition of human telomerase by rubromycins: implication of spiroketal system of the compounds as an active moiety. Biochemistry. 2000;39:5995–6002. doi: 10.1021/bi992661i. [DOI] [PubMed] [Google Scholar]
- 37.Pascolo E, Wenz C, Lingner J, Hauel N, Priepke H, Kauffmann I, Garin-Chesa P, Rettig WJ, Damm K, Schnapp A. Mechanism of human telomerase inhibition by BIBR1532, a synthetic, non-nucleosidic drug candidate. J. Biol. Chem. 2002;277:15566–15572. doi: 10.1074/jbc.M201266200. [DOI] [PubMed] [Google Scholar]
- 38.Francis R, West C, Friedman SH. Targeting telomerase via its key RNA/DNA heteroduplex. Bioorg. Chem. 2001;29:107–117. doi: 10.1006/bioo.2000.1196. [DOI] [PubMed] [Google Scholar]
- 39.Rangarajan S, Friedman SH. Design, synthesis, and evaluation of phenanthridine derivatives targeting the telomerase RNA/DNA heteroduplex. Bioorg. Med. Chem. Lett. 2007;17:2267–2273. doi: 10.1016/j.bmcl.2007.01.070. [DOI] [PubMed] [Google Scholar]
- 40.Kondo S, Tanaka Y, Kondo Y, Hitomi M, Barnett GH, Ishizaka Y, Liu J, Haqqi T, Nishiyama A, Villeponteau B, Cowell JK, Barna BP. Antisense telomerase treatment: induction of two distinct pathways, apoptosis and differentiation. FASEB J. 1998;12:801–811. doi: 10.1096/fasebj.12.10.801. [DOI] [PubMed] [Google Scholar]
- 41.Herbert B, Pitts AE, Baker SI, Hamilton SE, Wright WE, Shay JW, Corey DR. Inhibition of human telomerase in immortal human cells leads to progressive telomere shortening and cell death. Proc. Natl. Acad. Sci. U.S.A. 1999;96:14276–14281. doi: 10.1073/pnas.96.25.14276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hahn WC, Stewart SA, Brooks MW, York SG, Eaton E, Kurachi A, Beijersbergen RL, Knoll JH, Meyerson M, Weinberg RA. Inhibition of telomerase limits the growth of human cancer cells. Nat. Med. 1999;5:1164–1170. doi: 10.1038/13495. [DOI] [PubMed] [Google Scholar]
- 43.Zhang X, Mar V, Zhou W, Harrington L, Robinson MO. Telomere shortening and apoptosis in telomerase-inhibited human tumor cells. Genes Dev. 1999;13:2388–2399. doi: 10.1101/gad.13.18.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yi X, White DM, Aisner DL, Baur JA, Wright WE, Shay JW. An alternate splicing variant of the human telomerase catalytic subunit inhibits telomerase activity. Neoplasia. 2000;2:433–440. doi: 10.1038/sj.neo.7900113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim MM, Rivera MA, Botchkina IL, Shalaby R, Thor AD, Blackburn EH. A low threshold level of expression of mutant-template telomerase RNA inhibits human tumor cell proliferation. Proc. Natl. Acad. Sci. U.S.A. 2001;98:7982–7987. doi: 10.1073/pnas.131211098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Damm K, Hemmann U, Garin-Chesa P, Hauel N, Kauffmann I, Priepke H, Niestroj C, Daiber C, Enenkel B, Guilliard B, Lauritsch I, Müller E, Pascolo E, Sauter G, Pantic M, Martens UM, Wenz C, Lingner J, Kraut N, Rettig WJ, Schnapp A. A highly selective telomerase inhibitor limiting human cancer cell proliferation. EMBO J. 2001;20:6958–6968. doi: 10.1093/emboj/20.24.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zahler AM, Williamson JR, Cech TR, Prescott DM. Inhibition of telomerase by G-quartet DNA structures. Nature. 1991;350:718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- 48.Sun D, Thompson B, Cathers BE, Salazar M, Kerwin SM, Trent JO, Jenkins TC, Neidle S, Hurley LH. Inhibition of human telomerase by a G-quadruplex-interactive compound. J. Med. Chem. 1997;40:2113–2116. doi: 10.1021/jm970199z. [DOI] [PubMed] [Google Scholar]
- 49.Read M, Harrison RJ, Romagnoli B, Tanious FA, Gowan SH, Reszka AP, Wilson WD, Kelland LR, Neidle S. Structure-based design of selective and potent G quadruplex-mediated telomerase inhibitors. Proc. Natl. Acad. Sci. U.S.A. 2001;98:4844–4849. doi: 10.1073/pnas.081560598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Riou JF, Guittat L, Mailliet P, Laoui A, Renou E, Petitgenet O, Mégnin-Chanet F, Hélène C, Mergny JL. Cell senescence and telomere shortening induced by a new series of specific G-quadruplex DNA ligands. Proc. Natl. Acad. Sci. U.S.A. 2002;99:2672–2677. doi: 10.1073/pnas.052698099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim MY, Vankayalapati H, Shin-ya K, Wierzba K, Hurley LH. Telomestatin, a potent telomerase inhibitor that interacts quite specifically with the human telomeric intramolecular G-quadruplex. J. Am. Chem. Soc. 2002;124:2098–2099. doi: 10.1021/ja017308q. [DOI] [PubMed] [Google Scholar]
- 52.Gowan SM, Heald R, Stevens MF, Kelland LR. Potent inhibition of telomerase by small-molecule pentacyclic acridines capable of interacting with G-quadruplexes. Mol. Pharmacol. 2001;60:981–988. doi: 10.1124/mol.60.5.981. [DOI] [PubMed] [Google Scholar]
- 53.Binz N, Shalaby T, Rivera P, Shin-Ya K, Grotzer MA. Telomerase inhibition, telomere shortening, cell growth suppression and induction of apoptosis by telomestatin in childhood neuroblastoma cells. Eur. J. Cancer. 2005;41:2873–2881. doi: 10.1016/j.ejca.2005.08.025. [DOI] [PubMed] [Google Scholar]
- 54.El-Daly H, Kull M, Zimmermann S, Pantic M, Waller CF, Martens UM. Selective cytotoxicity and telomere damage in leukemia cells using the telomerase inhibitor BIBR1532. Blood. 2005;105:1742–1749. doi: 10.1182/blood-2003-12-4322. [DOI] [PubMed] [Google Scholar]
- 55.Tauchi T, Shin-ya K, Sashida G, Sumi M, Nakajima A, Shimamoto T, Ohyashiki JH, Ohyashiki K. Activity of a novel G-quadruplex-interactive telomerase inhibitor, telomestatin (SOT-095), against human leukemia cells: involvement of ATM-dependent DNA damage response pathways. Oncogene. 2003;22:5338–5347. doi: 10.1038/sj.onc.1206833. [DOI] [PubMed] [Google Scholar]
- 56.Tahara H, Shin-Ya K, Seimiya H, Yamada H, Tsuruo T, Ide T. G-Quadruplex stabilization by telomestatin induces TRF2 protein dissociation from telomeres and anaphase bridge formation accompanied by loss of the 3' telomeric overhang in cancer cells. Oncogene. 2006;25:1955–1966. doi: 10.1038/sj.onc.1209217. [DOI] [PubMed] [Google Scholar]
- 57.Miyazaki T, Pan Y, Joshi K, Purohit D, Hu B, Demir H, Mazumder S, Okabe S, Yamori T, Viapiano M, Shin-ya K, Seimiya H, Nakano I. Telomestatin impairs glioma stem cell survival and growth through the disruption of telomeric G-quadruplex and inhibition of the proto-oncogene, c-Myb. Clin. Cancer Res. 2012;18:1268–1280. doi: 10.1158/1078-0432.CCR-11-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gomez D, O'Donohue MF, Wenner T, Douarre C, Macadré J, Koebel P, Giraud-Panis MJ, Kaplan H, Kolkes A, Shin-ya K, Riou JF. The G-quadruplex ligand telomestatin inhibits POT1 binding to telomeric sequences in vitro and induces GFP-POT1 dissociation from telomeres in human cells. Cancer Res. 2006;66:6908–6912. doi: 10.1158/0008-5472.CAN-06-1581. [DOI] [PubMed] [Google Scholar]
- 59.Gomez D, Wenner T, Brassart B, Douarre C, O'Donohue MF, El Khoury V, Shin-ya K, Morjani H, Trentesaux C, Riou JF. Telomestatin-induced telomere uncapping is modulated by POT1 through G-overhang extension in HT1080 human tumor cells. J. Biol. Chem. 2006;281:38721–38729. doi: 10.1074/jbc.M605828200. [DOI] [PubMed] [Google Scholar]
- 60.Brassart B, Gomez D, De Cian A, Paterski R, Montagnac A, Qui KH, Temime-Smaali N, Trentesaux C, Mergny JL, Gueritte F, Riou JF. A new steroid derivative stabilizes g-quadruplexes and induces telomere uncapping in human tumor cells. Mol. Pharmacol. 2007;72:631–640. doi: 10.1124/mol.107.036574. [DOI] [PubMed] [Google Scholar]
- 61.Salvati E, Leonetti C, Rizzo A, Scarsella M, Mottolese M, Galati R, Sperduti I, Stevens MF, D'Incalci M, Blasco M, Chiorino G, Bauwens S, Horard B, Gilson E, Stoppacciaro A, Zupi G, Biroccio A. Telomere damage induced by the G-quadruplex ligand RHPS4 has an antitumor effect. J. Clin. Invest. 2007;117:3236–3247. doi: 10.1172/JCI32461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gunaratnam M, Greciano O, Martins C, Reszka AP, Schultes CM, Morjani H, Riou JF, Neidle S. Mechanism of acridine-based telomerase inhibition and telomere shortening. Biochem. Pharmacol. 2007;74:679–689. doi: 10.1016/j.bcp.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 63.Rodriguez R, Müller S, Yeoman JA, Trentesaux C, Riou JF, Balasubramanian S. A novel small molecule that alters shelterin integrity and triggers a DNA-damage response at telomeres. J. Am. Chem. Soc. 2008;130:15758–15759. doi: 10.1021/ja805615w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rizzo A, Salvati E, Porru M, D'Angelo C, Stevens MF, D'Incalci M, Leonetti C, Gilson E, Zupi G, Biroccio A. Stabilization of quadruplex DNA perturbs telomere replication leading to the activation of an ATR-dependent ATM signaling pathway. Nucleic Acids Res. 2009;37:5353–5364. doi: 10.1093/nar/gkp582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kondo T, Oue N, Yoshida K, Mitani Y, Naka K, Nakayama H, Yasui W. Expression of POT1 is associated with tumor stage and telomere length in gastric carcinoma. Cancer Res. 2004;64:523–529. doi: 10.1158/0008-5472.can-03-1196. [DOI] [PubMed] [Google Scholar]
- 66.Yang Q, Zhang R, Horikawa I, Fujita K, Afshar Y, Kokko A, Laiho P, Aaltonen LA, Harris CC. Functional diversity of human protection of telomeres 1 isoforms in telomere protection and cellular senescence. Cancer Res. 2007;67:11677–11686. doi: 10.1158/0008-5472.CAN-07-1390. [DOI] [PubMed] [Google Scholar]
- 67.Vega F, Cho-Vega JH, Lennon PA, Luthra MG, Bailey J, Breeden M, Jones D, Medeiros LJ, Luthra R. Splenic marginal zone lymphomas are characterized by loss of interstitial regions of chromosome 7q, 7q31.32 and 7q36.2 that include the protection of telomere 1 (POT1) and sonic hedgehog (SHH) genes. Br. J. Haematol. 2008;142:216–226. doi: 10.1111/j.1365-2141.2008.07176.x. [DOI] [PubMed] [Google Scholar]
- 68.Incles CM, Schultes CM, Kelland LR, Neidle S. Acquired cellular resistance to flavopiridol in a human colon carcinoma cell line involves up-regulation of the telomerase catalytic subunit and telomere elongation. Sensitivity of resistant cells to combination treatment with a telomerase inhibitor. Mol. Pharmacol. 2003;64:1101–1108. doi: 10.1124/mol.64.5.1101. [DOI] [PubMed] [Google Scholar]
- 69.Tang T, Zhou FX, Lei H, Yu HJ, Xie CH, Zhou YF, Liu SQ. Increased expression of telomere-related proteins correlates with resistance to radiation in human laryngeal cancer cell lines. Oncol. Rep. 2009;21:1505–1509. doi: 10.3892/or_00000381. [DOI] [PubMed] [Google Scholar]
- 70.Armbruster BN, Linardic CM, Veldman T, Bansal NP, Downie DL, Counter C. Rescue of an hTERT mutant defective in telomere elongation by fusion with hPOT1. Mol. Cell. Biol. 2004;24:3552–3561. doi: 10.1128/MCB.24.8.3552-3561.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Barrientos KS, Kendellen MF, Freibaum BD, Armbruster BN, Etheridge K, Counter CM. Distinct functions of POT1 at telomeres. Mol. Cell. Biol. 2008;28:5251–5264. doi: 10.1128/MCB.00048-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Croy JE, Wuttke DS. Themes in ssDNA recognition by telomere-end protection proteins. Trends Biochem. Sci. 2006;31:516–525. doi: 10.1016/j.tibs.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 73.Lei M, Baumann P, Cech TR. Cooperative binding of single-stranded telomeric DNA by the Pot1 protein of Schizosaccharomyces pombe. Biochemistry. 2002;41:14560–14568. doi: 10.1021/bi026674z. [DOI] [PubMed] [Google Scholar]
- 74.Lei M, Podell ER, Baumann P, Cech TR. DNA self-recognition in the structure of Pot1 bound to telomeric single-stranded DNA. Nature. 2003;426:198–203. doi: 10.1038/nature02092. [DOI] [PubMed] [Google Scholar]
- 75.Lei M, Podell ER, Cech TR. Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat. Struct. Mol. Biol. 2004;11:1223–1229. doi: 10.1038/nsmb867. [DOI] [PubMed] [Google Scholar]
- 76.Croy JE, Altschuler SE, Grimm NE, Wuttke DS. Nonadditivity in the recognition of single-stranded DNA by the Schizosaccharomyces pombe protection of telomeres 1 DNA-binding domain, Pot1-DBD. Biochemistry. 2009;48:6864–6875. doi: 10.1021/bi900307x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Altschuler SE, Dickey TH, Wuttke DS. Schizosaccharomyces pombe protection of telomeres 1 utilizes alternate binding modes to accommodate different telomeric sequences. Biochemistry. 2011;50:7503–7513. doi: 10.1021/bi200826a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zappulla DC, Roberts JN, Goodrich KJ, Cech TR, Wuttke DS. Inhibition of yeast telomerase action by the telomeric ssDNA-binding protein, Cdc13p. Nucleic Acids Res. 2009;37:354–367. doi: 10.1093/nar/gkn830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in Eealuation and validation of high throughput screening assays. J. Biomol. Screening. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 80.Stockley PG. Filter-binding assays. Methods Mol. Biol. 2009;543:1–14. doi: 10.1007/978-1-60327-015-1_1. [DOI] [PubMed] [Google Scholar]
- 81.Pedersen MØ, Mikkelsen K, Behrens MA, Pedersen JS, Enghild JJ, Skrydstrup T, Malmendal A, Nielsen NC. NMR reveals two-step association of Congo Red to amyloid β in low-molecular-weight aggregates. J. Phys. Chem. B. 2010;114:16003–16010. doi: 10.1021/jp108035y. [DOI] [PubMed] [Google Scholar]
- 82.Groenning M, Olsen L, van de Weert M, Flink JM, Frokjaer S, Jørgensen FS. Study on the binding of Thioflavin T to beta-sheet-rich and non-beta-sheet cavities. J. Struct. Biol. 2007;158:358–369. doi: 10.1016/j.jsb.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 83.Foderà V, Groenning M, Vetri V, Librizzi F, Spagnolo S, Cornett C, Olsen L, van de Weert M, Leone M. Thioflavin T hydroxylation at basic pH and its effect on amyloid fibril detection. J. Phys. Chem. B. 2008;112:15174–15181. doi: 10.1021/jp805560c. [DOI] [PubMed] [Google Scholar]
- 84.Pease AP. Master's thesis. Virginia Polytechnical Institute and State University; 2000. Novel approaches to evaluate osteoarthritis in the rabbit lateral meniscectomy model. [Google Scholar]
- 85.Phoacharern P. Master's thesis. Kasetsart University; 2006. Photocatalytic degradation of Trypan Blue using gold/titanium dioxide. [Google Scholar]
- 86.Croy JE, Podell ER, Wuttke DS. A new model for Schizosaccharomyces pombe telomere recognition: the telomeric single-stranded DNA-binding activity of Pot11-389. J. Mol. Biol. 2006;361:80–93. doi: 10.1016/j.jmb.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 87.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 88.Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 89.Bazin H, Préaudat M, Trinquet E, Mathis G. Homogeneous time resolved fluorescence resonance energy transfer using rare earth cryptates as a tool for probing molecular interactions in biology. Spectrochim. Acta, Part A. 2001;57:2197–2211. doi: 10.1016/s1386-1425(01)00493-0. [DOI] [PubMed] [Google Scholar]
- 90.Degorce F, Card A, Soh S, Trinquet E, Knapik GP, Xie B. HTRF: A technology tailored for drug discovery - a review of theoretical aspects and recent applications. Curr. Chem. Genomics. 2009;3:22–32. doi: 10.2174/1875397300903010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sierk ML, Kleywegt GJ. Déjà vu all over again. Structure. 2004;12:2103–2111. doi: 10.1016/j.str.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 92.Schrodinger LLC. The PyMOL Molecular Graphics System. Version 1.3r1. 2010.
- 93.Croy JE, Fast JL, Grimm NE, Wuttke DS. Deciphering the mechanism of thermodynamic accommodation of telomeric oligonucleotide sequences by the Schizosaccharomyces pombe protection of telomeres 1 (Pot1pN) protein. Biochemistry. 2008;47:4345–4358. doi: 10.1021/bi701778x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Macarrón R, Hertzberg RP. Design and implementation of high throughput screening assays. Mol. Biotechnol. 2011;47:270–285. doi: 10.1007/s12033-010-9335-9. [DOI] [PubMed] [Google Scholar]
- 95.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Delivery Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 96.Dallmann HG, Fackelmayer OJ, Tomer G, Chen J, Wiktor-Becker A, Ferrara T, Pope C, Oliveira MT, Burgers PM, Kaguni LS, McHenry CS. Parallel multiplicative target screening against divergent bacterial replicases: identification of specific inhibitors with broad spectrum potential. Biochemistry. 2010;49:2551–2562. doi: 10.1021/bi9020764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ungermannova D, Parker SJ, Nasveschuk CG, Chapnick DA, Phillips AJ, Kutcha RD, Liu X. Identification and mechanistic studies of a novel ubiquitin E1 inhibitor. J. Biomol. Screening. 2012;17:421–434. doi: 10.1177/1087057111433843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wong I, Chao KL, Bujalowski W, Lohman TM. DNA-induced dimerization of the Escherichia coli rep helicase. Allosteric effects of single-stranded and duplex DNA. J. Biol. Chem. 1992;267:7596–7610. [PubMed] [Google Scholar]
- 99.Wong I, Lohman TM. A double-filter method for nitrocellulose-filter binding: application to protein-nucleic acid interactions. Proc. Natl. Acad. Sci. U.S.A. 1993;90:5428–5432. doi: 10.1073/pnas.90.12.5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Skowronek M, Stopa B, Konieczny L, Rybarska J, Piekarska B, Szneler E, Bakalarski G, Roterman I. Self-assembly of Congo Red—A theoretical and experimental approach to identify its supramolecular organization in water and salt solutions. Biopolymers. 1998;46:267–281. [Google Scholar]
- 101.Lendel C, Bolognesi B, Wahlström A, Dobson CM, Gräslund A. Detergent-like interaction of Congo red with the amyloid beta peptide. Biochemistry. 2010;49:1358–1360. doi: 10.1021/bi902005t. [DOI] [PubMed] [Google Scholar]
- 102.Mourtzis N, Cordoyiannis G, Nounesis G, Yannakopoulou K. Single and Double Threading of Congo Red into γ-Cyclodextrin. Solution Structures and Thermodynamic Parameters of 1:1 and 2:2 Adducts, as Obtained from NMR Spectroscopy and Microcalorimetry. Supramol. Chem. 2003;15:639–649. [Google Scholar]
- 103.Edwards RA, Woody RW. Induced circular dichroism as a probe of Cibacron Blue and Congo Red bound to dehydrogenases. Biochem. Biophys. Res. Commun. 1977;79:470–476. doi: 10.1016/0006-291x(77)90181-4. [DOI] [PubMed] [Google Scholar]
- 104.Frid P, Anisimov SV, Popovic N. Congo red and protein aggregation in neurodegenerative diseases. Brain Res. Rev. 2007;53:135–160. doi: 10.1016/j.brainresrev.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 105.Vassar PS, Culling CF. Fibrosis of the breast. AMA Arch. Pathol. 1959;67:128–133. [PubMed] [Google Scholar]
- 106.Croy JE, Wuttke DS. Insights into the dynamics of specific telomeric single-stranded DNA recognition by Pot1pN. J. Mol. Biol. 2009;387:935–948. doi: 10.1016/j.jmb.2009.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Goodrich JA, Kugel JF. Binding and Kinetics for Molecular Biologists. Cold Spring Harbor Laboratory Press; New York: 2007. Using Competition to Assess Affinity: IC50; pp. 33–39. [Google Scholar]
- 108.Moyzis RK, Buckingham JM, Cram LS, Dani M, Deaven LL, Jones MD, Meyne J, Ratliff RL, Wu JR. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc. Natl. Acad. Sci. U.S.A. 1988;85:6622–6626. doi: 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ashburn TT, Han H, McGuinness BF, Lansbury PT. Amyloid probes based on Congo Red distinguish between fibrils comprising different peptides. Chem. Biol. 1996;3:351–358. doi: 10.1016/s1074-5521(96)90118-0. [DOI] [PubMed] [Google Scholar]
- 110.Groenning M, Norrman M, Flink JM, van de Weert M, Bukrinsky JT, Schluckebier G, Frokjaer S. Binding mode of Thioflavin T in insulin amyloid fibrils. J. Struct. Biol. 2007;159:483–497. doi: 10.1016/j.jsb.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 111.Krebs MRH, Bromley EHC, Donald AM. The binding of Thioflavin-T to amyloid fibrils: localisation and implications. J. Struct. Biol. 2005;149:30–37. doi: 10.1016/j.jsb.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 112.Mitton-Fry RM, Anderson EM, Hughes TR, Lundblad V, Wuttke DS. Conserved structure for single-stranded telomeric DNA recognition. Science. 2002;296:145–147. doi: 10.1126/science.1068799. [DOI] [PubMed] [Google Scholar]
- 113.Mitton-Fry RM, Anderson EM, Theobald DL, Glustrom LW, Wuttke DS. Structural basis for telomeric single-stranded DNA recognition by yeast Cdc13. J. Mol. Biol. 2004;338:241–255. doi: 10.1016/j.jmb.2004.01.063. [DOI] [PubMed] [Google Scholar]
- 114.Skowronek M, Roterman, Konieczny L, Stopa B, Rybarska J, Piekarska B, Górecki A, Król M. The conformational characteristics of Congo red, Evans blue and Trypan blue. Comput. Chem. 2000;24:429–450. doi: 10.1016/s0097-8485(99)00089-3. [DOI] [PubMed] [Google Scholar]
- 115.Feng BY, Toyama BH, Wille H, Colby DW, Collins SR, May BC, Prusiner SB, Weissman J, Shoichet BK. Small-molecule aggregates inhibit amyloid polymerization. Nat. Chem. Biol. 2008;4:197–199. doi: 10.1038/nchembio.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Stopa B, Górny M, Konieczny L, Piekarska B, Rybarska J, Skowronek M, Roterman I. Supramolecular ligands: monomer structure and protein ligation capability. Biochimie. 1998;80:963–968. doi: 10.1016/s0300-9084(99)80001-7. [DOI] [PubMed] [Google Scholar]
- 117.Stopa B, Piekarska B, Konieczny L, Rybarska J, Spólnik P, Zemanek G, Roterman I, Król M. The structure and protein binding of amyloid-specific dye reagents. Acta Biochim. Pol. 2003;50:1213–1227. [PubMed] [Google Scholar]
- 118.Seki T, Takahashi H, Yamamoto K, Ogawa K, Onji T, Adachi N, Tanaka S, Hide I, Saito N, Sakai N. Congo red, an amyloid-inhibiting compound, alleviates various types of cellular dysfunction triggered by mutant protein kinase cγ that causes spinocerebellar ataxia type 14 (SCA14) by inhibiting oligomerization and aggregation. J. Pharmacol. Sci. 2010;114:206–216. doi: 10.1254/jphs.10170fp. [DOI] [PubMed] [Google Scholar]
- 119.Gray LE, Ostby JS. The effects of prenatal administration of azo dyes on testicular development in the mouse: a structure activity profile of dyes derived from benzidine, dimethylbenzidine, or dimethoxybenzidine. Fundam. App. Toxicol. 1993;20:177–183. doi: 10.1006/faat.1993.1024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.