Abstract
Here we describe the Immunogenetic Management Software (IMS) system, a novel web-based application that permitsmultiplexed analysis of complex immunogenetic traits that are necessary for the accurate planning and execution of experiments involving large animal models, including nonhuman primates. IMS is capable of housing complex pedigree relationships, microsatellite-based MHC typing data, as well as MHC pyrosequencing expression analysis of class I alleles. It includes a novel, automated MHC haplotype naming algorithm and has accomplished an innovative visualization protocol that allows users to view multiple familial and MHC haplotype relationships through a single, interactive graphical interface. Detailed DNA and RNA-based data can also be queried and analyzed in a highly accessible fashion, and flexible search capabilities allow experimental choices to be made based on multiple, individualized and expandable immunogenetic factors. This web application is implemented in Java, MySQL, Tomcat, and Apache, with supported browsers including Internet Explorer and Firefox onWindows and Safari on Mac OS. The software is freely available for distribution to noncommercial users by contacting Leslie. kean@emory.edu. A demonstration site for the software is available at http://typing.emory.edu/typing_demo, user name: imsdemo7@gmail.com and password: imsdemo.
Keywords: Genetics, Visualization, Web services, Next-generation sequencing, MHC
Nonhuman primate (NHP) models represent the preclinical gold standard for both transplantation (Wiseman and O'Connor 2007; Miller et al. 2010; Singh et al. 2010) and HIV translational research (Sawai et al. 1996; Barouch et al. 2000; Igarashi et al. 2001; Wu et al. 2006; Wiseman et al. 2007). Despite their importance, the lack of knowledge about the immunogenetics of NHP (Thomas et al. 1989, 2000, 2006; Kenyon et al. 1999; Kawai et al. 2004; Lechler et al. 2005; Kean et al. 2006) has impacted both the reproducibility of results obtained with these models and the degree to which NHP studies can predict clinical success or failure. This is especially true for the field of transplantation, in which the degree of relatedness and MHC genetics can play a defining role in transplantation outcome (McGiffin et al. 1997; Malkki et al. 2007; Petersdorf et al. 2007). Given this limitation, our group has led a major NIH-funded initiative in the past 5 years that has resulted in the creation of the first fully pedigreed, MHC-defined, directed breeding NHP colonies. Through this initiative, thousands of animals have been subjected to microsatellite-based genetic analysis of their parentage and pedigree, medium-density microsatellite analysis of their MHC chromosomal structure (Penedo et al. 2005; Kanthaswamy et al. 2010), as well as detailed MHC expression sequencing of class I alleles using massively multiplexed pyrosequencing techniques (Wiseman et al. 2009). The first studies using this new national resource have now been completed (Larsen et al. 2010; Miller et al. 2010; Page et al. 2011) and have established detailed immunogenetically mapped macaques as the new gold standard for primate translational research.
While the MHC genotyping initiative has been a major advance, it has led to significant challenges in managing the resulting genetic database. Experiments now require the discernment of multiple genetic factors for their proper execution, including the degree of relatedness between multiple animals within complex polygamous social structures, theirMHC haplotype genetics, as well as the expression patterns of numerous MHC class I alleles (Wiseman and O'Connor 2007; Miller et al. 2010). Traditional family tree analysis software is inadequate to display even the pedigree information for these animals, given the highly complex mating patterns observed in NHPs. In order to most efficiently make experimental choices, researchers require a visual display that integrates multiple relational databases in a platform that is also capable of effectively accessing and managing detailed DNA and RNA-based immunogenetic data. To address this unmet need, we created a novel webbased application that permits multiplexed analysis of several complex immunogenetic traits for experimental NHP. It is capable of housing complex pedigree relationships, microsatellite-based MHC typing data, as well as MHC pyrosequencing expression analysis of class I alleles. It includes an automated, robust microsatellite-based MHC haplotype naming algorithm and has accomplished an innovative visualization protocol that allows users to view multiple familial and MHC haplotype relationships through a single, interactive graphical interface. Detailed DNA and RNA-based data can also be queried and analyzed in a highly accessible fashion, and flexible search capabilities allow experimental choices to be made based on multiple, individualized and expandable immunogenetic factors. We believe that this software will be a critical component of the evolution of immunologic research, as it will permit translational researchers to optimally utilize multiple sources of immunogenetic data now available on their prospective research subjects, and required for precise experimental planning and analysis. In what follows and in the Electronic supplementary materials, we describe the features and the implementation of the Immunogenetic Management Software system (“IMS”), highlighting both the visualization and search modalities.
Features
Novel multiparameter pedigree visualization tool
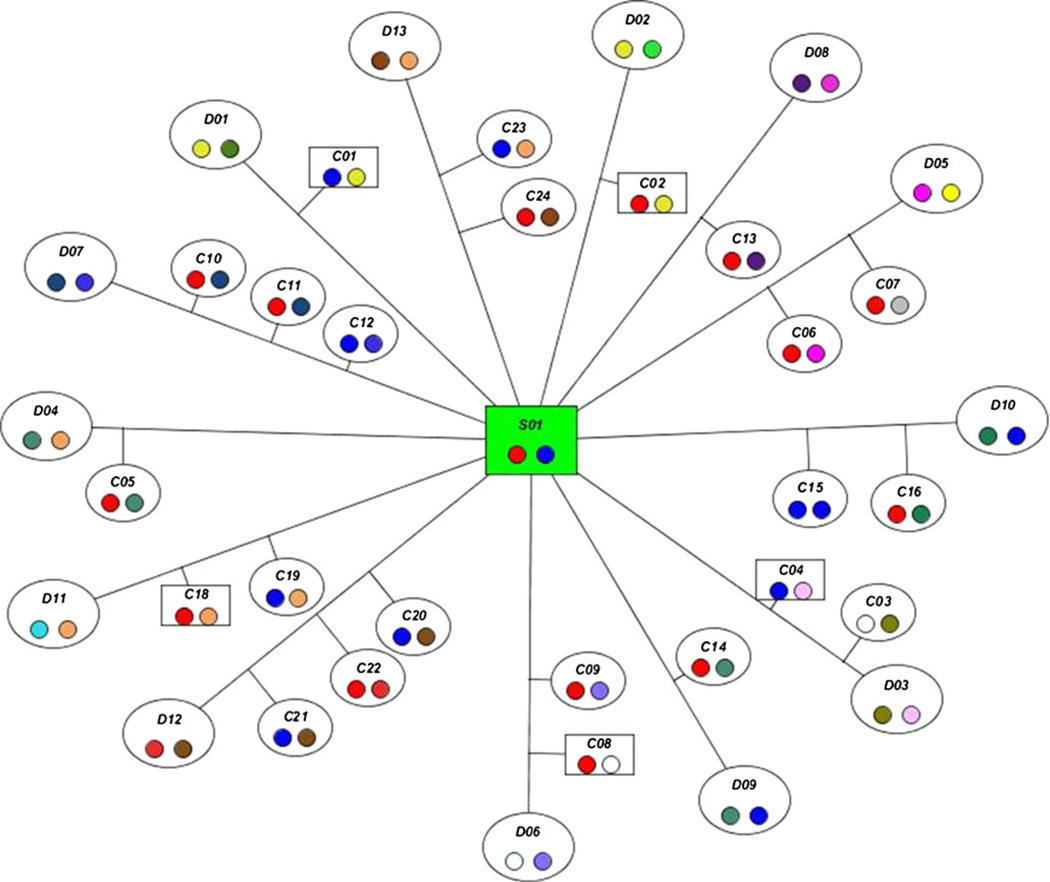
This application is designed to handle multicolony, multispecies data sets, each composed of numerous categories of genetic information. One of the major advances is the implementation of a novel visualization tool for viewing complex pedigrees along with associated immunogenetic data. The structural complexity of primate mating patterns makes traditional commercially available pedigree software tools both cumbersome and uninformative for primate family structural display. To address this, IMS analyzes relationships among animals within the colony and constructs the pedigree chart relative to a selected animal using a modified polar coordinate system (see Fig. 1). Coded microsatellite-based MHC haplotype patterns are overlaid onto the pedigree information using uniquely colored symbols, which enables the simultaneous visualization of multiple familial and MHC-derived relationships. The visualization tool also includes an interactive link to all source data, including DNA microsatellite-based MHC typing, detailed MHC expression databases, and the demographic database.
Fig. 1.
A sire centric view of a complex pedigree with the sire highlighted in green and each spoke representing a dam with whom the sire has produced at least one offspring.
MHC haplotype assignment tool
This application has implemented a novel algorithm that allows the de novo identification of MHC chromosomal haplotypes based on microsatellite genotypes. After uploading primary genotyping data that has been previously parsed [using utilities such as BEAGLE (Browning and Browning 2007) or PHASE (Stephens and Donnelly 2003)] into distinct inheritance patterns, the application allows a flexible core group of microsatellites to be queried in order to identify MHC haplotype patterns and assigns a colony-specific MHC haplotype name to each unique core sequence. To accomplish this, the entire MHC chromosome is divided into three regions, including the core group of microsatellite markers and two sets of flanking markers located on the telomeric and centromeric ends of the chromosome. Core sequence patterns are compared across the colony, and a counter is used to assign pattern names sequentially. Starting with 1, the counter is incremented every time a new core pattern sequence is found, and the value of the counter is assigned as the colonyspecific name of the pattern sequence. The process also identifies and flags sequence length discrepancies (which could be due to either recombination events or genotyping errors) through a comparison algorithm. For every animal, the flanking markers are compared with the matching haplotype patterns of its parents, and if differences are found in the base pair lengths of the corresponding patterns found on the parents, the pattern name on the offspring is flagged with an asterisk to indicate a discrepancy. This is a critical utility for genetic analysis of the MHC, given the significant rate of recombination that occurs within this chromosomal region (Doxiadis et al. 2001; Otting et al. 2005) The microsatellitebased MHC haplotypes are also coded with unique color codes, which are overlaid onto the pedigree visualization tool, described above. This allows rapid visual assessment of both degree of relatedness and degree of MHC similarity between animals considered for transplantation.
MHC expression sequencing search tool
Perhaps the most important technical advance to occur to NHP MHC typing over the last decade has been the advent of massively multiplexed MHC expression sequencing (Karl et al. 2009; Wiseman et al. 2009). Given the highly complexMHC chromosomal structure that exists in OldWorld monkeys, where the MHC has undergone significant DNA level duplication (Penedo et al. 2005; Bontrop 2006) and where multiple MHC genes are transcribed, in a hierarchical, rather than a codominant manner (Otting et al. 2005), a complete MHC analysis of NHP has been exceedingly difficult, time consuming, and costly to complete. Massively multiplexed pyrosequencing of class I alleles has fundamentally changed the landscape of NHP MHC typing and now affords NHP researchers extremely detailed MHC expression information on all potential experimental animals (Wiseman et al. 2009). However, the ability to mine this data continues to represent a major challenge for researchers.
IMS provides functionalities that enable transplant planning based on MHC expression data and which facilitates the integration of this data with pedigree and STR-based haplotype information. After primary MHC expression sequencing data are uploaded into the application, IMS calculates relative frequencies of the expressed MHC alleles to facilitate interanimal comparisons of MHC expression patterns. Animals can then be chosen based on complex patterns of both expressed and unexpressed MHC alleles. Integration of the microsatellite-based haplotype designations with pyrosequencing-based expression data will ultimately allow users to infer MHC expression patterns from microsatellite-based haplotype designations, such that the more inexpensive microsatellite surrogate can predict both the functional as well as the structural MHC of potential experimental animals. It should be noted, however, that transcription of an allele does not always result in its cell surface expression. Indeed, in rhesus macaques, some highly transcribed alleles have been found to be expressed at low levels on the cell surface (Rosner et al. 2010). Thus, the development of tools to rigorously assess cell surface MHC expression remains an unmet need in the field.
Flexibility to incorporate future genotyping strategies
While IMS was written to best meet the existing needs of the primate transplant/immunology community, with minor modification, this program can be used by investigators to meet very different goals. These include using the IMS platform to organize data relevant to genetic associations for behavioral studies including pedigree structure, inbreeding coefficients, 5HTTLPR length, and genotypes at multiple functional SNPs, as well as studies of viral immunology and the protective immune response, where interactions of MHC allele expression patterns and haplotypes will be tied to viral clearance or disease. Because of this inherent flexibility, we believe that IMS will be an integral tool for investigators using NHP or other animal models in which the visualization and organization of complex data within a pedigree are paramount to research goals.
IMS implementation
Language
The IMS web application is written in Java, allowing it to be deployed on any platform. The downloadable application is packagedwith Tomcat and MySQL included for installation on Windows.
Search hierarchy
The application provides robust search functionality to permit the selection of a single animal or a subgroup of animals for further analysis. When a single animal is selected for further analysis, the software enables analysis of the selected animal compared with other animals in the colony from different perspectives, including the pedigree visualization; the identification of the selected animal’s sire, dam, and full or half siblings; the identification of animals which share two, one, or zero microsatellite-based MHC haplotypes; and the choice of animals expressing similar or divergent MHC alleles.
Pedigree visualization
The pedigree visualization tool uses a novel approach to depict the colony, which includes the drawing of complex pedigrees from three perspectives: sire, dam, and offspring. As shown in the example in Fig. 1, when displayed in a sire centric view, the center node represents the sire, the outer nodes represent the dams, and the intermediate nodes represent the offspring.
To create the sire centric view, the algorithm starts with a point of origin, which represents the sire node, and is based on the centered coordinate of the dimension parameter. It then calculates four points based on a polar coordinate system at 0°, 90°, 180°, and 270° of the polar axis. These four points represent placement coordinates of dam nodes. The algorithm splits the coordinate system into four quadrants and calculates the points where the remaining dam nodes will be placed in the polar coordinate system. The boundaries of the quadrant are represented as vectors that are added together to produce a resultant vector that bisects them. This follows a recursive process of adding the new resultant vector within each boundary vector up to 16 times per quadrant to produce the endpoints for each potential dam node coordinate. The coordinates for the dam nodes are staggered in order to prevent overlapping of nodes.
In order to provide enough spacing between the dam nodes to display offspring nodes, the dam nodes are staggered by scaling the resultant vector by dividing the magnitude of the vector with the unit vector and then scaling the result with an alternating constant of 1.5 to create the staggered layout. The placement of the offspring nodes is determined by calculating points along the edge from the sire to the dam. These points represent attachment sites from the edge of the offspring node to the edge of the sire-to-dam edge. The coordinate for the offspring node is calculated as the vector orthogonal to attachment site from the sire to the dam edge mentioned previously. Since this type of edge connection (edge to edge) is not typical of computer science graph theory implementations, which consist of a collection a vertices and edges that connect pairs of vertices, it was necessary to create a custom layout algorithm for this purpose.
The dam centric view was implemented using the same algorithm as the sire centric approach, but the nodes representing the dams and sires are reversed. The offspring centric view is more complex because it joins pedigrees of both parents and links them using the mating line attached to the selected offspring. Rather than using four quadrants to draw the view, the algorithm divides the canvas and creates two sets of half quadrants to draw the sire and dam pedigrees. It then joins them using the mating line attached to the selected offspring.
Because the pedigree visualization displays two generations of animals, in some instances, an animal may appear in the chart with two distinct roles (either as both offspring and sire or as both offspring and dam). Given the complexities associated with depicting these dual roles, the pedigree algorithm is designed to display animals that have two roles as two distinct nodes. These nodes are identified with a red outline to indicate their duplication.
The pedigree visualization uses color coding to enable a visual distinction between microsatellite-based haplotype patterns. The algorithm calculates the maximum number of colors needed for a single chart by calculating twice the number of sire and dam nodes in the pedigree. The software has a limited preset array of colors. If more are needed, the application autogenerates them and then assigns them to chromosome patterns found in the pedigree. For simplicity and to assure that colors are easily discernable within a pedigree, the color coding is regenerated with each new pedigree visualization—a unique color is not assigned to each microsatellite-based MHC haplotype. Thus, the same color may be used to represent two different haplotypes, if they appear in two separate pedigree visualizations.
The pedigree visualization is displayed in the web application as a Java applet. The process that launches the pedigree visualization for a selected animal queries the database for that animal’s sire and dam. Using these attributes, the process identifies roles of animals relative to the selected animal. The application generates XML output that provides information used by the pedigree visualization to draw the resulting graphic.
List mode search and display capabilities
While the unique visualization tool allows multiple layers of genetic information to be summarized succinctly and in a visually simplified manner, final experiment choices are often made by superimposing the information derived from the visualized pedigree with that derived from other search capabilities that are included within this application. Multiple genetic parameters can be searched simultaneously, and results are displayed in an exportable, tabular format, which permits detailed analysis of large sets of primary genetic information that is housed within IMS for each animal. Examples of list mode search capabilities include the following:
Search for the degree of microsatellite-based MHC haplotype similarity: One of the most powerful search functions allows the user to query for animals within a colony by degree of microsatellite-based MHC haplotype similarity. The system provides the researcher with the capability to search for animals that are matched for two, one, or zero MHC haplotypes.
Allele-specific PCR analysis: While RNA pyrosequencing has become the new gold standard for MHC analysis in NHP, many colonies and researchers also use allele-specific PCR-based MHC typing. IMS has the capability to house these data as well as the capacity to search for positive or negative status across an array of PCR alleles.
Pyrosequencing-based MHC expression analysis: The software provides the capability to search for the presence or absence of multiple MHC alleles identified through pyrosequencing techniques. The application allows primary sequencing data to be stored and calculates a frequency for each expressed allele to facilitate interanimal comparisons. Both inclusive and exclusive search criteria are included in the application, allowing the identification of animals according to detailed MHC expression specificities.
Relational data display: For any selected animal, the researcher can view detailed information for all related animals, including its sire, dam, and full and half siblings. When the data are displayed in this context, the selected animal’s chromosomes are color coded—orange for the sire chromosome and blue for the dam. This enables the researcher to quickly view in tabular form the siblings having matching and mismatched microsatellite-based MHC haplotypes. In addition, in the detailed microsatellite-based haplotype view, primary microsatellite data are displayed, along with the identification of probes for which a recombination event has likely occurred.
Algorithm for calculating patterns of alleles determined by allele-specific PCR: The IMS software implements an algorithm to calculate the degree of MHC sharing based on allele-specific PCR results. To do this, the algorithm first evaluates the total number of MHC class I and MHC class II alleles for which allele-specific PCR-based results are available for each selected animal. Next, it calculates the numbers of MHC class I and MHC class II alleles that were identified as positive by allele-specific PCR and the number of these alleles that were identified as negative for both animals. Finally, the total number of positive or negative intersections (both animals positive for a given MHC allele or both animals negative for a given MHC allele) is calculated.
Algorithm for calculating shared MHC expressed alleles: The IMS software also implements an algorithm to calculate the degree of sharing of MHC alleles after pyrosequence-based expression analysis. To accomplish this, the algorithm calculates the total number of sequences having frequencies greater than 1% (coded as “positive” by MHC expression analysis) and the total number of sequences having frequencies less than or equal to 1%(coded as “negative” by expression analysis). Finally, the algorithm calculates the intersection of positive and negative MHC allele sequences between two selected animals.
Data management
Data management involves three key components, including the following:
Excel template file
Upload method
Algorithm for assigning microsatellite-based MHC haplotype pattern names
The downloadable distribution of IMS includes an Excel template file for uploading data into the IMS database. The upload method uses Apache POI to parse the Excel file and write to the database. Details about the upload method are included in the user manual.
Algorithm for assigning microsatellite-based haplotype pattern names
The algorithm that assigns microsatellite-based MHC haplotype pattern names is performed in two steps, first assigning names to unique core pattern sequences and then identifying and flagging recombination events.
The algorithm begins by dividing the sequence of microsatellite markers into three segments—(1) the core region, (2) the flanking markers on the telomeric end of the chromosome (called the “prefix”), and (3) the flanking markers on the centromeric end of the chromosome (called the “suffix”). The core, prefix, and suffix pattern sequences are represented as strings such that the base pair lengths in each of the regions are concatenated into three separate patterns representing the core, prefix, and suffix. The core pattern representation is used for comparing, identifying, and naming distinct haplotype patterns.
Three hashmaps are used for storing the core, prefix, and suffix pattern sequences described above. A hashmap is a data structure that maps that stores key/value pairs, and given a key, the value can be found. Keys must be unique, but values may be duplicated. Each of the hashmaps implemented contains entries for every animal chromosome in the data set. The entries are keyed on a combination of animal name and MHC haplotype number so that the algorithm can recall the pattern sequences when needed.
To generate the microsatellite-based haplotype pattern names, a counter is used and initialized to one. The first core pattern sequence is assigned the haplotype pattern name of 1. The distinct core pattern sequences and their names are stored in another hashmap, and after the first core pattern sequence is identified and named, it is added to the sequence name hashmap, with the entry keyed on the core pattern sequence. For every animal’s haplotypes, the algorithm compares the core pattern sequence by checking to see if it exists in the sequence name hashmap. If the core pattern sequence is found in the hashmap, the pattern name is recalled and assigned to the animal’s haplotype. If the core pattern sequence is not found in the sequence name hashmap, the counter is incremented by 1, assigned to the pattern sequence, an entry added to the sequence name hashmap, and the new haplotype pattern name is assigned to the animal. Finally, another hashmap is used for tracking animals with assigned pattern names.
After the pattern names are assigned, the algorithm iterates over the animals haplotypes to identify and flag genotyping discrepancies. For every animal, the algorithm checks for the animal’s sire and dam, and if both are present, compares the core sequence of the current animal with core sequences from both pairs of haplotypes found on the parents. If a match for the core sequence is found on one of the parent haplotypes, the corresponding prefix and suffix sequences of the current animal’s haplotype are compared with prefix and suffix patterns found on the matching parent haplotype. If the base pair lengths in either region are different, the genotyping discrepancy flag field in the database is marked with a “Y” to indicate that a genotyping discrepancy is detected within the haplotype pattern. If the current animal is missing a sire or dam, the genotyping discrepancy flag in the database is marked with a “U” indicating “unknown.” When the discrepancy flag is rendered in the web application, only the known genotyping discrepancies (i.e., those marked with a “Y” in the database) are displayed, using an asterisk appended at the end of the haplotype pattern name.
Haplotype pattern naming convention
To date, no single naming convention exists for microsatellite-based MHC haplotypes within the rhesus macaque (Evans et al. 1999; Penedo et al. 2005). This is due to the fact that individual investigators have used and published slightly different panels of microsatellites that take advantage of colonyspecific patterns of allelic frequency. To accommodate these differences, this application has implemented a colony-specific convention to naming microsatellite-based MHC haplotype patterns. As such, for each colony of origin, the application assigns a name and pattern number to patterns of base pair sequences found within the MHC region. The naming convention includes four parts: (1) the microsatellite-based haplotype being named (the MHC), (2) the species undergoing microsatellite-based MHC typing, (3) the colony to which the individual animal belongs, and (4) a unique six-digit number assigned to the designated microsatellite pattern. For example, for the Yerkes rhesus macaque colony, the first microsatellite-based MHC haplotype pattern would thus be named “MHC-Rhesus-Yerkes-000001.” It must be noted however that two animals with the same biological haplotype housed in different colonies would not be identified as matched using this current naming convention, given that the enumeration and naming of microsatellite-based MHC haplotype patterns occur independently for each colony. Once a universal microsatellite-based MHC haplotype naming convention is established, the IMS application can be easily converted to utilize these universal haplotype identifiers.
If a genotyping discrepancy is observed when comparing an offspring animal to his or her two parents, the existence of this discrepancy is shown with the “(*)” designation at the end of the alphanumeric designation. The ability to code for these discrepancies, which can arise either due to recombination events or to genotyping errors, is critical when naming within the MHC, given that this chromosomal region is one of the most gene-rich, polymorphic regions of the mammalian genome and is a hot spot for recombination (Trowsdale 1995; Gruen and Weissman 1997). Given the propensity for recombination, microsatellites are routinely genotyped both within the MHC and outside the core MHC region. Microsatellite patterns and recombination events in the flanking regions (which occur in as many as 2–3% of meiosis, Penedo et al. 2005) are identified with the “(*)” designation. While recombinations are also possible within the core MHC region, these are rarer than those occurring in the flanking regions and can also result in alterations in the expression patterns of individual MHC alleles. Given these issues, the current naming convention is founded on the identification of the core pattern; thus, if discrepancies occur within the core, they will not be designated with an “*” but rather with a new microsatellite-based MHC haplotype identification number. These events would be noted on pedigree analysis, which would reveal an apparent Mendelian error during reproduction.
Supplementary Material
Acknowledgments
The authors thank Kristy Kraemer, Ph.D. for her administrative and scientific leadership of the MHC typing initiative for the Nonhuman Primate Transplantation Tolerance Cooperative Steering Group and Isabelle Lussier for her significant contributions to the MHC typing initiative.
Funding This work was supported by Yerkes National Primate Research Center Base Grant, #RR00165. CPL was supported by NIH grant #s 2U19 AI051731, and 2P01 AI044644. LSK was supported by grant #s 5K08 AI065822, 2U19 AI051731, 1R01 HL095791 and 2U24 RR018109, and by a Burroughs Wellcome Fund Career Award in the Biomedical Sciences. Jim Else was supported by NIH grant # 2U24 RR018109. MCTP was supported by NIH grants # U42-RR16023 and U24 RR18144. Pyrosequencing efforts by DHO, RWW, and SML were supported by NIAID contract # HHSN266200400088C/N01-A1-40088.
Footnotes
ZP Johnson and RD Eady are co-first authors.
Electronic supplementary material The online version of this article (doi:10.1007/s00251-011-0587-8) contains supplementary material, which is available to authorized users.
Contributor Information
Z. P. Johnson, Yerkes National Primate Research Center, Emory University, Atlanta, GA 30322, USA
R. D. Eady, Research and Health Sciences Information Technology, Emory University, Atlanta, GA 30322, USA
S. F. Ahmad, Research and Health Sciences Information Technology, Emory University, Atlanta, GA 30322, USA
S. Agravat, Research and Health Sciences Information Technology, Emory University, Atlanta, GA 30322, USA
T Morris, Research and Health Sciences Information Technology, Emory University, Atlanta, GA 30322, USA.
J Else, Yerkes National Primate Research Center, Emory University, Atlanta, GA 30322, USA.
S. M. Lank, Wisconsin National Primate Research Center, University of Wisconsin–Madison, Madison, WI 53715, USA
R. W. Wiseman, Wisconsin National Primate Research Center, University of Wisconsin–Madison, Madison, WI 53715, USA
D. H. O’Connor, Wisconsin National Primate Research Center, University of Wisconsin–Madison, Madison, WI 53715, USA
M. C. T. Penedo, Veterinary Genetics Laboratory, University of California, Davis, CA 95616, USA
C. P. Larsen, The Emory Transplant Center, Department of Surgery, Emory University School of Medicine, Room 5203 WMB, 101 Woodruff Circle NE, Atlanta, GA 30322, USA
L. S. Kean, Email: Leslie.kean@emory.edu, The Emory Transplant Center, Department of Surgery, Emory University School of Medicine, Room 5203 WMB, 101 Woodruff Circle NE, Atlanta, GA 30322, USA
References
- Barouch DH, Craiu A, Kuroda MJ, Schmitz JE, Zheng XX, Santra S, Frost JD, Krivulka GR, Lifton MA, Crabbs CL, Heidecker G, Perry HC, Davies ME, Xie H, Nickerson CE, Steenbeke TD, Lord CI, Montefiori DC, Strom TB, Shiver JW, Lewis MG, Letvin NL. Augmentation of immune responses to HIV-1 and simian immunodeficiency virus DNA vaccines by IL-2/Ig plasmid administration in rhesus monkeys. Proc Natl Acad Sci U S A. 2000;97(8):4192–4197. doi: 10.1073/pnas.050417697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bontrop RE. Comparative genetics of MHC polymorphisms in different primate species: duplications and deletions. Hum Immunol. 2006;67(6):388–397. doi: 10.1016/j.humimm.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 2007;81(5):1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxiadis GG, Otting N, de Groot NG, Bontrop RE. Differential evolutionary MHC class II strategies in humans and rhesus macaques: relevance for biomedical studies. Immunol Rev. 2001;183:76–85. doi: 10.1034/j.1600-065x.2001.1830106.x. [DOI] [PubMed] [Google Scholar]
- Evans DT, Knapp LA, Jing P, Mitchen JL, Dykhuizen M, Montefiori DC, Pauza CD, Watkins DI. Rapid and slow progressors differ by a single MHC class I haplotype in a family of MHC-defined rhesus macaques infected with SIV. Immunol Lett. 1999;66(1–3):53–59. doi: 10.1016/s0165-2478(98)00151-5. [DOI] [PubMed] [Google Scholar]
- Gruen JR, Weissman SM. Evolving views of the major histocompatibility complex. Blood. 1997;90(11):4252–4265. [PubMed] [Google Scholar]
- Igarashi T, Brown CR, Endo Y, Buckler-White A, Plishka R, Bischofberger N, Hirsch V, Martin MA. Macrophage are the principal reservoir and sustain high virus loads in rhesus macaques after the depletion of CD4+ T cells by a highly pathogenic simian immunodeficiency virus/HIV type 1 chimera (SHIV): implications for HIV-1 infections of humans. Proc Natl Acad Sci U S A. 2001;98(2):658–663. doi: 10.1073/pnas.021551798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanthaswamy S, Kou A, Satkoski J, Penedo MC, Ward T, Ng J, Gill L, Lerche NW, Erickson BJ, Smith DG. Genetic characterization of specific pathogen-free rhesus macaque (Macaca mulatta) populations at the California National Primate Research Center (CNPRC) Am J Primatol. 2010;72(7):587–599. doi: 10.1002/ajp.20811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karl JA, Wiseman RW, O'Connor DH. Cost-effective sequence-based nonhuman primate MHC class I genotyping from RNA. Methods. 2009;49(1):11–17. doi: 10.1016/j.ymeth.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Sogawa H, Boskovic S, Abrahamian G, Smith RN, Wee SL, Andrews D, Nadazdin O, Koyama I, Sykes M, Winn HJ, Colvin RB, Sachs DH, Cosimi AB. CD154 blockade for induction of mixed chimerism and prolonged renal allograft survival in nonhuman primates. Am J Transplant. 2004;4(9):1391–1398. doi: 10.1111/j.1600-6143.2004.00523.x. [DOI] [PubMed] [Google Scholar]
- Kean LS, Gangappa S, Pearson TC, Larsen CP. Transplant tolerance in non-human primates: progress, current challenges and unmet needs. Am J Transplant. 2006;6(5 Pt 1):884–893. doi: 10.1111/j.1600-6143.2006.01260.x. [DOI] [PubMed] [Google Scholar]
- Kenyon NS, Chatzipetrou M, Masetti M, Ranuncoli A, Oliveira M, Wagner JL, Kirk AD, Harlan DM, Burkly LC, Ricordi C. Long-term survival and function of intrahepatic islet lografts in rhesus monkeys treated with humanized anti-CD154. Proc Natl Acad Sci U S A. 1999;96(14):8132–8137. doi: 10.1073/pnas.96.14.8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen CP, Page A, Linzie KH, Russell M, Deane T, Stempora L, Strobert E, Penedo MC, Ward T, Wiseman R, O'Connor D, Miller W, Sen S, Singh K, Kean LS. An MHC-defined primate model reveals significant rejection of bone marrow after mixed chimerism induction despite full MHC matching. Am J Transplant. 2010;10(11):2396–2409. doi: 10.1111/j.1600-6143.2010.03272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechler RI, Sykes M, Thomson AW, Turka LA. Organ transplantation—how much of the promise has been realized? Nat Med. 2005;11(6):605–613. doi: 10.1038/nm1251. [DOI] [PubMed] [Google Scholar]
- Malkki M, Gooley TA, Horowitz MM, Absi L, Christiansen FT, Cornelissen JJ, Dormoy A, Dubois V, Gagne K, Gluckman E, Haagenson MD, Oudshoorn M, Spellman S, Petersdorf EW. Mapping MHC-resident transplantation determinants. Biol Blood Marrow Transplant. 2007;13(8):986–995. doi: 10.1016/j.bbmt.2007.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGiffin DC, Naftel DC, Kirklin JK, Morrow WR, Towbin J, Shaddy R, Alejos J, Rossi A. Predicting outcome after listing for heart transplantation in children: comparison of Kaplan–Meier and parametric competing risk analysis. ***Pediatric Heart Transplant Study Group. J Heart Lung Transplant. 1997;16(7):713–722. [PubMed] [Google Scholar]
- Miller WP, Srinivasan S, Panoskaltsis-Mortari A, Singh K, Sen S, Hamby K, Deane T, Stempora L, Beus J, Turner A, Wheeler C, Anderson DC, Sharma P, Garcia A, Strobert E, Elder E, Crocker I, Crenshaw T, Penedo MC, Ward T, Song M, Horan J, Larsen CP, Blazar BR, Kean LS. GVHD after haploidentical transplantation: a novel, MHC-defined rhesus macaque model identifies CD28-CD8+ T cells as a reservoir of breakthrough T-cell proliferation during costimulation blockade and sirolimus-based immunosuppression. Blood. 2010;116(24):5403–5418. doi: 10.1182/blood-2010-06-289272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otting N, Heijmans CM, Noort RC, de Groot NG, Doxiadis GG, van Rood JJ, Watkins DI, Bontrop RE. Unparalleled complexity of theMHC class I region in rhesus macaques. ProcNatl Acad Sci U S A. 2005;102(5):1626–1631. doi: 10.1073/pnas.0409084102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page A, Srinivasan S, Singh K, Russell M, Hamby K, Deane T, Sen S, Stempora L, Leopardi F, Price AA, Strobert E, Reimann KA, Kirk AD, Larsen CP, Kean LS. CD40 blockade combines with CTLA4Ig and sirolimus to produce mixed chimerism in an MHC-defined rhesus macaque transplant model. Am J Transplant. 2011 doi: 10.1111/j.1600-6143.2011.03737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penedo MC, Bontrop RE, Heijmans CM, Otting N, Noort R, Rouweler AJ, de Groot N, de Groot NG, Ward T, Doxiadis GG. Microsatellite typing of the rhesus macaque MHC region. Immunogenetics. 2005;57(3–4):198–209. doi: 10.1007/s00251-005-0787-1. [DOI] [PubMed] [Google Scholar]
- Petersdorf EW, Malkki M, Gooley TA, Martin PJ, Guo Z. MHC haplotype matching for unrelated hematopoietic cell transplantation. PLoS Med. 2007;4(1):e8. doi: 10.1371/journal.pmed.0040008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosner C, Kruse PH, Lubke T, Walter L. Rhesus macaque MHC class I molecules show differential subcellular localizations. Immunogenetics. 2010;62(6):409–418. doi: 10.1007/s00251-010-0447-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawai ET, Khan IH, Montbriand PM, Peterlin BM, Cheng-Mayer C, Luciw PA. Activation of PAK by HIV and SIV Nef: importance for AIDS in rhesus macaques. Curr Biol. 1996;6(11):1519–1527. doi: 10.1016/s0960-9822(96)00757-9. [DOI] [PubMed] [Google Scholar]
- Singh K, Srinivasan S, Page A, Larsen C, Kean LS. The pitfalls of myeloid-only mixed-chimerism: lack of t cell chimerism correlates with transplant rejection after immunosuppression withdrawal in a novel, MHC-defined primate model: 2551. Transplantation. 2010;90:234. [Google Scholar]
- Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73(5):1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JM, Carver M, Cunningham P, Sash C, Park K, Thomas F. Promotion of incompatible allograft acceptance in rhesus monkeys given posttransplant antithymocyte globulin and donor bone marrow. II. Effects of adjuvant immunosuppressive drugs. Transplantation. 1989;47(2):209–215. doi: 10.1097/00007890-198902000-00002. [DOI] [PubMed] [Google Scholar]
- Thomas JM, Eckhoff DE, Contreras JL, Lobashevsky AL, Hubbard WJ, Moore JK, Cook WJ, Thomas FT, Neville DM., Jr Durable donor-specific T and B cell tolerance in rhesus macaques induced with peritransplantation anti-CD3 immunotoxin and deoxyspergualin: absence of chronic allograft nephropathy. Transplantation. 2000;69(12):2497–2503. doi: 10.1097/00007890-200006270-00007. [DOI] [PubMed] [Google Scholar]
- Thomas JM, Eckhoff DE, Contreras JL, Thomas FT, Neville DM, Jr, Lobashevsky A, Hubbard WJ, Cook WJ. Retraction: durable donor-specific T and B cell tolerance in rhesus macaques induced with peritransplantation anti-CD3 immunotoxin and deoxyspergualin: absence of chronic allograft nephropathy. Transplantation. 2006;82(4):577. doi: 10.1097/01.tp.0000239527.00075.60. [DOI] [PubMed] [Google Scholar]
- Trowsdale J. "Both man & bird & beast": comparative organization of MHC genes. Immunogenetics. 1995;41(1):1–17. doi: 10.1007/BF00188427. [DOI] [PubMed] [Google Scholar]
- Wiseman RW, Karl JA, Bimber BN, O'Leary CE, Lank SM, Tuscher JJ, Detmer AM, Bouffard P, Levenkova N, Turcotte CL, Szekeres E, Jr, Wright C, Harkins T, O'Connor DH. Major histocompatibility complex genotyping with massively parallel pyrosequencing. Nat Med. 2009;15(11):1322–1326. doi: 10.1038/nm.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiseman RW, O'Connor DH. Major histocompatibility complex-defined macaques in transplantation research. Transplant Rev. 2007;21(1):17–25. [Google Scholar]
- Wiseman RW, Wojcechowskyj JA, Greene JM, Blasky AJ, Gopon T, Soma T, Friedrich TC, O'Connor SL, O'Connor DH. Simian immunodeficiency virus SIVmac239 infection of major histocompatibility complex-identical cynomolgus macaques from Mauritius. J Virol. 2007;81(1):349–361. doi: 10.1128/JVI.01841-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Anderson JL, Campbell EM, Joseph AM, Hope TJ. Proteasome inhibitors uncouple rhesus TRIM5alpha restriction of HIV-1 reverse transcription and infection. Proc Natl Acad Sci U S A. 2006;103(19):7465–7470. doi: 10.1073/pnas.0510483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.