Introduction
The potential integration of whole genome sequencing into standard healthcare practice presents an array of challenges for clinicians [1]. Among these are concerns about whether clinicians with limited time are prepared to effectively communicate about complex multi-disease genetic risk results and what key information patients need to comprehend [2–4]. In narrowing down the essential topics to be discussed with patients, it will be necessary to determine whether genomic testing should be treated as a single entity (i.e. patients would decide whether to have the test and receive all or none of the genetic risk information) or provision of each health condition on the test should be explored independently (i.e. patients would make disease-specific decisions).
Although much research has examined patient decision-making for single-gene genetic tests, studies on how people choose among different diseases on a multiplex (i.e. multi-genetic variant) test are lacking. Even for newborn screening, the most widely used application of multi-disease genetic testing, research and practice guidelines focus on whether parents should be able to decline the screening test as a whole [5]. While one study did find that few healthcare providers (18.6%) support offering parents the option to choose among diseases on newborn screening tests, little consideration is typically given to this option due to pragmatic concerns [5, 6].
It is likely that there will be pressure to provide options among conditions offered on genomic tests. Typically, it is recommended that informed consent should be sought for genetic testing in the absence of overwhelming public health benefit [7]. Also, people typically prefer choice in controlling their health information, as can be seen in their support for control of what types of genetic research results are returned and desire for informed consent in biobanking research [8, 9]. How patients would make decisions among genetic tests for multiple diseases is unclear, although some research and theoretical work in health behavior suggests that they would attempt to develop an overall `gist' understanding of the test information [10]. While people have a preference for using the broadest gist representation possible, evidence indicates that nonexperts tend to use more details in their decision-making.
This report is an ancillary study of a parent project, the Multiplex Initiative, where participants were offered a genetic test for 8 common health conditions [11–14]. Although the Multiplex Initiative did not allow participants to choose which health conditions would be tested, it did present an opportunity for this ancillary study to assess what disease tests participants would have chosen if given an option. The purpose of this study is to describe whether people select among individual diseases on the multiplex genetic test and, if so, characterize the correlates of those preferences.
Methods
Recruitment
Participants in this study comprise a subset of those enrolled in the Multiplex Initiative, which is described in detail elsewhere [11–15]. The Multiplex Initiative was a collaborative effort among the National Human Genome Research Institute (Bethesda, Md.) and 2 members of the Cancer Research Network; the Group Health Cooperative (Seattle, Wash.), and the Henry Ford Health System (Detroit, Mich.). The participants were recruited from a sample of 350,000 members of the Henry Ford Health System who were 25–40 years old, Caucasian or African American, and not affected with type 2 diabetes, atherosclerotic cardiovascular disease, osteoporosis, or cancer. A phone survey collected baseline information for a total of 1,959 participants. Survey completers were sent a brochure that discussed the multiplex genetic test and referred participants to a website for more information [16]. The 612 participants who accessed the website could complete additional surveys, including the ancillary survey particular to this study (which had a USD 20 financial incentive). Participants who completed relevant questions on the ancillary survey were eligible for this study. On the website participants could also elect to visit the Henry Ford Health System clinic and decide whether to have blood drawn for testing. The test assessed 15 genetic variants contributing to modest risk (most OR <2.0) for 8 conditions: heart disease, hypercholesterolemia, hypertension, type 2 diabetes, osteoporosis, colon cancer, lung cancer, and skin cancer [14].
Measures
Baseline Measures
Gender and age were obtained from the Henry Ford Health System electronic records. Data on race and education was obtained from the baseline survey. Information about participants' family health histories was obtained for the diseases on the test, although only family history of cancer in general was addressed, rather than lung cancer, colon cancer and skin cancer specifically.
Ancillary Study Web-Based Measures: Disease Choice
A question developed for this study asked: `Multiplex Testing offers risk information on all of the conditions listed below. It was not an option for you to choose among the conditions, but we are interested in knowing what you might have done if you had been able to choose. Please check the conditions for which you would have chosen to receive risk information. Please leave blank those for which you would have chosen not to learn risk information.'
Outcome Measure at Clinic Visit: Test Uptake
Participants who had a clinic visit could learn about testing, go through a consent process and have a blood sample drawn for testing. Uptake was defined as having completed the blood draw.
Data Analysis
Data analysis was conducted using PASW Statistics software (version 18.0.3). The association between the number of diseases selected and test uptake was assessed via logistic regression, where a 10% change-in-estimate strategy was used to ensure a parsimonious model [17]. Associations between demographic factors and the number of diseases wanted were explored with linear regression. Pearson's χ2 tests were used to determine whether each of the health conditions were associated with demographic factors, family health history and test uptake (except for age, where logistic regression was applied).
Results
Sample Population
A total of 286 participants completed all relevant items and qualified for inclusion in the Multiplex Initiative ancillary study sample. Demographic and uptake information is presented in table 1. Forty-seven percent (n = 134) of participants took the multiplex genetic test. The characteristics of study participants in this sample did not differ significantly from the participants who accessed the website in the parent Multiplex Initiative study, except that this sample included more men (50% vs. 44%).
Table 1.
Factors associated with diseases selected
| Study demographics (n = 286) | Participants selecting disease |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| HD | HC | HTN | T2D | OS | LC | CC | SC | ||
| Mean age | 34.6 (SD = 4.0) | 34.8 | 34.8 | 34.8 | 35.0 | 34.6 | 34.9 | 35.2 | 34.9 |
| Sex | |||||||||
| Female | 142 (50%) | 76% | 67% | 69% | 65% | 52% | 52% | 68% | 46% |
| Male | 144 (50%) | 77% | 69% | 57% | 69% | 26% | 50% | 66% | 49% |
| Education | |||||||||
| HS or less | 56 (20%) | 63% | 50% | 61% | 64% | 25% | 50% | 63% | 38% |
| Some college | 98 (34%) | 79% | 75% | 65% | 74% | 42% | 52% | 66% | 49% |
| College or higher | 132 (46%) | 81% | 71% | 62% | 64% | 43% | 51% | 69% | 50% |
| Race | |||||||||
| Caucasian | 135 (47%) | 77% | 64% | 56% | 59% | 44% | 52% | 68% | 54% |
| Afr. Am. or other | 151 (53%) | 76% | 72% | 69% | 74% | 34% | 50% | 66% | 41% |
| Family history of disease | |||||||||
| Absent | N/Aa | 71% | 65% | 43% | 57% | 33% | 50%a | 60% a | 44%a |
| Present | N/Aa | 81% | 69% | 68% | 74% | 69% | 51%a | 73% a | 50%a |
Bold results indicate differences of statistical significance (p < 0.05).
HD = Heart disease; HC = high cholesterol; HTN = hypertension; T2D = type 2 diabetes; OS = osteoporosis; LC = lung cancer; CC = colon cancer; SC = skin cancer; Afr. Am. = African American; HS = high school.
Family health history data was not combined for an overall statistic. It should be noted that family history was obtained for cancer in general, rather than lung, colorectal, and skin cancer separately.
The table reads: The mean age of participants who chose heart disease was 34.8, and age was not significantly associated with choosing this disease. Seventy-six percent of females chose heart disease and 77% of males chose heart disease; there was no significant difference observed.
Total Number of Health Conditions Selected and Multiplex Test Uptake
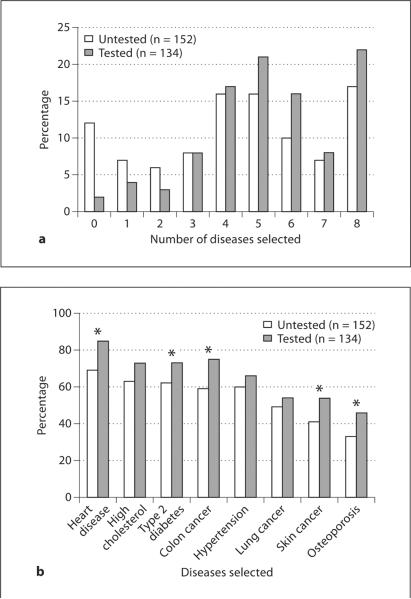
If given a choice, most participants indicated that they would select a subset of the available health conditions rather than selecting all or none of the diseases on the test (fig. 1a). On average, participants in the study would have opted to obtain genetic testing for 4.79 (SD = 2.41) of the 8 health conditions. Untested and tested participants differed in the mean number of conditions selected (untested: 4.36 (SD = 2.60) versus tested: 5.27 (SD = 2.07)). This association with test uptake was statistically significant, with an OR of 1.16 (CI = 1.04–1.29; p = 0.005) per health condition selected when controlled for education. Eighty-eight percent of those who declined testing would have wanted testing for one or more health conditions, while 78% of those who were tested would have declined testing for at least one health condition. Older age was the only demographic factor significantly associated with selecting more health conditions (β = 0.09; 95% CI = 0.02–0.16; p = 0.014).
Fig. 1.
Disease selection on the multiplex genetic test. a The total number of diseases for which participants would have wanted testing, separated by test uptake. b Percentage of participants indicating that they would prefer testing for each of the health conditions, separated by test uptake. *Significant differences (p < 0.05).
Differences between Participants Based on Health Conditions Selected
Differences in participant interest in genetic testing for the 8 health conditions is reported in figure 1b and table 1. Overall desire for testing ranged from 77% of participants interested in heart disease testing to 40% wanting testing for osteoporosis, and varied significantly among tested and untested participants for 5 health conditions (fig. 1b).
In some cases, desiring testing for specific health conditions varied with participant characteristics (table 1). Older participants were significantly more likely to want testing for type 2 diabetes and colorectal cancer, while female participants more often selected hypertension and osteoporosis. Those identifying as African American or `other' chose hypertension and type 2 diabetes more frequently and skin cancer less frequently. Participants that had lower educational levels (high school or less) were less likely to express interest in heart disease and hypercholesterolemia testing.
Although participants who had a family health history of a disease had higher frequencies of wanting testing for the disease, this was only significant for hypertension, type 2 diabetes and osteoporosis. Additionally, a family health history of cancer in general was associated with interest in testing for colorectal cancer.
Discussion
As genomic approaches to testing become increasingly available, questions remain about how to ensure that test decision-making is informed and leads to valuable outcomes. This study describes how a subset of participants in the Multiplex Initiative would choose among health conditions offered on a multiplex test if given that option. Participants who wanted testing for more diseases had a greater likelihood of actually taking the test. However, regardless of whether participants chose testing, they did not form a gist mental model of the multiplex test as a monolithic entity, where all diseases were accepted or refused together. Rather, the range in total number of diseases selected and differences noted across individual diseases suggest that participants engaged in active information processing for the 8 health conditions included on the multiplex genetic test.
The findings of our study are consistent with participants using personal risk information to select health conditions of interest. In particular, several of the significant differences in diseases selected across socio-demographic groups coincided with established population-based risks (e.g. lower interest in osteoporosis testing among male participants and skin cancer testing among African American participants [18, 19]). Additionally, family health history of a disease was associated with a significantly higher interest in 4 conditions, suggesting that increased salience and risk of disease may be associated with a stronger desire for genetic risk information.
The study results suggest that in addition to offering meaningful pre-test information, providers of genomic information should consider allowing people to select which types of risk information will be included in their report. Failure to do so could result in some people choosing not to be tested at all because they do not want risk information for certain diseases, while people who are tested could receive genetic risk information that they would have preferred not to know [20]. Both concerns are supported by the health condition preference data in this study.
There are study limitations, including a descriptive design and the use of a single question that explores hypothetical preferences among diseases (albeit in the context of an actual genetic test including those conditions). Additionally, while our study demonstrated that participants would want to select among diseases if given that option, we did not assess how important that choice was to them.
As genomic tests examine increasing numbers of health conditions, customizing results for individual needs would require careful research on effective communication strategies and the impact that complex or unanticipated information can have on people. Ultimately, if genomic testing is shown to have a public health benefit, it will be necessary to determine whether these tests should be designed as a population screen where options are minimized for efficiency or a personal choice where careful informed decision-making must be supported for each health condition on the test.
Acknowledgements
This research was supported by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health. The proposed research was made possible through a collaboration with the Cancer Research Network funded by the National Cancer Institute (U19CA 079689). Group Health Research Institute and Henry Ford Hospital provided additional resources. Genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University (HHSN268200782096C). Additionally, this research was supported in part by an appointment to the Senior Fellowship Program at the National Institutes of Health. This program is administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the US Department of Energy and the National Institutes of Health.
We would like to thank the study participants, members of the Henry Ford Health System and the Multiplex Initiate Steering Committee for their efforts in making the Multiplex Initiative Ancillary Studies possible.
References
- 1.Ormond KE, Wheeler MT, Hudgins L, Klein TE, Butte AJ, Altman RB, Ashley EA, Greely HT. Challenges in the clinical application of whole-genome sequencing. Lancet. 2010;375:1749–1751. doi: 10.1016/S0140-6736(10)60599-5. [DOI] [PubMed] [Google Scholar]
- 2.McBride CM, Wade CH, Kaphingst KA. Consumers' views of direct-to-consumer genetic information. Annu Rev Genomics Hum Genet. 2010;11:427–446. doi: 10.1146/annurev-genom-082509-141604. [DOI] [PubMed] [Google Scholar]
- 3.Powell KP, Cogswell WA, Christianson CA, Dave G, Verma A, Eubanks S, Henrich VC. Primary care physicians' awareness, experience and opinions of direct-to-consumer genetic testing. J Genet Couns. 2012;21:113–126. doi: 10.1007/s10897-011-9390-9. [DOI] [PubMed] [Google Scholar]
- 4.Leighton JW, Valverde K, Bernhardt BA. The general public's understanding and perception of direct-to-consumer genetic test results. Public Health Genomics. 2012;15:11–21. doi: 10.1159/000327159. [DOI] [PubMed] [Google Scholar]
- 5.Ross LF. Mandatory versus voluntary consent for newborn screening? Kennedy Inst Ethics J. 2010;20:299–328. [PubMed] [Google Scholar]
- 6.Miller FA, Hayeems RZ, Carroll JC, Wilson B, Little J, Allanson J, Bytautas JP, Paynter M, Christensen R, Chaktraborty P. Consent for newborn screening: the attitudes of health care providers. Public Health Genomics. 2010;13:181–190. doi: 10.1159/000240966. [DOI] [PubMed] [Google Scholar]
- 7.Task Force on Genetic Testing . Promoting safe and effective genetic testing in the United States. National Institutes of Health and the Department of Energy; Bethesda: 1997. http://www.genome.gov/10001733. [Google Scholar]
- 8.Murphy J, Scott J, Kaufman D, Geller G, Le-Roy L, Hudson K. Public perspectives on informed consent for biobanking. Am J Public Health. 2009;99:2128–2134. doi: 10.2105/AJPH.2008.157099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murphy J, Scott J, Kaufman D, Geller G, Le-Roy L, Hudson K. Public expectations for return of results from large-cohort genetic research. Am J Bioeth. 2008;8:36–43. doi: 10.1080/15265160802513093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reyna VF. A theory of medical decision making and health: fuzzy trace theory. Med Decis Making. 2008;28:850–865. doi: 10.1177/0272989X08327066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McBride CM, Alford SH, Reid RJ, Larson EB, Baxevanis AD, Brody LC. Putting science over supposition in the arena of personalized genomics. Nat Genet. 2008;40:939–942. doi: 10.1038/ng0808-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McBride CM, Alford SH, Reid RJ, Larson EB, Baxevanis AD, Brody LC. Characteristics of users of online personalized genomic risk assessments: implications for physician-patient interactions. Genet Med. 2009;11:582–587. doi: 10.1097/GIM.0b013e3181b22c3a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hensley Alford S, McBride CM, Reid RJ, Larson EB, Baxevanis AD, Brody LC. Participation in genetic testing research varies by social group. Public Health Genomics. 2011;14:85–93. doi: 10.1159/000294277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wade CH, McBride CM, Kardia SL, Brody LC. Considerations for designing a prototype genetic test for use in translational research. Public Health Genomics. 2010;13:155–165. doi: 10.1159/000236061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wade CH, Shiloh S, Woolford SW, Roberts JS, Alford SH, Marteau TM, Biesecker BB. Modelling decisions to undergo genetic testing for susceptibility to common health conditions: an ancillary study of the Multiplex Initiative. Psychol Health. 2012;27:430–444. doi: 10.1080/08870446.2011.586699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaphingst KA, McBride CM, Wade C, Alford SH, Brody LC, Baxevanis AD. Consumers' use of web-based information and their decisions about multiplex genetic susceptibility testing. J Med Internet Res. 2010;12:e41. doi: 10.2196/jmir.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hosmer DW, Lameshow S. Applied Logistic Regression. Wiley; New York: 2000. [Google Scholar]
- 18.Melton LJ, 3rd, Chrischilles EA, Cooper C, Lane AW, Riggs BL. Perspective. How many women have osteoporosis? J Bone Miner Res. 1992;7:1005–1010. doi: 10.1002/jbmr.5650070902. [DOI] [PubMed] [Google Scholar]
- 19.Howlader N, Noone AM, Krapcho M, Neyman N, Aminou R, Waldron W, Altekruse SF, Kosary CL, Ruhl J, Tatalovich Z, Cho H, Mariotto A, Eisner MP, Lewis DR, Chen HS, Feuer EJ, Cronin KA, Edwards BK. SEER Cancer Statistics Review, 1975-2008. National Cancer Institute; Bethesda: 2010. [Google Scholar]
- 20.Wilson J. To know or not to know? Genetic ignorance, autonomy and paternalism. Bioethics. 2005;19:492–504. doi: 10.1111/j.1467-8519.2005.00460.x. [DOI] [PubMed] [Google Scholar]