Abstract
Choline monooxygenase (CMO) catalyzes the committing step in the synthesis of glycine betaine, an osmoprotectant accumulated by many plants in response to salinity and drought. To investigate how these stresses affect CMO expression, a spinach (Spinacia oleracea L., Chenopodiaceae) probe was used to isolate CMO cDNAs from sugar beet (Beta vulgaris L., Chenopodiaceae), a salt- and drought-tolerant crop. The deduced beet CMO amino acid sequence comprised a transit peptide and a 381-residue mature peptide that was 84% identical (97% similar) to that of spinach and that showed the same consensus motif for coordinating a Rieske-type [2Fe-2S] cluster. A mononuclear Fe-binding motif was also present. When water was withheld, leaf relative water content declined to 59% and the levels of CMO mRNA, protein, and enzyme activity rose 3- to 5-fold; rewatering reversed these changes. After gradual salinization (NaCl:CaCl2 = 5.7:1, mol/mol), CMO mRNA, protein, and enzyme levels in leaves increased 3- to 7-fold at 400 mm salt, and returned to uninduced levels when salt was removed. Beet roots also expressed CMO, most strongly when salinized. Salt-inducible CMO mRNA, protein, and enzyme activity were readily detected in leaves of Amaranthus caudatus L. (Amaranthaceae). These data show that CMO most probably has a mononuclear Fe center, is inducibly expressed in roots as well as in leaves of Chenopodiaceae, and is not unique to this family.
Like other organisms, many plants accumulate betaines, polyols, or Pro in response to dry or saline conditions (Yancey, 1994; Bohnert et al., 1995). These compounds, termed compatible solutes or osmoprotectants, serve as nontoxic solutes for cytoplasmic osmoregulation and can also partly reverse the damaging effects of salts on proteins and membranes (Yancey, 1994). The metabolic engineering of osmoprotectant accumulation has therefore attracted interest as a way to improve crop stress resistance (McCue and Hanson, 1990; Bartels and Nelson, 1994; Bohnert and Jensen, 1996). One target for such engineeering is Gly betaine, a potent osmoprotectant that occurs widely among flowering plants, including the economically important families Chenopodiaceae, Amaranthaceae, Malvaceae, Compositae, and Gramineae (Gorham, 1995).
Plants synthesize Gly betaine via a two-step oxidation of choline: choline → betaine aldehyde → Gly betaine (Rhodes and Hanson, 1993). Bacteria use the same route, which has led several groups to express bacterial genes for choline oxidation (choline dehydrogenase or choline oxidase) in plants that lack Gly betaine, in some cases targeting the enzyme to chloroplasts because this is the site of Gly betaine synthesis in Chenopodiaceae (Rhodes and Hanson, 1993). The transformants made small amounts of Gly betaine and were significantly more salt tolerant (e.g. Lilius et al., 1996; Hayashi et al., 1997).
It is now possible to use plant genes for such engineering experiments, since cDNAs for both enzymes of Gly betaine synthesis have been cloned from Chenopodiaceae (McCue and Hanson, 1992a; Rathinasabapathi et al., 1997). The enzyme mediating the second reaction is BADH, an NAD-linked dehydrogenase that is known also from Amaranthaceae and Gramineae (Ishitani et al., 1993; Valenzuela-Soto and Muñoz-Clares, 1994). In Chenopodiaceae, BADH is predominantly (≥ 90%) located in the chloroplast stroma (Rathinasabapathi et al., 1994). Salinity and water deficit raise BADH mRNA and enzyme levels in leaves and roots, coincident with the accumulation of Gly betaine (McCue and Hanson, 1992a; Ishitani et al., 1995; Wood et al., 1996). BADH cDNAs from Chenopodiaceae and Gramineae have recently been functionally expressed in tobacco (Rathinasabapathi et al., 1994; Ishitani et al., 1995).
The enzyme catalyzing the first and committing step of Gly betaine synthesis is not as well known, having so far been found only in spinach (Chenopodiaceae). The spinach enzyme (CMO) is a stromal, Fd-dependent monooxygenase with a Rieske-type [2Fe-2S] center, and it is completely unrelated to the bacterial choline dehydrogenase and oxidase enzymes (Burnet et al., 1995; Rathinasabapathi et al., 1997). Little is known about CMO expression except that it increases in salinized spinach leaves (Brouquisse et al., 1989; Rathinasabapathi et al., 1997).
We cannot yet answer three questions about the natural expression of CMO that bear on its use in metabolic engineering. First, is it induced by drought as well as by salinity? Second, is it expressed in roots as well as in leaves? Third, is it unique to Chenopodiaceae or is it found in other Gly betaine-accumulating plants? To address the first two questions we chose sugar beet (Chenopodiaceae) because it can accumulate high levels of Gly betaine in both roots and shoots (≥ 300 μmol g−1 dry weight), it is a halophyte, and it withstands drought well (Hanson and Wyse, 1982; Dunham, 1993). This led us to isolate beet CMO cDNAs for use as homologous probes; analysis of their deduced amino acid sequences revealed a mononuclear Fe-binding motif.
MATERIALS AND METHODS
Plants and Growing Conditions
Sugar beet (Beta vulgaris L., cv Great Western D-2) seed was obtained from Hilleshög Mono-hy, Inc. (Longmont, CO). Beet plants from which mRNA was isolated for library construction were grown in a growth chamber (12-h day, 25°C, 350 μE m−2 s−1 PPFD/12-h night, 22°C) and salinized with 400 mm NaCl as described previously (McCue and Hanson, 1992a). For salinity experiments, beet plants were grown hydroponically (four per 18-L tank) in aerated one-half-strength Hoagland nutrient solution in a growth chamber (8-h day, 25°C, 350 μE m−2 s−1 PPFD/16-h night, 20°C). Salinization began when plants were 4 weeks old, using an NaCl:CaCl2 mixture (5.7:1 mol/mol, hereafter termed salt), and the concentration was raised by 50 mm every third day to final values of 100, 250, or 400 mm salt. Water was added daily to replace evapotranspiration and the medium was replaced every 2 weeks. Plants were harvested 3 d after the final concentration was reached in the 400 mm salt treatment, collecting four young leaves (blade length about 5–20 cm) and the taproot from each. At the same time, one batch of four plants was transferred from 250 mm salt to nutrient solution alone, and harvested 3 d later. Unsalinized controls were harvested at the same time as the 400 mm salt treatment or at a similar stage of development to the plants in this treatment; since the data were similar, only the latter are presented.
For drought experiments, beet plants were grown (one per 4-L pot) in heavy potting soil (Southland Importers, Greensboro, NC) supplemented with 5 g per pot of Osmocote 14:14:14 (N:P:K) (Scotts-Sierra, Maysville, OH) in a greenhouse in natural daylight during November and December, 1996; minimum temperature was 18°C. Irrigation was with one-half-strength Hoagland nutrient solution. Seven weeks after planting, irrigation was withheld for 7 d, by which time the expanded leaves had been wilted continuously for 3 d. One-half of the plants was then harvested, taking young leaves as above; the others were rewatered and harvested 3 d later. Control leaves were harvested just before irrigation was withheld. In all experiments, samples from individual plants were frozen in liquid N2 and stored at −80°C.
Amaranth (Amaranthus caudatus L. cv RRC 1036) seeds were obtained from U.S. Department of Agriculture, North Central Regional Plant Introduction Station (Ames, IA). Plants were grown (two per 2.5-L pot) in Metro-Mix 300 (Grace Sierra, Milpitas, CA), supplemented with Osmocote, in the 8-h day/16-h night conditions given above. Salinization was with NaCl:CaCl2 in nutrient solution as above, starting at 5 weeks, applying 1.5 L per pot daily, and raising the salt level by 50 mm every third day to a final value of 300 mm. Controls were irrigated with nutrient solution. After 3 d at 300 mm salt, young, fully expanded, and expanding leaves (≥ 50% final size) were harvested, frozen, and stored as above.
Measurement of Ψs and RWC
The fourth youngest leaf was sampled for beet, and the first fully expanded leaf was sampled for amaranth. Ψs was determined on frozen-thawed 0.8-cm diameter leaf discs using a thermocouple psychrometer (HR-33T, Wescor, Logan, UT) equipped with C-52 sample chambers. RWC was measured on sets of nine 1.6-cm discs from the same leaves, as described by Grumet and Hanson (1986).
Gly Betaine Analysis
Gly betaine was extracted from 50-mg dry weight leaf samples with a methanol/chloroform/water procedure and isolated by ion-exchange chromatography (Hanson et al., 1991). [Methyl-2H3]Gly betaine (448 nmol/sample) was added at the start of the extraction as internal standard. Gly betaine was converted to its n-butyl ester and determined by fast atom bombardment MS as described by Rhodes et al. (1987).
RNA Isolation and Analysis
For beet cDNA library construction, total RNA was isolated as described previously (Rathinasabapathi et al., 1997) and poly(A+) RNA was prepared using poly(U) Sephadex (Hondred et al., 1987). For expression studies, total RNA was isolated by a single-step method (Puissant and Houdebine, 1990). Poly(A+) RNA for expression studies was purified using polyA Spin Kits (New England Biolabs). RNA was separated on 1.2% formaldehyde gels and blotted to Duralon membranes (Stratagene); molecular size standards (0.24- to 9.5-kb RNA ladder, Gibco-BRL) were included. Probes were labeled (> 109 cpm μg−1) by the random primer method using [α-32P]dCTP. Autoradiographs of northern blots were scanned using a digital imaging system (IS-1000, Alpha Innotech, San Leandro, CA) to quantify signals. Signal strengths were normalized with respect to the amount of rRNA in each track, determined by reprobing blots with an 18S rRNA probe (Nairn and Ferl, 1988). The quality of amaranth poly(A+) RNA preparations was checked by probing blots with the 1.6-kb XhoI fragment of amaranth PEP carboxylase (Rydzik and Berry, 1996).
cDNA Cloning and DNA Sequence Analysis
A cDNA library (3.1 × 106 plaque-forming units) was constructed in the UniZap XR vector (Stratagene). Screening and in vivo excision were carried out according to the supplier's protocols. Filters were hybridized at 42°C and washed at 55°C in 2× SSC containing 0.1% (v/v) SDS. The amplified library was screened with a 1150-bp AccI-EcoRV fragment of spinach CMO cDNA clone pRS3 (Rathinasabapathi et al., 1997) labeled as described above. This yielded a near-full-length CMO cDNA (SB2), of which the 5′-terminal 265-bp EcoRI fragment was used to isolate clone SB30 from the unamplified library. Clones were sequenced in both strands using the fluorescent chain-terminating dideoxynucleotides method. Sequences were analyzed using the Wisconsin GCG Sequence Analysis Package (Version 8.0, Genetics Computer Group, Madison, WI).
Protein Isolation and Analysis
Samples (2 g fresh weight) were pulverized in liquid N2 and extracted with 8 mL of ice-cold buffer containing 100 mm Tris, 1 mm Na2EDTA, 10 mm DTT, and 4% (w/v) PVP, adjusted to pH 8.0 with HCl; for all samples except the drought-stressed beet leaves, 20 mm Na borate, 50 mm ascorbic acid, 1 mm PMSF, 1 μg mL−1 leupeptin, and 1 μg mL−1 antipain were also included. After centrifuging at 12,000g (10 min, 4°C), proteins were precipitated from the supernatant by adding 1 volume of ice-cold 50% (w/v) PEG 8000 in 25 mm Tris-HCl, pH 8.0 (without or with borate, ascorbate, leupeptin, and antipain as above), holding on ice for 2 h, and centrifuging at 9,000g (15 min, 4°C). This procedure was shown to precipitate 80% of the protein present in beet leaf extracts. The pellet was redissolved in a buffer (adjusted to pH 8.0 with HCl) containing Tris 50 mm, 0.5 mm Na2EDTA, 2 mm DTT, 10% (v/v) glycerol, and, except for drought-stressed beet leaves, 1 μg mL−1 leupeptin and 1 μg mL−1 antipain. The solution was then desalted on Sephadex G-25 equilibrated in the same buffer and used for CMO assays and western analyses. CMO activity was measured by the coupled radiometric assay described previously (Burnet et al., 1995) using an incubation time of 10 or 20 min, and adjusting the amount of extract so that product formation was proportional to time and protein concentration. Proteins were separated by SDS-PAGE and transferred to nitrocellulose as described previously (Tokuhisa et al., 1985). Blots were probed with mouse polyclonal antibodies to spinach CMO (Rathinasabapathi et al., 1997).
RESULTS
Isolation and Characterization of Beet CMO cDNAs
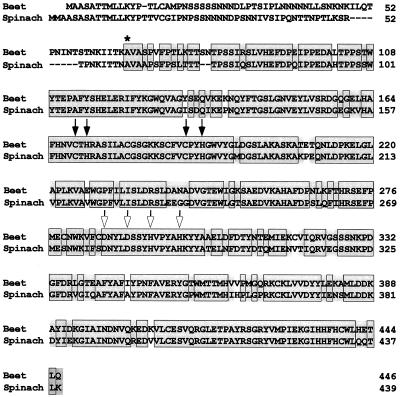
A cDNA library was prepared using mRNA from salinized beet leaves, and screened with a spinach CMO cDNA (Rathinasabapathi et al., 1997). This yielded a 5′-truncated CMO clone, SB2 (1709 bp). A 5′ fragment of SB2 was then used to isolate SB30, a 3′-truncated clone that included 36 bp of 5′-untranslated region and 468 bp of coding sequence, of which 462 bp overlapped with SB2. Together, SB2 and SB30 represent a full-length cDNA of 1751 bp that encodes a polypeptide of 446 amino acids and has a 377-bp 3′-untranslated region. The composite amino acid sequence is shown in Figure 1, aligned with that of spinach CMO. Comparison of the beet and spinach N-terminal regions suggests that beet CMO has a 65-residue stromal targeting peptide, similar in length to that of spinach CMO (60 residues) and of typical amino acid composition (Cline and Henry, 1996). The deduced amino acid sequence of the mature beet CMO polypeptide has 84% identity and 97% similarity to that of spinach, which are comparable values to those for their BADHs (McCue and Hanson, 1992a). Beet CMO contains the consensus sequence (Cys-X-His-X15–17-Cys-X2-His) for coordinating a Rieske-type [2Fe-2S] cluster, as found in spinach CMO (Rathinasabapathi et al., 1997).
Figure 1.
Comparison of the deduced amino acid sequences for sugar beet and spinach CMO cDNA clones. The beet sequence is a composite of two overlapping clones (SB2 and SB30, encoding amino acids 3 to 446 and 1 to 156, respectively). Identical amino acid residues are boxed and shaded; conservative substitutions are shaded. The asterisk shows the N-terminal residue of the mature spinach CMO polypeptide (Rathinasabapathi et al., 1997). Solid arrows mark the conserved Cys-His pairs of the Rieske-type [2Fe-2S] cluster-binding region, and open arrows indicate the conserved residues of the mononuclear Fe-binding motif. The accession number for the composite beet CMO nucleotide sequence is AF 023132.
Positioned 94 residues along the protein from the Rieske cluster-binding motif, beet CMO has a sequence very like the recently proposed consensus for coordination sites for mononuclear nonheme Fe: Glu/Asp-X3–4-Asp-X2-His-X4–5-His (Jiang et al., 1996; Gray et al., 1997). Exactly the same motif is present in spinach CMO (Fig. 1); the only departure from the consensus is that the central Asp and His are three residues apart, not two. Otherwise, the amino acids flanking the Asp and His residues in CMO are similar or identical to those in other mononuclear Fe-binding sites, and the location of these conserved residues relative to the Rieske cluster-binding motif is the same as in other oxygenases (Mason and Cammack, 1992; Jiang et al., 1996). Together, these observations strongly imply that beet and spinach CMOs contain mononuclear Fe.
CMO Expression in Beet Leaves during and after Water Deficit
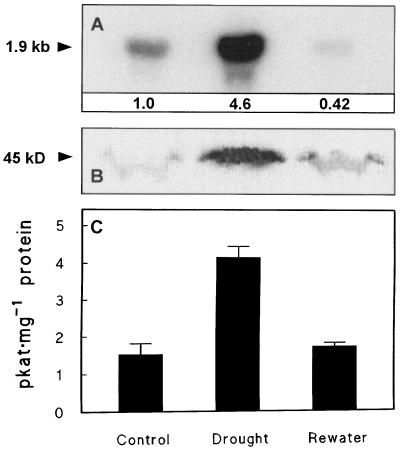
Prolonged withholding of irrigation resulted in marked declines in RWC and Ψs, which were fully reversed upon rewatering (Table I). As expected, this drought treatment promoted Gly betaine accumulation, although it did not lead to measurable overall osmotic adjustment (i.e. a net increase in solutes), since Ψs values corrected to 100% RWC did not fall. Consistent with the accumulation of Gly betaine, during the stress period the level of CMO mRNA rose 5-fold (Fig. 2A), CMO protein accumulated (Fig. 2B), and CMO enzyme activity tripled (Fig. 2C). Within 3 d of rewatering, CMO mRNA, protein, and enzyme activity had all fallen to or below their prestress levels (Fig. 2). The size of the CMO mRNA observed in all treatments (1.9 kb) was slightly above that of spinach (1.7 kb; Rathinasabapathi et al., 1997).
Table I.
Responses of sugar beet leaves to a cycle of drought stress and relief
| Treatment | RWCa | −Ψsa | Gly Betaine |
|---|---|---|---|
| % | MPa | μmol g−1 fresh wt | |
| Control | 91.1a | 1.14a | 11.8 |
| Drought stress | 58.5b | 1.69b | 23.1 |
| Drought/rewater | 89.4a | 1.12a | 17.0 |
Irrigation was withheld for 7 d, by which time the mature leaves had been wilted for 3 d, and then restored for 3 d. Control plants were irrigated daily. The Ψs and RWC measurements were made between 10:00 am and 1:00 pm; data are means of four replicates and were subjected to analysis of variance. The Ψs values shown are not corrected to 100% RWC. Gly betaine was determined using leaf discs pooled from four plants and is expressed on the basis of fresh weight at harvest.
Means within columns not followed by the same letter are significantly different at the 5% probability level.
Figure 2.
Expression of CMO in sugar beet leaves during a cycle of drought stress and relief. Irrigation was withheld until mature leaves had been continuously wilted for 3 d (Drought), and then restored for 3 d (Rewater). Controls were irrigated daily. A, Northern analysis. Lanes contained 10 μg of total RNA; the blot was probed with two EcoRI fragments comprising the 5′-terminal 815 bp from beet clone SB2. The numbers beneath each lane show the CMO mRNA abundance relative to the control (1.0), normalized for loading using 18S rRNA as a benchmark. B, Western analysis. Lanes contained 130 μg of protein (precipitated with 25% PEG); the blot was probed with mouse antibodies raised against spinach CMO. C, CMO activity in 25% PEG-precipitated protein (means and se for three replicates). The experiment was repeated with similar results.
CMO Expression in Salinized Beet Leaves and Roots
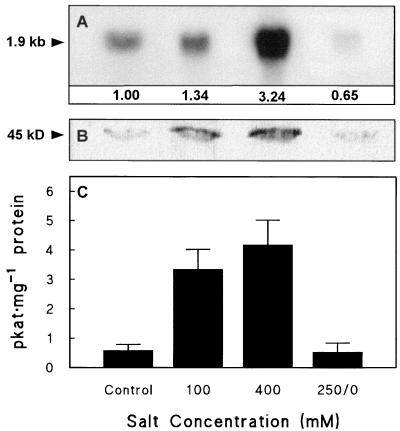
Adding salt to the hydroponic medium lowered its Ψs by about 0.55 MPa at 100 mm and 1.9 MPa at 400 mm, as predicted (Wyn Jones and Gorham, 1983). The declines in leaf Ψs in salinized plants were close to these values, and resulted from both osmotic adjustment and, to a lesser extent, a drop in RWC (Table II). In leaves the steady-state levels of CMO mRNA, protein, and enzyme activity all rose as salinity was increased, and at 400 mm salt they were 4- to 7-fold higher than in unsalinized controls (Fig. 3). Three days after relief of salt stress, RWC and Ψs had both risen markedly (Table II), and CMO mRNA, protein, and enzyme activity had fallen back to levels no higher than those in control leaves (Fig. 3).
Table II.
RWC and Ψs of leaves from salinized beet plants
| Salt Treatment | RWCa | −Ψsa |
|---|---|---|
| % | MPa | |
| Control | 93.5a | 0.70a |
| 100 mm | 78.4b | 1.20b |
| 400 mm | 73.4c | 2.36c |
| 250 mm/0 mm | 85.6d | 1.08b |
Plants were salinized gradually and then maintained at the various salinity levels for at least 3 d. One batch of plants was salinized to a salt level of 250 mm, then transferred to nutrient solution for 3 d before harvest. Controls received nutrient solution alone. The Ψs and RWC measurements were made 3 to 6 h after the start of the light period; Ψs values were not corrected to 100% RWC. Data are means of four or five replicates and were subjected to analysis of variance.
Means within columns not followed by the same letter are significantly different at the 5% probability level.
Figure 3.
CMO expression in sugar beet leaves after long-term salinization. Plants were salinized gradually and then maintained at the salinity levels shown for at least 3 d. One batch of plants (250/0) was salinized to an intermediate salt level (250 mm), then transferred to nutrient solution for 3 d before harvest; a prior experiment had demonstrated that CMO induction at 250 mm salt was the same as at 400 mm. Controls received nutrient solution alone. A, Northern analysis; numbers below the lanes show the CMO mRNA abundance relative to the unsalinized control (1.0), normalized for RNA loading. B, Western analysis; lanes contained 50 μg of protein. C, CMO activity (means and se for three replicates). Other experimental conditions were as in Figure 2. The experiment was repeated with similar results.
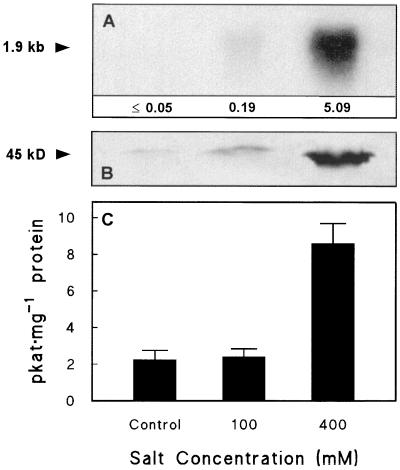
In taproots CMO mRNA levels were undetectable in the control and low in the 100 mm salt treatment, but at 400 mm salt they were comparable to those in leaves (Fig. 4A). CMO protein level and activity (Fig. 4, B and C) were also far higher at 400 mm salt, but were readily detectable in both other treatments. Expressed per unit protein, CMO activities in roots were higher than in leaves (Figs. 3C and 4C). SDS-PAGE analysis of the PEG-precipitated fractions used to assay CMO indicated that this reflected the high proportion of Rubisco in leaf samples.
Figure 4.
CMO expression in sugar beet taproots after long-term salinization. The plants were the same as those used for the leaf data of Figure 3. A, Northern analysis; numbers beneath the lanes show the CMO mRNA abundance relative to that in unsalinized leaves (1.0; Fig. 3), normalized for RNA loading. B, Western analysis; lanes contained 100 μg of protein (precipitated with 14% PEG). C, CMO activity (means and se for three replicates). Other experimental conditions were as in Figure 2. The experiment was repeated with similar results.
CMO Expression in Amaranth
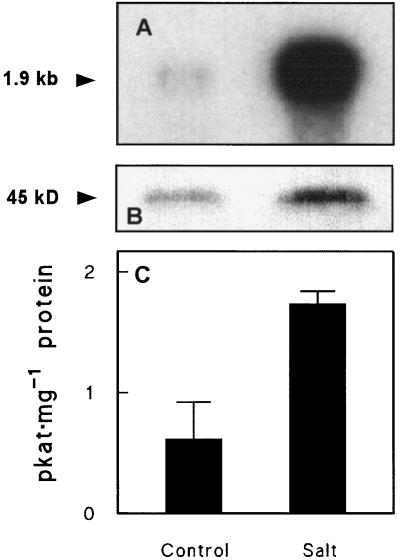
The families Amaranthaceae and Chenopodiaceae belong to the order Caryophyllales and are considered to stand close to each other within it (Takhtajan, 1969). Consistent with such a relationship, the beet CMO cDNA hybridized to a salt-inducible 1.9-kb CMO mRNA from amaranth leaves (Fig. 5A), and antibodies to spinach CMO recognized an amaranth CMO polypeptide (Fig. 5B) of the same mass (about 45 kD) as the CMO monomer from spinach and beet. CMO enzyme activity was also found in control and salinized amaranth leaves (Fig. 5C), at levels comparable to those in leaves of beet (Fig. 3C) and spinach (Rathinasabapathi et al., 1997).
Figure 5.
Evidence for salinity-inducible CMO in amaranth leaves. Plants were grown without salinization (Control) or gradually salinized to a final level of 300 mm (Salt). A, Northern analysis; lanes contained 3 μg of poly(A+) RNA; the blot was probed with the two 5′ EcoRI fragments from beet clone SB2 and washed at 30°C in 2× SSC containing 0.1% SDS (v/v). B, Western analysis; lanes contained 150 μg of protein (precipitated with 14% PEG). C, CMO activity (means and se for three replicates). Other experimental conditions were as in Figure 2.
DISCUSSION
We have isolated and characterized cDNAs encoding CMO from a second plant species, with spinach being the first. The deduced amino sequence of beet CMO contained the Rieske-type [2Fe-2S] cluster-binding motif found in the spinach sequence (Rathinasabapathi et al., 1997), as well as a newly recognized consensus motif for a mononuclear Fe-binding site (Jiang et al., 1996; Gray et al., 1997). The presence of a mononuclear Fe center in CMO fits with findings for other oxygenases. Thus, various bacterial oxygenases with Rieske [2Fe-2S] clusters are known also to have mononuclear Fe centers (whence the consensus motif) that are thought to be the site of dioxygen activation and catalysis (Mason and Cammack, 1992; Jiang et al., 1996).
If CMO contains mononuclear Fe, this could help to explain why CMO loses activity during purification (Burnet et al., 1995), since mononuclear Fe is essential for catalysis but is sometimes readily lost (Suen and Gibson, 1993; Jiang et al., 1996). Consistent with this possibility, purified CMO had an Fe:S ratio of 1.06:1 (Rathinasabapathi et al., 1997), well below the expected value of 1.5:1 for a protein with a Rieske-type [2Fe-2S] cluster and a mononuclear Fe center. A systematic study of the effects of Fe2+ treatment on the activity of purified CMO would consequently be of interest, although it could be complicated by the effect of Fe2+ on superoxide-driven Fenton chemistry (hydroxyl radical formation) in the CMO assay itself (Asada, 1996).
Subjecting beet to a cycle of water stress and relief showed that drought leads to Gly betaine accumulation in leaves and that this is associated with up-regulated CMO gene expression manifest in higher levels of CMO mRNA, protein, and enzyme activity. The latter increases were completely reversed within 3 d of stress relief, indicating that both CMO mRNA and protein can be turned over quite rapidly. In this experiment, as well as in those involving salinity, the increases and decreases in CMO protein as judged from western blots were roughly comparable to those in enzyme activity. This suggests that stress does not affect CMO activity by posttranslational mechanisms, which fits with mass spectral evidence against CMO having covalent posttranslational modifications (Rathinasabapathi et al., 1997).
The data for salinized beet confirm observations made with spinach that CMO is salt inducible in leaves (Rathinasabapathi et al., 1997) and extend them in two ways. First, the beet results show that the induction is fully reversed within 3 d of salt removal at both the mRNA and enzyme levels. This contrasts with the response of BADH to relief of salt stress: beet BADH mRNA levels decay rapidly but enzyme activity does not (McCue and Hanson, 1992b). This difference in poststress behavior between CMO and BADH is consistent with CMO being more important in controlling flux through the Gly betaine synthesis pathway. The second novel facet of the beet data is that they show that CMO is expressed in roots. Since beet roots also express BADH (McCue and Hanson, 1992a) and produce choline (Hanson and Wyse, 1982), they are most probably able to synthesize Gly betaine. The reduced Fd requirement for CMO could presumably be met by electron transfer from NADPH to nonphotosynthetic Fd, as it is for the glutamate synthase and nitrite reductase of root plastids (Bowsher et al., 1989, 1992). Consistent with this, nongreen cell cultures of Atriplex spp. (Chenopodiaceae) have been shown to produce Gly betaine (Koheil et al., 1992). Although little conversion of [14C]ethanolamine or [14C]formate to Gly betaine was seen in beet root tissues (Hanson and Wyse, 1982), 14C accumulated in choline and phosphorylcholine and it is possible that the endogenous pools of these compounds were large enough to act as traps for the label.
That amaranth has CMO mRNA, protein, and enzyme is, to our knowledge, the first evidence that CMO occurs outside the family Chenopodiaceae. However, since the Amaranthaceae are phylogenetically close to the Chenopodiaceae, it would be premature to assume that CMO also occurs in more distant families such as Malvaceae, Compositae, and Gramineae. No work has yet been done on the biochemistry of choline oxidation in these groups, but it was recently reported that BADH in Gramineae may be located mainly in peroxisomes rather than chloroplasts (Nakamura et al., 1997). If Gly betaine synthesis in Gramineae occurs in the peroxisome, then CMO is unlikely to be the choline-oxidizing enzyme, because its Fd requirement would not be met in this compartment. It may also be significant that we have been unable to detect CMO mRNA or activity in cotton (Malvaceae), although reprobing the RNA blots with a cotton α-tubulin cDNA gave satisfactory signals, and when cotton and beet tissues were mixed and extracted together, the beet CMO was not inactivated (B.L. Russell and A.D. Hanson, unpublished data).
ACKNOWLEDGMENTS
We thank J.O. Berry, R.J. Ferl, and T.A. Wilkins, respectively, for the gifts of the PEP carboxylase, 18S rRNA, and cotton α-tubulin cDNA clones; D.A. Gage for carrying out the mass spectral analyses of Gly betaine; and K.D. Nolte for help in preparing figures.
Abbreviations:
- BADH
betaine aldehyde dehydrogenase
- CMO
choline monooxygenase
- Ψs
solute potential
- RWC
relative water content
Footnotes
This work was supported in part by a grant from the U.S. Department of Agriculture National Research Initiative Competitive Grants Program (95-37100-1596) and by an endowment from the C.V. Griffin, Sr. Foundation. This is Florida Agricultural Experiment Station journal series no. R-06188.
LITERATURE CITED
- Asada K. Radical production and scavenging in the chloroplasts. In: Baker NR, editor. Photosynthesis and the Environment. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1996. pp. 123–150. [Google Scholar]
- Bartels D, Nelson D. Approaches to improve stress tolerance using molecular genetics. Plant Cell Environ. 1994;17:659–667. [Google Scholar]
- Bohnert HJ, Jensen RG. Strategies for engineering water-stress tolerance in plants. Trends Biotechnol. 1996;14:89–97. [Google Scholar]
- Bohnert HJ, Nelson DE, Jensen RG. Adaptations to environmental stresses. Plant Cell. 1995;7:1099–1011. doi: 10.1105/tpc.7.7.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowsher CG, Boulton EL, Rose J, Nayagam S, Emes MJ. Reductant for glutamate synthase is generated by the oxidative pentose phosphate pathway in non-photosynthetic root plastids. Plant J. 1992;2:893–898. [Google Scholar]
- Bowsher CG, Hucklesby DP, Emes MJ. Nitrite reduction and carbohydrate metabolism in plastids purified from roots of Pisum sativumL. Planta. 1989;177:359–366. doi: 10.1007/BF00403594. [DOI] [PubMed] [Google Scholar]
- Brouquisse R, Weigel P, Rhodes D, Yocum CF, Hanson AD. Evidence for a ferredoxin-dependent choline monooxygenase from spinach chloroplast stroma. Plant Physiol. 1989;90:322–329. doi: 10.1104/pp.90.1.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnet M, Lafontaine PJ, Hanson AD. Assay, purification, and partial characterization of choline monooxygenase from spinach. Plant Physiol. 1995;108:581–588. doi: 10.1104/pp.108.2.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline K, Henry R. Import and routing of nucleus-encoded chloroplast proteins. Annu Rev Cell Dev Biol. 1996;12:1–26. doi: 10.1146/annurev.cellbio.12.1.1. [DOI] [PubMed] [Google Scholar]
- Dunham RJ. Water use and irrigation. In: Cooke DA, Scott RK, editors. The Sugar Beet Crop. London: Chapman and Hall; 1993. pp. 279–309. [Google Scholar]
- Gorham J. Betaines in higher plants: biosynthesis and role in stress metabolism. In: Wallsgrove RM, editor. Amino Acids and Their Derivatives in Higher Plants. Cambridge, UK: Cambridge University Press; 1995. pp. 173–203. [Google Scholar]
- Gray J, Close PS, Briggs SP, Johal GS. A novel suppressor of cell death in plants encoded by the Lls1gene of maize. Cell. 1997;89:25–31. doi: 10.1016/s0092-8674(00)80179-8. [DOI] [PubMed] [Google Scholar]
- Grumet R, Hanson AD. Genetic evidence for an osmoregulatory function of glycinebetaine accumulation in barley. Aust J Plant Physiol. 1986;13:353–364. [Google Scholar]
- Hanson AD, Rathinasabapathi B, Chamberlin B, Gage DA. Comparative physiological evidence that β-alanine betaine and choline-O-sulfate act as compatible osmolytes in halophytic Limoniumspecies. Plant Physiol. 1991;97:1199–1205. doi: 10.1104/pp.97.3.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson AD, Wyse R. Biosynthesis, translocation, and accumulation of betaine in sugar beet and its progenitors in relation to salinity. Plant Physiol. 1982;70:1191–1198. doi: 10.1104/pp.70.4.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi H, Alia, Mustardy L, Deshnium P, Ida M, Murata N. Transformation of Arabidopsis thaliana with the codAgene for choline oxidase; accumulation of glycinebetaine and enhanced tolerance to salt and cold stress. Plant J. 1997;12:133–142. doi: 10.1046/j.1365-313x.1997.12010133.x. [DOI] [PubMed] [Google Scholar]
- Hondred D, Wadle D-M, Titus DE, Becker WM. Light-stimulated accumulation of the peroxisomal enzymes hydroxypyruvate reductase and serine: glyoxylate aminotransferase and their translatable mRNAs in cotyledons of cucumber seedlings. Plant Mol Biol. 1987;9:259–275. doi: 10.1007/BF00166462. [DOI] [PubMed] [Google Scholar]
- Ishitani M, Arakawa K, Mizuno K, Kishitani S, Takabe T. Betaine aldehyde dehydrogenase in the Gramineae: levels in leaves of both betaine-accumulating and nonaccumulating cereal plants. Plant Cell Physiol. 1993;34:493–495. [Google Scholar]
- Ishitani M, Nakamura T, Han SY, Takabe T. Expression of the betaine aldehyde dehydrogenase gene in barley in response to osmotic stress and abscisic acid. Plant Mol Biol. 1995;27:307–315. doi: 10.1007/BF00020185. [DOI] [PubMed] [Google Scholar]
- Jiang H, Parales RE, Lynch NA, Gibson DT. Site-directed mutagenesis of conserved amino acids in the alpha subunit of toluene dioxygenase: potential mononuclear non-heme iron coordination sites. J Bacteriol. 1996;178:3133–3139. doi: 10.1128/jb.178.11.3133-3139.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koheil MAH, Hilal SH, El-Alfy TS, Leistner E. Quaternary ammonium compounds in intact plants and cell suspension cultures of Atriplex semibaccata and A. halimusduring osmotic stress. Phytochemistry. 1992;31:2003–2008. [Google Scholar]
- Lilius G, Holmberg N, Bülow L. Enhanced NaCl stress tolerance in transgenic tobacco expressing bacterial choline dehydrogenase. Bio/Technology. 1996;14:177–180. [Google Scholar]
- Mason JR, Cammack R. The electron-transport proteins of hydroxylating bacterial oxygenases. Annu Rev Microbiol. 1992;46:277–305. doi: 10.1146/annurev.mi.46.100192.001425. [DOI] [PubMed] [Google Scholar]
- McCue KF, Hanson AD. Drought and salt tolerance: towards understanding and application. Trends Biotechnol. 1990;8:358–362. [Google Scholar]
- McCue KF, Hanson AD. Salt-inducible betaine aldehyde dehydrogenase from sugar beet: cDNA cloning and expression. Plant Mol Biol. 1992a;18:1–11. doi: 10.1007/BF00018451. [DOI] [PubMed] [Google Scholar]
- McCue KF, Hanson AD. Effects of soil salinity on the expression of betaine aldehyde dehydrogenase in leaves: investigation of hydraulic, ionic and biochemical signals. Aust J Plant Physiol. 1992b;19:555–564. [Google Scholar]
- Nairn CJ, Ferl RJ. The complete nucleotide sequence of the small-subunit ribosomal RNA coding region for the cycad Zamia pumila: phylogenetic implications. J Mol Evol. 1988;27:133–141. doi: 10.1007/BF02138373. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Yokota S, Muramoto Y, Tsutsui K, Oguri Y, Fukui K, Takabe T. Expression of a betaine aldehyde dehydrogenase gene in rice, a glycine betaine nonaccumulator, and possible localization of its protein in peroxisomes. Plant J. 1997;11:1115–1120. doi: 10.1046/j.1365-313x.1997.11051115.x. [DOI] [PubMed] [Google Scholar]
- Puissant C, Houdebine L-M. An improvement of the single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. BioTechniques. 1990;8:148–149. [PubMed] [Google Scholar]
- Rathinasabapathi B, Burnet M, Russell BL, Gage DA, Liao P-C, Nye GJ, Scott P, Golbeck JH, Hanson AD. Choline monooxygenase, an unusual iron-sulfur enzyme catalyzing the first step of glycine betaine synthesis in plants: prosthetic group characterization and cDNA cloning. Proc Natl Acad Sci USA. 1997;94:3454–3458. doi: 10.1073/pnas.94.7.3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinasabapathi B, McCue KF, Gage DA, Hanson AD. Metabolic engineering of glycine betaine synthesis: plant betaine aldehyde dehydrogenases lacking typical transit peptides are targeted to tobacco chloroplasts where they confer betaine aldehyde resistance. Planta. 1994;193:155–162. doi: 10.1007/BF00192524. [DOI] [PubMed] [Google Scholar]
- Rhodes D, Hanson AD. Quaternary ammonium and tertiary sulfonium compounds in higher plants. Annu Rev Plant Physiol Plant Mol Biol. 1993;44:357–384. [Google Scholar]
- Rhodes D, Rich PJ, Myers AC, Reuter CC, Jamieson GC. Determination of betaines by fast atom bombardment mass spectrometry. Plant Physiol. 1987;84:781–788. doi: 10.1104/pp.84.3.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydzik E, Berry JO. The C4 phosphoenolpyruvate carboxylase (PEPCase) from grain amaranth (accession no. L49175) (PGR 95-135) Plant Physiol. 1996;110:713. [Google Scholar]
- Suen W-J, Gibson DT. Isolation and preliminary characterization of the subunits of the terminal component of naphthalene dioxygenase from Pseudomonas putidaNCIB 9816–4. J Bacteriol. 1993;175:5877–5881. doi: 10.1128/jb.175.18.5877-5881.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takhtajan A (1969) Flowering Plants: Origin and Dispersal. Smithsonian Institution Press, Washington, D.C.
- Tokuhisa JG, Daniels SM, Quail PH. Phytochrome in green tissue: spectral and immunochemical evidence for two distinct molecular species of phytochrome in light grown Avena sativaL. Planta. 1985;164:321–332. doi: 10.1007/BF00402943. [DOI] [PubMed] [Google Scholar]
- Valenzuela-Soto EM, Muñoz-Clares RA. Betaine-aldehyde dehydrogenase from leaves of Amaranthus hypochondriacusL. exhibits an Iso Ordered Bi Bi steady state mechanism. J Biol Chem. 1994;268:23818–23824. [PubMed] [Google Scholar]
- Wood AJ, Saneoka H, Rhodes D, Joly RJ, Goldsbrough PB. Betaine aldehyde dehydrogenase in sorghum. Molecular cloning and expression of two related genes. Plant Physiol. 1996;110:1301–1308. doi: 10.1104/pp.110.4.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyn Jones RG, Gorham J. Osmoregulation. In: Lange OL, Osmond CB, Ziegler H, editors. Encyclopaedia of Plant Physiology, Vol 12C. Berlin: Springer-Verlag; 1983. pp. 35–58. [Google Scholar]
- Yancey PH. Compatible and counteracting solutes. In: Strange K, editor. Cellular and Molecular Physiology of Cell Volume Regulation. Boca Raton, FL: CRC Press; 1994. pp. 81–109. [Google Scholar]