Abstract
Backgroud and Objective
Genetic factors may influence the colonization of pathogenic bacteria, therefore increasing the risk for the initiation and development of periodontal disease. The present study was carried out to investigate the association of CD14-260 polymorphisms, subgingival microbiota, and gingival crevicular fluid (GCF) cytokine levels with cyclosporine A (CsA)-induced gingival overgrowth (GO) in renal transplant patients.
Material and Methods
204 patients were dichotomized into two groups: 124 with GO and 80 without GO. The CD14-260 polymorphisms were measured using an allele-specific PCR method. The levels of periodontal pathogens were determined by real-time PCR of subgingival samples. GCF levels of IL-1β and sCD14 were detected by ELISA.
Results
The frequency of CD14-260 genotype CT + TT was found to be similar in both groups. Patients with GO presented increased prevalence of Pg, Td, and Tf (red complex) and significantly higher levels of IL-1β than those without GO. GO patients carrying CT+TT genotypes were found to have higher frequencies of Pg, Td, and Tf than those carrying CC genotype. Furthermore, in the presence of red complex, CT+TT genotypes were associated with higher IL-1β levels and severe GO. Multiple logistic regression analysis demonstrated that the severity of GO is not dependent on age, gender and pharmacological variables, being only associated with CD14-260 genotype and red complex periodontopathogens.
Conclusions
No association between CD14-260 polymorphisms and the prevalence of GO was revealed in renal transplant patients administered CsA. However, CD14-260 CT+TT genotypes are associated with the prevalence of red complex periodontopathogens in GO patients, and may thus play some role in the development of severe CsA-induced GO.
Keywords: cyclosporine A, gingival overgrowth, polymorphism, CD14, cytokines, periodontal pathogens
The pathogenesis of cyclosporine (CsA)-induced gingival overgrowth (GO) is multifactorial. Plaque-induced gingival inflammation has been shown to play a pivotal role in the development of CsA-induced GO (1, 2). CsA may override the inhibitory effect of lipopolysaccharide(LPS) on cell proliferation and maintain a capacity to stimulate fibroblast DNA synthesis (3, 4). In co-cultures of gingival fibroblasts and marcophages, CsA can inhabit the activities of matrix metalloproteinase in the presence of Porphyromonas gingivalis(Pg) LPS, promoting abnormal accumulation of extracellular matrix components in the gingival lamina propria (5). LPS induces the proliferation of periodontal epithelial cells via the CD14 and Toll-like receptor (TLR) signaling pathway (6). CsA positively regulates TLR-mediated inflammatory responses of gingival fibroblasts to microbial components, enhancing the production of pro-inflammatory cytokines and potentially augmenting the proliferation of gingival fibroblasts(7). These results suggest that complex interactions between the LPS signaling pathway and tissue metabolism might be involved in the pathogenesis of CsA-induced GO.
CD14, a pattern recognition receptor on monocyte and macrophage, plays a critical role in innate immunity through recognition of bacterial LPS (8). Through interaction with both CD14 and LPS-binding protein, signal transduction on effector cells is then transferred via the TLR/MD-2 signaling complex (9), resulting in the activation of innate immune response and the release of pro-inflammatory cytokines, such as interleukin (IL)-1β, IL-6, and transforming growth factor-β1, that act synergistically with CsA in the modulation of gingival fibroblast metabolism (10–12).
A -260C>T polymorphism in the CD14 gene promoter results in decreased affinity of DNA/protein interaction at a GC box that contains a binding site for SP proteins and modulates the activity of the promoter(13). Carriage of the TT genotype has been associated with significantly higher serum levels of the soluble CD14 receptor (sCD14) and an increased density of CD14 in monocytes (14). Individuals carrying the TT genotype are more susceptible to developing periodontal infections induced by opportunistic pathogens (15). The carriers of the T-containing genotype of CD14-260(CT+TT) have been shown to have a higher extent of periodontal disease compared to those carrying the CC genotype (16), while other studies did not find associations (17, 18).
The concept of periodontal infectogenomics is defined as the relationship between host genetic factors and the composition of the subgingiva microbiota (19). Genetic factors may influence the colonization of pathogenic bacteria, therefore increasing the risk for the initiation and development of periodontal disease. Subjects positive for the composite IL-1 genotype had increased counts of red and orange complex species (20). Other studies also demonstrated an association between IL-6 gene and Fcr gene receptor variants and subgingival detection of Pg and Aggregatibacter actinomycetemcomitans (Aa) (21–24). Therefore, we hypothesized that CD14-260 genotype may influence the composition of subgingival microbiota, contributing to the increase of pro-inflammatory cytokines within gingival tissues and hence be associated with the severityof CsA-induced GO.
The aim of this study was to investigate the possible association of CD14-260 polymorphisms, subgingival microbiota, and gingival crevicular fluid (GCF) cytokine levels of IL-1β and sCD14 with CsA-induced GO in renal transplant patients.
Materials and methods
Study population and clinical examination
The cross-sectional study was conducted in the Department of Urology and Department of Stomatology in Zhongshan Hospital, Fudan University, Shanghai, China, where renal transplant recipients were seen on a regular basis to monitor drug therapy and graft survival. The immunosuppressive therapy consisted of a triple regimen including CsA, mycophenolate mofetil and prednisolone. At the time of the study, all patients had been followed for a minimum of six months. No subjects had received periodontal therapy and no subjects had taken medication known to affect periodontal status (e.g., antibiotics and anticonvulsants) within three months prior to enrolment. Radiographic examination was also carried out to detect alveolar bone destruction. Subjects receiving CsA without any sign of alveolar bone loss were selected for the present study. Demographic and pharmacologic data were obtained from the medical documentation and recorded by a single investigator. The research protocol was approved by the Ethics Committee of Fudan University.
Clinical measurements included plaque index (PLI) (25), papillary bleeding index (PBI) (26) and probing depths (PD). The subjects were assessed using a clinical scoring method according to Pernu et al (27). The subjects were ascribed a general whole-mouth score of between 0 and 3:0= no overgrowth seen; 1= mild GO (thickening of the marginal gingival and /or lobular granulation of the gingival pocket as well as overgrowth covering the gingival third of the crown or less); 2= moderate GO (overgrowth extending to the middle of the crown); and 3= severe GO (overgrowth covering two-thirds of the crown or affecting the whole attached gingival). The patients were dichotomized into a gingival overgrowth-negative (GO−) group, those with no signs of GO (score 0), and a gingival overgrowth-positive (GO+) group, those with signs of overgrowth (score 1–3) for analysis. All subjects were examined by two trained and calibrated examiners with good to excellent intra- and inter-examiner agreement.
Genotyping
DNA was extracted from epithelial buccal cells with a sequential phenol-chloroform solution and precipitated with a salt/ethanol solution. The CD14-260 polymorphisms were measured using an allele-specific PCR method, as previously described (14). Amplification reactions were performed in a total volume of 25 μl containing 50–100 ng of DNA, 1×PCR buffer (Mg2+ Plus), 0.2 mM of each dNTP, 0.75 U of HotStart Taq™ DNA polymerase (Takara, Dalian, China), and 5 pmol of each primer (Invitrogen, Shanghai, China). PCR was performed in a Mastercycle 5333 PCR system (Eppendorf, Wesseling-Berzdorf, Germany) as follows: 35 cycles at 94°C for 30s, at 60 °C for 30s, and at 72°C for 1 min. The final extension step was at 72°C for 5 min. The amplified samples were visualized by electrophoresis in 2% (w/v) agarose gel stained with ethidium bromide. The assay thus yielded a 227-bp band for the C allele and a 381-bp band for the T allele. In each PCR series, samples with known genotypes were also included to show that the PCR works. Whenever the results were not clear, the analysis was repeated. If the result was still uncertain after repetition, no result was recorded for that polymorphism.
GCF and subgingival plaque collection
GCF samples and subgingival plaque samples were collected from one proximal site of one tooth in the lower anterior arch from each subject after measuring PLI and before measuring PBI and PD. For GO+ patients, one site presenting moderate to severe GO (score 2–3) was chosen to be the sample site, and for GO-patients, one clinically uninflamed site without overgrowth was chosen.
Following the careful removal of all supragingival plaque, areas were washed with a water spray, isolated with cotton rolls, and gently dried for 30 seconds. Subgingival plaque samples were taken with one sterile paper point #30 that was inserted into the bottom of the periodontal pocket for 30 seconds. Points with blood marks were discarded. The paper points were placed in sterile polypropylene tubes containing 1.5 ml of phosphate buffered solution.
The teeth were washed again; the area was then isolated and gently dried. GCF was collected with paper strips (Whatman International Ltd, Maidstone UK) inserted into the gingival crevice and maintained for 30 seconds. Strips with blood marks were discarded. The GCF flow volume was measured by weighing the strips in sterile polypropylene tubes before and after sample collection (Mettler AE240 balance, Mettler-Toledo, Swissland). This method was used as an alternative method to direct GCF volume estimation with Periotron measurement (28). The absorbed fluid was eluted from each strip into 350 μl of phosphate buffered solution (PH 7.2). All samples (subgingival plaque and GCF) were stored at −20°C until assayed.
Microbiological evaluation
The real-time PCR method is based on the amplification of variable regions of the 16S rRNA genes of Pg, Aa, Prevotella intermedia (Pi), Treponema denticola (Td) and Tannerella forsythia (Tf). Species-specific primers were selected using software (Primer Premier 5.0) based on the published 16S rRNA sequences (Table. 1).
Table 1.
Species-specific primers used for real-time PCR
| Primer | Sequence (5′-3′) | Size of amplicon(bp) | Accession Numbers |
|---|---|---|---|
| Pg | F1:GGAATAACGGGCGATACGA | 155 | X73964 |
| F2:CACCGCTGACTTACCGAACA | |||
| Aa | F1:ATTGGGCATAAAGGGCATCT | 204 | X90833 |
| F2:TTCGCACATCAGCGTCAGTA | |||
| Pi | F1:GCCTAATACCCGATGTTGTCC | 237 | L16468 |
| F2:ACTTGGCTGGTTCAGACTTCC | |||
| Td | F1:CTGAGGACTCTGGCGGAACT | 228 | D85438 |
| F2:ACCGTGCTGATGTGTGCGATTA | |||
| Tf | F1:AGAGCCTGAACCGGCCAAGT | 208 | L16495 |
| F2:ACAGCCCCACCTACGCACC |
Pg, Porphyromonas gingivalis; Aa, Aggregatibacter actinomycetemcomitans; Pi, Prevotella intermedia; Td, Treponema denticola; Tf, Tannerella forsythia.
Bacterial DNA was extracted as described previously (29). A primer concentration of 0.25μM was ultimately used for the five species. Real-time PCR reaction was carried out using a Mastercycler system (Eppendorf, Wesseling-Berzdorf, Germany) with SYBR Green Mix (Ruicheng Biotech, Shanghai, China). Samples were assayed in duplicate in 25-μl reaction mixtures containing 2μl template DNA, 0.5μl forward primer and reverse primer, and 12.5μl SYBR Green Mix. The cycling conditions used were as follows: (1) 95°C/15 min, 95°C/30 s, 53°C/30 s, 72°C/30 s and 40 cycles for Pg and Tf and (2) 95°C/15 min, 95°C/30 s, 54°C/30 s, 72°C/30 s and 40 cycles for Aa, Pi, and Td. Melting peaks were used to determine the specificity of the PCR.
The absolute quantification of target bacteria in subgingival samples was performed using Pg (ATCC 33277), Aa (Y4), Pi (ATCC 23256), Td (ATCC 33520), and Tf (ATCC 43037) as controls. Standard curves were established with the controls, which could be used to convert cycle threshold values into the number of bacterial cells using controls with known amounts of bacterial-specific DNA. The level of detection was set to103 bacteria/subgingival plaque sample for the real-time PCR. The determination of DNA content in the controls was based on the genome size of each bacteria and the mean weight of one nucleotide pair (30, 31)
Immunological analysis
GCF IL-1β and sCD14 levels were analyzed after centrifugal elution, by using a human IL-1β enzyme-linked immunosorbent assay (ELISA) kit (Invitrogen, CA, USA) and a human sCD14 ELISA kit (Yusen, Shanghai, China) according to the manufacturer’s instructions. The results for IL-1β and sCD14 were expressed as pg/site for total cytokine levels, which has been suggested to be as a better indicator of relative GCF constituent activity rather than the GCF volume, which might result in a decrease of the cytokine concentration (32).
Statistical analysis
A comparison of the demographic, pharmacological, and periodontal data between the two groups was made using an independent sample t-test, the χ2 test, or the Mann-Whitney U test as appropriate.
The distribution of genotypes and allele frequencies were compared by the Fisher’s exact test. The odds-ratio (OR) and 95% confidence interval (95% CI) were also assessed. Genotypes were grouped according to the allele T carrier status, as this allele is known to be associated with a higher level of sCD14 (14). The Mann-Whitney U test was used to assess statistical significance in the levels of GCF cytokine between GO status, as well as between CD14-260 genotypes.
The association between bacterial prevalence and GO status and CD14-260 genotypes were analyzed by Fisher’s exact test. Logarithmic transformations were performed for the amounts of bacteria to improve normality. Bacterial amounts in both groups were analyzed by the independent t test. The spearman rank correlation coefficient was used to explore associations between levels of GCF cytokines and bacterial amounts. A multiple logistic regression analysis was performed to evaluate possible associations between these variables and the severity of GO. The p-value <0.05 were considered statistically significant. All data analysis was performed using a statistical package (SPSS 13.0, SPSS Inc., Chicago, IL, USA).
Results
Clinical parameters and genotype profiles in the GO+ and GO− groups
A total of 204 renal transplant patients(132 males and 72 females) aged 16 to 72 years (mean of 45.0 ±11.0 years) were recruited for statistical analysis (3 subjects were excluded because of their unclear genotype results, even after analysis was repeated). Eighty subjects were classified as score 0 of GO, 69 patients were ascribed score 1, 39 subjects score 2 and 16 score 3. Table 2 shows that GO+ patients presented significantly higher PLI, PBI and PD levels compared with GO− patients(p< 0.001); Concomitant CCB use was also higher in GO+ patients; however, the differences were statistically not significant (p> 0.05). The cause of end-stage renal disease, such as glomerulonephritis, diabetes, hypertension, and chronic pyelonephritis, did not show significant difference in patients with and without GO (Table 3). The proportion of polycystic kidney disease was significantly higher in GO+ patients than in GO− patients (p< 0.05).
Table 2.
Demographic, periodontal and bacterial variables of CsA-treated patients in GO+ and GO− groups
| GO+(n=124) | GO−(n=80) | p-value | |
|---|---|---|---|
| Age(years) | 45.0±10.1 | 45.0±12.2 | 0.98 |
| Gender distribution(F:M) | 40:84 | 32:48 | 0.26 |
| Duration of CsA therapy (years) | 4.2±4.0 | 4.1±4.6 | 0.74 |
| CsA dosage(mg/day) | 190±48 | 189±46 | 0.84 |
| CsA serum concentration(μg/l) | 705±233 | 764±269 | 0.10 |
| Calcium channel blocker use (%) | 64(52%) | 36(45%) | 0.36 |
| PLI | 1.44±0.65 | 0.58±0.41 | <0.001 |
| PBI | 1.21±0.75 | 0.42±0.36 | <0.001 |
| PD(mm) | 3.44±0.87 | 1.90±0.49 | <0.001 |
|
| |||
| Pg | 106(85.5) | 45(56.3) | <0.001 |
| Aa | 92(74.2) | 57(71.3) | 0.747 |
| Pi | 72(58.1) | 36(45.0) | 0.085 |
| Td | 108(87.1) | 43(53.8) | <0.001 |
| Tf | 97(78.2) | 49(61.3) | 0.011 |
| Pg+ Td+ Tf | 81(65.3) | 20(25.0) | <0.001 |
Bold represents a significant difference (p<0.05).
CsA, cyclosporine A; GO, gingival overgrowth; PLI, plaque index; PBI, papillary bleeding index; PD, probing depth; Pg, Porphyromonas gingivalis; Aa, Aggregatibacter actinomycetemcomitans; Pi, Prevotella intermedia; Td, Treponema denticola; Tf, Tannerella forsythia.
Table 3.
Cause of end-stage renal disease in CsA-treated patients with and without GO
| Cause of end-stage renal disease | GO+(n=124) | GO−(n=80) | p-value |
|---|---|---|---|
| Glomerulonephritis | 34(27.4) | 26(32.5) | 0.437 |
| Diabetes | 38(30.6) | 27(33.8) | 0.648 |
| Hypertension | 23(18.5) | 13(16.3) | 0.711 |
| Chronic pyelonephritis | 6(4.8) | 4(5.0) | 1.000 |
| Polycystic kidney disease | 9(7.3) | 2(2.5) | 0.008 |
| Other | 14(11.3) | 8(10.0) | - |
Bold represents a significant difference (p<0.05).
CsA, cyclosporine A; GO, gingival overgrowth.
The distribution of CD14-260 genotypes in the two groups did not differ significantly from the Hardy-Weinberg equilibrium. The frequency of genotypes CT + TT and of the allele T was found to be similar in the GO+ and GO− group (Table 4).
Table 4.
Genotypes and allele frequencies of CD14-260 polymorphisms in CsA-treated patients with and without GO
| GO+ group (n=124) | GO− group (n=80) | p-value | |
|---|---|---|---|
| Genotype | |||
| -260CC | 49(39..5) | 28(35.0) | |
| -260CT | 50(40.3) | 39(48.8) | p=0.413, OR=1.365, CI=0.730–2.551 |
| -260TT | 25(20.2) | 13(16.3) | p=0.984, OR=0.910, CI=0.403–2.057 |
| -260CT+TT | 75(60.4) | 52(65.0) | p=0.616, OR=1.213, CI=0.677–2.175 |
| allele | |||
| -260C allele | 148(59.7) | 95(59.4) | |
| -260T allele | 100(40.3) | 65(40.6) | p=1.000, OR=1.013, CI=0.676–1.518 |
CsA, cyclosporine A; ; GO, gingival overgrowth; OR, odds ratio; CI, confidence interval;
Association of genotype and GCF cytokine levels with periodontal parameters
The total amount and concentration of GCF IL-1β, but not sCD14, were statistically higher in the GO+ group than those in the GO− group (Fig. 1A, B). No difference were found between IL-1β, sCD14 levels and periodontal parameters in the different genotype of the GO− group, whereas CT+TT carriers from the GO+ group presented significantly increased GCF IL-1β levels(Fig. 1G). We found that in the GO+ group, CT+TT carriers were also associated with higher levels of PLI, PD and GO scores (Fig. 1C,E,F). In view of the absence of any association of sCD14 with genotype and/or clinical parameters in both groups, sCD14 was not included in the subsequent figures.
Fig. 1.
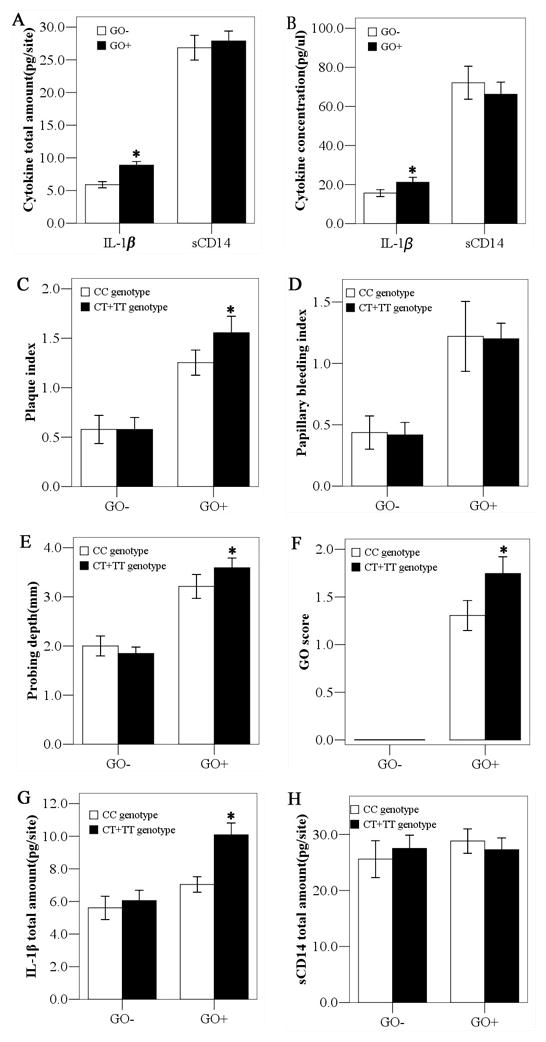
Association of genotype with GCF cytokine levels and periodontal parameters in patients with and without GO. * denotes p< 0.05 by Mann-Whitney U test. GCF, gingival crevicular fluid; GO, gingival overgrowth; sCD14, soluble CD14; IL-1β, interleukin-1β.
Association of periodontopathogens and GCF IL-1β levels with GO status
The frequencies of Pg, Td, and Tf, and the prevalence of the simultaneous occurrence of red complex pathogens (Pg+Td+Tf) were significantly more prevalent in the GO+ group than in the GO− group (Table 2). The amounts of red complex and Pi were found to be markedly higher in the GO+ group (Fig 2A). In the GO+ group, the presence of red complex and Pi were associated with significantly high levels of GCF IL-1β (Fig 2B–G). The amounts of red complex and Pi were also positively associated with the total amounts of IL-1β in GCF (Fig 3).
Fig. 2.
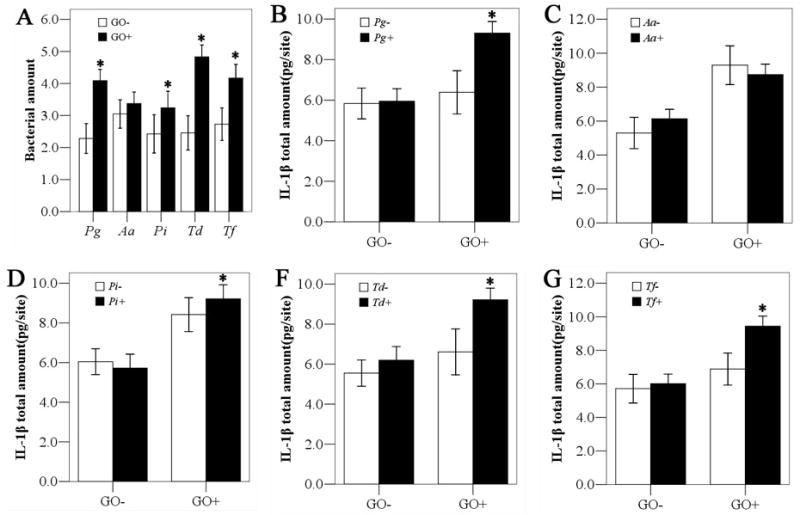
Association of the presence and amount of periodontopathogens and GCF IL-1β levels with GO status. * denotes p< 0.05 by Mann-Whitney U test or independent t test. IL-1β, interleukin-1β; GO, gingival overgrowth; Pg, Porphyromonas gingivalis; Aa, Aggregatibacter actinomycetemcomitans; Pi, Prevotella intermedia; Td, Treponema denticola; Tf, Tannerella forsythia;
Fig. 3.
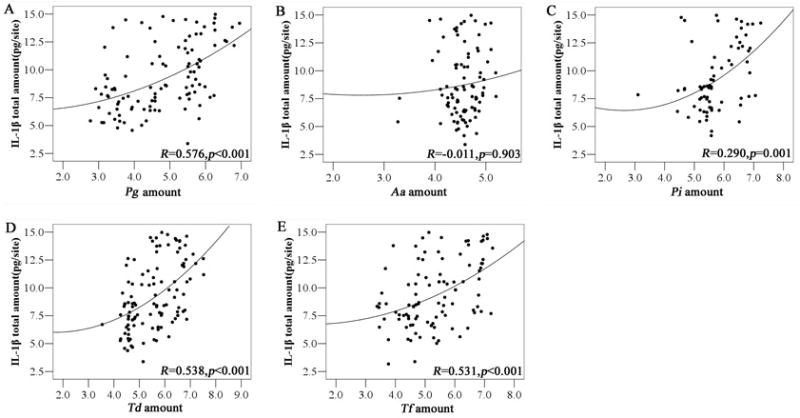
Association between the amounts of periodontal pathogens and GCF IL-1β in patients with GO. * denotes p< 0.05 by spearman rank correlation coefficient. GCF, gingival crevicular fluid; IL-1β, interleukin-1β; GO, gingival overgrowth. Pg, Porphyromonas gingivalis; Aa, Aggregatibacter actinomycetemcomitans; Pi, Prevotella intermedia; Td, Treponema denticola; Tf, Tannerella forsythia;
Association of genotype, periodontopathogens, and GCF IL-1β levels with the severity of GO
In the GO− group, no differences were found in the frequency of the periodontopathogens in the different CD14 -260 genotype groups. However, in the GO+ group, CT+TT genotype carriers presented higher frequencies of Pg, Td, and Tf and red complex (Table 5). Interestingly, in the absence of Pg, Td, and Tf, GO patients bearing CT+TT genotypes showed similar levels of GCF IL-1β levels, whereas in the presence of Pg, Td, and Tf, CT+TT genotype groups presented significantly higher levels of IL-1β (Fig 4). Additionally, in the presence of red complex, CT+TT carriers were also found to be associated with higher GO scores (p<0.05) (data not shown).
Table 5.
Frequencies of periodontal pathogens in the GO+ and GO-group with regard to CD14-260 genotype
| Bacteria | GO+ group
|
p-value | GO− group
|
p-value | ||
|---|---|---|---|---|---|---|
| CC(n=49) | CT+TT(n=75) | CC(n=28) | CT+TT(n=52) | |||
| Pg | 36(73.4) | 70(93.3) | 0.003 | 15(53.6) | 30(57.7) | 0.815 |
| Aa | 38(77.6) | 54(72.0) | 0.535 | 22(78.6) | 35(67.3) | 0.316 |
| Pi | 32(65.3) | 40(53.3) | 0.199 | 15(53.6) | 21(40.4) | 0.347 |
| Td | 39(79.6) | 69(92.0) | 0.042 | 16(57.1) | 27(51.9) | 0.815 |
| Tf | 32(65.3) | 65(86.7) | 0.007 | 18(64.3) | 31(59.6) | 0.811 |
| Red complex | 24(49.0) | 57(76.0) | 0.004 | 6(21.4) | 14(26.9) | 0.787 |
Fisher’s Exact Test was performed to determine the association of the prevalence of the five periodontal pathogens and different genotype subgroups (CC vs. CT+TT). Bold represents a significant difference (p<0.05).
GO, gingival overgrowth; Pg, Porphyromonas gingivalis; Aa, Aggregatibacter actinomycetemcomitans; Pi, Prevotella intermedia; Td, Treponema denticola; Tf, Tannerella forsythia.
Fig. 4.
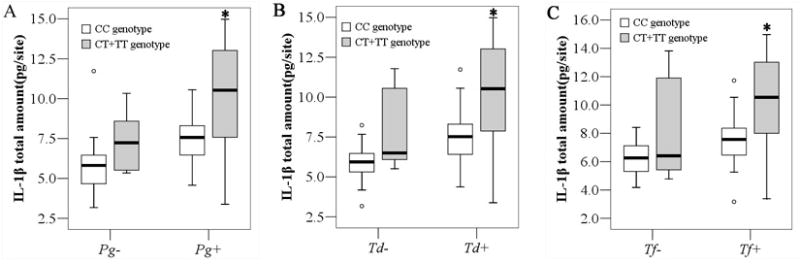
Association between genotype, the prevalence of red complex periodontopathogens, and GCF IL-1β levels in patients with GO. The box represents the first and the third quartiles (rectangular boxes); the line within the box is the median and the little circles represent atypical values that were plotted separately. * denotes p< 0.05 by Mann-Whitney U test. GCF, gingival crevicular fluid; IL-1β, interleukin-1β. Pg, Porphyromonas gingivalis; Td, Treponema denticola; Tf, Tannerella forsythia;
For further comparison, subjects in the GO+ group were categorized as mild GO patients (score 1) and moderate to severe GO patients (score 2–3). Moderate to severe GO patients were found to have significantly higher levels of GCF IL-1β and red complex than mild GO patients (p<0.05) (data not shown). The frequency of genotypes CT+TT was also significantly higher in the moderate to severe GO patients (76.4%) than that in the mild GO patients (47.8%, p=0.001, OR=3.5, 95%CI=1.6–7.7). Multiple logistic regression analysis demonstrated that the severity of GO is not dependent on age, gender and pharmacological variables including duration of CsA therapy, CsA dosage, CsA serum concentration(peak) and concomitant calcium channel blocker use, being only associated with CD14 -260 genotype(p=0.012, OR=3.0, 95% CI=1.3–7.0) and red complex periodontopathogens (p=0.009, OR=3.3, 95%CI=1.3–8.2).
Discussion
Limited data were available to delineate the effect of microbial profiles on the pathogenesis of CsA-induced GO, and even these findings are varied. Romito et al. demonstrated a positive association between Micromonas micros and the GO positive group in heart transplant patients receiving CsA (33). Animal research indicated a higher proportion of Pg in the test sites of CsA-medicated beagle dogs (34). A recent study showed that GO subjects presented a higher frequency of Tf in the salivary samples of immunosuppressed patients under the administration of CsA, tacrolimus, or sirolimus (35). The results of the present study demonstrated a relationship between the prevalence of red complex and CsA induced GO. These findings differed from the study of Vieira et al. showing that renal transplant patients taking immunosuppressive medications had a lower frequency of Pg, Td, and Tf in their dental plaque using a BANA test (36). The reason for such contrary results is not clear. It may be due to different methods of sample collection and analysis.
As a result of gingival enlargement, gingival swelling and edema cause pseudopockets that accelerate the accumulation of dental plaque, which can act as a reservoir, slowly releasing CsA and sustaining the deleterious effects on gingival tissues (37). The red complex periodontal pathogens detected in the present study have been shown to coexist as a consortium in deeper periodontal pockets (38). Enlarged gingival tissues may disrupt the normal symbiotic relationship between the host and its resident microbes, creating a more appropriate environment for the proliferation of these specifies (39). In addition, the host-impaired response due to immunosuppressive medications may also interfere in the alteration of microbial flora, favoring the growth of strictly anaerobic periodontal pathogens (40, 41).
Pg, Td, and Tf have been associated with IL-1β production in vitro and in vivo (42). The levels of IL-1β are thought to be a critical determinant of periodontal disease outcome (43). IL-1β is capable of synergizing with CsA to up-regulate the secretion of IL-6 in human gingival fibroblasts, exerting a positive modulation on collagen and glycoaminoglycan synthesis (11). Atilla et al. reported that subjects with CsA-induced GO had higher levels of GCF IL-1β than those without overgrowth (10), which is confirmed by our results. In the GO+ group, red complex-positive patients presented high levels of IL-1β than red complex-negative patients, whereas no association could be found in the GO− group. Furthermore, the amounts of red complex were also positively associated with the levels of IL-1β in the GO+ group, suggesting these elevated red complex periodontal pathogens may represent an important role in the modulation of IL-1β, which in turn synergizes with CsA to up-regulate fibroblast proliferation and extracellular matrix production.
Several studies have identified specific polymorphisms involved in the inflammatory immune responses as risk factors for CsA-induced GO, including IL-1A, IL-6, TGF-β1, and CTLA-4 (44–47). However, their findings are inconsistent. In the present study, we failed to demonstrate an association between CD14-260 genotype and the prevalence of GO, which is inconsistent with some of previous studies that the CD14-260 polymorphism was associated with chronic periodontitis in Caucasians (48, 49). The ethnic background of the study populations and the different clinical selection criteria may explain these discrepancies. However, our data demonstrated that moderate to severe GO patients presented higher proportion of CT+TT genotype compared with mild GO patients. Multiple logistic regression analysis also demonstrated an association between the CT+TT genotype and GO severity. Our data is partly in agreement with the study of Tervonen et al. that carriers of the T-containing genotype of CD14-260 have a higher extent of periodontal disease compared with those carrying the CC genotype(16), although the aetiology of drug induced GO and periodontitis are quite different. In contrast, there are also reports which demonstrated the presence of the T allele, associated with increased expression of CD14, may be protective in periodontal disease (50).
Complex interactions between the microbiota and host genetic factor are at the basis of susceptibility to periodontal disease. In the present study, GO patients carrying CT+TT genotype presented higher frequencies of Pg, Td, and Tf compared with CC genotype carriers, whereas no difference could be found in the patients without GO. Moreover, CT+TT genotype and the presence of red complex were shown to be associated with the severity of GO by multiple logistic regression analysis after adjusting for age, gender and pharmacological variables. Considering the multifactorial aetiology of periodontal disease, the two pathways for periodontal infectogenomics can explain why the predominant periodontal pathogens preferably develop in GO subjects with the T allele of CD14-260. First, genetic factor coding for pattern recognition receptors (TLRs and CD14) involved in recognizing and killing bacteria may affect bacterial clearance (19). In vitro studies have shown that TLR4 polymorphisms can affect responsiveness to Pg from gingival epithelial cells (51). Secondly, subjects carrying the T allele may increase the chance of overgrowth of periodontal pathogens due to increased inflammatory response. Monocytes with TT genotype of CD14-260 have been shown to present elevated tumor necrosis factor-α production in response to Pg LPS stimulation (18). A high concentration of IL-1β and IL-10 in culture supernatants were observed in the peripheral blood mononuclear cells of asthmatic children with the TT genotype on the response to endotoxin (52). The results of our study also demonstrate a close relationship between CT+TT genotype and higher levels of IL-1β in patients with GO. These elevated pro-inflammatory cytokines might directly lead to an increased inflammatory response and affect the proliferation of red complex pathogens in GO patients carrying CT+TT genotype.
When the putative role of these variables were evaluated individually, our results demonstrated that in the absence of red complex pathogens, CT+TT carriers showed similar levels of GCF IL-1β levels, whereas in the presence of red complex pathogens,, CT+TT genotype groups presented significantly higher levels of IL-1β and were positively associated with the severity of GO. These positive associations between CT+TT genotype and red complex pathogens in severe GO patients reinforce the concept of periodontal infectogenomics, and may help uncover the complex aetiology of CsA induced GO, possibly assisting in the prevention and management of this disease.
There are some limitations to this study that need to be considered. With the nature of cross-sectional study, it is difficult to indentify whether the gingival inflammation was present before onset of GO or was a consequence of the gingival changes. On the other hand, some susceptible individuals without gingival changes may develop severe overgrowth in the future. This may result in selection biases in the interpretation of the results (53). Additionally, in cross-sectional studies, it is hard to identify whether the gingival inflammation was present before onset of GO or was a consequence of the gingival changes. The increased levels of red complex and IL-1β in GO patients might be more a consequence of deeper and more inflamed pockets than a cause of GO. Further extensive studies are needed to analyze this putative relevance.
Conclusions
Taken together, no association between CD14-260 polymorphisms and the prevalence of GO was revealed in renal transplant patients administered CsA. However, CD14-260 CT+TT genotypes are found to be associated with the prevalence of red complex periodontopathogens in patients with GO, and may thus play some role in the development of severe CsA-induced GO.
Acknowledgments
We are grateful to Drs. Ming Xu and Ruiming Rong, Department of Urology, Zhongshan Hospital, Fudan University, for their clinical service.
Source of funding
The work is supported by the Scientific Research Foundation Project of Shanghai Health Bureau (2009089) to Y.G. and by National Institutes of Health grants DE 14537 and DE 16710 to J.C.
References
- 1.Costa FO, Ferreira SD, Lages EJ, Costa JE, Oliveira AM, Cota LO. Demographic, pharmacologic, and periodontal variables for gingival overgrowth in subjects medicated with cyclosporin in the absence of calcium channel blockers. J Periodontol. 2007;78:254–261. doi: 10.1902/jop.2007.050445. [DOI] [PubMed] [Google Scholar]
- 2.Dannewitz B, Kruck E-M, Staehle HJ, et al. Cyclosporine-induced gingival overgrowth correlates with NFAT regulated gene expression: a pilot study. J Clin Periodontol. 2011;38:984–991. doi: 10.1111/j.1600-051X.2011.01773.x. [DOI] [PubMed] [Google Scholar]
- 3.Bartold PM. Regulation of human gingival fibroblast growth and synthetic activity by cyclosporine-A in vitro. J Periodont Res. 1989;24:314–321. doi: 10.1111/j.1600-0765.1989.tb00876.x. [DOI] [PubMed] [Google Scholar]
- 4.Barber MT, Savage NW, Seymour GJ. The effect of cyclosporin and lipopolysaccharide on fibroblasts: implications for cyclosporin-induced gingival overgrowth. J Periodontol. 1992;63:397–404. doi: 10.1902/jop.1992.63.5.397. [DOI] [PubMed] [Google Scholar]
- 5.Kuo PJ, Tu HP, Chin YT, Lu SH, Chiang CY, Chen RY, Fu E. Cyclosporine-A inhibits MMP-2 and -9 activities in the presence of Porphyromonas gingivalis lipopolysaccharide: an experiment in human gingival fibroblast and U937 macrophage co-culture. J Periodontal Re. 2012 doi: 10.1111/j.1600-0765.2011.01450.x. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 6.Putnins EE, Sanaie AR, Wu Q, Firth JD. Induction of keratinocyte growth factor 1 Expression by lipopolysaccharide is regulated by CD-14 and toll-like receptors 2 and 4. Infect Immun. 2002;70:6541–6548. doi: 10.1128/IAI.70.12.6541-6548.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suzuki AM, Yoshimura A, Ozaki Y, Kaneko T, Hara Y. Cyclosporin A and phenytoin modulate inflammatory responses. J Dent Res. 2009;88:1131–1136. doi: 10.1177/0022034509350566. [DOI] [PubMed] [Google Scholar]
- 8.Wang PL, Ohura K. Porphyromonas gingivalis lipopolysaccharide signaling in gingival fibroblasts-CD14 and Toll-like receptors. Crit Rev Oral Biol Med. 2002;13:132–142. doi: 10.1177/154411130201300204. [DOI] [PubMed] [Google Scholar]
- 9.da Silva Correia J, Soldau K, Christen U, Tobias PS, Ulevitch RJ. Lipopolysaccharide is in close proximity to each of the proteins in its membrane receptor complex transfer from CD14 to TLR4 and MD-2. J Biol Chem. 2001;276:21129–21135. doi: 10.1074/jbc.M009164200. [DOI] [PubMed] [Google Scholar]
- 10.Atilla G, Kütükçüler N. Crevicular fluid interleukin-1beta, tumor necrosis factor-alpha, and interleukin-6 levels in renal transplant patients receiving cyclosporine A. J Periodontol. 1998;69:784–790. doi: 10.1902/jop.1998.69.7.784. [DOI] [PubMed] [Google Scholar]
- 11.Morton RS, Dongari-Bagtzoglou AI. Regulation of gingival fibroblast interleukin-6 secretion by cyclosporine A. J Periodontol. 1999;70:1464–1471. doi: 10.1902/jop.1999.70.12.1464. [DOI] [PubMed] [Google Scholar]
- 12.Cotrim P, Martelli-Junior H, Graner E, Sauk JJ, Coletta RD. Cyclosporin A induces proliferation in human gingival fibroblasts via induction of transforming growth factor-beta1. J Periodontol. 2003;74:1625–1633. doi: 10.1902/jop.2003.74.11.1625. [DOI] [PubMed] [Google Scholar]
- 13.LeVan TD, Bloom JW, Bailey TJ, et al. A common single nucleotide polymorphism in the CD14 promoter decreases the affinity of Sp protein binding and enhances transcriptional activity. J Immunol. 2001;167:5838–5844. doi: 10.4049/jimmunol.167.10.5838. [DOI] [PubMed] [Google Scholar]
- 14.Karhukorpi J, Yan Y, Niemela S, et al. Effect of CD14 promoter polymorphism and H. pylori infection and its clinical outcomes on circulating CD14. Clin Exp Immunol. 2002;128:326–332. doi: 10.1046/j.1365-2249.2002.01837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laine ML, Morré SA, Murillo LS, van Winkelhoff AJ, Peña AS. CD14 and TLR4 gene polymorphisms in adult periodontitis. J Dent Res. 2005;84:1042–1046. doi: 10.1177/154405910508401114. [DOI] [PubMed] [Google Scholar]
- 16.Tervonen T, Raunio T, Knuuttila M, Karttunen R. Polymorphisms in the CD14 and IL-6 genes associated with periodontal disease. J Clin Periodontol. 2007;34:377–383. doi: 10.1111/j.1600-051X.2007.01067.x. [DOI] [PubMed] [Google Scholar]
- 17.James JA, Poulton KV, Haworth SE, et al. Polymorphisms of TLR4 but not CD14 are associated with a decreased risk of aggressive periodontitis. J Clin Periodontol. 2007;35:111–117. doi: 10.1111/j.1600-051X.2006.01030.x. [DOI] [PubMed] [Google Scholar]
- 18.Yamazaki K, Ueki-Maruyama K, Oda T, et al. Single-nucleotide polymorphism in the CD14 promoter and periodontal disease expression in a Japanese population. J Dent Res. 2003;82:612–6. doi: 10.1177/154405910308200808. [DOI] [PubMed] [Google Scholar]
- 19.Nibali L, Donos N, Henderson B. Periodontal infectogenomics. J Med Microbiol. 2009;58:1269–1274. doi: 10.1099/jmm.0.012021-0. [DOI] [PubMed] [Google Scholar]
- 20.Socransky SS, Haffajee AD, Smith C, Duff GW. Microbiological parameters associated with IL-1 gene polymorphisms in periodontitis patients. J Clin Periodontol. 2000;27:810–818. doi: 10.1034/j.1600-051x.2000.027011810.x. [DOI] [PubMed] [Google Scholar]
- 21.Nibali L, Ready DR, Parkar M, et al. Gene polymorphisms and the prevalence of key periodontal pathogens. J Dent Res. 2007;86:416–420. doi: 10.1177/154405910708600505. [DOI] [PubMed] [Google Scholar]
- 22.Nibali L, Madden I, Franch Chillida F, Heitz-Mayfield L, Brett P, Donos N. IL6 -174 genotype associated with Aggregatibacter actinomycetemcomitans in Indians. Oral Dis. 2011;17:232–237. doi: 10.1111/j.1601-0825.2010.01731.x. [DOI] [PubMed] [Google Scholar]
- 23.Nibali L, Donos N, Farrell S, et al. Association between interleukin-6 -174 polymorphism and Aggregatibacter actinomycetemcomitans in chronic periodontitis. J Periodontol. 2010;81:1814–1819. doi: 10.1902/jop.2010.100084. [DOI] [PubMed] [Google Scholar]
- 24.Nibali L, Tonetti MS, Ready D, et al. Interleukin-6 polymorphisms are associated with pathogenic bacteria in subjects with periodontitis. J Periodontol. 2008;79:677–683. doi: 10.1902/jop.2008.070453. [DOI] [PubMed] [Google Scholar]
- 25.Silness J, Loe H. Periodontal disease in pregnancy. II. Correlation between oral hygiene and periodontal condtion. Acta Odontol Scand. 1964;22:121–135. doi: 10.3109/00016356408993968. [DOI] [PubMed] [Google Scholar]
- 26.Saxer UP, Mühlemann HR. Motivation and education. SSO Schweiz Monatsschr Zahnheilkd. 1975;85:905–919. [PubMed] [Google Scholar]
- 27.Pernu HE, Pernu LM, Huttunen KR, Nieminen PA, Knuuttila ML. Gingival overgrowth among renal transplant recipients related to immunosuppressive medication and possible local background factors. J Periodontol. 1992;63:548–53. doi: 10.1902/jop.1992.63.6.548. [DOI] [PubMed] [Google Scholar]
- 28.Kuru L, Griffiths GS, Petrie A, Olsen I. Changes in transforming growth factor-beta1 in gingival crevicular fluid following periodontal surgery. J Clin Periodontol. 2004;31:527–533. doi: 10.1111/j.1600-051x.2004.00521.x. [DOI] [PubMed] [Google Scholar]
- 29.Morillo JM, Lau L, Sanz M, Herrera D, Silva A. Quantitative real-time PCR based on single copy gene sequence for detection of Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis. J Periodont Res. 2003;38:518–524. doi: 10.1034/j.1600-0765.2003.00684.x. [DOI] [PubMed] [Google Scholar]
- 30.Del Peloso Ribeiro E, Bittencourt S, Sallum EA, Nociti FH, Jr, Gonçalves RB, Casati MZ. Periodontal debridement as a therapeutic approach for severe chronic periodontitis: a clinical, microbiological and immunological study. J Clin Periodontol. 2008;35:789–798. doi: 10.1111/j.1600-051X.2008.01292.x. [DOI] [PubMed] [Google Scholar]
- 31.Dolezel J, Bartos J, Voglmayr H, Greilhuber J. Nuclear DNA content and genome size of trout and human. Cytometry Part A. 2003;51:127–128. doi: 10.1002/cyto.a.10013. [DOI] [PubMed] [Google Scholar]
- 32.Gamonal J, Acevedo A, Bascones A, Jorge O, Silva A. Levels of interleukin-1 beta, -8, and -10 and RANTES in gingival crevicular fluid and cell populations in adult periodontitis patients and the effect of periodontal treatment. J Periodontol. 2000;71:1535–1545. doi: 10.1902/jop.2000.71.10.1535. [DOI] [PubMed] [Google Scholar]
- 33.Romito GA, Pustiglioni FE, Saraiva L, Pustiglioni AN, Lotufo RF, Stolf NA. Relationship of subgingival and salivary microbiota to gingival overgrowth in heart transplant patients following cyclosporin A therapy. J Periodontol. 2004;75:918–924. doi: 10.1902/jop.2004.75.7.918. [DOI] [PubMed] [Google Scholar]
- 34.Berglundh T, Lindhe J, Tarkowski A. Effects of cyclosporine A on plasma cells in experimental gingivitis in dogs. J Clin Periodontol. 1996;23:507–511. doi: 10.1111/j.1600-051x.1996.tb01817.x. [DOI] [PubMed] [Google Scholar]
- 35.Cota LO, Aquino DR, Franco GC, Cortelli JR, Cortelli SC, Costa FO. Gingival overgrowth in subjects under immunosuppressive regimens based on cyclosporine, tacrolimus, or sirolimus. J Clin Periodontol. 2010;37:894–902. doi: 10.1111/j.1600-051X.2010.01601.x. [DOI] [PubMed] [Google Scholar]
- 36.Vieira ML, Martins WJ, Grisi MF, Novaes AB, Souza SL, Salvador SL. Clinical and microbiological analysis of periodontally diseased sites after renal transplant. Spec Care Dentist. 2002;22:115–120. doi: 10.1111/j.1754-4505.2002.tb01173.x. [DOI] [PubMed] [Google Scholar]
- 37.Niimi A, Tohnai I, Kaneda T, Takeuchi M, Nagura H. Immunohistochemical analysis of effects of cyclosporin A on gingival epithelium. J Oral Pathol Med. 1990;19:397–403. doi: 10.1111/j.1600-0714.1990.tb00867.x. [DOI] [PubMed] [Google Scholar]
- 38.Socransky SS, Haffajee AD, Ximenez-Fyvie LA, Feres M, Mager D. Ecological considerations in the treatment of Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis periodontal infections. Periodontology 2000. 1999;20:341–362. doi: 10.1111/j.1600-0757.1999.tb00165.x. [DOI] [PubMed] [Google Scholar]
- 39.Fischer RG, Edwardsson S, Klinge B, Attström R. The effect of cyclosporin-A on the oral microflora at gingival sulcus of the ferret. J Clin Periodontol. 1996;23:853–860. doi: 10.1111/j.1600-051x.1996.tb00623.x. [DOI] [PubMed] [Google Scholar]
- 40.Leung WK, Yau JY, Jin LJ, et al. Subgingival microbiota of renal transplant recipients. Oral Microbiol Immunol. 2003;18:37–44. doi: 10.1034/j.1399-302x.2003.180106.x. [DOI] [PubMed] [Google Scholar]
- 41.Saraiva L, Lotufo RF, Pustiglioni AN, Silva HT, Jr, Imbronito AV. Evaluation of subgingival bacterial plaque changes and effects on periodontal tissues in patients with renal transplants under immunosuppressive therapy. Oral Surg Oral Med Oral Pathol Oral Radio Endod. 2006;101:457–462. doi: 10.1016/j.tripleo.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 42.Bodet C, Chandad F, Grenier D. Inflammatory responses of a macrophage/epithelial cell co-culture model to mono and mixed infections with Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia. Microbes Infect. 2006;8:27–35. doi: 10.1016/j.micinf.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 43.Ferreira SB, Jr, Trombone AP, Repeke CE, et al. An interleukin-1beta (IL-1beta) single-nucleotide polymorphism at position 3954 and red complex periodontopathogens independently and additively modulate the levels of IL-1beta in diseased periodontal tissues. Infect Immun. 2008;76:3725–34. doi: 10.1128/IAI.00546-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Linden GJ, Haworth SE, Maxwell AP, et al. The influence of transforming growth factor-beta1 gene polymorphisms on the severity of gingival overgrowth associated with concomitant use of cyclosporin A and a calcium channel blocker. J Periodontol. 2001;72:808–814. doi: 10.1902/jop.2001.72.6.808. [DOI] [PubMed] [Google Scholar]
- 45.Bostanci N, Ilgenli T, Pirhan DC, et al. Relationship between IL-1A polymorphisms and gingival overgrowth in renal transplant recipients receiving Cyclosporin A. J Clin Periodontol. 2006;33:771–778. doi: 10.1111/j.1600-051X.2006.00990.x. [DOI] [PubMed] [Google Scholar]
- 46.Kusztal M, Radwan-Oczko M, Kościelska-Kasprzak K, Boratyńska M, Patrzałek D, Klinger M. Possible association of CTLA-4 gene polymorphism with cyclosporine-induced gingival overgrowth in kidney transplant recipients. Transplant Proceedings. 2007;39:2763–2765. doi: 10.1016/j.transproceed.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 47.Radwan-Oczko M, Boratyńska M, Zietek M, Zołedziewska M, Jonkisz A. The relationship of transforming growth factor-beta1 gene polymorphism, its plasma level, and gingival overgrowth in renal transplant recipients receiving different immunosuppressive regimens. J Periodontol. 2006;77:865–873. doi: 10.1902/jop.2006.050086. [DOI] [PubMed] [Google Scholar]
- 48.Donati M, Berglundh T, Hytönen AM, Hahn-Zoric M, Hanson LA, Padyukov L. Association of the -159 CD14 gene polymorphism and lack of association of the -308 TNFA and Q551R IL-4RA polymorphisms with severe chronic periodontitis in Swedish Caucasians. J Clin Periodontol. 2005;32:474–9. doi: 10.1111/j.1600-051X.2005.00697.x. [DOI] [PubMed] [Google Scholar]
- 49.Folwaczny M, Glas J, Török HP, Fricke K, Folwaczny C. The CD14 -159C-to-T promoter polymorphism in periodontal disease. J Clin Periodontol. 2004;31:991–5. doi: 10.1111/j.1600-051X.2004.00600.x. [DOI] [PubMed] [Google Scholar]
- 50.Sahingur SE, Xia XJ, Gunsolley J, Schenkein HA, Genco RJ, De Nardin E. Single nucleotide polymorphisms of pattern recognition receptors and chronic periodontitis. J Periodontal Res. 2011;46:184–92. doi: 10.1111/j.1600-0765.2010.01327.x. [DOI] [PubMed] [Google Scholar]
- 51.Kinane DF, Shiba H, Stathopoulou PG, et al. Gingival epithelial cells heterozygous for Toll-like receptor 4 polymorphisms Asp299Gly and Thr399Ile are hypo-responsive to Porphyromonas gingivalis. Genes Immun. 2006;7:190–200. doi: 10.1038/sj.gene.6364282. [DOI] [PubMed] [Google Scholar]
- 52.Keskin O, Birben E, Saçkesen C, et al. The effect of CD14-c159T genotypes on the cytokine response to endotoxin by peripheral blood mononuclear cells from asthmatic children. Ann Allergy Asthma Immunol. 2006;97:321–8. doi: 10.1016/S1081-1206(10)60796-X. [DOI] [PubMed] [Google Scholar]
- 53.Schäfer AS, Jepsen S, Loos BG. Periodontal genetics: a decade of genetic association studies mandates better study designs. J Clin Periodontol. 2011;38:103–107. doi: 10.1111/j.1600-051X.2010.01653.x. [DOI] [PubMed] [Google Scholar]


