Abstract
Mutations in DLX3 in humans lead to defects in craniofacial and appendicular bones, yet the in vivo activity related to Dlx3 function during normal skeletal development have not been fully elucidated. Here we used a conditional knockout approach to analyze the effects of neural crest deletion of Dlx3 on craniofacial bones development. At birth, mutant mice exhibit a normal overall positioning of the skull bones, but a change in the shape of the calvaria was observed. Molecular analysis of the genes affected in the frontal bones and mandibles from these mice identified several bone markers known to affect bone development, with a strong prediction for increased bone formation and mineralization in vivo. Interestingly, while a subset of these genes were similarly affected in frontal bones and mandibles (Sost, Mepe, Bglap, Alp, Ibsp, Agt), several genes, including Lect1 and Calca, were specifically affected in frontal bones. Consistent with these molecular alterations, cells isolated from the frontal bone of mutant mice exhibited increased differentiation and mineralization capacities ex vivo, supporting cell autonomous defects in neural crest cells. However, adult mutant animals exhibited decreased bone mineral density in both mandibles and calvaria, as well as a significant increase in bone porosity. Together, these observations suggest that mature osteoblasts in the adult respond to signals that regulate adult bone mass and remodeling. This study provides new downstream targets for Dlx3 in craniofacial bone, and gives additional evidence of the complex regulation of bone formation and homeostasis in the adult skeleton.
Keywords: Dlx3, Neural crest, Craniofacial, development bone
INTRODUCTION
Dlx homeobox genes are known to play crucial roles in craniofacial development. In mammals, there are six Dlx genes organized into three pairs of inverted and convergently transcribed genes: Dlx1/2, Dlx3/4 and Dlx5/6 (McGuinness et al., 1996; Nakamura et al., 1996; Sumiyama et al., 2002). All members of the Dlx family are expressed in the neural crest (NC) and branchial arches. At E9.5, the expression patterns of Dlx1/2, Dlx5/6 and Dlx3/4 overlap in the distal part of the arches but expand to different degrees towards the proximal side of the arches (Depew et al., 2005; Minoux and Rijli, 2010). The effects of Dlx1, Dlx2, Dlx5 and Dlx6 deletions on craniofacial development have been studied, with double knockout mice (Dlx1/2 and Dlx5/6) exhibiting more pronounced defects than single knockout mice (Qiu et al., 1997; Thomas et al., 1997). The craniofacial defects observed in these mouse models are various, affecting skull and jaw morphology, as well as inner ear and tooth development. Among all Dlx knockout studies, Dlx5−/−/Dlx6−/− mice exhibit the most severe craniofacial defects. However, there has been no report showing the effects of Dlx3 or Dlx4 ablation on craniofacial bone development.
Mutations in DLX3 in humans lead to Tricho-Dento-Osseous syndrome (TDO), an ectodermal dysplasia characterized by defects in hair, teeth and bone (Price et al., 1998; Nieminen et al., 2011). Craniofacial and appendicular bones in TDO patients exhibit increased bone mineral density, and patients often have prognathic mandible and aberrant skull shape (Lichtenstein et al., 1972; Nieminen et al., 2011; Nguyen et al., 2012). Early embryonic lethality (E9.5) caused by placental defects precluded the analysis of the craniofacial phenotype in Dlx3−/− mice. In a recent study, we used Wnt1-cre mice and floxed Dlx3 mice (Dlx3F/F) to delete Dlx3 in the NC. With this approach, we showed that Dlx3 expression in the dental mesenchyme was essential for normal odontoblast differentiation and dentin formation during tooth development (Duverger et al., 2012).
In the present study, we have characterized alterations in the craniofacial skeleton of these conditional knockout mice during embryogenic bone formation and in the adult skeleton, and identified pathways affected by the absence of Dlx3 in NC-derived craniofacial bones.
MATERIALS AND METHODS
Mice breeding and genotyping
Dlx3LacZ/WT mice and Dlx3F/F mice were generated and genotyped as previously described (Hwang et al., 2008). Wnt1-cre mice (Jax3829) were used to delete Dlx3 in the NC and the activity of the cre recombinase in Wnt1-cre mice was traced by mating with R26RLacZ (Jax3474) mice (The Jackson Laboratories, Bar Harbor, ME). All animal work was approved by the NIAMS Animal Care and Use Committee.
Whole mount LacZ staining
Whole mount LacZ staining of Wnt1-cre:R26RLacZ embryos was performed according to (Danielian et al., 1998). Staining procedure for Dlx3LacZ/WT embryos was described previously (Hwang et al., 2008). Images were acquired using an Olympus SZX9 dissecting microscope (Olympus, Center Valley, PA).
Skeletal staining
Embryos were collected and kept in ice cold 1X PBS for one hour. They were dipped in hot water (60–70°C) for one minute to facilitate the removal of the skin. Skinned embryos were fixed in absolute ethanol at room temperature for one day with gentle shaking, and subsequently incubated for two days at room temperature in acetone with gentle shaking to dissolve fat tissues. Skeletal staining was performed in 75% ethanol, 5% acetic acid, 0.02% Alcian Blue 8GX (Sigma-Aldrich, Saint-Louis, MO, A3157), 0.01% Alizarin Red S (Sigma-Aldrich, Saint-Louis, MO, A5533), with Thymol (Sigma-Aldrich, Saint-Louis, MO, T0501) to prevent fungi contamination. After two days of staining at 37°C with gentle shaking, specimens were washed once with water and cleared in 1% KOH for several days. Clearing was performed at room temperature without shaking and 1% KOH solution was changed every day, until all soft tissues appeared transparent. Cleared specimens were incubated in 20% glycerol for one day and transferred to 50% glycerol for long term storage at room temperature. Images were acquired using an Olympus SZX9 dissecting microscope (Olympus, Center Valley, PA).
Microarray analysis
Mandibles and calvaria from newborn (P0) heads were dissected in RNA later. Muscle tissues surrounding the mandibles were removed. Frontal and parietal bones were separated using a scalpel and sutures were removed. Dissected tissues were immediately immerged in Trizol® reagent (Invitrogen, Carlsbad, CA) and homogenized using a tissue homogenizer with disposable plastic probes (OMNI international, Kennsaw, GA). Total RNA was extracted following the manufacturer’s protocol (Invitrogen, Carlsbad, CA). Microarray analysis was performed on three control and three mutant animals by the NIH NIDDK Genomics Core Facility (Yadav et al., 2011). Samples were hybridized with affymetrix Mouse 430.2 arrays (Affymetrix Inc., Santa-Clara, CA) and processed using Affymetrix 450 Fluidic stations. Chips were scanned using Affymetrix GeneChip scanner 3000 running Affymetrix (GeneChip Operating Software) GCOS 1.4 version software. Wild type samples were averaged and used as a baseline to mutant samples. The significantly affected genes (p < 0.05 and fold change ≥ 1.5) were selected based on ANOVA analysis by Partek Pro software (Partek, St. Charles, MO, USA). Ingenuity Pathway Analysis (Ingenuity Systems, Redwood City, CA) was used to identify affected functions.
Quantitative real-time PCR
One microgram of total RNA was reverse transcribed into cDNA using the ImProm-II™ Reverse Transcription System (Promega, Madison, WI). Quantitative real-time PCR analysis (qPCR) was performed on a MyiQ™ Single Color Real-Time PCR Detection System, using iQ™ Sybr® Green Supermix (Bio-Rad, Hercules, CA). Quantifications were performed on four control and four mutant animals. Primers used: Rs15 (Forward) CTTCCGCAAGTTCACCTACC and (Reverse) GGCTTGTAGGTGATGGAGAA; Dlx3 (Forward) ATTACAGCGCTCCTCAGCAT and (Reverse) CTTCCGGCTCCTCTTTCAC; Sost (Forward) TCCTCCTGAGAACAACCAGACC and (Reverse) TCTGTCAGGAAGCGGGTGTAGT; Mepe (Forward) ATGCAGGGAGAGCTGGTTAC and (Reverse) TGGTTCCCTTTGGACTCTTC; Bglap (Forward) CTTGGTGCACACCTAGCAGA and (Reverse) ACCTTATTGCCCTCCTGCTT; Col1a1 (Forward) AAGATGTGCCACTCTGACTG and (Reverse) ATAGGTGATGTTCTGGGAGG; Alp (Forward) CCAGCAGGTTTCTCTCTTGG and (Reverse) CTGGGAGTCTCATCCTGAGC; Dlx5 (Forward) CTGGCCGCTTTACAGAGAAG and (Reverse) CTGGTGACTGTGGCGAGTTA; Lect1 (Forward) TCCTTGAACTCTGTGGCGACCT and (Reverse) GGAGCACTGTTTCTCACGACTTC; Calca Forward) GCACTGGTGCAGGACTATATGC and (Reverse) CTCAGATTCCCACACCGCTTAG; Ibsp (Forward) CTTACCGAGCTTATGAGGATGAATA and (Reverse) AAATGGTAGCCAGATGATAAGAC.
Primary cell cultures
Osteoblastic cells were enzymatically isolated from frontal bones dissected from calvaria of control and mutant neonates (from P2 to P3) and maintained as described previously (Owen et al., 1990). Briefly, frontal and parietal bones of calvaria were aseptically dissected, sutures were removed and frontal bone fragments were digested at 37°C in 0.25% trypsin (Invitrogen, Carlsbad, CA) and 0.1% collagenase P (Roche Diagnostics, Indianapolis, IN). Cells were centrifuged for 5 minutes at 1000g, plated at a density of 1× 104 cells/cm2 in 12-well-plates and maintained in α-MEM (Invitrogen, Carlsbad, CA) with 10% fetal bovine serum (Atlanta biologicals, Lawrenceville, GA), 100 UI/ml penicillin, and 0.25mg/ml streptomycin (Sigma-Aldrich, Saint-Louis, MO) at 37°C in a fully humidified atmosphere in air with 5% CO2. After 5 days of culture, attached cells from each well were collected by trypsinization (0.25% trypsin (Invitrogen, Carlsbad, CA)), cells from the same genotype were pooled and seeded at a density of 1×104 cell/cm2. To induce differentiation and mineralization (80% confluency), regular growth medium (α-MEM supplemented with 10% FBS) was replaced with osteogenic medium (α-MEM supplemented with 10% FBS, 280μM ascorbic acid and 10 mM β-glycerophosphate disodium (Sigma-Aldrich, Saint-Louis, MO)).
Cell differentiation and mineralization assay
Alkaline phosphatase (ALP) and Von Kossa stainings were performed after 0, 7, 14, 21 and 28 days of culture in osteogenic medium. At each time point, culture media was removed and the cells were washed twice with phosphate-buffered saline (PBS). The cells were fixed on ice for 15 minutes in 10% neutral-buffered formalin (Sigma-Aldrich, Saint-Louis, MO). Subsequently, the cells were washed with deionized water and kept in 0.2M Tris pH 9.2 for 15 minutes. The cells were stored in PBS at 4°C and protected from light until all the time-points were collected. For ALP staining, cells were stained for 20 minutes with a diazonium salt solution composed of 0.1% Fast red AS TR Salt and 0.05% Naphthol AS-MX-PO4 solution (Sigma-Aldrich, Saint-Louis, MO). Afterward, the cells were rinsed with deionized water. Cells positive for ALP were stained red. For mineralization assay (Von Kossa), cells were stained with 5% silver nitrate (Sigma-Aldrich, Saint-Louis, MO) solution in bright light until mineralized areas (calcium phophate deposits) turned brown/black. After extensive washes, plates stained for ALP or Von Kossa were imaged using a flat-bed scanner. Using ImageJ, the Von Kossa stained mineralized areas were quantified as a percentage of total well surface at 14, 21 and 28 days.
High resolution X-ray, micro-CT analysis and 3-D reconstructions
Mandibles and calvaria were fixed in 4% PFA in 1X PBS overnight at 4°C. High-resolution X-rays were performed using a Faxitron MX-20 and Kodak XTL2 films (5×, 90 seconds, 40 kV). Micro-CT analysis was performed using a Skyscan 1172 desktop X-ray microfocus CT scanner (Skyscan, Aartselaar, Belgium). The scanning procedure was completed at 40 kV, at 12 μm per pixel and with a 0.5° rotation step. Bone phantoms of 0.25 and 0.75 g/cm3 were used for bone mineral density calibration. Reconstruction of raw images into axial cross-sections was performed using NRecon V1.4.0 software (Skyscan). For analysis of the data, measurements and 3D reconstruction, CT-an and CT-vol software were used (Skyscan).
Statistical analysis
All quantitative experiments were performed on at least three control and three mutant animals (Mean±SEM). Statistical analyses were performed on Prism 5 statistical software (GraphPad Software Inc., San Diego, CA), using T-test with a significance level of 0.05. *: p<0.05; **: p<0.01; ***: p<0.001.
RESULTS
Dlx3 is efficiently deleted from neural crest-derived craniofacial bones in Wnt1-cre:Dlx3F/LacZ mice
Previous studies have shown that Dlx3 is expressed in post migratory NC cells in the branchial arches as early as E9.5 (Robinson and Mahon, 1994). Wnt1-cre mice were mated with Dlx3LacZ/WT mice to generate Wnt1-cre:Dlx3LacZ/WT mice that were then mated with homozygous Dlx3F/F mice. We recently showed that this strategy led to efficient NC-specific deletion of Dlx3 in Wnt1-cre:Dlx3F/LacZ mice (cKO) that were compared to their control Dlx3F/WT littermates (WT) (Duverger et al., 2012). These conditional knockout animals are viable and live a normal lifespan when fed soft food.
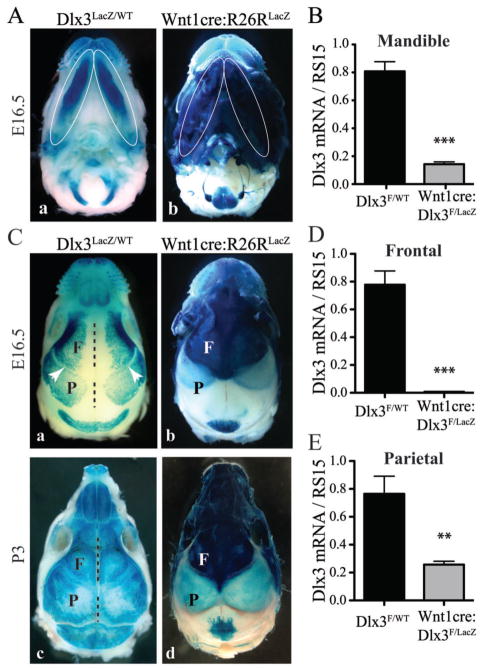
By analyzing LacZ staining in Dlx3LacZ/WT mice, we determined that Dlx3 was expressed in most developing bones in the craniofacial region at E16.5 (Figure 1, A and C). Strong Dlx3 expression was found in the mandible (Figure 1A, a), mostly derived from the NC (Chai et al., 2000), as corroborated by LacZ activity detected in Wnt1cre:R26RLacZ mice (lineage of cre-expressing cells) at E16.5 (Figure 1A, b). The level of Dlx3 mRNA in the whole mandible of cKO mice was significantly lower (7-fold) than in WT mice at P0 (Figure 1B).
Figure 1. Dlx3 expression during craniofacial development and targeted deletion using Wnt1-cre mice.
A) LacZ staining in the mandibles (bottom view of the head) of Dlx3LacZ/WT mice (a) and Wnt1-cre:R26RLacZ mice (b) at E16.5. Mandibles are marked by white ellipses. B) qPCR analysis of Dlx3 mRNA levels in mandibles at P0. T-test, n=4. C) LacZ staining in the calvaria (top view of the head) of Dlx3LacZ/WT mice (a and c) and Wnt1-cre:R26RLacZ mice (b and d), at E16.5 (a and b) and P3 (c and d). The black dotted lines and the white arrows are indicating the sagittal and coronal sutures, respectively. F: frontal bone; P: parietal bone. D) qPCR analysis of Dlx3 mRNA levels in frontal bones at P0. T-test, n=4. E) Same as D for parietal bones.
In Dlx3LacZ/WT mice, Dlx3 expression was also detected in frontal and parietal bones in the developing craniofacial region at E16.5 (Figure 1C, a). At this stage, a large area surrounding the sagittal sutures was devoid of LacZ staining, whereas the coronal sutures separating frontal and parietal bones exhibited a narrow line of LacZ-negative tissue (Figure 1C, a, white arrowhead). At P3, the LacZ staining in Dlx3LacZ/WT mice progressed centrally towards the sagittal sutures and covered the whole surface of all calvaria bones (Figure 1C, c). Previous studies have shown that while frontal bones are NC-derived, parietal bones are derived from the mesoderm (Jiang et al., 2002; Yoshida et al., 2008). This implies that cre is not expressed in the parietal bones of Wnt1-cre mice. Analyzing Wnt1-cre:R26RLacZ mice at E16.5 and P3, we concurred that frontal bones exhibited a very strong LacZ staining when compared to parietal bones (Figure 1C, b and d). The light blue staining observed in the parietal bone area has been previously shown to come from the underlying meninges and not from the bone (Jiang et al., 2002; Yoshida et al., 2008). The level of Dlx3 mRNA was reduced 100-fold in the frontal bones of Wnt1-cre:Dlx3F/LacZ mice at P0, confirming that most of the cells expressing Dlx3 in the frontal bones are NC-derived (Figure 1D). Parietal bones, in which cre is not expressed, exhibited a milder decrease in Dlx3 mRNA levels (Figure 1E). This reduction is likely due to the fact that parietal bones in Wnt1-cre:Dlx3F/LacZ mice only have one active copy of Dlx3 (Dlx3F is active; Dlx3LacZ is inactive), while two copies are active in the parietal bones of Dlx3F/WT mice.
These data indicate that Dlx3 is expressed in most craniofacial bones and can be efficiently deleted from NC-derived craniofacial bones using Wnt1-cre mice. The milder decrease in the mandibles compared to the frontal bones (7- vs 100-fold) is due to the fact that Dlx3 is also expressed in the epithelial compartment of the teeth where cre is not expressed (Zhao et al., 2000; Duverger et al., 2012). The robust Dlx3 expression in the frontal bone and mandible suggests that Dlx3 is a significant factor driving the intramembranous process of ossification from NC progenitor cells.
Deletion of Dlx3 in the neural crest alters the shape of the calvaria
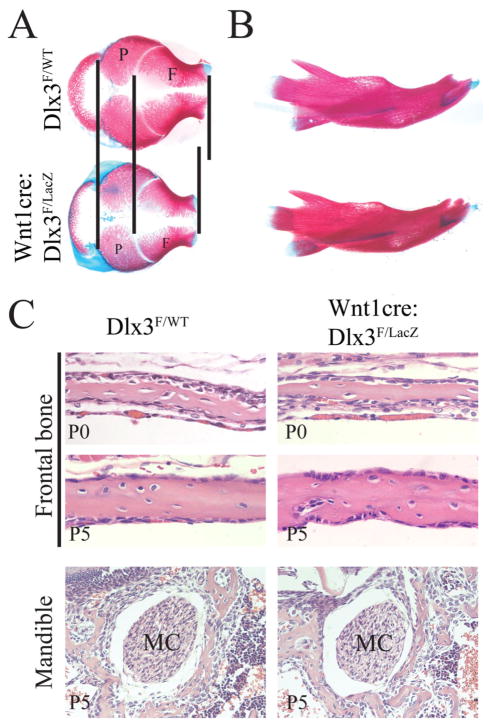
To analyze the effects of Dlx3 deletion on the development and patterning of the craniofacial skeleton, we performed a skeletal staining (Alizarin red and Alcian blue) of WT and cKO embryos at E18.5. The development of the skull appeared normal in cKO mice and no major patterning defects were observed (Figure 2). However, the calvaria appeared more rounded in the cKO than in the WT, with a visible shortening of the frontal bones along the sagittal axis (Figure 2A). At that stage the shape and structure of the mandibles also appeared normal in cKO animals (Figure 2B). All the bones of the basal part of the skull were present and no significant change in their shape was observed (data not shown). Additionally, the histology of the developing bone was not visibly altered in the frontal bone and the mandible of newborn animals (Figure 2C).
Figure 2. Structure of the developing skull of Wnt1-cre:Dlx3F/LacZ mice.
Skeletal staining (Alizarin red and Alcian blue) of the calvaria (A) and mandibles (B) from WT and cKO animals at E18.5. The vertical lines in A are indicating the limits of the frontal and parietal bones, and highlighting the relative shortening of the frontal bones in cKO animals. C) Histology of the developing craniofacial bone. H&E staining of coronal head sections showing the histology of the frontal bone from WT and cKO animals at P0 and P5 (upper panels), and the histology of the bone surrounding the Meckel’s cartilage (MC) in the area of the mandible adjacent to the first molar at P5 (lower panels).
These observations demonstrate that the absence of Dlx3 in the NC does not lead to major patterning defects (missing or transformed bone) in skull development but does alter the shape of the developing calvaria, possibly due to a disproportion between frontal and parietal bone development. This suggests that Dlx3 regulates the rate of frontal bone formation to maintain the normal pace of coordinate formation of parietal and frontal bones.
Changes in gene expression in frontal bones in Wnt1-cre:Dlx3F/LacZ mice predict an increase in bone formation and mineralization
To address mechanisms related to Dlx3 regulation of the frontal bone, genes affected by the absence of Dlx3 function in NC-derived craniofacial bones were identified by cDNA microarray analysis. We performed the study on frontal bones from cKO and WT animals at the P0 stage, in order to focus on early molecular effects of Dlx3 deletion on bone development. Using a threshold of >1.5 for the fold change, and <0.05 for the p value, 250 genes were found to be significantly affected (107 down-regulated; 143 up-regulated) in the frontal bones from cKO animals (Figure 3A and Supplemental Table S1). Ingenuity Pathway Analysis performed on this list of affected genes revealed significant alteration of functions related to bone development, with a predominant prediction for increased bone formation and mineralization. Table I presents the list of affected genes previously shown to have an effect on bone formation and/or mineralization in vivo. Based on the change in the level of expression of these genes in cKO mice and their function in bone development in vivo, we were able to predict the potential effects of these variations on bone formation and/or mineralization in our conditional knockout model. Consistent with the global prediction from Ingenuity Pathway Analysis, the majority of the genes listed in Table I are affected in a way that predicts an increase in bone formation and mineralization in cKO mice.
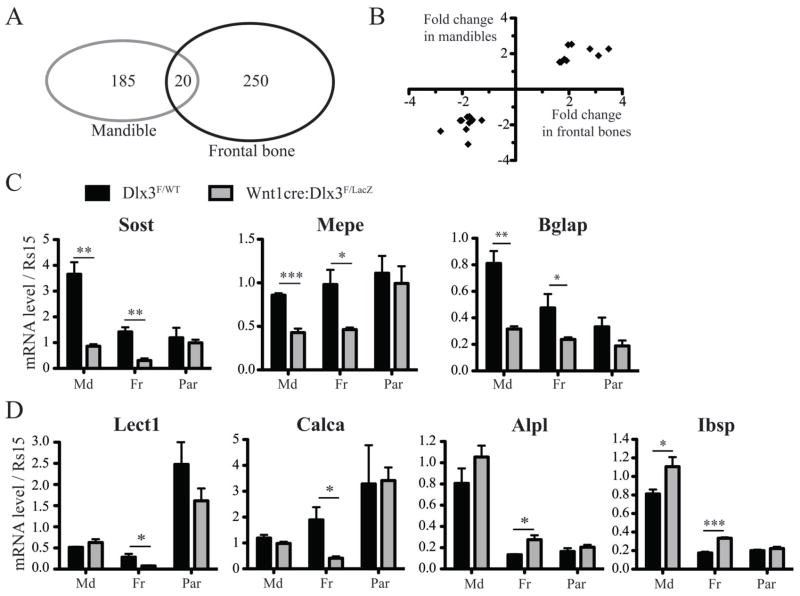
Figure 3. Bone markers affected in frontal bones and/or mandibles in Wnt1-cre:Dlx3F/LacZ mice.
A) Diagram showing the overlap between the genes significantly affected by NC-deletion of Dlx3 in mandibles and in frontal bones at P0. Including Dlx3, 21 genes were found to be affected in both tissues. B) Graph plotting the 20 genes affected in both mandibles and frontal bones, according to their fold change in both tissues. All these genes, listed in table I, exhibit the same trend in both tissues. C) qPCR analysis of the mRNA levels of Sost, Mepe and Bglap in mandibles, frontal bones and parietal bones from WT and cKO mice at P0. T-test, n=4. D) Same as C for Lect1, Calca, Alp and Ibsp.
Among the eighteen genes listed in Table I, one encodes a golgi protein (Galnt3), three encode nuclear proteins (Trps1, Mef2c and FosB), three encode membrane proteins or receptors (Dlk1, Pthr1 and Cd200), while the eleven remaining genes all encode secreted proteins. The three most strongly downregulated genes are Lect1, Calca and Sost. Lect1 (Leucocyte cell derived chemotaxin 1), also known as ChM-I (Chondromodulin-I), was revealed as a major bone mineralization factor in chondromodulin null mice that exhibit increased bone mineral density (Nakamichi et al., 2003). Tissue-specific alternative splicing of the Calca gene gives rise to two proteins, calcitonin and α-calcitonin gene-related peptide, that are distinctively involved in bone resorption and vascular regulation, respectively (Huebner et al., 2008). Sost (Sclerostin) is secreted by osteocytes and have been shown to have an inhibitory effect on bone formation and mineralization in vivo (Li et al., 2008). Among the most strongly upregulated genes encoding secreted proteins were Agt, Ibsp and Ihh. The Agt gene encodes angiotensinogen that accelerates osteoporosis when injected in mice (Shimizu et al., 2008). Ibsp, more commonly known as Bsp (Bone sialoprotein), is a member of the SIBLING (small, integrin-binding ligand N-linked glycoprotein) family and is a promoter of bone formation and mineralization (Malaval et al., 2008). Ihh (Indian Hedgehog) signaling has also been shown to promote osteoblast maturation and bone formation in vivo (St-Jacques et al., 1999; Lenton et al., 2011).
Table 1.
Genes affected in the frontal bones of Wnt1-cre:Dlx3F/LacZ mice with known oriented effects on bone formation and/or mineralization in vivo
| Gene Symbol | RefSeq Transcript ID | Fold- change | p-value | In vivo data related to gene function in bone | References | Prediction for cKO mice | ||
|---|---|---|---|---|---|---|---|---|
| Bone formation | Bone mineralization | |||||||
| Down-regulated | Lect1 | NM_010701 | −4.19 | 0.0006 | • Chondromodulin knockout mice exhibit increased bone mineral density | (Nakamichi et al., 2003) | - | Increased |
| Calca | NM_001033954/NM_007587 | −3.17 | 0.0429 | • Deletion of the Calcitonin gene in mice leads to increased bone mass | (Hoff et al., 2002) | Increased | Increased | |
| Sost | NM_024449 | −2.81 | 0.0119 | • Sclerostin knockout mice exhibit increased bone formation and mineral density | (Li et al., 2008) | Increased | Increased | |
| Dlk1 | NM_010052 | −2.64 | 0.0132 | • Overexpression of the Delta-like 1 homolog gene in bone leads to decreased bone formation and mineral density | (Abdallah et al., 2011) | Increased | Increased | |
| Nov | NM_010930 | −2.25 | 0.0496 | • Deletion of the nephroblastoma overexpressed gene in mice leads to increased bone formation and mineralization | (Heath et al., 2008) | Increased | Increased | |
| Hapln1 | NM_013500 | −1.97 | 0.0022 | • Mice lacking the hyaluronan and proteoglycan link protein 1 exhibit cartilage defects and delayed bone formation | (Watanabe and Yamada, 1999) | Decreased | - | |
| Mepe | NM_053172 | −1.85 | 0.0139 | • Deletion of the matrix extracellular phosphoglycoprotein gene in mice leads to increased bone mass in vivo and increased formation of mineralized nodules ex vivo | (Gowen et al., 2003; David et al., 2009) | Increased | Increased | |
|
| ||||||||
| Up-regulated | Pth1r | NM_011199 | 1.51 | 0.0369 | • Deletion of the parathyroid hormone 1 receptor in mice leads to reduced bone formation and mineralization | (Lanske et al., 1996; O’Brien et al., 2008) | Increased | Increased |
| Alpl | NM_007431 | 1.53 | 0.0109 | • Tissue-nonspecific Alkaline Phosphatase knockout mice exhibit defective skeletal mineralization | (Fedde et al., 1999) | - | Increased | |
| Trps1 | NM_032000 | 1.53 | 0.0234 | • Deletion of the GATA domain of the trichorhinophalangeal syndrome I gene in mice leads to decreased bone formation | (Malik et al., 2002) | Increased | - | |
| Galnt3 | NM_015736 | 1.55 | 0.0105 | • Mice lacking the UDP-N-acetyl-alpha-D- galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 gene exhibit hyperphosphatemia with increased bone mineral density | (Ichikawa et al., 2009) | Decreased | Decreased | |
| Cd200 | NM_010818 | 1.57 | 0.0111 | • Mice lacking the Cd200 molecule exhibit increased bone formation due to impaired osteoclast differentiation | (Cui et al., 2007) | Decreased | Decreased | |
| Wnt4 | NM_009523 | 1.66 | 0.0448 | • Grafting of MSCs expressing the wingless-related MMTV integration site 4 protein enhances bone formation during bone healing in vivo | (Chang et al., 2007) | Increased | Increased | |
| Mef2c | NM_025282 | 1.75 | 0.0133 | • Deletion of the myocyte enhancer factor 2C gene in mice leads to impaired chondrocyte hypertrophy and reduced bone formation | (Arnold et al., 2007) | Increased | - | |
| Ihh | NM_010544 | 1.83 | 0.0070 | • Indian hedgehog knockout mice exhibit reduced bone formation due to defective chondrocyte proliferation and failure of osteoblast maturation | (St-Jacques et al., 1999; Lenton et al., 2011) | Increased | - | |
| Ibsp | NM_008318 | 1.87 | 0.0040 | • Mice lacking the Integrin binding sialoprotein gene exhibit reduced bone formation and mineralization | (Malaval et al., 2008) | Increased | Increased | |
| Agt | NM_007428 | 1.91 | 0.0017 | • In vivo administration of angiotensinogen accelerates osteoporosis by activating osteoclasts via RANKL | (Shimizu et al., 2008) | Decreased | Decreased | |
| FosB | NM_008036 | 2.14 | 0.0494 | • Overexpression of the FBJ murine osteosarcoma viral oncogene homolog B gene increases bone formation and mineral density | (Sabatakos et al., 2000) | Increased | Increased | |
Altogether, these data suggest that the absence of Dlx3 in frontal bone osteoblasts leads to molecular changes that would predict an increase in bone formation and mineralization in Wnt1-cre:Dlx3F/LacZ mice.
A small subset of genes are similarly affected in the mandibles and frontal bones of Wnt1-cre:Dlx3F/LacZ mice
To determine if a similar gene signature was also found in other NC-derived bones, microarray analysis was performed on mandibles. We found 185 genes to be significantly affected (140 down-regulated; 45 up-regulated) in the mandibles of cKO animals at P0. By overlapping the data from frontal bones and mandibles, 20 genes were found to be affected in both tissues (Figure 3A). Importantly, all these genes were affected in a similar way (up- or down-regulated) in both tissues, as shown in Figure 3B. Table II shows the list of these genes with their fold change in mandibles and frontal bones. Among them, four genes were also listed in Table I (Sost, Mepe, Ihh and Agt), three of which were affected in a way that predicts increased bone formation and/or mineralization. Like Sost, Mepe (Matrix Extracellular Phosphoglycoprotein, also known as OF45) is secreted by osteoblasts and play an inhibitory role on bone formation and mineralization in mouse (Gowen et al., 2003). Bglap was significantly downregulated in both frontal bones and mandibles, even though it did not pass the threshold of 1.5-fold change for the frontal bones (not listed in Table I). Bglap encodes osteocalcin, the most abundant non-collagenous protein in bone extracellular matrix. Osteocalcin plays a crucial role in the regulation of bone formation, with Bglap−/− mice exhibiting increased bone formation (Ducy et al., 1996). Interestingly, Lect1 and Calca, the two bone markers that were most strongly downregulated in frontal bones were not affected in mandibles.
Table II.
Genes affected by Dlx3 deletion in both mandibles and frontal bones
| Gene Symbol | RefSeq Transcript ID | Mandible | Frontal bone | ||
|---|---|---|---|---|---|
|
| |||||
| p-value (cKO vs WT) | Fold-Change (cKO vs WT) | p-value (cKO vs WT) | Fold-Change (cKO vs. WT) | ||
| Dlx3 | NM_010055 | 2.41E-06 | −7.30 | 2.27E-05 | −16.77 |
| Sost | NM_024449 | 0.0122 | −2.36 | 0.0119 | −2.81 |
| H2-K1 | NM_001001892 | 0.0215 | −1.76 | 0.0115 | −2.08 |
| H2afv | NM_029938 | 0.0007 | −1.75 | 0.0005 | −2.02 |
| Mepe | NM_053172 | 0.0029 | −2.25 | 0.0139 | −1.85 |
| Fbln7 | NM_024237 | 0.0065 | −1.56 | 0.0172 | −1.81 |
| Lce1a2 | NM_028625 | 0.0040 | −3.09 | 0.0097 | −1.78 |
| Gal3st4 | NM_001033416 | 0.0327 | −1.89 | 0.0486 | −1.74 |
| Ahnak2 | NM_001033476 | 0.0483 | −1.53 | 0.0180 | −1.73 |
| Fetub | NM_001083904 | 0.0002 | −1.77 | 0.0027 | −1.64 |
| Magi2 | NM_001170745 | 3.32E-05 | −1.71 | 0.0170 | −1.62 |
| Bglap | NM_001032298 | 0.0034 | −1.74 | 0.0418 | −1.27* |
|
| |||||
| Cuedc1 | NM_001172099 | 0.0014 | 1.53 | 0.0128 | 1.65 |
| Rasgrp2 | NM_011242 | 0.0123 | 1.53 | 0.0007 | 1.71 |
| Ihh | NM_010544 | 0.0030 | 1.69 | 0.0071 | 1.83 |
| Agt | NM_007428 | 0.0097 | 1.61 | 0.0017 | 1.91 |
| Myl3 | NM_010859 | 0.0065 | 2.50 | 0.0173 | 1.97 |
| Hba-a1/2 | NM_001083955 | 4.13E-05 | 2.53 | 0.0014 | 2.10 |
| Chad | NM_007689 | 0.0056 | 2.27 | 4.73E-05 | 2.79 |
| Slc30a2 | NM_001039677 | 0.0024 | 1.89 | 0.0018 | 3.11 |
| Tns3 | NM_001083587 | 0.0004 | 2.27 | 0.0024 | 3.49 |
Note: Genes in shaded rows are known for their oriented effect on bone formation and mineralization in vivo. These were listed in table I, except for Bglap (* fold change lower than 1.5 in frontal bone).
These results show that, although a subset of genes were similarly affected by the absence of Dlx3 in the mandible and the calvaria, these two tissues exhibit a specific molecular signature in Wnt1-cre:Dlx3F/LacZ mice.
The osteoblast markers affected in the frontal bones and/or mandibles of Wnt1-cre:Dlx3F/LacZ mice are not affected in parietal bones
To validate the microarray data and demonstrate that the molecular changes observed in cKO mice are specific to NC-derived bones, we performed qPCR analysis for a subset of relevant genes and used parietal bones as a control tissue (not NC-derived). qPCR analysis confirmed that Sost and Mepe were downregulated in frontal bones and mandibles, while these two genes were not affected in parietal bones (Figure 3C). Similarly, Bglap expression was significantly reduced in frontal bones and mandibles, while no significant change was measured in parietal bones (Figure 3C).
qPCR analysis confirmed a significant downregulation of Lect1 and Calca expression in frontal bones while these two markers were unaffected in mandibles and parietal bones (Figure 3D). Interestingly, the basal expression level of Lect1 was significantly higher in parietal bones than in frontal bones (Figure 3D). The specific increase in Alpl expression in frontal bones was also confirmed by qPCR analysis (Figure 3D). Microarray analysis revealed a significant increase in Ibsp expression in frontal bones (Table I) but not in mandibles. qPCR analysis confirmed Ibsp up-regulation in frontal bones, but also showed a slight but significant increase in mandibles, while parietal bones showed no change in the expression of this marker (Figure 3D). We confirmed the specificity of these molecular effects by analyzing the expression of two common osteogenic-related genes that, according to the microarray analyses, showed no variation in expression: Collagen 1a1 (Col1a1) and Dlx5. The expression of these markers was unaffected in all the tissues analyzed (data not shown).
These data demonstrate that the molecular effects of the absence of Dlx3 in the NC are specific to NC-derived bones.
Calvaria osteoblasts isolated from frontal bones from Wnt1-cre:Dlx3F/LacZ mice exhibit increased differentiation and mineralization capacity in culture
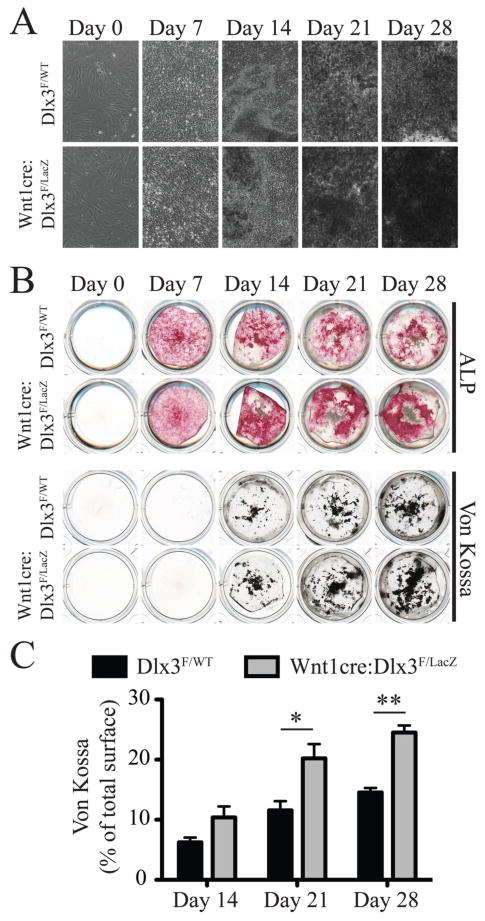
The gene signature obtained for Wnt1-cre:Dlx3F/LacZ mandibles and calvaria predicted an increase in bone formation in these mice. To check the validity of these predictions, we performed ex vivo cultures of primary osteoblasts isolated from dissected frontal bones from WT and cKO neonates. The cells were grown to subconfluence in proliferating medium before being induced to differentiate and mineralize using osteogenic medium. Cultures were analyzed 0, 7, 14, 21 and 28 days after induction. While no morphological differences could be observed at days 0 and 7 (Figure 4A), the mineralization nodules that were visible from day 14 onwards showed a different appearance in the cultures from WT and cKO mice. Indeed, we observed higher density and opacity of the nodules in cultures from cKO mice under a bright field microscope (Figure 4A). From day 14 onwards, Alkaline phosphatase (ALP) staining was stronger in cKO cultures when compared to WT (Figure 4B, upper panel), which is consistent with the increase in Alpl expression detected by microarray and qPCR analysis of frontal bones at P0 (Table I and Figure 3D). In order to measure the extent of mineralization in these cultures, we performed Von Kossa staining to detect calcium phosphate deposits. We showed that cKO cultures formed more minerals than WT cultures, with a significantly broader staining at days 21 and 28 (Figure 4, B and C).
Figure 4. Ex vivo differentiation of osteoblasts isolated from neonatal frontal bones.
A) Calvaria cell cultures isolated from frontal bones from WT and cKO animals. Images were acquired 0, 7, 14, 21 and 28 days after induction of osteoblast differentiation using osteogenic medium. Images at days 14, 21 and 28 were taken in areas showing the highest nodule density. B) Alkaline phosphatase (ALP, upper panel) and Von Kossa (upper panel) stainings of calvaria cell cultures described in A. C) Quantification of the relative surface of Von Kossa staining (percentage of total well surface) for WT and cKO cell cultures at 14, 21 and 28 days after induction of differentiation.
Taken together, these observations demonstrate that, in the absence of Dlx3, calvaria osteoblasts exhibit a higher cell autonomous mineralization capacity ex vivo. These results are consistent with the gene signature of Wnt1-cre:Dlx3F/LacZ frontal bones at P0.
Adult Wnt1-cre:Dlx3F/LacZ mice exhibit morphological defects and decreased bone mineral density in mandibles and calvaria
In light of the molecular and ex vivo data presented above, we analyzed the structure of adult craniofacial bones in Wnt1-cre:Dlx3F/LacZ mice using high resolution X-ray and micro-CT analysis.
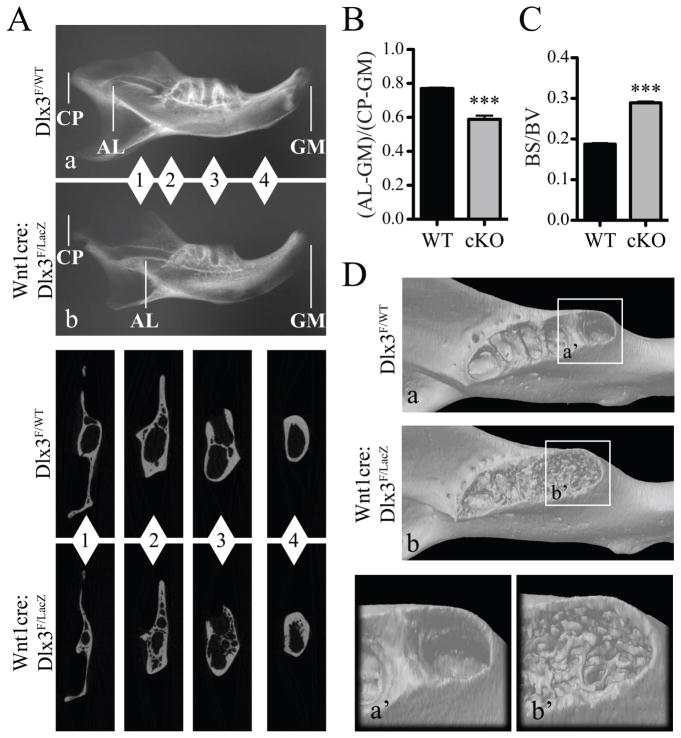
Mandibles at 8 weeks exhibited morphological and structural defects. High-resolution X-ray analysis showed a shortening of the distance between the incisor apical loop and the gingival margin (Figure 5, A and B). Micro-CT cross sections taken at different positions along the proximal-distal axis of the mandible (Figure 5A, insets 1–4) showed high porosity of the mandibular bone in cKO mice compared to controls. This was further supported by a significant increase in the ratio between bone surface and bone volume in mandibles from cKO mice (Figure 5C), and by 3D reconstruction images of the alveolar region (Figure 5D). Of interest, the average bone mineral density of the mandible was lower in cKO mice than in WT mice (Table III).
Figure 5. Structural defects in mandibles in adult Wnt1-cre:Dlx3F/LacZ mice.
A) High-resolution X-ray of mandibles from WT (a) and cKO (b) mice at 8 weeks. CP: condyloid process; AL: apical loop; GM: gingival margin. Insets show micro-CT section images at different levels (1–4) along the proximal-distal axis of the mandible. B) Measure of the distance between the apical loop and the gingival margin (AL-GM) relative to the total length (CP-GM) of the mandible. C) Measure of the ratio between bone surface (BS) and bone volume (BV), reflecting the increased porosity in mandibular bone in cKO mice. D) Micro-CT 3D-reconstruction of mandibles from WT (a) and cKO (b) mice at 8 weeks. Insets show a close-up view of the alveolar region.
Table III.
Bone mineral density measurements on Dlx3F/WT and Wnt1cre:Dlx3F/LacZ mice at 8 weeks
|
|
|||
|---|---|---|---|
| Mandible | Frontal bone | Parietal bone | |
| WT | 1.165 ± 0.008 (N=4) | 0.820 ± 0.007 (N=6) | 0.811 ± 0.007 (N=6) |
| Wnt1cre: Dlx3F/LacZ | 1.060 ± 0.008 (N=4) | 0.743 ± 0.009 (N=4) | 0.765 ± 0.010 (N=4) |
| p value | < 0.0001 (***) | 0.0002 (***) | 0.0056 (**) |
Values are in g.cm−3 ± SEM (standard error of the mean)
N: number of samples scanned.
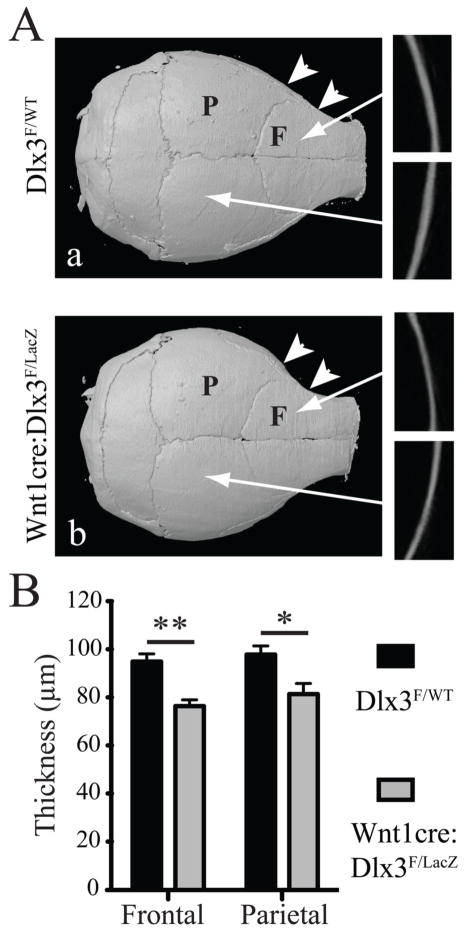
High-resolution X-ray and micro-CT analysis of the calvaria at 8 weeks confirmed a more rounded shape for the calvaria in cKO mice (Figure 6A). Micro-CT sections showed a significant reduction in the thickness of both frontal (76.4±2.6 μm for cKO versus 94.9±3.1 μm for WT; NcKO=4; NWT=6; p=0.008) and parietal bones (81.4±4.5 μm for cKO versus 97.8±3.7 μm for WT; NcKO=4; NWT=6; p=0.03) in cKO mice, with a stronger decrease for frontal bones (Figure 6A, insets, and 6B). Bone mineral density measurements showed a significant decrease in mutant animals for both frontal bones and parietal bones (Table III), with a stronger decrease in frontal bones.
Figure 6. Structural defects in calvaria in adult Wnt1-cre:Dlx3F/LacZ mice.
A) High-resolution X-ray of calvaria at 8 weeks. Insets show micro-CT cross sections of the frontal and parietal bones. The white arrowheads highlight the increased curvature of the frontal bone in cKO animals. C) Thickness measurement of the same bones. T-test, n=3. F: frontal bone; P: parietal bone.
These data indicate that, in contrast to our molecular and ex vivo observations, adult Wnt1-cre:Dlx3F/LacZ mice exhibit decreased bone mineralization. Also, despite their mesodermal origin, parietal bones exhibited decreased bone thickness and mineral density. This could potentially be due to a moderate reduction in Dlx3 expression levels in this bone (Figure 1E). However, the molecular data showing that the markers significantly affected in frontal bones and/or mandible are unaffected in parietal bones, suggest that Dlx3 haploinsufficiency has no significant effect on the expression of these genes. Therefore, the effects observed on parietal bones are likely due to a late systemic effect.
DISCUSSION
In the present study, we have analyzed the effects of NC-specific deletion of Dlx3 on craniofacial bone development. While Wnt1cre:Dlx3F/LacZ mice exhibited changes in the shape of the calvaria, they did not exhibit major patterning defects at birth. This might be due to functional redundancy with other Dlx genes expressed in the NC. As is the case in Dlx1−/−/Dlx2−/− and Dlx5−/−/Dlx6−/− double knockout animals (Qiu et al., 1997; Thomas et al., 1997), the inactivation of Dlx3 and Dlx4, could potentially lead to a stronger craniofacial phenotype. Indeed, since Dlx3 and Dlx4 follow the same expression pattern in the brachial arches (Depew et al., 2005), they could potentially play redundant functions during craniofacial development.
Microarray and qPCR analysis performed on frontal bones and mandibles from newborn animals identified molecular markers affected by the absence of Dlx3 in NC-derived bones. These markers are potential targets of Dlx3 in NC-derived osteoblasts. Among the genes that are known to modulate bone formation and mineralization in vivo, we identified Sost, Mepe, Bglap, Ibsp, Ihh and Agt as being similarly affected in frontal bones and mandibles. Interetingly, all the other makers affected in both tissues were also showing the same trend in frontal bones and mandibles. Even though these genes have not yet been implicated in bone development, the alteration of their expression could potentially have an influence on osteogenesis. Overall, the number of genes affected both in frontal bones and mandibles was relatively low (20 genes), and a substantial number of genes known to be involved in the regulation of bone formation and mineralization were affected exclusively in the mandible or the frontal bone. This demonstrates that, even though osteoblasts in frontal bones and mandibles are from the same lineage (NC), their later differentiation involves both overlaping and tissue-specific mechanisms. A fine tuning of Dlx3 function through the involvement of different transactivation partners and/or chromatin remodeling factors must be involved in the differential determination of its targets in distinct bone compartments. Such tuning has been shown for the direct regulation of Bglap and Runx2 expression by Dlx3, Dlx5, Msx2, Hoxa10 and other players during in vitro osteoblast differentiation (Hassan et al., 2004; Hassan et al., 2009). The differential response of specific craniofacial tissue sites to a same transcription factor was also shown in vivo for Msx2 (Molla et al., 2010). Further investigations will be required to determine more global/genomics mechanisms (e.g., epigenetics, micro-RNA control) that are involved in the regulation by Dlx3 of the new target genes identified in the Wnt1-cre:Dlx3F/LacZ mouse model.
Bone development and homeostasis involves both local and systemic signals that regulate the balance between bone formation and degradation. In the present study, we show that the molecular signature of neonatal frontal bones and mandibles devoid of Dlx3 would predict an increase in bone formation and mineralized in these two tissues. This prediction is further supported by ex vivo differentiation assays showing that osteobasts isolated from frontal bones of Wnt1-cre:Dlx3F/LacZ mice have higher differentiation/mineralization capacities when compared to frontal osteoblasts isolated from WT littermates. In contrast, what we observed in vivo by micro-CT analysis on adult mice was decreased bone mineral density. This indicates that, even though the absence of Dlx3 in the NC leads to the down-regulation of several inhibitors of bone formation and up-regulation of several activators of bone formation in calvaria and mandibles, later mechanisms that potentially involve peripheral signals may counteract this process and lead to a decrease in bone mineral density in adulthood. This hypothesis is supported by the fact that parietal bones, in which none of the bone markers analyzed were significantly affected, also exhibit a decrease in bone mineral density in the adult. It should also be noted that all the bones that are not NC-derived (parietal, appendicular, axial, etc) and still express Dlx3 may affect the overall balance of skeletal formation in Wnt1-cre:Dlx3F/LacZ mice. Further investigations will be required to identify the signals altering adult bone homeostasis in this model.
Patients with TDO syndrome have increased bone mineral density in both craniofacial and appendicular bones (Lichtenstein et al., 1972). A transgenic mouse model in which mutant Dlx3 was expressed under the Col1a1 promoter (Choi et al., 2009), exhibited increased formation and mineral density of trabecular bone in the femur through decreased osteoclasts number and activity due to increased IFN-γ levels in the plasma of transgenic mice expressing the TDO mutant. However, the mechanism by which the expression of the mutant Dlx3 isoform in osteoblasts leads to this effect on osteoclasts has not been elucidated. Even though the complete deletion of Dlx3 in bone cannot be directly compared to a situation where an autosomal dominant mutation is overexpressed, it is intriguing that most of the molecular alterations observed in NC-derived craniofacial bones lacking Dlx3 are predicting an increase in bone formation and mineralization. Therefore, we suggest that the osteoblast markers identified in this study will potentially be of interest for investigations on TDO syndrome.
This is the first in vivo study showing the structural and molecular effects of Dlx3 deletion on craniofacial bone development. In addition to providing new potential targets for Dlx3 in intramembranous bone formation, these data demonstrate that, during the development of different bone structures (e.g., mandibles and frontal bones) derived from the same lineage (e.g., neural crest), one transcription factor may be involve in the regulation of distinct sets of genes. The contrasting results obtained between young and adult bone formation and mineralization in this model also adds new evidence to the established complexity of bone homeostasis regulation by Dlx3.
Supplementary Material
Acknowledgments
MM is supported by the Intramural Research Program of the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health. JBL is supported by National Institute of Health grant R37 DE012528.
This research was supported by the Intramural Research Program of the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health. JBL is supported by NIH grant R37 DE012528. For the use of instruments, we thank Dr. Pamela G. Robey (X-ray). For microarray analysis, we thank George Poy and Weiping Chen. We thank the Mouse Imaging Facility of the NIH. We thank Dr. Jonathan Gordon for helpful direction and technical advice for the culture of primary osteblasts from mouse calvaria.
References
- Abdallah BM, Ditzel N, Mahmood A, Isa A, Traustadottir GA, Schilling AF, Ruiz-Hidalgo MJ, Laborda J, Amling M, Kassem M. DLK1 is a novel regulator of bone mass that mediates estrogen deficiency-induced bone loss in mice. J Bone Miner Res. 2011;26:1457–1471. doi: 10.1002/jbmr.346. [DOI] [PubMed] [Google Scholar]
- Arnold MA, Kim Y, Czubryt MP, Phan D, McAnally J, Qi X, Shelton JM, Richardson JA, Bassel-Duby R, Olson EN. MEF2C transcription factor controls chondrocyte hypertrophy and bone development. Dev Cell. 2007;12:377–389. doi: 10.1016/j.devcel.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Chai Y, Jiang X, Ito Y, Bringas P, Jr, Han J, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development. 2000;127:1671–1679. doi: 10.1242/dev.127.8.1671. [DOI] [PubMed] [Google Scholar]
- Chang J, Sonoyama W, Wang Z, Jin Q, Zhang C, Krebsbach PH, Giannobile W, Shi S, Wang CY. Noncanonical Wnt-4 signaling enhances bone regeneration of mesenchymal stem cells in craniofacial defects through activation of p38 MAPK. J Biol Chem. 2007;282:30938–30948. doi: 10.1074/jbc.M702391200. [DOI] [PubMed] [Google Scholar]
- Choi SJ, Roodman GD, Feng JQ, Song IS, Amin K, Hart PS, Wright JT, Haruyama N, Hart TC. In vivo impact of a 4 bp deletion mutation in the DLX3 gene on bone development. Dev Biol. 2009;325:129–137. doi: 10.1016/j.ydbio.2008.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui W, Cuartas E, Ke J, Zhang Q, Einarsson HB, Sedgwick JD, Li J, Vignery A. CD200 and its receptor, CD200R, modulate bone mass via the differentiation of osteoclasts. Proc Natl Acad Sci U S A. 2007;104:14436–14441. doi: 10.1073/pnas.0702811104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr Biol. 1998;8:1323–1326. doi: 10.1016/s0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- David V, Martin A, Hedge AM, Rowe PS. Matrix extracellular phosphoglycoprotein (MEPE) is a new bone renal hormone and vascularization modulator. Endocrinology. 2009;150:4012–4023. doi: 10.1210/en.2009-0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depew MJ, Simpson CA, Morasso M, Rubenstein JL. Reassessing the Dlx code: the genetic regulation of branchial arch skeletal pattern and development. Journal of Anatomy. 2005;207:501–561. doi: 10.1111/j.1469-7580.2005.00487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducy P, Desbois C, Boyce B, Pinero G, Story B, Dunstan C, Smith E, Bonadio J, Goldstein S, Gundberg C, Bradley A, Karsenty G. Increased bone formation in osteocalcin-deficient mice. Nature. 1996;382:448–452. doi: 10.1038/382448a0. [DOI] [PubMed] [Google Scholar]
- Duverger O, Zah A, Isaac J, Sun HW, Bartels AK, Lian JB, Berdal A, Hwang J, Morasso MI. Neural Crest Deletion of Dlx3 Leads to Major Dentin Defects through Down-regulation of Dspp. J Biol Chem. 2012;287:12230–12240. doi: 10.1074/jbc.M111.326900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedde KN, Blair L, Silverstein J, Coburn SP, Ryan LM, Weinstein RS, Waymire K, Narisawa S, Millan JL, MacGregor GR, Whyte MP. Alkaline phosphatase knock-out mice recapitulate the metabolic and skeletal defects of infantile hypophosphatasia. J Bone Miner Res. 1999;14:2015–2026. doi: 10.1359/jbmr.1999.14.12.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowen LC, Petersen DN, Mansolf AL, Qi H, Stock JL, Tkalcevic GT, Simmons HA, Crawford DT, Chidsey-Frink KL, Ke HZ, McNeish JD, Brown TA. Targeted disruption of the osteoblast/osteocyte factor 45 gene (OF45) results in increased bone formation and bone mass. J Biol Chem. 2003;278:1998–2007. doi: 10.1074/jbc.M203250200. [DOI] [PubMed] [Google Scholar]
- Hassan MQ, Javed A, Morasso MI, Karlin J, Montecino M, van Wijnen AJ, Stein GS, Stein JL, Lian JB. Dlx3 transcriptional regulation of osteoblast differentiation: temporal recruitment of Msx2, Dlx3, and Dlx5 homeodomain proteins to chromatin of the osteocalcin gene. Molecular & Cellular Biology. 2004;24:9248–9261. doi: 10.1128/MCB.24.20.9248-9261.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan MQ, Saini S, Gordon JA, van Wijnen AJ, Montecino M, Stein JL, Stein GS, Lian JB. Molecular switches involving homeodomain proteins, HOXA10 and RUNX2 regulate osteoblastogenesis. Cells Tissues Organs. 2009;189:122–125. doi: 10.1159/000151453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath E, Tahri D, Andermarcher E, Schofield P, Fleming S, Boulter CA. Abnormal skeletal and cardiac development, cardiomyopathy, muscle atrophy and cataracts in mice with a targeted disruption of the Nov (Ccn3) gene. BMC Dev Biol. 2008;8:18. doi: 10.1186/1471-213X-8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoff AO, Catala-Lehnen P, Thomas PM, Priemel M, Rueger JM, Nasonkin I, Bradley A, Hughes MR, Ordonez N, Cote GJ, Amling M, Gagel RF. Increased bone mass is an unexpected phenotype associated with deletion of the calcitonin gene. J Clin Invest. 2002;110:1849–1857. doi: 10.1172/JCI200214218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huebner AK, Keller J, Catala-Lehnen P, Perkovic S, Streichert T, Emeson RB, Amling M, Schinke T. The role of calcitonin and alpha-calcitonin gene-related peptide in bone formation. Arch Biochem Biophys. 2008;473:210–217. doi: 10.1016/j.abb.2008.02.013. [DOI] [PubMed] [Google Scholar]
- Hwang J, Mehrani T, Millar SE, Morasso MI. Dlx3 is a crucial regulator of hair follicle differentiation and cycling. Development. 2008;135:3149–3159. doi: 10.1242/dev.022202. Epub 2008 Aug 3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa S, Sorenson AH, Austin AM, Mackenzie DS, Fritz TA, Moh A, Hui SL, Econs MJ. Ablation of the Galnt3 gene leads to low-circulating intact fibroblast growth factor 23 (Fgf23) concentrations and hyperphosphatemia despite increased Fgf23 expression. Endocrinology. 2009;150:2543–2550. doi: 10.1210/en.2008-0877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Iseki S, Maxson RE, Sucov HM, Morriss-Kay GM. Tissue origins and interactions in the mammalian skull vault. Dev Biol. 2002;241:106–116. doi: 10.1006/dbio.2001.0487. [DOI] [PubMed] [Google Scholar]
- Lanske B, Karaplis AC, Lee K, Luz A, Vortkamp A, Pirro A, Karperien M, Defize LH, Ho C, Mulligan RC, Abou-Samra AB, Juppner H, Segre GV, Kronenberg HM. PTH/PTHrP receptor in early development and Indian hedgehog-regulated bone growth. Science. 1996;273:663–666. doi: 10.1126/science.273.5275.663. [DOI] [PubMed] [Google Scholar]
- Lenton K, James AW, Manu A, Brugmann SA, Birker D, Nelson ER, Leucht P, Helms JA, Longaker MT. Indian hedgehog positively regulates calvarial ossification and modulates bone morphogenetic protein signaling. Genesis. 2011;49:784–796. doi: 10.1002/dvg.20768. [DOI] [PubMed] [Google Scholar]
- Li X, Ominsky MS, Niu QT, Sun N, Daugherty B, D’Agostin D, Kurahara C, Gao Y, Cao J, Gong J, Asuncion F, Barrero M, Warmington K, Dwyer D, Stolina M, Morony S, Sarosi I, Kostenuik PJ, Lacey DL, Simonet WS, Ke HZ, Paszty C. Targeted deletion of the sclerostin gene in mice results in increased bone formation and bone strength. J Bone Miner Res. 2008;23:860–869. doi: 10.1359/jbmr.080216. [DOI] [PubMed] [Google Scholar]
- Lichtenstein J, Warson R, Jorgenson R, Dorst JP, McKusick VA. The tricho-dento-osseous (TDO) syndrome. Am J Hum Genet. 1972;24:569–582. [PMC free article] [PubMed] [Google Scholar]
- Malaval L, Wade-Gueye NM, Boudiffa M, Fei J, Zirngibl R, Chen F, Laroche N, Roux JP, Burt-Pichat B, Duboeuf F, Boivin G, Jurdic P, Lafage-Proust MH, Amedee J, Vico L, Rossant J, Aubin JE. Bone sialoprotein plays a functional role in bone formation and osteoclastogenesis. J Exp Med. 2008;205:1145–1153. doi: 10.1084/jem.20071294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik TH, Von Stechow D, Bronson RT, Shivdasani RA. Deletion of the GATA domain of TRPS1 causes an absence of facial hair and provides new insights into the bone disorder in inherited tricho-rhino-phalangeal syndromes. Mol Cell Biol. 2002;22:8592–8600. doi: 10.1128/MCB.22.24.8592-8600.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuinness T, Porteus MH, Smiga S, Bulfone A, Kingsley C, Qiu M, Liu JK, Long JE, Xu D, Rubenstein JL. Sequence, organization, and transcription of the Dlx-1 and Dlx-2 locus. Genomics. 1996;35:473–485. doi: 10.1006/geno.1996.0387. [DOI] [PubMed] [Google Scholar]
- Minoux M, Rijli FM. Molecular mechanisms of cranial neural crest cell migration and patterning in craniofacial development. Development. 2010;137:2605–2621. doi: 10.1242/dev.040048. [DOI] [PubMed] [Google Scholar]
- Molla M, Descroix V, Aioub M, Simon S, Castaneda B, Hotton D, Bolanos A, Simon Y, Lezot F, Goubin G, Berdal A. Enamel protein regulation and dental and periodontal physiopathology in MSX2 mutant mice. Am J Pathol. 2010;177:2516–2526. doi: 10.2353/ajpath.2010.091224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi Y, Shukunami C, Yamada T, Aihara K, Kawano H, Sato T, Nishizaki Y, Yamamoto Y, Shindo M, Yoshimura K, Nakamura T, Takahashi N, Kawaguchi H, Hiraki Y, Kato S. Chondromodulin I is a bone remodeling factor. Mol Cell Biol. 2003;23:636–644. doi: 10.1128/MCB.23.2.636-644.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S, Stock DW, Wydner KL, Bollekens JA, Takeshita K, Nagai BM, Chiba S, Kitamura T, Freeland TM, Zhao Z, Minowada J, Lawrence JB, Weiss KM, Ruddle FH. Genomic analysis of a new mammalian distal-less gene: Dlx7. Genomics. 1996;38:314–324. doi: 10.1006/geno.1996.0634. [DOI] [PubMed] [Google Scholar]
- Nguyen T, Phillips C, Frazier-Bower S, Wright T. Craniofacial variations in the tricho-dento-osseous syndrome. Clin Genet. 2012;9999 doi: 10.1111/j.1399-0004.2012.01907.x. [DOI] [PubMed] [Google Scholar]
- Nieminen P, Lukinmaa PL, Alapulli H, Methuen M, Suojarvi T, Kivirikko S, Peltola J, Asikainen M, Alaluusua S. DLX3 homeodomain mutations cause tricho-dento-osseous syndrome with novel phenotypes. Cells Tissues Organs. 2011;194:49–59. doi: 10.1159/000322561. [DOI] [PubMed] [Google Scholar]
- O’Brien CA, Plotkin LI, Galli C, Goellner JJ, Gortazar AR, Allen MR, Robling AG, Bouxsein M, Schipani E, Turner CH, Jilka RL, Weinstein RS, Manolagas SC, Bellido T. Control of bone mass and remodeling by PTH receptor signaling in osteocytes. PLoS One. 2008;3:e2942. doi: 10.1371/journal.pone.0002942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen TA, Aronow M, Shalhoub V, Barone LM, Wilming L, Tassinari MS, Kennedy MB, Pockwinse S, Lian JB, Stein GS. Progressive development of the rat osteoblast phenotype in vitro: reciprocal relationships in expression of genes associated with osteoblast proliferation and differentiation during formation of the bone extracellular matrix. J Cell Physiol. 1990;143:420–430. doi: 10.1002/jcp.1041430304. [DOI] [PubMed] [Google Scholar]
- Price JA, Bowden DW, Wright JT, Pettenati MJ, Hart TC. Identification of a mutation in DLX3 associated with tricho-dento-osseous (TDO) syndrome. Human Molecular Genetics. 1998;7:563–569. doi: 10.1093/hmg/7.3.563. [DOI] [PubMed] [Google Scholar]
- Qiu M, Bulfone A, Ghattas I, Meneses JJ, Christensen L, Sharpe PT, Presley R, Pedersen RA, Rubenstein JL. Role of the Dlx homeobox genes in proximodistal patterning of the branchial arches: mutations of Dlx-1, Dlx-2, and Dlx-1 and -2 alter morphogenesis of proximal skeletal and soft tissue structures derived from the first and second arches. Developmental Biology. 1997;185:165–184. doi: 10.1006/dbio.1997.8556. [DOI] [PubMed] [Google Scholar]
- Robinson GW, Mahon KA. Differential and overlapping expression domains of Dlx-2 and Dlx-3 suggest distinct roles for Distal-less homeobox genes in craniofacial development. Mechanisms of Development. 1994;48:199–215. doi: 10.1016/0925-4773(94)90060-4. [DOI] [PubMed] [Google Scholar]
- Sabatakos G, Sims NA, Chen J, Aoki K, Kelz MB, Amling M, Bouali Y, Mukhopadhyay K, Ford K, Nestler EJ, Baron R. Overexpression of DeltaFosB transcription factor(s) increases bone formation and inhibits adipogenesis. Nat Med. 2000;6:985–990. doi: 10.1038/79683. [DOI] [PubMed] [Google Scholar]
- Shimizu H, Nakagami H, Osako MK, Hanayama R, Kunugiza Y, Kizawa T, Tomita T, Yoshikawa H, Ogihara T, Morishita R. Angiotensin II accelerates osteoporosis by activating osteoclasts. FASEB J. 2008;22:2465–2475. doi: 10.1096/fj.07-098954. [DOI] [PubMed] [Google Scholar]
- St-Jacques B, Hammerschmidt M, McMahon AP. Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev. 1999;13:2072–2086. doi: 10.1101/gad.13.16.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumiyama K, Irvine SQ, Stock DW, Weiss KM, Kawasaki K, Shimizu N, Shashikant CS, Miller W, Ruddle FH. Genomic structure and functional control of the Dlx3-7 bigene cluster. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:780–785. doi: 10.1073/pnas.012584999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas BL, Tucker AS, Qui M, Ferguson CA, Hardcastle Z, Rubenstein JL, Sharpe PT. Role of Dlx-1 and Dlx-2 genes in patterning of the murine dentition. Development. 1997;124:4811–4818. doi: 10.1242/dev.124.23.4811. [DOI] [PubMed] [Google Scholar]
- Watanabe H, Yamada Y. Mice lacking link protein develop dwarfism and craniofacial abnormalities. Nat Genet. 1999;21:225–229. doi: 10.1038/6016. [DOI] [PubMed] [Google Scholar]
- Yadav H, Quijano C, Kamaraju AK, Gavrilova O, Malek R, Chen W, Zerfas P, Zhigang D, Wright EC, Stuelten C, Sun P, Lonning S, Skarulis M, Sumner AE, Finkel T, Rane SG. Protection from obesity and diabetes by blockade of TGF-beta/Smad3 signaling. Cell Metab. 2011;14:67–79. doi: 10.1016/j.cmet.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Vivatbutsiri P, Morriss-Kay G, Saga Y, Iseki S. Cell lineage in mammalian craniofacial mesenchyme. Mech Dev. 2008;125:797–808. doi: 10.1016/j.mod.2008.06.007. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Stock D, Buchanan A, Weiss K. Expression of Dlx genes during the development of the murine dentition. Dev Genes Evol. 2000;210:270–275. doi: 10.1007/s004270050314. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.