Abstract
Lung cancer is a highly heterogeneous disease in terms of both underlying genetic lesions and response to therapeutic treatments. We performed deep whole-genome sequencing and transcriptome sequencing on 19 lung cancer cell lines and three lung tumor/normal pairs. Overall, our data show that cell line models exhibit similar mutation spectra to human tumor samples. Smoker and never-smoker cancer samples exhibit distinguishable patterns of mutations. A number of epigenetic regulators, including KDM6A, ASH1L, SMARCA4, and ATAD2, are frequently altered by mutations or copy number changes. A systematic survey of splice-site mutations identified 106 splice site mutations associated with cancer specific aberrant splicing, including mutations in several known cancer-related genes. RAC1b, an isoform of the RAC1 GTPase that includes one additional exon, was found to be preferentially up-regulated in lung cancer. We further show that its expression is significantly associated with sensitivity to a MAP2K (MEK) inhibitor PD-0325901. Taken together, these data present a comprehensive genomic landscape of a large number of lung cancer samples and further demonstrate that cancer-specific alternative splicing is a widespread phenomenon that has potential utility as therapeutic biomarkers. The detailed characterizations of the lung cancer cell lines also provide genomic context to the vast amount of experimental data gathered for these lines over the decades, and represent highly valuable resources for cancer biology.
Lung cancer is the most common cause of cancer-related death (Siegel et al. 2012). There are two broad categories of lung cancer, small-cell lung cancer (SCLC) and non-small-cell lung cancer (NSCLC), and the latter is further divided into three major types, squamous cell carcinoma, adenocarcinoma, and large cell carcinoma (Herbst et al. 2008). Significant heterogeneity exists even within a given subtype of lung cancer (Sun et al. 2007; Herbst et al. 2008), thus necessitating personalized treatment based on the underlying causal genetic events (Salgia et al. 2011). Indeed, key drivers such as EGFR and EML4–ALK have become attractive therapeutic targets for subsets of lung cancer patients. A more comprehensive understanding of genomic alterations in lung cancers is critical for identifying new therapeutic targets as well as for identifying suitable patients who might respond to a given targeted agent.
Recent advances in high-throughput sequencing have enabled the systematic analysis of genomic and transcriptomic alterations in cancer samples on the nucleotide level (Ley et al. 2008; Maher et al. 2009; Zhao et al. 2009; Lee et al. 2010; Parsons et al. 2010; Pleasance et al 2010a,b; Wu et al. 2012). For example, whole-genome sequencing of the NCI-H209 SCLC cell line identified a total of 22,910 somatic substitutions (Pleasance et al. 2010b). We previously performed whole-genome sequencing on the tumor and normal pair from a lung adenocarcinoma patient with a history of smoking (Lee et al. 2010), and identified 50,675 high-confidence somatic point mutations and extensive structural variations (SVs) throughout the cancer genome. In both the lung cancer cell line and lung tumor studies, mutational selection pressure was found to be associated with various genomic and transcriptomic features such as the expression status, and the overall mutation spectra were consistent with tobacco exposure (Lee et al. 2010; Pleasance et al. 2010b). The broad similarities suggest that cancer cell lines might retain fundamental genomic features of the original tumor samples and therefore should recapitulate some of the phenotypes of original tumors. Indeed, breast cancer cell lines exhibit strikingly similar patterns of DNA copy number alteration to those observed in breast tumors (Neve et al. 2006). Cell lines are pivotal to drug screening and response studies (Gazdar et al. 2010), so it is important to establish whether cell lines retain key mutational features observed in tumors. Detailed understanding of genomic features of a large number of cell lines also helps selecting appropriate cell lines as model systems with particular genetic lesions. Recently, two research teams used a combination of SNP array, expression array, and targeted sequencing technologies to interrogate several hundred cancer cell lines, uncovering significant links between drug activities and the functional complexity of cancer genomes (Barretina et al. 2012; Garnett et al. 2012). Although these cell lines were not characterized at the whole genome or transcriptome levels, it is clear that projects like Cancer Cell Line Encyclopedia (CCLE) provide highly useful resources for the generation and testing of hypotheses related to the grand goals of personalizing cancer medicine (Weinstein 2012).
In this study, we have applied next generation sequencing technologies to multiple lung cancer cell lines and tissue samples (Supplemental Tables 1, 2, 3). For 19 lung cancer cell lines, we performed whole-genome sequencing, RNA sequencing, SNP array analysis, and spectral karyotyping (SKY) in order to obtain detailed and comprehensive views of genomic and transcriptomic alterations. For three lung cancer patients, including the smoker lung cancer patient we previously described (Lee et al. 2010), we performed whole-genome sequencing, RNA sequencing, and SNP array analysis on tumor/normal pairs in order to compare smoker and never-smoker tumor genomes, and to compare tumors with cell lines. The combined genome and transcriptome analyses also provide opportunities to characterize mutations that might have an effect on transcription and splicing. To this end, we performed a genome-wide survey of cancer-associated splicing events that might be caused by mutations in essential splice sites. In addition, we examined differential isoform expression between cancer and normal samples from the transcriptome data, and found that expression of RAC1b, which is up-regulated in tumors, is associated with cell line response to a MAP2K (MEK) inhibitor. It is therefore interesting to consider such genomic features as potentially unconventional biomarkers found from the combined genome and transcriptome information of cancer cell lines.
Results
Mutation landscape in lung cancer genomes
We first performed whole-genome sequencing (∼60×) for paired lung adenocarcinoma and adjacent normal lung tissue samples from two patients with no history of cigarette smoking, and compared the results with our previous study on a smoker genome. Somatic mutations in tumors were identified by comparing the variant calls in the tumor with the corresponding normal genomes, and a subset of candidate mutations was validated using mass spectrometry-based genotyping (Supplemental Table 4; Supplemental Fig. 1). In the two never-smoker tumor genomes, we identified 1802 and 1169 novel, high-confidence somatic single base substitutions, which reflect an approximate rate of 0.62 and 0.40 mutations per Mb. Among these, 16 are nonsynonymous coding mutations in each genome. In comparison, the number of somatic mutations and the consequent mutation rate were 20-fold higher in the previous smoker genome, underscoring the profound DNA damage caused by cigarette smoking. Among the somatic missense mutations are some previously reported in lung cancer. While the previously sequenced smoker tumor genome contains a mutant KRAS and wild-type EGFR, both never-smoker tumor genomes contain wild-type KRAS but mutated EGFR.
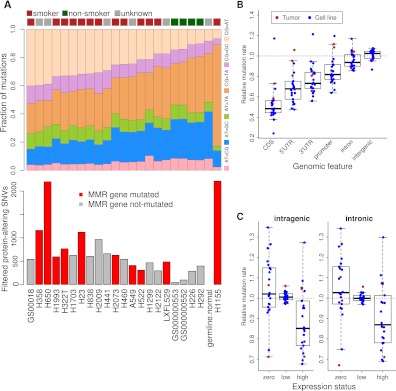
A collection of 19 lung cancer cell lines was sequenced in the same fashion. Although the definitive list of somatically mutated genes cannot be obtained due to the lack of matched normal samples for most cell lines, we approximated this list by removing all known germline variants from the cell line single nucleotide variant (SNV) collection (Supplemental Table 5). The germline variants include those in the dbSNP database, the 1000 Genomes Project, the 69 fully sequenced genomes by Complete Genomics, and the NHLBI GO Exome Sequencing Project (ESP). The numbers of filtered protein-altering SNVs vary dramatically among the cell lines. Cell lines derived from smoker patients tend to have more SNVs than those from never-smoker patients. We also noticed that the four cell lines with the most protein-altering SNVs all have mutations in at least one of the mismatch repair (MMR) genes (Fig. 1A, bottom panel).
Figure 1.
The mutation spectra and mutation rate of lung cancer genomes. (A) Lung cancer genomes have a few distinct patterns of mutation composition. Genomes from smokers tend to have a large fraction of C:G > A:T transversions, a known tobacco-related DNA-damage pattern (top panel), and have larger number of filtered variations (bottom panel). In addition, the four cell lines with the most protein-altering SNVs all have mutations in at least one of the mismatch repair (MMR) genes. (B) CDS and UTRs have the lowest mutation rates among genomic features. Mutation rates (i.e., number of mutations per mega base) for different genomic features were calculated, and then normalized by the genome-wide mutation rates to obtain the relative mutation rates. Each dot in the plot represents one genome (blue, cell lines; red, tumors). The boxes in the box-and-whisker plots represent the interquartile range between the first and third quartiles; the dashed lines (whiskers) extend to the most extreme data points which is no more than 1.5 times the interquartile range from the box. (C) Mutation rates are negatively correlated with expression level. In each genome, genes were divided into three groups according to their expression level based on the RNA-seq data from the same sample: zero (RPKM = 0), low (0 < RPKM ≤ 1), and high (RPKM > 1). Mutation rate for each group was calculated, and then normalized by average mutation rate for all genes in the genome to obtain relative mutation rates. Genes with high expression levels tend to have the lowest mutation rates in either the entire intragenic region (i.e., exons and introns, left panel) or introns only (right panel). This suggests that transcription-coupled DNA-repair mechanisms may play a role. The boxes in the box-and-whisker plots represent the interquartile range between the first and third quartiles; the dashed lines (whiskers) extend to the most extreme data points which are no more than 1.5 times the interquartile range from the box.
We examined the spectra of filtered variations from tumors and cell lines, and compared them with those of germline variations in the normal genomes. In our sequenced lung sample panel, the previously published smoker genome (GS00018) has the largest fraction of C:G > A:T transversions, the tobacco-related DNA-damage signature (Fig. 1A, top panel). In contrast, the normal genomes and our two never-smoker tumor genomes (GS000000553 and GS000000552) are among those with the lowest proportion, suggesting that never-smoker related lung cancer is distinct from the smoking-related disease. Consistent with this, most cell lines derived from known smokers (Supplemental Table 1) had significantly higher C:G > A:T fraction than somatic mutations from the never-smoker-derived tumor and cell line genomes (P = 4 × 10−5, Student's t-test) (Fig. 1A). Interestingly, the fraction of C:G > A:T transversions is negatively correlated with that of C:G > T:A transitions (Supplemental Fig. 2). In contrast, prior to filtering, all called variants have base change patterns similar to germline variants (data not shown). This suggests that our variation filtering strategy largely preserves the mutation spectra of cell lines. The consistency between the mutation spectra and the smoking history in our panel of samples also indicates that the mutation data can be used to annotate the smoking history of unannotated samples, or even to resolve conflicting annotations.
The analysis of mutation rates of different genomic features revealed that the CDS and UTR regions have the lowest mutation rates, suggesting strong selective pressure (Fig. 1B). Interestingly, when genes are grouped by their expression values derived from our transcriptome sequencing data, we found that the mutation rate was inversely correlated with expression status (Fig. 1C), consistent with the previous observations related to the transcription-coupled DNA-repair mechanism (Lee et al. 2010; Pleasance et al. 2010a,b). The reduced mutation rate in highly expressed genes cannot be solely explained by selective constraints in the coding region, since intronic regions display the same trend as the entire gene regions (Fig. 1C).
Based on mutation effect predictors including SIFT (Kumar et al. 2009), PolyPhen (Ramensky et al. 2002), and mCluster (Yue et al. 2010), we found that a significantly higher proportion of somatic nonsynonymous mutations in the tumors are predicted to be deleterious compared with germline variations (Supplemental Fig. 3; Supplemental Table 7). Overall, 41% of the somatic mutations are predicted to have an impact on protein function, based on at least two of three prediction methods, in contrast to only ∼12% in the germline set (P = 4.4 × 10−61, χ2 test).
We also investigated the RNA and DNA sequence differences (RDDs) in our genomes. We started with exonic positions that have high-confidence nonreference calls in the RNA-seq data, but homozygous calls in the genomic sequence from the same sample. Among such 12,476 positions in our 25 genomes, we identified sequence differences between RNA and DNA in 624 positions (Supplemental Table 8). The most prevalent RDDs in our data set are A > G changes, suggesting that the observed differences are enriched for RNA-editing events by ADARs (adenosine deaminases that act on RNA) that deaminate adenosine to inosine. We observed a highly recurrent A > G RDD at position chr1:225974614, appearing in 14 of our samples, including normal tissues, tumor tissues, and cell lines. This potential editing event results in an amino acid substitution (I64M) in SRP9, the same gene that was reported to undergo RNA-editing in breast cancer (Shah et al. 2009). It is also interesting to note that, in all three tissue samples, the tumor genomes have a higher percentage of RNA–DNA differences than their matched normal genomes, and two of three tumors have a notably higher ADAR expression level than matched normal; nevertheless, the differences did not reach statistical significance.
Analysis of DNA copy number for our 19 lung cancer cell lines revealed significantly recurrent gain and loss at regions typical for NSCLC (Weir et al. 2007), including gain at CCND1, CCNE1, EGFR, MYC, and chromosome 1q and loss at CDKN2B and PTPRD (Supplemental Fig. 4). The most significant region of gain at 1q was focused precisely on the RNA-editing gene ADAR. This gene has been implicated in altered patterns of RNA editing in cancer, resulting in activation of proto-oncogenes and inactivation of tumor suppressor genes (Dominissini et al. 2011). ADAR expression and copy number, by RNA-seq and SNP Array, are significantly correlated (Spearman's ρ = 0.65, P = 2.82 × 10−3) (Supplemental Fig. 5). FHIT and ADAM3A were the foci of other significantly recurrent deletions (Supplemental Fig. 4). FHIT is frequently inactivated in many tumor types, and reintroduction of FHIT leads to reduced tumor cell growth in in vitro and animal models (Ishii et al. 2001) and apoptosis in SCLC cells (Zandi et al. 2011). Focal, homozygous deletion of ADAM3A has been reported in pediatric high-grade glioma (Barrow et al. 2011).
Frequently altered genes in lung cancer cell line genomes
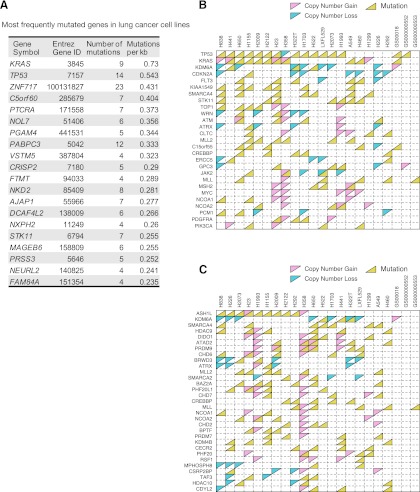
We also found that frequently mutated genes from lung cancer cell lines are similar to what was found in lung tumors (Ding et al. 2008). Such frequently mutated genes were identified by calculating the number of protein-altering single-nucleotide mutations per kilobase of coding sequences. Genes that are not expressed based on transcriptome data are excluded from this list. The KRAS proto-oncogene and the TP53 tumor suppressor exhibited the highest mutation rates (Fig. 2A; Supplemental Table 9). Also among the highly mutated genes is STK11, a gene frequently inactivated in lung cancers (Sanchez-Cespedes et al. 2002). These findings are consistent with previous genome-wide analyses of lung tumor samples (Ding et al. 2008), suggesting that cell lines exhibit mutations in genes similar to those of tumor tissues when extensive germline variant filtering is applied. It is worth noting that the mutation rate of known cancer genes, as defined by the Cancer Gene Census (Futreal et al. 2004), is significantly higher than that of other protein-coding genes in the genome (P = 0.0004, one-sided Wilcoxon rank sum test). We further examined genes that are affected by either protein-altering mutations or substantial copy number alterations. Such integrated analysis reveals 27 known cancer-related genes that are altered in at least five cell lines (Fig. 2B). Interestingly, these commonly altered cancer-related genes include several genes involved in epigenetic regulation, SMARCA4, CREBBP, MLL, and MLL2.
Figure 2.
Frequently altered genes in lung cancer genomes. (A) The top 20 genes with the highest mutation rates in lung cancer cell lines include three frequently mutated genes (KRAS, TP53, and STK11) in lung tumors from previous studies (Ding et al. 2008). (B) Profiles of single-nucleotide variation and copy number alterations in lung cancer cell lines and tumors for a subset of known cancer-related genes. (C) Profiles of single-nucleotide variation and copy number alterations in lung cancer cell lines and tumors for a subset of genes in epigenetic pathways.
To examine if epigenetic regulators as a class are frequently mutated as compared with other classes of genes, we examined the mutation status of all genes known or predicted to be involved in writing, erasing, and reading histone modifications (Chi et al. 2010). Indeed, such genes are mutated at a higher rate than others (P = 0.004, one-sided Wilcoxon rank sum test). Combined with DNA copy number data, we found that 31 epigenetic regulators have either mutations or copy number alterations in at least five cell lines (Fig. 2C). For example, SMARCA4 (BRG1), a gene encoding a SWI/SNF family member that is part of an ATP-dependent chromatin remodeling complex, is mutated in seven distinct cell lines, and two additional cell lines harbor SMARCA4 copy number gains. KDM6A, a gene encoding a JmjC domain-containing protein that catalyzes the demethylation of K27 of histone H3, has a point mutation or copy number loss in nine cell lines. The ASH1L gene is mutated in nine cell lines. ASH1L is a bromodomain-containing protein that also contains a SET domain and is predicted to methylate K4 of histone H3. ATAD2, another bromodomain-containing protein that has been shown to co-activate E2F1, CCND1, and MYC in breast cancers (Kalashnikova et al. 2010; Revenko et al. 2010), has DNA copy number gain or mutations in seven cell lines. It is clear that such epigenetic modifiers are significantly altered in these lung cancer cell lines.
Structural variations and cytogenetic characterization
We screened for SVs and translocations based on discordant paired-end genomic sequencing data. Normal SVs called from a pool of normal samples were used as a filter to remove germline SVs and technical artifacts, resulting in a list of candidate SVs. A subset of these alterations was experimentally validated using PCR and sequencing (Supplemental Tables 10, 11; 57% successfully validated at the base pair level).
Among the 22,030 candidate SVs in the tumors and cell lines, 812 have both ends mapped to intragenic regions of two different genes. We examined our transcriptome sequencing data for reads consistent with the fusion gene pairs in these SVs, and found 62 putative gene fusion events (Supplemental Table 12). We also attempted to obtain a list of candidate chimera transcripts from our transcriptome sequencing data alone using ChimeraScan (Iyer et al. 2011). There are 1097 predicted chimeras in our tumors and cell lines, 291 of which have at least one read spanning the fusion junction. Among these predicted chimeras, 44% have more than one genomic sequencing read supporting the same gene pair (Supplemental Fig. 6), suggesting that they may result from genomic-level SVs, rather than read-through at the transcript level.
Among the SVs supported by both genomic and transcriptomic evidence, we selected four cases involving genes in the cancer-related pathways for further experimental validation. In three out of four cases, we were able to confirm the existence of the chimera transcripts and the exact sequences spanning the breakpoints by RT-PCR (Supplemental Fig. 7) and Sanger sequencing. They are CLTC–VMP1, HIF1A–SNAPC1, and MLLT3–TMIGD1 fusions in two cell lines. The VMP1 (vacuole membrane protein 1) locus is fused to the CLTC (clathrin heavy chain gene) locus in H1229. The fusion is supported by 81 discordant RNA-seq reads, more than 60 of which can be perfectly mapped across the breakpoint (Supplemental Fig. 8A,B). Sequence analysis based on this fusion junction indicated a putative fusion protein product containing the first 1609 amino acids from CLTC and the last 141 amino acids from VMP1 (Supplemental Fig. 8C). CLTC has been previously implicated in oncogenic gene fusions in inflammatory myofibroblastic tumor (Bridge et al. 2001), pediatric renal adenocarcinoma (Argani et al. 2003), and large B-cell lymphoma (De Paepe et al. 2003). Interestingly, the breakpoint within the CLTC protein in the CLTC–VMP1 fusion is very close to what was reported in the CLTC–ALK fusion in the case of inflammatory myofibroblastic tumor (Bridge et al. 2001). The HIF1A (hypoxia inducible factor 1, alpha subunit) locus is juxtaposed to the SNAPC1 (small nuclear RNA activating complex, polypeptide 1) locus through a 50-kb deletion in H1299 (Supplemental Fig. 9A). The resulting potentially in-frame fusion protein product is 284 amino acids in length, missing the critical C-terminal transactivation domain of the wild-type HIF1A protein. The MLLT3–TMIGD1 fusion in H838 resulted from an inter-chromosomal translocation between chromosome 9 and chromosome 17 (Supplemental Fig. 9B). The fusion was predicted to be out-of-frame, therefore no productive fusion protein product was expected; nevertheless, we were able to detect the fusion transcript from both RNA-seq reads and RT-PCR (Supplemental Fig. 7).
For the 19 cell lines, we performed SKY analysis to characterize the cytogenetic abnormalities (Supplemental Fig. 10A,B,C). Although the resolution of SKY is limited and cannot be used for detection of all breakpoints in complex samples, the SKY results allow us to assess the overall ploidy level of the cell lines and to observe large chromosomal abnormalities, along with the rate of occurrence to determine clonality. Ten to 20 cells were examined in detail for each cell line. Within a given cell line, different cells exhibit largely consistent chromosomal patterns, but minor deviations from the main pattern were frequently observed, suggesting the heterogeneous nature of the cell lines. In one cell line (NCI-H226, Supplemental Fig. 10A), ∼50% of cells have diploid genome (2n) while the remaining cells are tetraploid (4n), but the overall translocation pattern clearly suggests that they have the same origin. Based on the SKY results, copy number increases in the form of an increase in the number of chromosomes are apparent. For example, in the 4n cell line H1993 there were seven to eight distinct copies of chromosome 8q, which contains the MYC gene (Supplemental Fig. 10C). This demonstrates that the MYC amplification was primarily caused by the presence of multiple chromosomes rather than by tandem duplication of the genomic region. Such spectral karyotyping data also revealed a large number of translocations for each cell line. Although detailed comparison with WGS-based translocation data is not feasible due to significant differences in resolution, we were able to identify cases where SKY-supported translocations are clearly missed by whole-genome sequencing. Surprisingly, sequencing data did not support about half of the cytogenetic abnormalities reported by SKY results. Such discrepancies likely result from the fact that many translocations occur at low complexity genomic regions, and therefore cannot be detected by our WGS approach that uses only uniquely mapped sequence reads.
Splice site mutations in tumors and cell lines
The combined whole-genome and RNA-seq data provide opportunities for global analysis of mutations potentially affecting the mRNA splicing process. Among the 133,738 somatic mutations in the three tumor genomes and 2,953,975 filtered variations in the cell lines, 438 were found to affect essential splice acceptor and donor sites (i.e., the first two or the last two base pairs of introns) in 433 genes. To evaluate the potential functional significance of these splice site mutations, we examined transcriptome sequencing data in these samples for splicing events (exon skipping or inclusion, aberrant 5′ or 3′ splice sites) that are inconsistent with current exon models from all RefSeq and Ensembl transcripts. By associating splice site mutations with aberrant splice junctions observed in the same sample, we identified 101 genes with potential mutation-associated aberrant splicing (Supplemental Table 13). This constitutes about one-third of the 321 genes containing splice site mutations that were expressed. We did not detect any recurrent splice site mutations among the examined samples. Among the mutations with no detectable aberrant splicing, 106 do not have any RNA-seq reads covering the adjacent exons, and 30 additional mutations have fewer than three reads covering the adjacent exons. Therefore, of mutated genes that have sufficient read coverage, the majority exhibit evidence for altered splicing patterns.
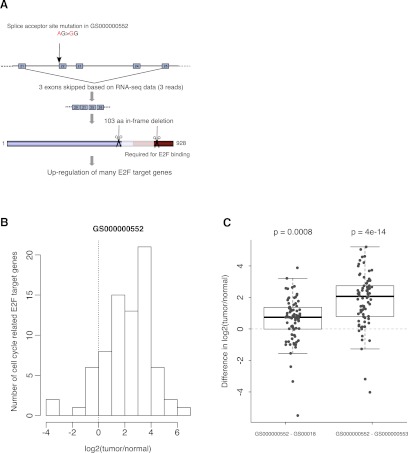
Of the 101 genes with splice site mutations and evidence of aberrant splicing, several are known cancer-related genes including RB1, EP300, ABL1, and AKT3 (Supplemental Table 14). In tumor genome GS000000552, we identified a novel mutation in the tumor suppressor gene RB1 affecting the AG splice acceptor sequence of exon 22 (Fig. 3A). From the RNA-seq data, we observed three reads spanning a novel exon–exon junction connecting exons 21 and 25, skipping the three exons in between. The resulting protein product has a 103-amino-acid in-frame deletion close to the C terminus of RB1, which is essential for the binding of RB1 to the E2F–DP transcription factor complexes (Rubin et al. 2005). Therefore, this mutation likely leads to a RB1 protein product that is unable to bind E2F to block the G1-S cell cycle transition. Consistent with this finding, many E2F target genes involved in the G1-S transition are up-regulated in this tumor, compared with both the adjacent normal tissue and the other two tumors (Fig. 3B,C). Another example is a mutation in a splice donor site of a serine/threonine–protein kinase AKT3. Four reads support the expression of a novel isoform of AKT3 that skips exons 6 and 7, which would lead to an in-frame deletion of 45 residues, thereby disrupting the essential protein kinase domain (Supplemental Fig. 11).
Figure 3.
Splice mutation in RB1 and the associated aberrant splicing event. (A) Splice site mutation is associated with an exon skipping event in RB1 in the tumor genome GS000000552. A novel mutation in the tumor suppressor gene RB1 alters the AG essential splice acceptor sequence just before exon 22. There are three RNA-seq reads spanning a novel exon–exon junction between exons 21 and 25, skipping the three exons in between. The resulting protein product has a 103-amino-acid in-frame deletion close to the C terminus of RB1, which is essential for the binding of RB1 to the E2F–DP transcription factor complexes. (B) Most cell cycle-related E2F target genes (Bracken et al. 2004) are up-regulated in sample GS000000552. For each gene, we obtained the expression values in matched normal and tumor samples from the patient, and calculated the log2 fold-change between tumor and normal. (C) Among the three tumor samples, E2F target genes are up-regulated the most in sample GS000000552. Each dot represents one of the known cell cycle-related E2F target genes (Bracken et al. 2004). P-values shown on the plot are derived from paired t-tests between GS000000552 and one of the other tumors. The result is consistent with the hypothesis that the truncated RB1 in this sample resulting from aberrant splicing is unable to bind to E2F and suppress the expression of its target genes. The boxes in the box-and-whisker plots represent the interquartile range between the first and third quartiles; the dashed lines (whiskers) extend to the most extreme data points which are no more than 1.5 times the interquartile range from the box.
Differential isoform usage revealed by RNA-seq
The comprehensive transcriptome sequence data also allowed us to directly identify differential isoform usages related to cancer. We compared the tumor transcriptomes with their corresponding normal transcriptomes using Cufflinks (Trapnell et al. 2010) and complemented the analysis by checking exon-level expression values and by manual inspection. We identified four genes, ENAH, OSBPL8, PSD3, and RAC1, with splicing products that were expressed at significantly higher levels in all three tumors compared with the corresponding normal samples (Supplemental Fig. 12). Similar analysis was performed for the cell lines using pooled transcriptome data from the three normal lung tissue samples as baseline, resulting in 153 genes with splicing isoforms differentially expressed between the cancer cell lines and normal samples across at least four different cell lines. We investigated whether these cancer-related alternative splicing events are related to any splice site mutations. Among these events, we found only ∼5% of them have splice site variations (including germline variations and variations located at nonessential splice sites). Very similar percentages were found in genes without cancer-related alternative splicing events. Therefore, we found no evidence in our data suggesting the systematic role of cis-acting mutations in cancer-related alternative splicing.
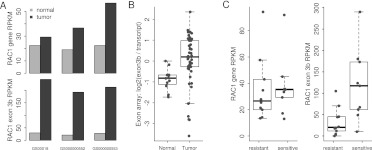
Overall, 13 out of these 153 genes were involved in cancer-related pathways, including RAC1, KRAS, CHEK2, and FBXW11 (Supplemental Table 15). RAC1 (ras-related C3 botulinum toxin substrate 1), which is differentially spliced in both tumors and cell lines (Supplemental Fig. 12; Supplemental Table 15), encodes a rho family small GTPase that is involved in regulating several cancer-related pathways including the PI3K/AKT pathway, the mitogen-activated protein kinase (MAPK) cascades, and the JUN NH2-terminal kinase (JNK) pathway (Bosco et al. 2009). RAC1b, a splice variant of RAC1 that predominantly exists in the GTP-bound active form, has been previously shown to be highly expressed in breast and colon cancers (Jordan et al. 1999; Schnelzer et al. 2000). More recently, two papers reported that RAC1b promotes K-ras-induced lung tumorigenesis, and that RAC1b stimulates epithelial–mesenchymal transition and spontaneous tumor formation (Stallings-Mann et al. 2012; Zhou et al. 2012). We observed near twofold up-regulation of total RAC1 expression levels in all three lung tumors. In contrast, the expression level of exon 3b, which is unique to the RAC1b isoform, is about 10 times higher in the tumors than in the corresponding normal samples, suggesting preferential up-regulation of the RAC1b isoform in the tumors (Fig. 4A). To investigate the prevalence of RAC1b up-regulation in tumors, we examined an Affymetrix exon array data set (GEO accession: GSE16534) (Lin et al. 2009) containing 12 normal lung tissues and 49 lung tumor samples. Tumor samples expressed significantly higher levels of exon 3b, even after subtracting the gene-level difference (Fig. 4B, P = 0.00026, Student's t-test). We did not detect any mutations in RAC1 in our tumors or cell lines, suggesting that expression of RAC1b in these samples may be regulated by trans-acting splice factors, similar to what was reported previously in a colorectal cancer cell line (Gonçalves et al. 2009).
Figure 4.
Alternative splicing of RAC1 in lung cancer. (A) The RAC1b isoform, which includes exon 3b, is preferentially up-regulated in all three lung tumors in our study. (B) Up-regulation of the RAC1b isoform is confirmed by exon array data from an independent data set (GSE16534). Each dot represents a tissue sample. To account for differences in the expression of total RAC1, we calculated the difference between the exon 3b expression and total RAC1 expression, and compared the differences between normal and tumor samples. The analysis showed that RAC1b is significantly up-regulated in tumors (P = 0.00026, Student's t-test). (C) Cell lines sensitive to PD-0325901 have significantly higher expression of the RAC1b isoform (P = 0.019, Student's t-test), while no difference in RAC1 total expression was observed between resistant and sensitive cell lines. Each dot represents one cell line. The boxes in the box-and-whisker plots represent the interquartile range between the first and third quartiles; the dashed lines (whiskers) extend to the most extreme data points which are no more than 1.5 times the interquartile range from the box.
Differential isoform usage as potential biomarkers
Cancer cell lines have been extensively used as in vitro systems and in preclinical animal models to study disease progression and drug response. We hypothesize that expression of the RAC1b isoform, given its potential to influence the RAS–MEK–ERK pathway activity, may influence the response of cell lines to drugs that target this pathway. To this end, we tested how this panel of cell lines responds to PD-0325901, a small molecule inhibitor of mitogen-activated protein kinase kinase (MAP2K or MEK, including MAP2K1 and MAP2K2). Interestingly, cell lines sensitive to PD-0325901 have significantly higher expression of the RAC1b isoform (P = 0.019, Student's t-test), while no difference in RAC1 total expression was observed between resistant and sensitive cell lines (Fig. 4C). This suggests that expression of the specific RAC1b isoform can potentially serve as a predictive biomarker for response to PD-0325901.
Discussion
Although cell lines are widely used in cancer biology study and drug screening, it is still controversial whether they provide a suitable system for studying cancer genomics (Borrell 2010). There are two major challenges associated with cell line sequencing. One is the concern over spontaneous accumulation of mutations during the cell culturing and passaging process; and the second is the typical lack of matched normal samples. Both factors lead to difficulties in data interpretation. To our best knowledge, whole-genome sequencing has been reported on only three cancer cell lines, NCI-H209 (Pleasance et al. 2010b), COLO-829 (Pleasance et al. 2010a), and U87MG (Clark et al. 2010), although exome sequencing is more frequently applied (Chang et al. 2011; Wang et al. 2012). While H209 and COLO-829 have matched normal cell lines, the lack of a noncancer counterpart for U87MG limited the biological interpretation of such a large collection of variations throughout the cancer genome. Regardless, the U87MG study showed that cell lines can be genetically stable since genotype results across different groups were remarkably consistent (Clark et al. 2010). The key question with cell lines is how to extract meaningful mutation information despite the lack of matched normal samples.
Our study suggests that, in absence of matched normal samples, it is reasonable to use a large pool of germline variants as a filter to obtain a list of likely somatic mutations. With the ever-increasing collection of germline variants in the public domain, this strategy is gaining traction for cell line genome studies. To estimate the effectiveness of the filtering strategy, we took our three normal lung tissue genomes and went through the same filtering steps that remove known SNPs. We found that the three normal genomes have an average of 73,700 private SNPs, or ∼2.5% of the entire SNP collection per genome. Restricting to the protein altering private SNPs, we observed an average of 315 per normal genome. These private SNPs form the basis for false positive mutation calls. In contrast, most of our cell line samples harbor a much higher number of variants not already represented by known SNP collections (Supplemental Fig. 13; Supplemental Table 5). These variants are a combination of private SNPs and true mutations. Assuming the number of private SNPs after our filtering procedure is relatively constant, our false positive rate of “somatic” mutation calls vary substantially, depending on the total number of filtered SNVs in the genomes. Subtracting the expected number of private SNPs from all cell lines, we estimate that ∼55% of reported variants are true somatic mutations. By that count, the fraction of somatic mutations in our filtered set is enriched by more than 50-fold compared with the unfiltered set. We anticipate that the percentage of true somatic mutations will gradually increase as we include more comprehensive germline variants in our filtering step.
We also demonstrated the effectiveness of such a germline filtering process in two other aspects: the overall mutation pattern and the most frequently mutated genes. Our previous study showed that compositions of germline and somatic variants are different in a smoker lung cancer genome, where C:G > A:T transversions are enriched in the somatic mutation group (Lee et al. 2010). For the cell lines we analyzed in this study, while called variants prior to filtering resemble the germline SNP collection (data not shown), the candidate mutations after filtering show an obvious presence of the C:G > A:T smoker signature in those smoker samples (Fig. 1A). More importantly, the most frequently mutated genes in these cell lines (such as KRAS, TP53, STK11) are consistent with previously reported frequent somatic mutations from a group of lung tumors (Ding et al. 2008), even though the precise list of commonly mutated genes is dependent on the specific lung tumor cohort. Therefore, the vast majority of germline variants can be filtered out using the current data sets and strategies, revealing patterns of somatic mutations in these cell lines. The similarities in the mutation spectra and frequently mutated genes between cell lines and tumors are also quite remarkable considering that many of these cell lines were derived ∼30 yr ago, so they have stably maintained their presumed genetic alterations in cultured life.
Our integrated mutation and DNA copy number analysis reveals that several epigenetic regulators are frequently mutated in these lung cancer cell lines (Fig. 2C). In recent years, advances in both cancer genomics and epigenomics have led to discoveries that many epigenetic regulators are frequently mutated in various cancer types, and many of those mutations have been shown to be driver mutations and thus could serve as a new class of anti-cancer targets (Esteller 2007; Jones and Baylin 2007; Baylin and Jones 2011; Rodriguez-Paredes and Esteller 2011; Wang et al. 2011). Our study shows that, as in other cancer types, epigenetic regulators tend to be mutated, suggesting epigenetic distortion in these lung cancer cell lines. In fact, all of the cell lines we have studied so far have at least one mutation or copy number alteration in ASH1L, KDM6A, SMARCA4, or ATAD2 (Fig. 2C). The combined mutation and copy number loss profile of KDM6A is reminiscent of the status of tumor suppressors like TP53 or CDKN2A in our study. In contrast, the mutation and copy number gain of ATAD2 is similar to the profiles of oncogenes like KRAS and PIK3CA. The prevalence of alterations in epigenetic regulators in lung tumors still needs to be established in a bigger data set, since the three tumors in our data (two were derived from never-smokers with a very small number of mutations) do not appear to harbor many of these alterations. In addition, the definitive functional roles of these epigenetic regulators in lung cancer still require further biochemical validation, but our study provides the needed genomic evidence and choices of cell lines with various genetic backgrounds for further experimental work.
The combined genome and transcriptome studies also provide ample opportunities to study the functional effect of mutations on different aspects of transcription: expression level, splicing, and allele-specific expression. In this study, we focused on the relationship between splice site mutations and splicing differences by examining two complementary angles. One is to examine the frequency of observing aberrant splicing events in genes with splicing site mutations; the other is to determine the contribution of cis-acting mutations to differential isoform usage. On the first aspect, our data showed that for genes with an essential splicing site mutation, if adequate RNA-seq reads are available, the majority of these genes show aberrant splicing around the mutation. We have identified relevant splice site mutations in cancer, including a novel mutation in the RB1 gene (Fig. 3A), which results in a truncated protein predicted to deregulate E2F targets (Fig. 3B). On the second aspect, for all differential splicing isoform usage cases detected by RNA-seq analysis, only ∼5% have splice site variations. Since a majority of alternative splicing in this study cannot be explained by cis-acting variants within a gene, we argue that alternative splicing primarily results from other independent factors. The finding of the RAC1b splicing isoform, for example, is independent of the mutation data. Regardless of mutation-dependent aberrant splicing or mutation-independent alternative splicing, it is important to characterize the expression of particular splicing isoforms like RAC1b, whose expression status is associated with cell line response to MEK inhibition (Fig. 4C). This is of particular interest in biomarker discovery since the abundant genome and transcriptome data significantly broaden the search space for these biomarkers.
In summary, we used multiple genomic, transcriptomic, and cytogenetic technologies to characterize more than twenty lung cancer samples (Supplemental Fig. 14). Our comprehensive and integrative analysis showed that cancer cell lines can be useful models for finding mutations of interest, uncovering functional splice site mutations, and exploring events like splicing isoforms as unconventional predictive biomarkers. Such data on a much wider collection of cancer cell lines should prove to be extremely valuable resources for cancer biology study and therapeutic drug development.
Methods
Sample descriptions and preparation
Frozen tissues samples were obtained from Indivumed. Four-micron thick frozen sections were obtained from both primary lung adenocarcinoma and the matched normal lung tissues. Sections were H&E stained and examined by a pathologist to verify diagnosis and evaluate tumor content. Both tumor samples had a tumor percentage >80%. The DNA and RNA were extracted from frozen tissues and cell lines by a standard protocol using DNA/RNA extraction kit (Qiagen).
Since reports on smoking history of the patients from which the cell lines were derived can be inconsistent (such as those from the ATCC database), we obtained such information from the original report describing these cell lines (Phelps et al. 1996). In the case of H460, we based the information on personal communications with the doctor who treated the original donor patient.
Whole genome sequencing
Whole genome DNA sequencing (DNA-seq) was performed by “unchained combinatorial probe anchor ligation sequencing,” as described previously (Drmanac et al. 2009). The resulting mate-paired reads with an expected intervening distance (∼400 bp) were mapped to the human reference genome (NCBI Build 37). First, both paired-end reads were aligned to the reference genome, resulting in an average of 185 billion base pairs of mapped sequences per sample. The average coverage was >60× (Supplemental Table 3). For locations with any evidence of differences from the reference genome, mapped reads were assembled into the best-fit diploid genome. This process results in single nucleotide variation, insertion, and deletion calls with associated variant quality scores (Drmanac et al. 2009). Overall, 92%–96% of the human reference genome was fully called.
Mutation detection, filtering, and validation
Variations were called, with respect to the reference genome, for each of the sequenced genomes as described previously (Drmanac et al. 2009). Loci that were called as variant in the tumor and reference in the normal genome were considered as somatic mutations. Somatic scores were assigned to the mutation calls using calldiff-1.3, where a higher score indicates a lower likelihood that the called variation in the tumor genome is false positive and the reference call in the normal genome is false negative. Mutations that were present in dbSNP (v131) were filtered out to obtain novel mutation calls. We further filtered out any variations that were found in 1000 Genomes (Nov 2010 release), variations present in 69 complete human genomes release by Complete Genomics Inc. (http://www.completegenomics.com/sequence-data/download-data/), and SNPs present in the ESP2500 release of the NHLBI Exome Sequencing Project (http://www.ncbi.nlm.nih.gov/projects/SNP/snp_viewBatch.cgi?sbid=1055207). Mutations that were also present in COSMIC v55 (http://www.sanger.ac.uk/genetics/CGP/cosmic/) were retained, even if also present in any of the previously mentioned filtering sets.
Mutations were annotated for their effect on transcripts using the variant effect predictor tool (McLaren et al. 2010). The different types of consequences predicted are intergenic, regulatory region, upstream (within 5 kb), 5′ UTR, complex indel (spans intron/exon border), splice site (1–3 bp into exon, 3–8 bp into intron), synonymous coding, nonsynonymous coding, intronic, frame-shift coding, stop gained, stop lost, 3′ UTR and downstream (within 5 kb).
We experimentally tested a selective subset of somatic single base substitutions for tumor-normal comparisons from all three patients using Sequenom (223 mutations from patient 21214, 187 from 34560, and 995 mutations from patient 39876). For the purpose of validation, we filtered out mutations that were annotated as intergenic, intronic, or downstream. We also filtered out mutations in pseudogenes or hypothetical genes, based on their description in the Entrez Gene database. The score performed well in all the patients (Supplemental Fig. 1; AUC for never-smokers were 0.92 each and 0.79 for the smoker). The optimal somatic score threshold was determined this way: A set of scores was chosen such that the positive predictive value (PPV) was ≥80%. Among these scores, the score with the highest true positive rate (TPR) was chosen, and its PPV and TPR were reported. For the two never-smokers, the data were pooled and an optimal score threshold of 0.064 was determined, at a PPV of 82.3% and a TPR of 68.9%. For the smoker, a score threshold of 0.034 was used, which gave a PPV as well as a TPR value of 92%.
A selected list of mutations is shown in Supplemental Table 4 satisfying all of these criteria, that the mutation is:
novel
high-confidence (somatic score is greater than determined threshold and mutation was not invalidated experimentally)
transcript-associated (i.e., it does not have the consequences “intergenic”, “intronic”, “downstream”, “upstream”, or “within_non_coding_gene”).
We also attempted to validate our lists of filtered variations using our transcriptome sequence data. The number of SNVs that can be validated is limited by the RNA-seq coverage at the SNV positions, which vary greatly among the genomes (Supplemental Fig. 15, top panel). Nevertheless, among SNV positions with sufficient RNA-seq coverage (10 or more reads), ∼75% of SNVs are supported by the presence of variant RNA-seq reads (Supplemental Fig. 15, bottom panel; Supplemental Table 6). To estimate the false negative rate of our SNV calling, we compared our results with recently published targeted-sequencing data from the CCLE (Barretina et al. 2012). Fourteen cell lines in our collection were also analyzed in the CCLE project. On average 94% of all the mutations published by CCLE were also called by our Complete Genomics data, representing a possible 6% false negative rate (Supplemental Fig. 16).
Predicting the effects of amino acid substitutions on protein function
SIFT 4.04 (Kumar et al. 2009), PolyPhen-2 (Ramensky et al. 2002; Adzhubei et al. 2010), and mCluster (Yue et al. 2010) were applied to predict the effects of nonsynonymous mutations on protein function. To compare the proportions of deleterious mutations in both the somatic and the germline mutation set, we applied SIFT and PolyPhen using default parameters. Mutations, which were predicted to be “deleterious” by SIFT or “probably damaging” by PolyPhen, were defined as deleterious in our analysis. For the derivation of mutations that were predicted to affect protein function based on at least two methods, mutations classified as “possibly damaging” by PolyPhen were also defined to have an impact on function. Nonsynonymous mutations that could not be scored (e.g., because of the lack of homologous proteins) were excluded from our analysis.
Transcriptome sequencing
Total RNA was subject to enrichment using the Ribo-minus Eukaryote kit (Invitrogen), and the resulting RNA fraction was used to construct complementary DNA libraries. Transcriptome sequencing (RNA-seq) was performed on the Illumina GAIIx Platform using the standard paired-end protocol. The RNA-seq reads were first aligned to ribosomal RNA sequences to remove potential ribosomal reads. The remaining reads were aligned to the human reference genome (NCBI Build 37) using GSNAP (Wu and Nacu 2010), allowing a maximum of five mismatches per 75-bp sequencing end. To quantify the gene expression level, the number of reads mapped to the exons of each RefSeq gene was calculated, and the corresponding RPKM value (reads mapping to the genome per kilobase of transcript per million reads sequenced) (Mortazavi et al. 2008) was also derived.
Structural variations (SV) detection and validation
All uniquely mapped reads from whole-genome sequencing were used for the estimation of normal pair-span. The set of mate pairs was further refined by aligning each read within a normal range of pair-span with penalties for mismatches and indels. Each read with fewer than four penalty units was retained. The orientation of the reads that were mapped to the forward strand of the reference genome was designated as plus (“+”); otherwise it was assigned minus (“−”). The discordant mate pairs were defined as either (1) mate-span beyond normal range (500 bp) or (2) with discordant orientation. Adjacent discordant reads (within 500 bp) with the same orientation were then merged to make a discordant reads cluster. The clusters that contained too few discordant mate pairs (<3) after merging were discarded.
We then applied a filtering process to define high confidence somatic SVs as (1) SVs supported by a sufficient number of discordant mate pairs (pair_count ≥ 10), and (2) a discordant span (≥5 kb) for an intrachromosomal deletion call. We also excluded those SVs that were also present in matched non-neoplastic samples or other unrelated normal samples that we sequenced. A subset of putative somatic SVs that overlapped with RefSeq genes were subjected to a further experimental validation by PCR. PCR primers were designed to flank putative SV breakpoints. PCR amplification was performed on both tumor and nontumor lung tissues and cell lines. PCR conditions and validation criteria were described previously (Jiang et al. 2012). About 57% (51/89) breakpoints were confirmed at base-pair resolution.
Spectral karyotyping (SKY)
Metaphase slides were prepared from 20 lung cancer cell lines that were cultured, harvested, and fixed with methanol:acetic acid (3:1), according to standard cytogenetic procedures. Seven microliters of denatured SkyPaint probe (Applied Spectral Imaging [ASI]) was added to each denatured metaphase slide, which then was covered by a glass coverslip and incubated overnight in a 37°C humidified chamber. Slide pretreatment and post-hybridization washes were performed according to the standard supplied protocol (ASI) with slight modifications. Image acquisition was performed with a COOL-1300 SpectraCube camera (ASI) mounted on an Olympus BX43 microscope using a SKY optical filter (ASI). For each sample, a minimum of 10–15 metaphases were analyzed using the HiSKY v6.0 software (ASI).
Copy number variations (CNV) and loss of heterozygosity (LOH) detection from sequencing data
DNA copy number variation (CNV) and allele-imbalance (AIB/LOH) in tumor samples was defined by read depth coverage and B-allele frequency analysis. For each sample, DNA sequencing reads were binned at 50-Kbp intervals along the genome and counted. The ratio of counts per bin in the tumor and its matched normal sample, log2 transformed, was calculated as the raw measure of copy number (log2[tumor/normal]). This value was corrected for GC content bias using GC content information from UCSC (http://hgdownload.cse.ucsc.edu/goldenPath/hg19/gc5Base/hg19.gc5Base.txt.gz). These GC content data were averaged over 500-bp windows and then smoothed over a 1-Mb window using a running mean. The residuals of the regression of log2 ratio on GC content were taken as the measure of copy number. These log2 ratios were then shifted to have a mode of 0. These values were segmented into discrete blocks of uniform copy number using the CBS algorithm from the Bioconductor package DNAcopy (Venkatraman and Olshen 2007). The parameters for CBS were smooth.region = 2, outlier.SD.scale = 4, smooth.SD.scale = 2, and trim = 0.025. Segments with a log2 ratio less than or equal to −0.15 were considered as regions of copy loss whereas segments with a log2 ratio ≥0.15 were defined as copy gain regions.
Genome-wide allelic imbalance (AI) was assessed using the counts of A, C, G, and T nucleotides in the tumor at positions called heterozygous in the matched normal sample. The most common nucleotide was called “Allele B” and the sum of the counts for the other three nucleotides was taken as the frequency of the “Allele A.” The raw B-Allele Frequency (BAF) was calculated as BAF = 2/pi * atan(B counts/A counts) (Peiffer et al. 2006). BAF was converted to modified BAF (mBAF) by reflecting it around the value 0.5 (Diskin et al. 2008). mBAF values were averaged in the same 50-kb bins used for the copy number above. These binned mBAF values were segmented using CBS and the same parameters used for copy number (Diskin et al. 2008). Segments with a mBAF value ≥0.75 were considered as allelic imbalance. Segments with AI and without copy gain were said to have loss of heterozygosity (LOH).
Copy number variations (CNV) analysis from SNP array
Illumina HumanOmni2.5_4v1 arrays were used to assay 19 lung cancer cell lines for genotype, DNA copy number, and LOH. A subset of 2,295,239 high-quality SNPs was selected for all analyses. These SNPs were concordant in >17 cell lines assayed by both Illumina HumanOmni2.5_4v1 array and Complete Genomics full-genome sequencing.
We applied a modified version of the PICNIC (Greenman et al. 2010) algorithm to estimate total copy number and allele-specific copy number/LOH. PICNIC was modified to work with Illumina arrays as described previously (Seshagiri et al. 2012).
Genomic regions with recurrent DNA copy gain and loss were identified using GISTIC (Mermel et al. 2011), version 2.0. Segmented integer total copy number values obtained from PICNIC, c, were converted to log2 ratio values, y, as y = log2(c + 0.1) – 1. Cutoffs of ±0.2 were used to categorize log2 ratio values as gain or loss, respectively. A minimum segment length of 20 SNPs and a log2 ratio “cap” value of 3 were used.
Association between essential splice site variations and aberrant splicing junctions
Variations at essential splice sites (the first and last two base pairs of introns in RefSeq transcripts) were extracted from the list of all filtered mutations. Variations in genes with low expression level (RPKM < 0.01) were excluded from the analysis. The aberrant splicing junctions were obtained from the spliced reads in transcriptome sequencing data by removing splicing junctions which match any exon junction in RefSeq or Ensembl transcripts (downloaded from http://genome.ucsc.edu/). Aberrant splicing junctions were excluded if they do not completely fall within the gene boundaries. Filtered essential splice site mutations that are located within the ranges of introns spanned by aberrant splicing junctions were reported.
Identification of differential isoform expression between cancer and normal samples
We used a two-step procedure to identify differential isoform expression between cancer and normal samples. In both steps, three tumor transcriptomes were compared with corresponding paired normal genomes, and 19 cancer cell line transcriptomes were compared with data pooled from three normal transcriptomes. In the first step, cancer transcriptomes were compared with normal transcriptomes using the Cufflinks method (Trapnell et al. 2010) to identify genes which are expressed (average transcript FPKM > 0.1, Fragments Per Kilobase of exon per Million fragments mapped) and have significant changes in splicing (P < 0.01) across at least a number of transcriptomes (three for tumor and four for cell lines) with consistent (>70%) top expressed transcripts. In order to pinpoint the specific isoforms that were differentially expressed, the gene list obtained in the first step was further filtered by comparing gene and exon level expression (RPKM) of the cancer transcriptomes to those of normal transcriptomes. Specifically, genes were kept if they are expressed (RPKMgene > 0.01) and have isoform-specific exon(s) which have at least twofold change of relative expression ratio (RPKMexon/RPKMgene) between cancer samples and normal samples across at least a number of cancer transcriptomes (three for tumor and four for cell lines).
Validation of fusion transcripts in cell lines
Cell culture
H1299, H441, and H838 cell lines were obtained from the American Type Culture Collection and were grown at 5% CO2 in RPMI 1640 supplemented with 10% Fetal Bovine Serum and 2 mM Glutimax (Life Technologies, Cat No. 35050061).
Nucleic acid extraction, PCR, and RT-PCR
Cell line DNA was extracted using DNeasy Blood & Tissue Kit (Qiagen, Cat. No. 69506) and RNA was extracted using TRIzol (Life Technologies, Cat No. 15596-026) according to the manufacturer's instructions. cDNA was generated using the ABI High Capacity cDNA Reverse Transcriptase Kit (Applied Biosystems, Cat No. 4368813). Both genomic DNA and cDNA were amplified with Platinum PCR Supermix (Life Technologies, Cat No. 11306-016) at 95°C for 5 min, then 35 cycles of 95°C for 30 sec, 55°C for 30 sec, and 72°C for 1 min, and then a final extension at 72°C for 8 min. PCR products were separated using 2% E-gels (Life Technologies, Cat No. G800802) and were imaged with a FluorChem 8900 gel imager (Alpha Innotech). PCR primers used are as follows: (F, forward primer; R, reverse primer; all sequences are 5′ to 3′) MLLT3_TMIGD1, F: CTGGGGAAATTGTGGATTTG, R: CCTGTACTGTGAGCCATGCT; HIF1A_SNAPC1, F: ATCTCCAAAATGCAGAACCG, R: AAAGCCACACACCTACGACC; CLTC_VMP1, F: GCTTTGGTTGAGAGACCAGC, R: CCTGGGTGACAAGAGGGAG; PCNXL3_RELA, F: CCCCTGTTCCACAGCACTAT, R: GACCCAGAGCTGTCCTTGAG. RT-PCR primers used are as follows (RT1, primer set 1; RT2, primer set 2; F, forward primer; R, reverse primer; all sequences are 5′ to 3′): MLLT3_TMIGD1, RT1F: TGTTTGGAAGTCGTTTCCACT, RT1R: GCTGGTTTCATTTTGCCAAT, RT2F: GCCTGCAGGTAAAGCTGATT, RT2R: CTTCCACTTGCACGAAAGC; HIF1A_SNAPC1, RT1F: CCTTCCTGCTGGTTTCAATC, RT1R: TGCGTGTGAGGAAACTTCTG, RT2F: CCCTTGACCAGATGCAGAAT, RT2R: TGCTCATCAGTTGCCACTTC; CLTC_VMP1, RT1F: CGTTGAGCCTCCAGGTACTC, RT1R: CAACAATCGCTGGAAACAGA, RT2F: CATTTCGCTTTTGTGGTGAA, RT2R: CAGCCTTTACAAGGATGCAA; PCNXL3_RELA, RT1F: GGCGAGAGGAGCACAGATAC, RT1R: TGAAGCCAAACACAGAGTGC, RT2F: TCTGCTTCCAGGTGACAGTG, RT2R: AAGCTCTTGCTCAGCTCCTG.
TA cloning and sequencing
PCR products were cloned using the TOPO TA Cloning Kit for Sequencing (Life Technologies, Cat No. K457540) according to manufacturer's instructions and plated on LB agar plates containing 50 μg/mL Carbenicillin. A minimum of 12 colonies per PCR product was selected and grown overnight at 37°C in LB media containing 50 μg/mL Carbenicillin. Plasmid DNA was extracted with a miniprep kit (QIAGEN, Cat No. 27361) and sequenced using a 3730 × 1 DNA Analyzer (Applied Biosystems). DNA sequence was analyzed using Sequencher v4.10.1.
Data access
The sequencing data used in this study have been submitted to the NCBI database of Genotypes and Phenotypes (dbGaP) (http://www.ncbi.nlm.nih.gov/gap) under accession number phs000299. The SNP array data for lung cancer cell lines have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE40908.
Competing interest statement
All Genentech authors are stockholders of Roche.
Acknowledgments
We thank Jens Reeder, Cory Barr, Melanie Huntley, Meg Green, Michael Lawrence, and Jeremiah Degenhardt for development of the transcriptome sequencing analysis pipeline and assistance in processing the transcriptome data; Richard Bourgon, Kiran Mukhyala, Oleg Mayba, Robert Yauch, David Dornan, Krishna Pant, and Dennis Ballinger for constructive discussions. J.D.M. is supported by NCI SPORE P50CA70907, CPRIT (RP101251, RP110709). A.F.G. is supported by U01 CA086402 from the Early Detection Research Network, NCI and the Canary Foundation.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.140988.112.
Freely available online through the Genome Research Open Access option.
References
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR 2010. A method and server for predicting damaging missense mutations. Nat Methods 7: 248–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argani P, Lui MY, Couturier J, Bouvier R, Fournet JC, Ladanyi M 2003. A novel CLTC-TFE3 gene fusion in pediatric renal adenocarcinoma with t(X;17)(p11.2;q23). Oncogene 22: 5374–5378 [DOI] [PubMed] [Google Scholar]
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, et al. 2012. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483: 603–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrow J, Adamowicz-Brice M, Cartmill M, MacArthur D, Lowe J, Robson K, Brundler MA, Walker DA, Coyle B, Grundy R 2011. Homozygous loss of ADAM3A revealed by genome-wide analysis of pediatric high-grade glioma and diffuse intrinsic pontine gliomas. Neuro-oncol 13: 212–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylin SB, Jones PA 2011. A decade of exploring the cancer epigenome—biological and translational implications. Nat Rev Cancer 11: 726–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrell B 2010. How accurate are cancer cell lines? Nature 463: 858 doi: 10.1038/463858a [DOI] [PubMed] [Google Scholar]
- Bosco EE, Mulloy JC, Zheng Y 2009. Rac1 GTPase: A “Rac” of all trades. Cell Mol Life Sci 66: 370–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken AP, Ciro M, Cocito A, Helin K 2004. E2F target genes: Unraveling the biology. Trends Biochem Sci 29: 409–417 [DOI] [PubMed] [Google Scholar]
- Bridge JA, Kanamori M, Ma Z, Pickering D, Hill DA, Lydiatt W, Lui MY, Colleoni GW, Antonescu CR, Ladanyi M, et al. 2001. Fusion of the ALK gene to the clathrin heavy chain gene, CLTC, in inflammatory myofibroblastic tumor. Am J Pathol 159: 411–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H, Jackson DG, Kayne PS, Ross-Macdonald PB, Ryseck RP, Siemers NO 2011. Exome sequencing reveals comprehensive genomic alterations across eight cancer cell lines. PLoS ONE 6: e21097 doi: 10.1371/journal.pone.0021097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi P, Allis CD, Wang GG 2010. Covalent histone modifications—miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer 10: 457–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MJ, Homer N, O'Connor BD, Chen Z, Eskin A, Lee H, Merriman B, Nelson SF 2010. U87MG decoded: The genomic sequence of a cytogenetically aberrant human cancer cell line. PLoS Genet 6: e1000832 doi: 10.1371/journal.pgen.1000832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Paepe P, Baens M, van Krieken H, Verhasselt B, Stul M, Simons A, Poppe B, Laureys G, Brons P, Vandenberghe P, et al. 2003. ALK activation by the CLTC-ALK fusion is a recurrent event in large B-cell lymphoma. Blood 102: 2638–2641 [DOI] [PubMed] [Google Scholar]
- Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan MB, et al. 2008. Somatic mutations affect key pathways in lung adenocarcinoma. Nature 455: 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diskin SJ, Li M, Hou C, Yang S, Glessner J, Hakonarson H, Bucan M, Maris JM, Wang K 2008. Adjustment of genomic waves in signal intensities from whole-genome SNP genotyping platforms. Nucleic Acids Res 36: e126 doi: 10.1093/nar/gkn556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Amariglio N, Rechavi G 2011. Adenosine-to-inosine RNA editing meets cancer. Carcinogenesis 32: 1569–1577 [DOI] [PubMed] [Google Scholar]
- Drmanac R, Sparks AB, Callow MJ, Halpern AL, Burns NL, Kermani BG, Carnevali P, Nazarenko I, Nilsen GB, Yeung G, et al. 2009. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science 327: 78–81 [DOI] [PubMed] [Google Scholar]
- Esteller M 2007. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet 8: 286–298 [DOI] [PubMed] [Google Scholar]
- Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, Stratton MR 2004. A census of human cancer genes. Nat Rev Cancer 4: 177–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, Greninger P, Thompson IR, Luo X, Soares J, et al. 2012. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature 483: 570–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazdar AF, Girard L, Lockwood WW, Lam WL, Minna JD 2010. Lung cancer cell lines as tools for biomedical discovery and research. J Natl Cancer Inst 102: 1310–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves V, Matos P, Jordan P 2009. Antagonistic SR proteins regulate alternative splicing of tumor-related Rac1b downstream of the PI3-kinase and Wnt pathways. Hum Mol Genet 18: 3696–3707 [DOI] [PubMed] [Google Scholar]
- Greenman CD, Bignell G, Butler A, Edkins S, Hinton J, Beare D, Swamy S, Santarius T, Chen L, Widaa S, et al. 2010. PICNIC: An algorithm to predict absolute allelic copy number variation with microarray cancer data. Biostatistics 11: 164–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbst RS, Heymach JV, Lippman SM 2008. Lung cancer. N Engl J Med 359: 1367–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii H, Dumon KR, Vecchione A, Fong LY, Baffa R, Huebner K, Croce CM 2001. Potential cancer therapy with the fragile histidine triad gene: Review of the preclinical studies. JAMA 286: 2441–2449 [DOI] [PubMed] [Google Scholar]
- Iyer MK, Chinnaiyan AM, Maher CA 2011. ChimeraScan: A tool for identifying chimeric transcription in sequencing data. Bioinformatics 27: 2903–2904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z, Jhunjhunwala S, Liu J, Haverty PM, Kennemer MI, Guan Y, Lee W, Carnevali P, Stinson J, Johnson S, et al. 2012. The effects of hepatitis B virus integration into the genomes of hepatocellular carcinoma patients. Genome Res 22: 593–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, Baylin SB 2007. The epigenomics of cancer. Cell 128: 683–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan P, Brazao R, Boavida MG, Gespach C, Chastre E 1999. Cloning of a novel human Rac1b splice variant with increased expression in colorectal tumors. Oncogene 18: 6835–6839 [DOI] [PubMed] [Google Scholar]
- Kalashnikova EV, Revenko AS, Gemo AT, Andrews NP, Tepper CG, Zou JX, Cardiff RD, Borowsky AD, Chen HW 2010. ANCCA/ATAD2 overexpression identifies breast cancer patients with poor prognosis, acting to drive proliferation and survival of triple-negative cells through control of B-Myb and EZH2. Cancer Res 70: 9402–9412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC 2009. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 4: 1073–1081 [DOI] [PubMed] [Google Scholar]
- Lee W, Jiang Z, Liu J, Haverty PM, Guan Y, Stinson J, Yue P, Zhang Y, Pant KP, Bhatt D, et al. 2010. The mutation spectrum revealed by paired genome sequences from a lung cancer patient. Nature 465: 473–477 [DOI] [PubMed] [Google Scholar]
- Ley TJ, Mardis ER, Ding L, Fulton B, McLellan MD, Chen K, Dooling D, Dunford-Shore BH, McGrath S, Hickenbotham M, et al. 2008. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature 456: 66–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin E, Li L, Guan Y, Soriano R, Rivers CS, Mohan S, Pandita A, Tang J, Modrusan Z 2009. Exon array profiling detects EML4-ALK fusion in breast, colorectal, and non–small cell lung cancers. Mol Cancer Res 7: 1466–1476 [DOI] [PubMed] [Google Scholar]
- Maher CA, Kumar-Sinha C, Cao X, Kalyana-Sundaram S, Han B, Jing X, Sam L, Barrette T, Palanisamy N, Chinnaiyan AM 2009. Transcriptome sequencing to detect gene fusions in cancer. Nature 458: 97–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F 2010. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics 26: 2069–2070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G 2011. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol 12: R41 doi: 10.1186/gb-2011-12-4-r41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, et al. 2006. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 10: 515–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons DW, Li M, Zhang X, Jones S, Leary RJ, Lin JC, Boca SM, Carter H, Samayoa J, Bettegowda C, et al. 2010. The genetic landscape of the childhood cancer medulloblastoma. Science 331: 435–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiffer DA, Le JM, Steemers FJ, Chang W, Jenniges T, Garcia F, Haden K, Li J, Shaw CA, Belmont J, et al. 2006. High-resolution genomic profiling of chromosomal aberrations using Infinium whole-genome genotyping. Genome Res 16: 1136–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelps RM, Johnson BE, Ihde DC, Gazdar AF, Carbone DP, McClintock PR, Linnoila RI, Matthews MJ, Bunn PA Jr, Carney D, et al. 1996. NCI-Navy Medical Oncology Branch cell line data base. J Cell Biochem Suppl 24: 32–91 [DOI] [PubMed] [Google Scholar]
- Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, Varela I, Lin ML, Ordonez GR, Bignell GR, et al. 2010a. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature 463: 191–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasance ED, Stephens PJ, O'Meara S, McBride DJ, Meynert A, Jones D, Lin ML, Beare D, Lau KW, Greenman C, et al. 2010b. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature 463: 184–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramensky V, Bork P, Sunyaev S 2002. Human non-synonymous SNPs: Server and survey. Nucleic Acids Res 30: 3894–3900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revenko AS, Kalashnikova EV, Gemo AT, Zou JX, Chen HW 2010. Chromatin loading of E2F-MLL complex by cancer-associated coregulator ANCCA via reading a specific histone mark. Mol Cell Biol 30: 5260–5272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Paredes M, Esteller M 2011. Cancer epigenetics reaches mainstream oncology. Nat Med 17: 330–339 [DOI] [PubMed] [Google Scholar]
- Rubin SM, Gall AL, Zheng N, Pavletich NP 2005. Structure of the Rb C-terminal domain bound to E2F1-DP1: A mechanism for phosphorylation-induced E2F release. Cell 123: 1093–1106 [DOI] [PubMed] [Google Scholar]
- Salgia R, Hensing T, Campbell N, Salama AK, Maitland M, Hoffman P, Villaflor V, Vokes EE 2011. Personalized treatment of lung cancer. Semin Oncol 38: 274–283 [DOI] [PubMed] [Google Scholar]
- Sanchez-Cespedes M, Parrella P, Esteller M, Nomoto S, Trink B, Engles JM, Westra WH, Herman JG, Sidransky D 2002. Inactivation of LKB1/STK11 is a common event in adenocarcinomas of the lung. Cancer Res 62: 3659–3662 [PubMed] [Google Scholar]
- Schnelzer A, Prechtel D, Knaus U, Dehne K, Gerhard M, Graeff H, Harbeck N, Schmitt M, Lengyel E 2000. Rac1 in human breast cancer: Overexpression, mutation analysis, and characterization of a new isoform, Rac1b. Oncogene 19: 3013–3020 [DOI] [PubMed] [Google Scholar]
- Seshagiri S, Stawiski EW, Durinck S, Modrusan Z, Storm EE, Conboy CB, Chaudhuri S, Guan Y, Janakiraman V, Jaiswal BS, et al. 2012. Recurrent R-spondin fusions in colon cancer. Nature 488: 660–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah SP, Morin RD, Khattra J, Prentice L, Pugh T, Burleigh A, Delaney A, Gelmon K, Guliany R, Senz J, et al. 2009. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature 461: 809–813 [DOI] [PubMed] [Google Scholar]
- Siegel R, Naishadham D, Jemal A 2012. Cancer statistics, 2012. CA Cancer J Clin 62: 10–29 [DOI] [PubMed] [Google Scholar]
- Stallings-Mann ML, Waldmann J, Zhang Y, Miller E, Gauthier ML, Visscher DW, Downey GP, Radisky ES, Fields AP, Radisky DC 2012. Matrix metalloproteinase induction of Rac1b, a key effector of lung cancer progression. Sci Transl Med 4: 142ra95 doi: 10.1126/scitranslmed.3004062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Schiller JH, Gazdar AF 2007. Lung cancer in never smokers—a different disease. Nat Rev Cancer 7: 778–790 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatraman ES, Olshen AB 2007. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics 23: 657–663 [DOI] [PubMed] [Google Scholar]
- Wang K, Kan J, Yuen ST, Shi ST, Chu KM, Law S, Chan TL, Kan Z, Chan AS, Tsui WY, et al. 2011. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nat Genet 43: 1219–1223 [DOI] [PubMed] [Google Scholar]
- Wang L, Tsutsumi S, Kawaguchi T, Nagasaki K, Tatsuno K, Yamamoto S, Sang F, Sonoda K, Sugawara M, Saiura A, et al. 2012. Whole-exome sequencing of human pancreatic cancers and characterization of genomic instability caused by MLH1 haploinsufficiency and complete deficiency. Genome Res 22: 208–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein JN 2012. Drug discovery: Cell lines battle cancer. Nature 483: 544–545 [DOI] [PubMed] [Google Scholar]
- Weir BA, Woo MS, Getz G, Perner S, Ding L, Beroukhim R, Lin WM, Province MA, Kraja A, Johnson LA, et al. 2007. Characterizing the cancer genome in lung adenocarcinoma. Nature 450: 893–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TD, Nacu S 2010. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26: 873–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Northcott PA, Dubuc A, Dupuy AJ, Shih DJ, Witt H, Croul S, Bouffet E, Fults DW, Eberhart CG, et al. 2012. Clonal selection drives genetic divergence of metastatic medulloblastoma. Nature 482: 529–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue P, Forrest WF, Kaminker JS, Lohr S, Zhang Z, Cavet G 2010. Inferring the functional effects of mutation through clusters of mutations in homologous proteins. Hum Mutat 31: 264–271 [DOI] [PubMed] [Google Scholar]
- Zandi R, Xu K, Poulsen HS, Roth JA, Ji L 2011. The effect of adenovirus-mediated gene expression of FHIT in small cell lung cancer cells. Cancer Invest 29: 683–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Caballero OL, Levy S, Stevenson BJ, Iseli C, de Souza SJ, Galante PA, Busam D, Leversha MA, Chadalavada K, et al. 2009. Transcriptome-guided characterization of genomic rearrangements in a breast cancer cell line. Proc Natl Acad Sci 106: 1886–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Licciulli S, Avila JL, Cho M, Troutman S, Jiang P, Kossenkov AV, Showe LC, Liu Q, Vachani A, et al. 2012. The Rac1 splice form Rac1b promotes K-ras-induced lung tumorigenesis. Oncogene. doi: 10.1038/onc.2012.99 [DOI] [PMC free article] [PubMed] [Google Scholar]