Abstract
The order of genes in eukaryotic genomes has generally been assumed to be neutral, since gene order is largely scrambled over evolutionary time. Only a handful of exceptional examples are known, typically involving deeply conserved clusters of tandemly duplicated genes (e.g., Hox genes and histones). Here we report the first systematic survey of microsynteny conservation across metazoans, utilizing 17 genome sequences. We identified nearly 600 pairs of unrelated genes that have remained tightly physically linked in diverse lineages across over 600 million years of evolution. Integrating sequence conservation, gene expression data, gene function, epigenetic marks, and other genomic features, we provide extensive evidence that many conserved ancient linkages involve (1) the coordinated transcription of neighboring genes, or (2) genomic regulatory blocks (GRBs) in which transcriptional enhancers controlling developmental genes are contained within nearby bystander genes. In addition, we generated ChIP-seq data for key histone modifications in zebrafish embryos, which provided further evidence of putative GRBs in embryonic development. Finally, using chromosome conformation capture (3C) assays and stable transgenic experiments, we demonstrate that enhancers within bystander genes drive the expression of genes such as Otx and Islet, critical regulators of central nervous system development across bilaterians. These results suggest that ancient genomic functional associations are far more common than previously thought—involving ∼12% of the ancestral bilaterian genome—and that cis-regulatory constraints are crucial in determining metazoan genome architecture.
The evolutionary and functional implications of high-order eukaryotic genome structures remain topics of contention, and have inspired some of the most ambitious evolutionary hypotheses of the genetic and genomic eras (e.g., Doolittle 1978; Lynch 2007; Koonin and Wolf 2010). Largely absent from these debates has been the question of the local ordering of genes themselves within a genome, generally reflecting the common assumption that gene position within the genome is mostly a secondary concern and/or is usually not constrained (Koonin and Wolf 2010).
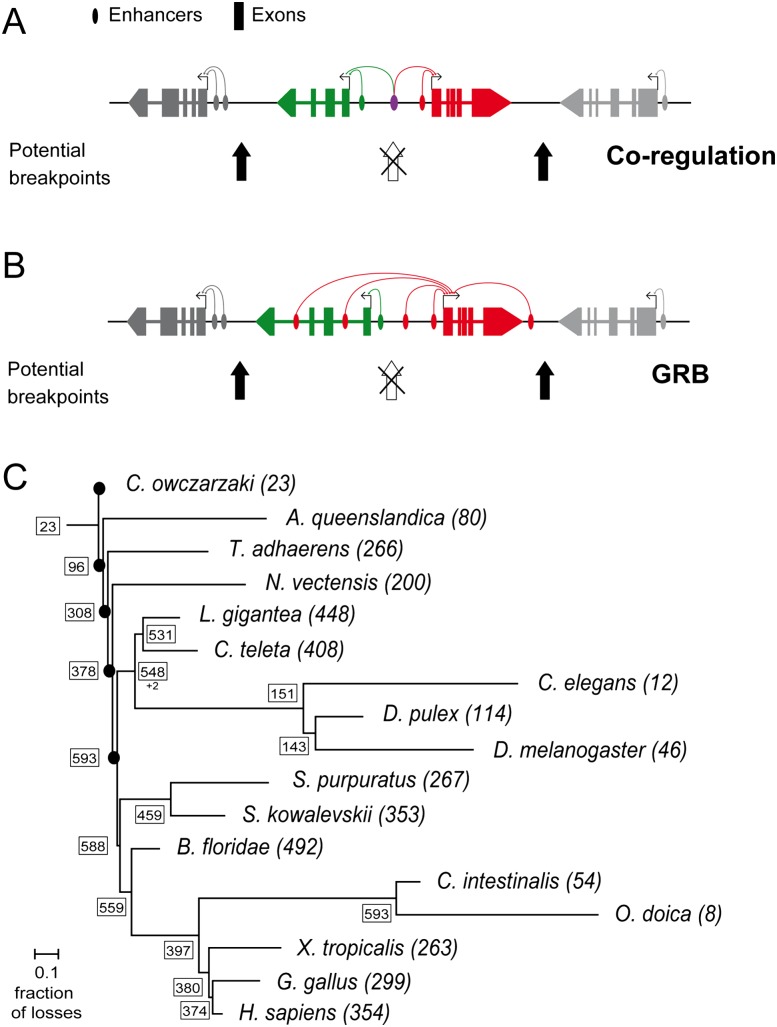
However, a variety of studies suggest that the situation is not so simple. Gene order is nonrandom (Hurst et al. 2004; Oliver and Misteli 2005; Michalak 2008), with clustering of genes in some species according to metabolic pathways and/or similarity of expression (Lee and Sonnhammer 2003; Fukuoka et al. 2004; Hurst et al. 2004). Particularly interesting are the genomic regulatory blocks (GRBs) (Becker and Lenhard 2007; Engstrom et al. 2007; Kikuta et al. 2007), which usually consist of (1) a trans-dev gene encoding a key transcriptional regulator with a complex spatiotemporal expression pattern, often involved in embryonic development (Woolfe et al. 2005; Kikuta et al. 2007); and (2) nearby functionally unrelated bystander gene(s), which contain cis-regulatory sequences for the trans-dev gene within their introns (Fig. 1B). Gene linkage is thus important in GRBs since breakage of the microsyntenic association would disrupt trans-dev-associated cis-regulatory functions.
Figure 1.
Functional causes of conserved microsynteny and phylogenetic distribution of conserved gene pairs. (A) (Coregulation) Two neighbor genes share one or more regulatory elements. Chromosomal breakpoints in the intergenic region of the two genes are opposed by selection since it would affect the coordinated expression of the two genes. (B) Genomic regulatory block (GRB): A bystander gene (green) contains regulatory elements in its introns that target a neighbor gene (red), often a trans-dev gene with key regulatory roles in animal development. The breakage of the association would result in affected regulation of the trans-dev gene. (Based on Becker and Lenhard [2007]). (C) Consensus phylogeny of the studied metazoans, showing the total number of CAMPs in each species (in parentheses), and the minimum number of pairs at each node inferred by parsimony (in boxes). Branch lengths correspond to the fraction of gene pairs lost out of the total pairs present in the last common ancestor. Black dots at basal nodes indicate that branch length could not be estimated.
Despite these instances of nonrandomness, questions remain about the generality and strength of natural selection in maintaining gene order. First, similarity of expression between neighboring genes could be the result rather than the cause of their proximity (e.g., because of shared chromatin structure). Second, direct studies of selection have produced ambiguous results, with evidence for purifying selection on maintaining gene order in hemiascomycetous yeasts (Hurst et al. 2002; Fischer et al. 2006; Poyatos and Hurst 2007), but not in Drosophila (Weber and Hurst 2011), even over short phylogenetic distances. Third, gene order is nearly completely scrambled between humans and model invertebrates (Putnam et al. 2008; Denoeud et al. 2010). Furthermore, although whole-genome analyses of slow-evolving species and ancestral karyotype reconstructions showed that the chromosome-scale organization (so-called macrosynteny) has been largely conserved since the last common metazoan ancestor, these same studies found almost no traces of preservation of the specific ancestral local gene order (microsynteny) across metazoans (Putnam et al. 2007, 2008; Srivastava et al. 2008, 2010).
Even in cases where selection appears to conserve microsynteny, questions remain about the generality and duration of these forces over evolutionary time. First, known GRBs are generally restricted to vertebrates (Kikuta et al. 2007) or insects (Engstrom et al. 2007), with only three transphyletic GRBs described to date (Wang et al. 2007; Irimia et al 2012; Maeso et al. 2012a). Second, while tandemly duplicated Drosophila genes tend to remain linked (Quijano et al. 2008), such duplicates usually reflect recent duplications (Thomas 2007; Irimia et al. 2008; Baldwin et al. 2009). This suggests that these associations are short-lived, again with only a few known exceptions (e.g., Hox genes and histone clusters). Third, while coexpression of linked genes is well-established (most strikingly, by usage of bidirectional promoters between genes) (Adachi and Lieber 2002), few gene linkages are conserved between eukaryotic kingdoms (Davila-Lopez et al. 2010). As such, the general paradigm holds that neutral processes dominate microsynteny, particularly over long evolutionary times (Srivastava et al. 2008; Koonin and Wolf 2010).
We report the first genome-wide analysis of microsynteny conservation across metazoans, analyzing 17 species spanning 1000 million years (MY) of evolution. We identified 795 groups of genes that are associated in four or more major animal taxa, 595 of which correspond to unrelated (nonparalogous) genes, which we term conserved ancestral microsyntenic pairs (CAMPs). Multiple lines of evidence suggest conservation of gene expression coregulation for some CAMPs, and of ancient GRBs involving key trans-dev genes for others.
Results
Identification of ancient conserved gene associations across metazoans
For each pair of neighboring genes in the genome of a given species, we assessed whether the two genes were also tightly genetically linked (with four or fewer intervening genes) in other lineages (see Methods for details). We identified hundreds of pairs in each genome that were conserved across at least four major lineages, ranging from 197 in the tunicate Ciona intestinalis to 842 in the cephalochordate Branchiostoma floridae, and 600 in human (Supplemental Table S1). To test this significance we randomized gene order within each chromosome/scaffold. Across 100 replicates per species, we found an average of 0.015 conserved pairs using the same criteria, a false discovery rate lower than 0.0002 for all species (Supplemental Table S1).
We then merged the data for each species into a single data set, and filtered for potential annotation errors (see Supplemental File S1), yielding a final set of 795 unique groups of gene pairs (or three or more gene clusters) (Supplemental Table S2). Due to high duplication rates, precise orthology and paralogy relationships could not be established for 89/795 groups. These included previously described examples, such as the histone clusters (Davila-Lopez et al. 2010) and cytochrome-P450 genes (Thomas 2007; Baldwin et al. 2009), as well as eight groups of clusters/pairs of important developmental genes (Hox, Wnt, Hes, En, Irx, Six, Tbx, and other homeobox genes).
Another 110 groups included paralogous gene pairs whose group orthology could be more confidently assigned by BLAST. Interestingly, using Bayesian phylogenetic inferences (see Supplemental File S1 for details), we found evidence that at least 68% of these pairs (Supplemental Table S3) likely arose by independent tandem duplications in the different lineages and are therefore not ancestrally linked (e.g., the homologs of the human gene C16orf5; Supplemental Fig. S1). This suggests a significant level of recurrent evolution of tandem gene duplicates across metazoan genomes (Maeso et al. 2012b). Alternatively, however, these patterns could also reflect events of gene conversion between ancestral duplicates within each species.
Deeply conserved phylogenetically unrelated gene pairs
We also identified 595 groups of nonparalogous gene pairs. These pairs ranged in degree of conservation, with 377 of them conserved in four out of the 11 studied major metazoan lineages, 153 in five, 46 in six, and 19 in seven or more. Since these gene pairs did not result from tandem duplications, it is very unlikely that they have become linked independently in different lineages, and therefore are likely to represent ancient gene associations. We refer to these gene pairs as conserved ancestral microsyntenic pairs (CAMPs).
To expand the phylogenetic coverage, we next assessed conservation of these 595 CAMPs in four additional phylogenetically key species whose incomplete genome assemblies or annotations precluded their inclusion in the initial analyses: the hemichordate Saccoglossus kowalevskii, the tunicate Oikopleura dioica, and two outgroups, the sponge Amphimedon queenslandica and the unicellular opistokont Capsaspora owczarzaki. With this information, we applied parsimony to reconstruct the evolutionary history of CAMPs. At least 378 CAMPs were already present at the origin of Eumetazoans (all animals but sponges and placozoans) and 593 at the origin of Bilateria (two pairs were specific to Protostomes). We also reconstructed the degree of CAMP disruption (fraction of ancestral CAMPs lost) on each branch. Notably, despite several potential sources of error (see Supplemental File S1), branches with high rates of CAMP disruption correspond closely to branches previously shown to have high rates of other genomic changes (e.g., intron loss, gene duplications, genome rearrangements, nucleotide substitutions, etc.) (Kent and Zahler 2000; Lynch and Conery 2000; Stein et al. 2003; Bourque et al. 2005; Bhutkar et al. 2008; Irimia and Roy 2008; Putnam et al. 2008; Denoeud et al. 2010) (Fig. 1C). For example, generally slow-evolving species such as amphioxus and the mollusk Lottia gigantea have retained 448–492 CAMPs, whereas fast-evolving lineages such as nematodes (12 CAMPs), flies (46), and C. intestinalis (54) have conserved far fewer (Fig. 1C). These results are also in full agreement with previous studies on lineage-specific losses of chromosomal-scale gene linkage (macrosysteny) (Putnam et al. 2007, 2008; Srivastava et al. 2008, 2010; Denoeud et al. 2010; Lv et al. 2011). These results thus indicate that CAMP evolution closely follows overarching (though still poorly understood) trends of genome evolution.
Functional causes for evolutionary conservation of ancient gene associations
We next sought independent tests that CAMPs have been specifically maintained by selection. We tested predictions made by each of the two known general hypotheses for functional roles of the conservation of microsynteny: coordinated transcriptional regulation (i.e., coregulation) and association of the genes as a GRB (Fig. 1).
Coregulation of gene pairs
Linked genes may share common cis-regulatory sequences, leading to coordinated gene expression (Fig. 1A). In this case, potential chromosomal breakpoints in the intergenic region between such linked pairs will disrupt expression. We tested six predictions of this scenario: (1) enrichment of divergently transcribed genes (i.e., in a head-to-head or 5′–5′ orientation) due, e.g., to a bidirectional promoter, (2) preferential conservation of this 5′–5′ orientation, (3) short intergenic distances, (4) correlated expression patterns, (5) few insulator elements, and (6) highly conserved intergenic sequences (due to functional transcriptional elements).
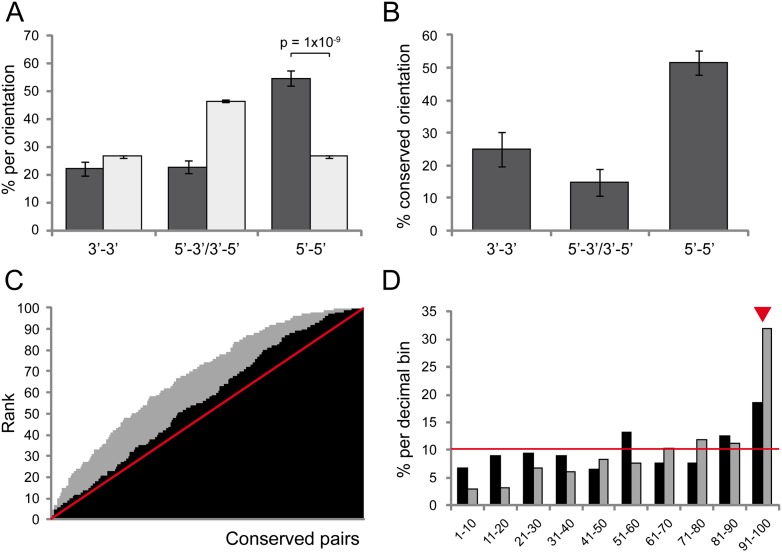
We found that most CAMPs were organized in a 5′–5′ orientation (prediction 1). In humans, 54.8% of CAMPs showed this orientation, compared with 26.7% of the control set (i.e., all nonparalogous neighboring gene pairs without conserved microsynteny; P = 1.37 × 10−9, χ2 test) (Fig. 2A). A similar pattern was found for all species (reaching 68.6% in L. gigantea) except for flies and tunicates (which have similar 3′–3′ and 5′–5′ orientations, see below). The 5′–5′ orientations are far more conserved (prediction 2): Orientation was conserved across all species sharing the CAMP for 51.4% of CAMPs with a 5′–5′ in human, compared with 15%–25% for other orientations (Fig. 2B); indeed, 5′–5′ CAMPs account for 75.8% of human CAMPs with fully conserved orientation. A similar result was observed for other species (again with the exceptions of flies and tunicates) and for highly conserved CAMPs (five or more lineages; data not shown).
Figure 2.
Predictive features of coregulated gene pairs. (A) Percentage of gene pairs oriented in 3′–3′ (convergently transcribed), 5′–3′ or 3′–5′ (codirectionally transcribed), or 5′–5′ (divergently transcribed) among human conserved gene pairs (dark gray) and control set (all nonconserved, nonparalogous gene pairs; light gray). Conserved pairs are highly enriched in 5′–5′ orientation (P = 1 × 10−9, χ2 test). (B) Percentage of pairs with fully conserved orientation across species for human conserved gene pairs; 5′–5′ pairs are more conserved. (C) Cumulative plot of ranks of ranked correlation coefficients of conserved pairs among the control data set (nonconserved neighbor pairs with the same orientation and similar intergenic distance; black) and random gene pairs (gray). The red line shows the expected cumulative pattern if coexpression was similar to the controls, and the area above the line the excess of highly coexpressed pairs. (D) Percentage of conserved pairs with ranks in each of the decimal bins. The red line (10%) indicates the expected values. Conserved pairs were significantly enriched in ranks of the last decimal bin (91–100, red arrowhead).
Next, we assessed whether CAMPs have shorter intergenic distances (prediction 3), which may facilitate coordinated expression (Davila-Lopez et al. 2010). In humans, intergenic regions of CAMPs were 2.3-fold shorter than the control set (mean of 48 vs. 113 Kbp, P = 2.3 × 10−5, KS test). This pattern was observed for all orientations, although it was strongest for 5′–5′ orientations (41 vs. 126 Kbp, P = 1.2 × 10−6). Moreover, 5′–5′ CAMPs with conserved orientation had the shortest intergenic regions of all subsets (29 Kbp). Finally, 5′–5′ CAMPs were twice as likely to have very short intergenic regions (<1 Kbp, a signature of bidirectional promoters) (Adachi and Lieber 2002) than the control set (12.5% vs 6%, P = 0.001, χ2 test).
To study coexpression of CAMPs (prediction 4), we calculated gene expression correlations across 23,941 different human microarray experiments (conducted using the Affymetrix U133 Plus 2.0 array). Since neighboring genes are known to have higher coexpression than unlinked genes (Fukuoka et al. 2004; Hurst et al. 2004), we compared each CAMP with a control set of 100 phylogenetically unrelated adjacent gene pairs with the same orientation and similar intergenic distance (see Methods). CAMPs showed significantly higher coexpression levels (average Spearman correlation coefficient, r = 0.30) than both the control sets (r = 0.21, P = 8.7 × 10−6, KS test) and random gene pairs (r = 0.14, P = 9.6 × 10−21). For each CAMP, we then ranked its coexpression value relative to its control set and also relative to 100 random gene pairs. The cumulative plot shows an excess of high ranks with respect to both the control and the random set (Fig. 2C, black and gray areas above the red line, respectively). This excess is mostly due to an enrichment in the most highly coexpressed decile (ranks 91–100, red arrowhead in Fig. 2D). This pattern was observed for all orientations, but was strongest for 5′–5′gene pairs (data not shown). Finally, to assess whether this may be a general pattern across metazoans, we performed a similar analysis for D. melanogaster, using 1909 published microarray experiments. These showed even stronger coexpression, with 81% of the pairs ranking in the top half of their 100 matched control pairs (50% expected) and 25% in the most highly coexpressed decile (10% expected; Supplemental Fig. S2). This excess of coexpression of CAMPs relative to other neighboring pairs is consistent with selection to maintain some CAMPs because of shared transcriptional regulatory elements.
We next investigated whether intergenic regions separating CAMPs show few insulators (prediction 5), which may impose a barrier for coordinated expression. Using insulators defined by chromatin signatures in nine human cell lines (Ernst et al. 2011), we found that CAMPs had significantly fewer intergenic insulators than their matched controls (0.062 vs. 0.076 insulators/Kbp, P = 6.4 × 10−5, KS test). This was more evident for 5′–5′ orientations (0.054 vs. 0.070 insulators/Kbp, P = 1.4 × 10−5) and for relatively short intergenic regions (<50 Kbp, 0.047 vs. 0.073 insulators/Kbp, P = 2.9 × 10−4). In contrast, CAMPs showed a higher density of transcriptional enhancers (Ernst et al. 2011) in intergenic regions (0.180 vs. 0.155 enhancers/Kbp, P = 0.022), in particular for highly coexpressed genes (see below). Finally, intergenic sequence conservation was also significantly higher in CAMPs than in nonconserved pairs (average mammalian phastCons score of 0.18 vs. 0.14, P = 2.9 × 10−6), fulfilling prediction 6.
In summary, coordination of gene expression is likely to explain the preservation of many CAMPs. One example involves the mitochondrial chaperonin genes Hspe1 and Hspd1, which are found together in nearly all studied species, from humans to the unicellular holozoan Capsaspora (and also in fungi) (Davila-Lopez et al. 2010), in a conserved 5′–5′ orientation. In humans, they have a very short intergenic distance (<1 Kbp) and are known to share a bidirectional promoter (Hansen et al. 2003). In agreement with this, both genes show high coexpression levels in our microarray analysis (r = 0.81, rank = 99). Some other strongly coexpressed pairs in humans include: UBA3-ARL6IP5 (r = 0.84, rank = 100), HNRNPA2B1-CBX3 (r = 0.84, rank = 100), HADHA-HADHB (r = 0.79, rank = 100), and ZMYM3-NONO (r = 0.83, rank = 100).
Genomic regulatory blocks (GRBs)
Another major cause of conserved gene linkage may be preservation of GRBs. The typical GRB comprises a trans-dev gene and one or more functionally unrelated bystander genes, whose introns harbor cis-regulatory sequences that act on the trans-dev gene promoter (Fig. 1B). Thus, separation of the genes in a GRB is likely to result in misregulation of the trans-dev gene. Several features of known GRBs are not expected from gene pairs with coordinated expression, yielding four specific predictions: (1) presence of a trans-dev gene and one or more non-trans-dev bystander genes, (2) extensive intronic sequence in the bystander gene, (3) evolutionary conservation of intronic sequence in the bystander gene, specifically in relatively short highly conserved noncoding regions (HCNRs), and (4) transcriptional enhancers (acting on the trans-dev gene) in bystander introns.
In order to test these predictions, we first identified trans-dev genes using Gene Ontology (see Methods). A total of 29.4% of conserved CAMPs in human were composed of a trans-dev and a non-trans-dev gene, compared with 19.5% in the control set (P = 6.0 × 10−4, χ2 test). Similar patterns were found in all species, with even higher proportions of trans-dev-plus-nondevelopmental CAMPs in fast-evolving species such as tunicates and flies (54.6% and 42.9%, respectively). This is consistent with the lower fraction of 5′–5′ (putatively coregulated) CAMPs in these lineages.
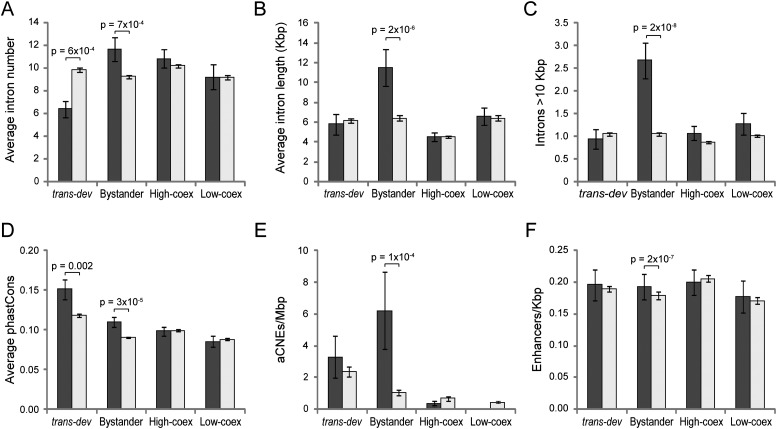
We next studied bystander gene's introns (prediction 2). We found higher intron number (P = 7 × 10−4, KS test) and average intron length (twice as long; P = 1.7 × 10−6) in bystander genes relative to controls (Fig. 3A,B). Greater intron length mostly reflected a subset of very long introns (a feature generally associated with the presence of HCNRs) (Irimia et al. 2011): Bystander genes have an average of 2.7 introns longer than 10 Kbp, compared with 0.9–1.1 in the other gene sets (P = 2.3 × 10−8) (Fig. 3C).
Figure 3.
Different functional and evolutionary features of intragenic sequences of conserved pairs. (A) Average intron number for each gene type. (Dark gray) Conserved pairs; (light gray) control set. Putative human bystander genes (nondevelopmental genes associated with trans-dev genes in conserved pairs) have significantly more introns on average than the control set (P = 7 × 10−4, KS test), whereas associated trans-dev genes have fewer (P = 6 × 10−4, KS test). Genes in highly coexpressed or lowly coexpressed conserved pairs have similar average intron numbers than the control set. (B) Average intron length (in Kbp). Putative bystander genes have significantly longer introns than the other genes (P = 2 × 10−6, KS test). (C) Average number of introns longer than 10 Kbp. Bystander genes have nearly three times more long introns than other genes (P = 2 × 10−8). (D) Average mammalian-wide phastCons score of intronic regions of different types of conserved gene pairs. Both trans-dev and bystander genes from conserved pairs show higher intronic sequence conservation than the nonconserved genes (P = 0.002 and P = 3 × 10−5, KS test), whereas conserved highly coexpressed nondevelopmental gene pairs show higher conservation than the control set at the intergenic region. (E) Density of ancient conserved noncoding elements (aCNEs) per Mbp in introns of different types of conserved gene pairs. (F) Density of functionally defined strong enhancers per Kbp in introns of different types of conserved gene pairs. Error bars correspond to standard errors.
We also found higher conservation of intron sequences in trans-dev and bystander genes than in the respective control sets (prediction 3; P = 0.0017 and P = 3.2 × 10−5, respectively, using mammalian phastCons scores) (Siepel et al. 2005). In contrast, CAMPs with no trans-dev gene showed no greater intronic conservation than their matched controls (Fig. 3D). Because enhancers likely constitute only a small fraction of all intronic sequence, we expect an even stronger enrichment of HCNRs than overall sequence conservation. Consistent with this, putative bystander genes in CAMPs had six to 10-fold more HCNRs than in nonconserved pairs, both for ancient conserved noncoding elements (aCNEs0) (Lee et al. 2011) (6.2 vs. 1.0 aCNEs/Mbp, P = 1.4 × 10−4) (Fig. 3E), and for VISTA HCNRs (Visel et al. 2007) (1.6 vs. 0.2 VISTAe/Mbp, P = 0.036). Finally, following prediction 4, CAMP's bystander introns had a higher density of functionally defined transcriptional enhancers (Ernst et al. 2011) than the control set (0.194 vs. 0.180 enhancers/Kbp, P = 1.5 × 10−7) (Fig. 3F). These four lines of evidence thus suggest that a subset of CAMPs are likely GRBs.
Experimental evidence for GRBs in vertebrate development
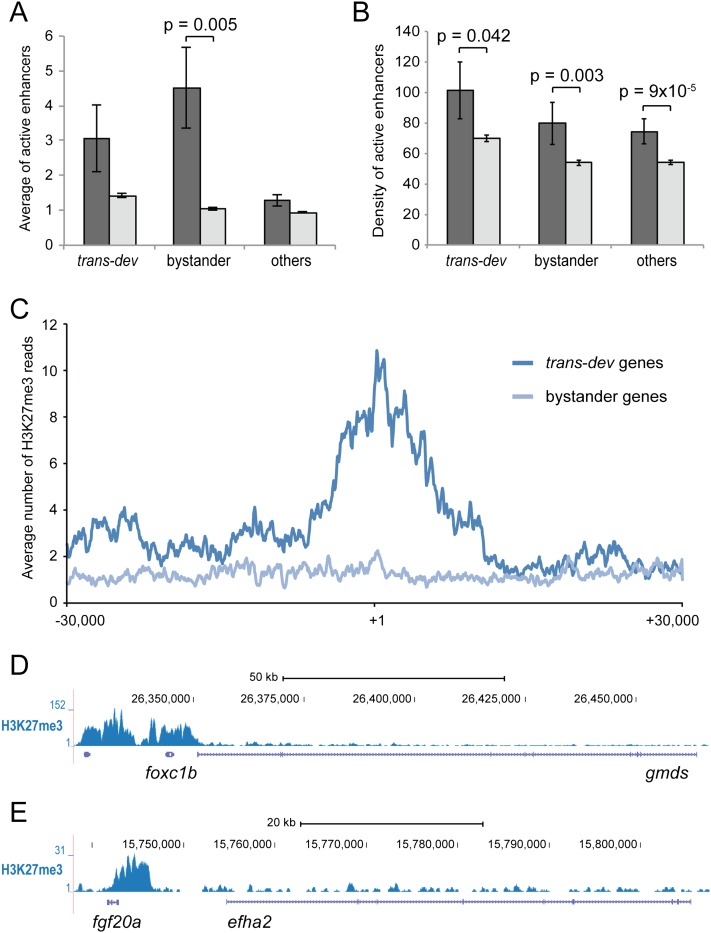
The ancient GRBs we identified typically involve genes from well known developmental gene families, such as Fox, Fgf, Tbx, Sox, Smad, etc. (Supplemental Table S4). Therefore, despite the significantly higher density of enhancers active in cell lines (Ernst et al. 2011), the major effect of bystander-contained enhancers is expected during embryonic development. In order to test this hypothesis, we used chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) for three key epigenetic marks in zebrafish embryos at 24-h post-fertilization (hpf): Histone 3 Lysine 4 trimethylation (H3K4me3, marks active promoters), Histone 3 Lysine 4 monomethylation (H3K4me1, often marks active enhancers when not overlapping with H3K4me3), and Histone 3 Lysine 27 trimethylation (H3K27me3, often marks inactive promoters, and indicates genes with tissue-specific expression patterns in whole embryos) (Turner 2007; Akkers et al. 2009; Margueron and Reinberg 2010). We searched for conservation of the 593 bilaterian CAMPs in the zebrafish genome, finding 260 conserved pairs of 205 unique groups, of which 29.2% included one trans-dev gene. Mapping of H3K4me1+/H3K4me3- peaks shows that putative bystanders for CAMPs contain approximately four times more active enhancers than for nonconserved pairs (P = 0.005, KS test) (Fig. 4A), and with a higher density (P = 0.003) (Fig. 4B). These results suggest that conserved GRBs have complex cis-regulatory landscapes in zebrafish development. Importantly, we found that H3K27me3 is significantly increased in trans-dev, but not in bystander genes in whole zebrafish embryos (Fig. 4C–E). This suggests that, globally, the trans-dev genes of GRBs, but not the enhancer-containing bystanders, have complex tissue-specific expression patterns (Akkers et al. 2009).
Figure 4.
Differential epigenetic marks in in bystander and trans-dev genes in early zebrafish development. (A,B) Average number (A) and density (peaks/Kbp, B) of H3K4me1+/H3K4me3− peaks (putative active enhancers) in 24-hpf zebrafish embryos within trans-dev, bystander, and other nondevelopmental genes for conserved pairs (dark gray) and for nonconserved pairs (light gray). Error bars correspond to standard errors. (C) Average number of reads for H3K27me3 ChIP-seq around transcription start sites show an specific deposition of this epigenetic mark in trans-dev genes. (D,E) Two examples of differential distribution of H3K27me3 in a trans-dev gene (foxc1b, D and fgf20a, E) and its nondevelopmental partner (gmds, D and efha2, E).
A striking case of conserved GRB involves the ISL LIM homeobox (Isl). Isl plays important conserved roles in animal development, in particular in neuron ontogeny in diverse phyla (Thor and Thomas 1997; Jackman et al. 2000; Voutev et al. 2009; Liang et al. 2011). We found that Isl genes are linked to Scaper/ssp3 (S-phase cyclin A-associated protein in the ER) genes in nearly all studied species, from sponges to humans, for over 1000 MY of evolution (Fig. 5A). In human, SCAPER spans ∼500 Kbp and contains 16 introns longer than 10 Kbp, as expected for a bystander. Furthermore, the expression patterns of these two genes are very different in flies (r = 0.08), humans (r = −0.07), and zebrafish (Fig. 5B). In 24-hpf zebrafish embryos, isl2a is expressed in specific cephalic neuronal domains and in discrete neurons in the spinal cord (arrow), and in hindgut (red arrow)—a pattern conserved in other animal lineages (Jackman et al. 2000; Gelbart and Emmert 2010; Graveley et al. 2011)—whereas scaper is not expressed at this stage.
Figure 5.
Functional characterization of the GRB of isl2a-scaper in zebrafish. (A) Phylogenetic distribution of the GRB across the studied metazoan species. The GRB was only not conserved in C. elegans and N. vectensis. (B) Top and middle panels are 24-h post fertilization (hpf) zebrafish embryos showing the expression patterns of isl2a and scaper. isl2a is expressed in discrete neurons in the spinal cord (arrow) and in the proctodeum (arrowhead), while scaper is not detectable at this stage. The third panel is an embryo at a similar developmental stage showing GFP expression promoted by an enhancer located within the intron of scaper (asterisk in C). This enhancer is active in isl2a-expressing territories. The bottom panel shows immunodetection of the transgenic GFP (green), the endogenous Islet proteins (red) and the overlay between the two showing coexpression. (C) Distribution of H3K4me3, H3K4me1, and H3K27me3 tracks along the isl2a-scaper GRB in 24-hpf zebrafish embryos. H3K4me1 peaks tested for enhancer activity in zebrafish stable transgenic lines are shaded in red. H3K4me3 and H3K27me3 distribution show that scaper is inactive and this stage and that isl2a is tissue specific, suggesting that the H3K4me1-positive enhancer may be acting on the active isl2a promoter. (Below) Conservation track from the UCSC Genome Browser.
In zebrafish, ChIP-seq data for H3K4me1 showed three potential enhancers within scaper at 24 hpf (red bars in Fig. 5C), whereas H3K4me3 indicated that isl2a's promoter is active but scaper's is not (Fig. 5C), suggesting that the active enhancers within scaper may be acting on isl2a. We thus used the GFP reporter vector ZED (Bessa et al. 2009) to examine the enhancer activity of the three H3K4me1+/H3K4me3− peaks in stable zebrafish transgenic lines. One of them (red asterisk in Fig. 5C) promoted GFP expression in different neuron populations and hindgut, two domains coexpressing the endogenous Isl2a protein, as shown by ISH and immunocolocalization (Fig. 5; Supplemental Fig. S3). These results strongly suggest that at least this scaper-contained enhancer is acting specifically on isl2a, and provide a plausible explanation for the conservation of this gene association.
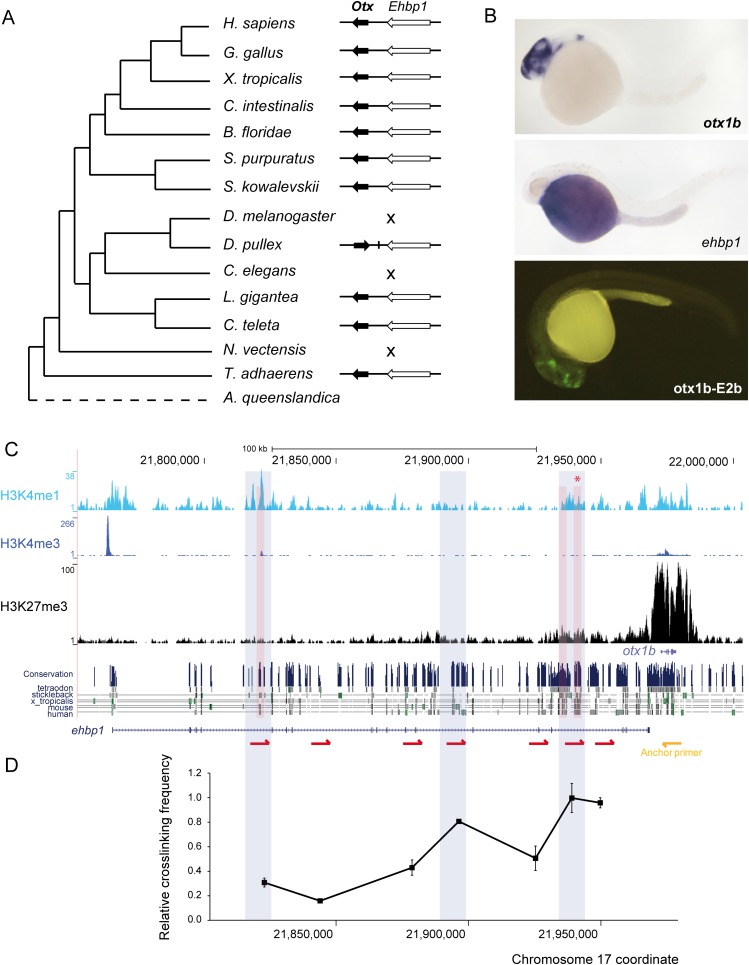
Another interesting case is the homeobox transcription factor Otx (Fig. 6). This gene is involved in establishing a deeply conserved early anterior–posterior (A–P) brain patterning (Simeone et al. 1992; Hirth et al. 2003; Castro et al. 2006; Irimia et al. 2010) and is expressed in the anterior nervous system of nearly all studied bilaterians (Simeone et al. 1992; Williams and Holland 1996; Hirth et al. 2003; Lowe et al. 2003; Scholpp et al. 2007). We found that Otx is linked to Ehbp1 (EH domain binding protein 1) in nearly all species, from placozoans to humans, spanning over 700 MY (Fig. 6A). As in the case of Isl-Scaper, the expression of OTX1 and EHBP1 is very different in human (r = −0.12) and in zebrafish (Fig. 6B). During early zebrafish development, only otx1b is expressed in the embryo, mainly in the cephalic region. In addition, zebrafish ehbp1 also has several long introns that contain potential enhancers, as indicated by high levels of H3K4me1 without H3K4me3 (Fig. 6C). We therefore generated stable transgenic zebrafish reporter lines for three of these H3K4me1 peaks, one of which drove strong and consistent expression to the classical conserved Otx anterior domain in the central nervous system. Next, to test whether the remaining ehbp1 intronic sequences may also contain other otx1b cis-regulatory elements, we performed chromosome conformation capture (3C) assays (Hagège et al. 2007) to explore physical interaction between several regions of the ehbp1 gene (red arrows in Fig. 6C) and the otx1b promoter. This assay confirmed the interaction of the identified enhancer, and revealed another potential cis-regulatory element within the ehbp1 intron that clearly contacted the otx1b promoter (Fig. 6C). Collectively, these results show that ehbp1 contains cis-regulatory elements that specifically regulate the expression of otx1b in deeply conserved domains.
Figure 6.
Functional characterization of the GRB of otx1b-ehbp1 in zebrafish. (A) Phylogenetic distribution of the GRB across the studied metazoan species. The GRB was not conserved in D. melanogaster, C. elegans, and N. vectensis. (B) Top and middle panels are zebrafish embryos at 24 hpf showing the expression of otx1b and ehbp1 genes. otx1b is detected in the anterior brain, while ehbp1 is expressed in the yolk. (Lower panel) GFP expression promoted by an enhancer located within the intron of ehbp1 (asterisk in C). This enhancer is active in most tissues expressing otx1b. (C) Distribution of H3K4me3, H3K4me1, and H3K27me3 tracks along the otx1b-ehbp1 GRB in 24-hpf zebrafish embryos. H3K4me1 peaks tested for enhancer activity in zebrafish stable transgenic lines are shaded in red. The regions that physically interact in 3C assays with the otx1b promoter are shaded in blue. H3K27me3 distribution indicates that otx1b, but not ehbp1, has tissue-specific expression; hence, H3K4me1 enhancers located in ehbp1 introns are likely acting on otx1b. (Below) conservation track from the UCSC Genome Browser. (D) Graph showing a 3C experiment to detect interaction between different ehbp1 intronic regions and the otx1b promoter in 24-hpf embryos. A fixed primer (yellow arrow) was set at the otx1b promoter, and seven regions were assayed for interaction with that promoter using different primers (red arrows) distributed along the ehbp1 intronic genomic area. The highest cross-linking frequency value is set to 1. Error bars indicate standard error (n = 3).
Discussion
We have reported a large number of genes belonging to conserved ancestral microsyntenic pairs (CAMPs) shared across several deeply diverged metazoan lineages. In contrast to the few previously described cases of deeply conserved microsynteny, nearly all of which involve tandemly duplicated gene pairs or clusters, we found nearly 600 pairs of unrelated genes that are closely physically linked in several major bilaterian lineages spanning over 600 MY of animal evolution (half of them already present in the nonbilaterian ancestors, 700–1000 MY ago). Moreover, these numbers are likely underestimates, since most genome sequence assemblies used in our study are highly fragmented into relatively small scaffolds (Supplemental Table S1). Thus, much of the microsynteny in most organisms could not be truly evaluated and was conservatively considered to be nonconserved.
These results are quite unexpected in light of previous studies of pairwise conservation. Current and previous comparisons between pairs of distantly related species show very little conservation of local microsynteny (e.g., only ∼1%–5% between humans and other lineages; Supplemental Fig. S4). Indeed, even studies of slow-evolving metazoans, which found conservation of chromosome-level linkage (e.g., genes on the same chromosome in humans also tend to be on the same chromosome in cnidarians) found very little or no microsyntenic conservation across several species (Putnam et al. 2007, 2008; Srivastava et al. 2008, 2010). Our finding that comparable fractions of gene linkages (e.g., including 1%–2% of genes in humans; Supplemental Table S1) have been independently conserved in several different lineages is therefore quite unexpected, and implies that the same specific subset of gene linkages have been actively retained in widely diverged lineages (including examples even in typically fast-evolving species) (Chavali et al. 2011; Lv et al. 2011; Weber and Hurst 2011). Accordingly, we have provided extensive evidence that many of these associations have been conserved due to functional reasons. First, many CAMPs show high coexpression levels in humans and/or flies, suggesting that they may share cis-regulatory inputs resulting in coordinated transcription. Second, we found more than 100 putative ancient GRBs, often involving important developmental regulators, substantially adding to the three previously known trans-phyletic GRB (Wang et al. 2007; Irimia et al. 2012; Maeso et al. 2012a). The deep conservation of GRBs suggests that they may underlie the remarkable conservation of some developmental genetic programs, such as the A–P patterning of the bilaterian central nervous system (in the case of Otx-Ehbp1) or neuronal ontogeny (Isl-Scaper). At the same time, the conservation of some GRBs in lineages with very different body plans or cell types is intriguing. For example, in the case of Isl, it is not clear which common regulatory role may be responsible for the conservation of the association between bilaterians and sponges, which have no proper neurons (Hooper and Van Soest 2002; Sakarya et al. 2007).
The phylogenetic distribution of CAMPs also shows that, despite their deep conservation, many of the associations have been repeatedly lost in different lineages. This was observed even for extremely conserved CAMPs such as the coexpressed mitochondrial chaperonins HSPE1-HSPD1 (lost in flies) and the GRBs described in detail in the present study (Figs. 5A, 6A). Different causes may underlie the loss of these associations. For example, major modification of body plans may render some of the regulatory constraints obsolete, and thus the microsynteny can be lost, presumably without selective cost (as in the case of Hox clusters) (Duboule 2007). Alternatively, associations within GRBs could be disentangled through the acquisition of genetic redundancy (McEwen et al. 2006; Kikuta et al. 2007; Navratilova et al. 2010; Goode et al. 2011; Maeso et al. 2012a). Complex genes are often regulated by a redundant set of cis-regulatory elements, which increase robustness, and are continuously evolving (Jeong et al. 2006; Hong et al. 2008; Frankel et al. 2010; Perry et al. 2010; Schmidt et al. 2010). For example, if a new, redundant cis-regulatory module arises outside of the bystander gene (e.g., in the intergenic region), the bystander gene could be now translocated without affecting the trans-dev gene's regulation. In a more complex scenario, duplication of the gene pair (either by whole-genome or segmental duplication) can aid this process. In coregulated gene pairs, each gene of the pair could be reciprocally lost from one of the duplicated regions while keeping the common cis-regulatory elements in both, therefore becoming two nonassociated genes with fully functional regulatory landscapes. In the case of GRBs, the coding sequences of extra bystander gene copies may be erased, while the trans-dev-associated regulatory elements are conserved (McEwen et al. 2006; Kikuta et al. 2007; Maeso et al. 2012a).
Finally, the extent of ancient conserved microsynteny we have uncovered is even more striking when considering that the ancestral bilaterian likely had fewer than 10,000 unique genes (Miller and Ball 2009): Thus, over 12% of these are involved in close microsyntenic relationships conserved in multiple lineages to this day. It can thus be argued that microsynteny is among the most conserved features of metazoan genomes, and that ancient cis-regulatory inputs may be far more common than currently appreciated (Royo et al. 2011). We expect that as metazoan genomes continue to be sequenced at an ever-faster rate, many more microsyntenic relationships will be discovered, and many more details surrounding their role as key components of the genome's architecture will be revealed.
Methods
Genome-wide search for deeply conserved gene pairwise associations
We downloaded full genome annotations for the initial 13 species (Supplemental File S1). For multiple-transcript genes we used only the longest protein isoform. Homologs were identified by pairwise BLASTP [E-value <10−5 without filtering for low complexity sequences (-F F)].
For each pair of immediately adjacent protein-coding genes (excluding noncoding RNA genes, pseudogenes, etc.) in each genome, we asked whether the first or second best BLAST hits for both proteins were also neighboring in each of the 12 other genomes. This was defined as on the same chromosome/scaffold with four or fewer intervening genes, a threshold chosen to deal with: (1) common annotation errors (e.g., spurious automatic gene models, split genes), and (2) the fact that GRBs often include nonadjoining genes (Kikuta et al. 2007). Cutoffs of three to 10 intervening genes yielded similar results; Supplemental Fig. S5). Based on randomization simulations (see below), we considered a gene pair conserved if it was linked in four total lineages (except vertebrates, each species was considered a distinct lineage, since pairwise species comparisons showed no enrichment for pair conservation due to phylogenetic proximity, except within vertebrates; Supplemental Fig. S4). To determine “conservation” by chance, we randomized gene order within each chromosome/scaffold for each of the 12 species, with 100 replicates. These simulations indicated that a cutoff of four or more lineages with four or fewer intervening genes yields a false-positive discovery rate of <0.0002 per gene pair for all species (Supplemental Table S1).
Generation of a unique data set of ancient microsyntenic gene pairs
Conserved pairs for each of the 13 species were then merged into a single data set of nonredundant groups. Each unique group contained the syntenic pairs for all species in which it was conserved, including duplicates of the pairs within species (e.g., paralogons in vertebrates resulted from the two rounds of whole-genome duplication), if any. Next, we assessed whether the genes were related (paralogous pairs), and filters were applied to account for reciprocal blast consistency and common annotation errors (see Supplemental File S1 for details).
To reconstruct the history of linked duplicate genes, we performed Bayesian phylogenetic inferences for genes for all species for each pair, to distinguish (1) independent tandem duplications (clustering of genes by species on phylogenetic trees) and (2) retained ancestral linkage. Statistical significance was tested by comparison to randomized trees with the same topology (see Supplemental File S1 for details).
We also studied disruption of CAMPs. First, we assessed conservation in four additional species: S. kowalevskii, O. dioica, A. queenslandica, and C. owczarzaki using tBLASTN against the assembled contigs (Supplemental File S1). Then, we applied parsimony to infer the number of CAMPs that were present at each node of a consensus phylogenetic tree, and estimated the fraction that were disrupted along each branch.
Coexpression of CAMPs using microarray data
We downloaded the full set of experiments from Affymetrix Human Genome U133 Plus 2.0 Array and Affymetrix Drosophila Genome 2.0 Array (GEO accession nos. GPL570 and GPL1322). After excluding experiments with missing probes, we obtained a matrix with 54,608 probes with data for 23,941 experiments in humans, and 13,935 probes with data for 1909 experiments in Drosophila. The expression levels were converted to ranks within each experiment to correct for different measurement and normalization methods.
Of the CAMPs with no intervening genes, 279 in humans and 27 in Drosophila had at least one probe for each gene. For these, we calculated the correlation coefficient for gene expression between the two genes. For comparisons, we generated two control sets for each conserved pair: (1) 100 nonparalogous, nonconserved gene pairs with the same orientation and the most similar intergenic distances; (2) 100 pairs of two randomly selected nonsyntenic genes. Then, the correlation coefficient of each conserved pair was ranked with respect to each of its two control sets. When multiple probes covered a gene, we used the combination of probes that gave the highest correlation (for both test and control sets). Using the average between probes gave the same pattern of higher coexpression between conserved pairs.
Study of genome structure
Orientation and intergenic distance were calculated from GFF/GTF annotation files, using the stable transcript for Ensembl genomes, and the best gene models for other genomes. We merged all gene isoforms into a single intron–exon structure to determine intron number/lengths. Global sequence conservation was calculated using the phastCons46wayPlacental scores from UCSC Comparative track (http://genome.ucsc.edu/). Only sites with scores >0 (confidently aligned across genomes) were used for calculations. HCNRs were obtained from two sources: (1) vistaEnhancers track at UCSC, and (2) ancient conserved noncoding elements (aCNEs) (Lee et al. 2011). Active strong enhancers and insulators for nine human cell lines were obtained from Ernst et al. (2011). Data for all cell lines were merged into a unique, nonredundant set of coordinates.
To study GRBs, we defined developmental (trans-dev) genes as genes with GO terms embryo development (GO:0009790) and/or organ development (GO:0048513). For comparison, we also defined two sets of CAMPs composed of two nondevelopmental genes, based on the analyses of coexpression presented above: pairs of highly coexpressed nondevelopmental genes (r > 0.40, likely enriched in gene pairs conserved due to coregulatory reasons) and of weakly coexpressed nondevelopmental genes (r < 0.05). In addition, each of these conserved pair sets had its corresponding control set, consisting of similar nonparalogous syntenic pairs (i.e., bystander plus trans-dev or highly/lowly coexpressed) that are not evolutionarily conserved.
Experimental analyses in zebrafish embryos
Chromatin immunoprecipitation (ChIP) was performed following the protocol described in Wardle et al. (2006) with minor modifications, and Genome Analyzer (Illumina) ChIP-seq was performed as described in Bogdanović et al. (2012) (see Supplemental File S1 for details). Highly enriched regions (peaks) of histone methylation were obtained by the MACS (v.1.3.3) algorithm (Zhang et al. 2008) using standard settings with one modification (mfold = 20). Twenty-five randomly selected peaks were verified by qPCR and compared with their random controls (false-positive discovery rate [FDR] <0.04). The PCRs were performed on 1:50 dilutions of the ChIP samples using the C1000 Thermal Cycler (BioRad).
For each H3K4me1-positive peak that was tested in stable zebrafish transgenic assays, we designed primers to span the whole region plus ∼100 nt at each side (primer sequences are available in Supplemental Table S5). Specific details for cloning, preparation, and injection of candidate enhancer sequences into zebrafish eggs, as well as for in situ hybridization and immunostaining, are provided in Supplemental File S1; in all cases, previously described protocols were followed with minor modifications (Kawakami 2004; Tena et al. 2007; Bessa et al. 2009). 3C assays were performed as referred in Hagège et al. (2007) and Tena et al. (2011), (see Supplemental File S1). A set of locus-specific primers (Supplemental Table S5) was designed to perform qPCRs to measure relative enrichment in each ligation product. Negative control primers were designed ∼30 Kbp upstream of and downstream from the regions of interest. PCR values were normalized with primers for Ercc3. ChIP-seq data for H3K4me1 and H3K4me3 were downloaded from the NCBI Gene Expression Omnibus (GEO) (GSE32483, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=vnyhbkqayaioczc&acc=GSE32483).
Data access
H3K27me3 short read data have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE35050.
Acknowledgments
M.I., M.S.A., S.W.R., and H.B.F. were funded by NIH grant 1R21HG005240-01A1. H.B.F. is an Alfred P. Sloan Fellow and Pew Scholar in the Biomedical Sciences. J.J.T., A.F-M., O.B., E.C-M., and J.L.G-S. were funded by grants BFU2010-14839, CSD2007-00008, and Proyecto de Excelencia CVI-3488.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.139725.112.
References
- Adachi N, Lieber MR 2002. Bidirectional gene organization: A common architectural feature of the human genome. Cell 109: 807–809 [DOI] [PubMed] [Google Scholar]
- Akkers RC, van Heeringen SJ, Jacobi UG, Janssen-Megens EM, Françoijs KJ, Stunnenberg HG, Veenstra GJ 2009. A hierarchy of H3K4me3 and H3K27me3 acquisition in spatial gene regulation in Xenopus embryos. Dev Cell 17: 425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin W, Marko P, Nelson D 2009. The cytochrome P450 (CYP) gene superfamily in Daphnia pulex. BMC Genomics 10: 169 doi: 10.1186/1471-2164-10-169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker TS, Lenhard B 2007. The random versus fragile breakage models of chromosome evolution: A matter of resolution. Mol Genet Genomics 278: 487–491 [DOI] [PubMed] [Google Scholar]
- Bessa J, Tena JJ, de la Calle-Mustienes E, Fernández-Miñán A, Naranjo S, Fernández A, Montoliu L, Akalin A, Lenhard B, Casares F, et al. 2009. Zebrafish enhancer detection (ZED) vector: A new tool to facilitate transgenesis and the functional analysis of cis-regulatory regions in zebrafish. Dev Dyn 238: 2409–2417 [DOI] [PubMed] [Google Scholar]
- Bhutkar A, Schaeffer SW, Russo SM, Xu M, Smith TF, Gelbart WM 2008. Chromosomal rearrangement inferred from comparisons of 12 Drosophila genomes. Genetics 179: 1657–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanović O, Fernandez-Miñán A, Tena JJ, de la Calle-Mustienes E, Hidalgo C, van Kruysbergen I, van Heeringen SJ, Veenstra GJ, Gómez-Skarmeta JL 2012. Dynamics of enhancer chromatin signatures mark the transition from pluripotency to cell specification during embryogenesis. Genome Res 22: 2043–2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque G, Zdobnov EM, Bork P, Pevzner PA, Tesler G 2005. Comparative architectures of mammalian and chicken genomes reveal highly variable rates of genomic rearrangements across different lineages. Genome Res 15: 98–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro LFC, Rasmussen SLK, Holland PWH, Holland ND, Holland LZ 2006. A Gbx homeobox gene in amphioxus: Insights into ancestry of the ANTP class and evolution of the midbrain/hindbrain boundary. Dev Biol 295: 40–51 [DOI] [PubMed] [Google Scholar]
- Chavali S, Morais DA, Gough J, Babu MM 2011. Evolution of eukaryotic genome architecture: Insights from the study of a rapidly evolving metazoan, Oikopleura dioica: Non-adaptive forces such as elevated mutation rates may influence the evolution of genome architecture. Bioessays 33: 592–601 [DOI] [PubMed] [Google Scholar]
- Davila-Lopez M, Martinez-Guerra JJ, Samuelsson T 2010. Analysis of gene order conservation in eukaryotes identifies transcriptionally and functionally linked genes. PLoS ONE 5: e10654 doi: 10.1371/journal.pone.0010654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denoeud F, Henriet S, Mungpakdee S, Aury JM, Da Silva C, Brinkmann H, Mikhaleva J, Olsen LC, Jubin C, Cañestro C, et al. 2010. Plasticity of animal genome architecture unmasked by rapid evolution of a pelagic tunicate. Science 330: 1381–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle WF 1978. Genes in pieces: Were they ever together? Nature 272: 581–582 [Google Scholar]
- Duboule D 2007. The rise and fall of Hox gene clusters. Development 134: 2549–2560 [DOI] [PubMed] [Google Scholar]
- Engstrom PG, Ho Sui SJ, Drivenes O, Becker TS, Lenhard B 2007. Genomic regulatory blocks underlie extensive microsynteny conservation in insects. Genome Res 17: 1898–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. 2011. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473: 43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer G, Rocha EP, Brunet F, Vergassola M, Dujon B 2006. Highly variable rates of genome rearrangements between hemiascomycetous yeast lineages. PLoS Genet 2: e32 doi: 10.1371/journal.pgen.0020032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel N, Davis GK, Vargas D, Wang S, Payre F, Stern DL 2010. Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature 466: 490–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuoka Y, Inaoka H, Kohane IS 2004. Inter-species differences of co-expression of neighboring genes in eukaryotic genomes. BMC Genomics 5: 4 doi: 10.11861471-2164-5-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelbart WM, Emmert DB 2010. 2010 FlyBase Reference Report: Gelbart and Emmert, 2010.10.13, FlyBase high throughput expression pattern data beta version. http://flybase.org/reports/FBrf0212041.html
- Goode DK, Callaway HA, Cerda GA, Lewis KE, Elgar G 2011. Minor change, major difference: Divergent functions of highly conserved cis-regulatory elements subsequent to whole genome duplication events. Development 138: 879–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, van Baren MJ, Boley N, Booth BW, et al. 2011. The developmental transcriptome of Drosophila melanogaster. Nature 471: 473–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagège H, Klous P, Braem C, Splinter E, Dekker J, Cathala G, de Laat W, Forné T 2007. Quantitative analysis of chromosome conformation capture assays (3C-qPCR). Nat Protoc 2: 1722–1733 [DOI] [PubMed] [Google Scholar]
- Hansen JJ, Bross P, Westergaard M, Nielsen MN, Eiberg H, Børglum AD, Mogensen J, Kristiansen K, Bolund L, Gregersen N 2003. Genomic structure of the human mitochondrial chaperonin genes: HSP60 and HSP10 are localised head to head on chromosome 2 separated by a bidirectional promoter. Hum Genet 112: 71–77 [DOI] [PubMed] [Google Scholar]
- Hirth F, Kammermeier L, Frei E, Walldorf U, Noll M, Reichert H 2003. An urbilaterian origin of the tripartite brain: Developmental genetic insights from Drosophila. Development 130: 2365–2373 [DOI] [PubMed] [Google Scholar]
- Hong JW, Hendrix DA, Levine MS 2008. Shadow enhancers as a source of evolutionary novelty. Science 321: 1314 doi: 10.1126/science.116631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper JNA, Van Soest RWM 2002. Systema Porifera: A guide to the classification of sponges. Kluwer Academic/Plenum Publishers, New York [Google Scholar]
- Hurst LD, Williams EJ, Pal C 2002. Natural selection promotes the conservation of linkage of co-expressed genes. Trends Genet 18: 604–606 [DOI] [PubMed] [Google Scholar]
- Hurst LD, Pal C, Lercher MJ 2004. The evolutionary dynamics of eukaryotic gene order. Nat Rev Genet 5: 299–310 [DOI] [PubMed] [Google Scholar]
- Irimia M, Roy SW 2008. Spliceosomal introns as tools for genomic and evolutionary analysis. Nucleic Acids Res 36: 1703–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia M, Maeso I, Garcia-Fernandez J 2008. Convergent evolution of clustering of Iroquois homeobox genes across metazoans. Mol Biol Evol 25: 1521–1525 [DOI] [PubMed] [Google Scholar]
- Irimia M, Piñeiro C, Maeso I, Gómez-Skarmeta JL, Casares F, Garcia-Fernàndez J 2010. Conserved developmental expression of Fezf in chordates and Drosophila and the origin of the Zona Limitans Intrathalamica (ZLI) brain organizer. EvoDevo 1: 7 doi: 10.1186/2041-9139-1-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia M, Maeso I, Burguera D, Hidalgo-Sánchez M, Puelles L, Garcia-Fernàndez J, Roy SW, Ferran JL 2011. Contrasting 5′ and 3′ evolutionary histories and frequent evolutionary convergence in meis/hth gene structures. Genome Biol Evol 3: 551–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia M, Royo JL, Burguera D, Maeso I, Gómez-Skarmeta JL, Garcia-Fernandez J 2012. Comparative genomics of the Hedgehog loci in chordates and the origins of Shh regulatory novelties. Sci Rep 2: 433 doi: 10.1038/srep00433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman WR, Langeland JA, Kimmel CB 2000. islet reveals segmentation in the Amphioxus hindbrain homolog. Dev Biol 220: 16–26 [DOI] [PubMed] [Google Scholar]
- Jeong Y, El-Jaick K, Roessler E, Muenke M, Epstein DJ 2006. A functional screen for sonic hedgehog regulatory elements across a 1 Mb interval identifies long-range ventral forebrain enhancers. Development 133: 761–772 [DOI] [PubMed] [Google Scholar]
- Kawakami K 2004. Transgenesis and gene trap methods in zebrafish by using the Tol2 transposable element. Methods Cell Biol 77: 201–222 [DOI] [PubMed] [Google Scholar]
- Kent WJ, Zahler AM 2000. Conservation, regulation, synteny, and introns in a large-scale C. briggsae–C. elegans genomic alignment. Genome Res 10: 1115–1125 [DOI] [PubMed] [Google Scholar]
- Kikuta H, Laplante M, Navratilova P, Komisarczuk AZ, Engstrom PG, Fredman D, Akalin A, Caccamo M, Sealy I, Howe K, et al. 2007. Genomic regulatory blocks encompass multiple neighboring genes and maintain conserved synteny in vertebrates. Genome Res 17: 545–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Wolf YI 2010. Constraints and plasticity in genome and molecular-phenome evolution. Nat Rev Genet 11: 487–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JM, Sonnhammer EL 2003. Genomic gene clustering analysis of pathways in eukaryotes. Genome Res 13: 875–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AP, Kerk SY, Tan YY, Brenner S, Venkatesh B 2011. Ancient vertebrate conserved noncoding elements have been evolving rapidly in teleost fishes. Mol Biol Evol 28: 1205–1215 [DOI] [PubMed] [Google Scholar]
- Liang X, Song MR, Xu Z, Lanuza GM, Liu Y, Zhuang T, Chen Y, Pfaff SL, Evans SM, Sun Y 2011. Isl1 is required for multiple aspects of motor neuron development. Mol Cell Neurosci 47: 215–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe CJ, Wu M, Salic A, Evans L, Lander E, Stange-Thomann N, Gruber CE, Gerhart J, Kirschner M 2003. Anteroposterior patterning in hemichordates and the origins of the chordate nervous system. Cell 113: 853–865 [DOI] [PubMed] [Google Scholar]
- Lv J, Havlak P, Putnam NH 2011. Constraints on genes shape long-term conservation of macro-synteny in metazoan genomes. BMC Bioinformatics 12: S11 doi: 10.1186/1471-2105-12-S9-S11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. 2007 The origins of genome architecture. Sinauer Associates, Sunderland, MA. [Google Scholar]
- Lynch M, Conery JS 2000. The evolutionary fate and consequences of duplicate genes. Science 290: 1151–1155 [DOI] [PubMed] [Google Scholar]
- Maeso I, Irimia M, Tena JJ, González-Pérez E, Tran D, Ravi V, Venkatesh B, Campuzano S, Gómez-Skarmeta JL, Garcia-Fernàndez J 2012a. An ancient genomic regulatory block conserved across bilaterians and its dismantling in tetrapods by retrogene replacement. Genome Res 22: 642–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeso I, Roy SW, Irimia M 2012b. Widespread recurrent evolution of genomic features. Genome Biol Evol 4: 486–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D 2010. Chromatin structure and the inheritance of epigenetic information. Nat Rev Genet 11: 285–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen GK, Woolfe A, Goode D, Vavouri T, Callaway H, Elgar G 2006. Ancient duplicated conserved noncoding elements in vertebrates: A genomic and functional analysis. Genome Res 16: 451–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalak P 2008. Coexpression, coregulation, and cofunctionality of neighboring genes in eukaryotic genomes. Genomics 91: 243–248 [DOI] [PubMed] [Google Scholar]
- Miller DJ, Ball EE 2009. The gene complement of the ancestral bilaterian—was Urbilateria a monster? J Biol 8: 89 doi: 10.1186/jbiol192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navratilova P, Fredman D, Lenhard B, Becker TS 2010. Regulatory divergence of the duplicated chromosomal loci sox11a/b by subpartitioning and sequence evolution of enhancers in zebrafish. Mol Genet Genomics 283: 171–184 [DOI] [PubMed] [Google Scholar]
- Oliver B, Misteli T 2005. A non-random walk through the genome. Genome Biol 6: 214 doi: 10.1186/gb-2005-6-4-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry MW, Boettiger AN, Bothma JP, Levine M 2010. Shadow enhancers foster robustness of Drosophila gastrulation. Curr Biol 20: 1562–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyatos JF, Hurst LD 2007. The determinants of gene order conservation in yeasts. Genome Biol 8: R233 doi: 10.1186/gb-2007-8-11-r233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam NH, Srivastava M, Hellsten U, Dirks B, Chapman J, Salamov A, Terry A, Shapiro H, Lindquist E, Kapitonov VV, et al. 2007. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 317: 86–94 [DOI] [PubMed] [Google Scholar]
- Putnam N, Butts T, Ferrier DEK, Furlong RF, Hellsten U, Kawashima T, Robinson-Rechavi M, Shoguchi E, Terry A, Yu JK, et al. 2008. The amphioxus genome and the evolution of the chordate karyotype. Nature 453: 1064–1071 [DOI] [PubMed] [Google Scholar]
- Quijano C, Tomancak P, Lopez-Marti J, Suyama M, Bork P, Milan M, Torrents D, Manzanares M 2008. Selective maintenance of Drosophila tandemly arranged duplicated genes during evolution. Genome Biol 9: R176 doi: 10.1186/gb-2008-9-12-r176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royo JL, Maeso I, Irimia M, Gao F, Peter IS, Lopes CS, D'Aniello S, Casares F, Davidson EH, Garcia-Fernández J, et al. 2011. Transphyletic conservation of developmental regulatory state in animal evolution. Proc Natl Acad Sci 108: 14186–14191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakarya O, Armstrong KA, Adamska M, Adamski M, Wang IF, Tidor B, Degnan BM, Oakley TH, Kosik KS 2007. A post-synaptic scaffold at the origin of the animal kingdom. PLoS ONE 6: e506 doi: 10.1371/journal.pone.0000506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Wilson MD, Ballester B, Schwalie PC, Brown GD, Marshall A, Kutter C, Watt S, Martinez-Jimenez CP, Mackay S, et al. 2010. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science 328: 1036–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholpp S, Foucher I, Staudt N, Peukert D, Lumsden A, Houart C 2007. Otx1l, Otx2 and Irx1b establish and position the ZLI in the diencephalon. Development 134: 3167–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res 15: 1034–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeone A, Acampora D, Gulisano M, Stornaiuolo A, Boncinelli E 1992. Nested expression domains of four homeobox genes in developing rostral brain. Nature 358: 687–690 [DOI] [PubMed] [Google Scholar]
- Srivastava M, Begovic E, Chapman J, Putnam NH, Hellsten U, Kawashima T, Kuo A, Mitros T, Salamov A, Carpenter ML, et al. 2008. The Trichoplax genome and the nature of placozoans. Nature 454: 955–960 [DOI] [PubMed] [Google Scholar]
- Srivastava M, Simakov O, Chapman J, Fahey B, Gauthier MEA, Mitros T, Richards GS, Conaco C, Dacre M, Hellsten U, et al. 2010. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature 466: 720–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein LD, Bao Z, Blasiar D, Blumenthal T, Brent MR, Chen N, Chinwalla A, Clarke L, Clee C, Coghlan A, et al. 2003. The genome sequence of Caenorhabditis briggsae: A platform for comparative genomics. PLoS Biol 1: E45 doi: 10.1371/journal.pbio.0000045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tena JJ, Neto A, de la Calle-Mustienes E, Bras-Pereira C, Casares F, Gomez-Skarmeta JL 2007. Odd-skipped genes encode repressors that control kidney development. Dev Biol 301: 518–531 [DOI] [PubMed] [Google Scholar]
- Tena JJ, Alonso ME, de la Calle-Mustienes E, Splinter E, de Laat W, Manzanares M, Gómez-Skarmeta JL 2011. An evolutionarily conserved three-dimensional structure in the vertebrate Irx clusters facilitates enhancer sharing and co-regulation. Nat Commun 2: 310 doi: 10.1038/ncomms1301 [DOI] [PubMed] [Google Scholar]
- Thomas JH 2007. Rapid birth-death evolution specific to xenobiotic cytochrome p450 genes in vertebrates. PLoS Genet 3: e67 doi: 10.1371/journal.pgen.0030067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thor S, Thomas JB 1997. The Drosophila islet gene governs axon pathfinding and neurotransmitter identity. Neuron 18: 397–409 [DOI] [PubMed] [Google Scholar]
- Turner BM 2007. Defining an epigenetic code. Nat Cell Biol 9: 2–6 [DOI] [PubMed] [Google Scholar]
- Visel A, Minovitsky S, Dubchak I, Pennacchio LA 2007. VISTA Enhancer Browser–a database of tissue-specific human enhancers. Nucleic Acids Res 35: D88–D92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voutev R, Keating R, Hubbard EJ, Vallier LG 2009. Characterization of the Caenorhabditis elegans Islet LIM-homeodomain ortholog, lim-7. FEBS Lett 583: 456–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Zhong J, Su B, Zhou Y, Wang YQ 2007. Comparison of Pax1/9 locus reveals 500-myr-old syntenic block and evolutionary conserved noncoding regions. Mol Biol Evol 24: 784–791 [DOI] [PubMed] [Google Scholar]
- Wardle FC, Odom DT, Bell GW, Yuan B, Danford TW, Wiellette EL, Herbolsheimer E, Sive HL, Young RA, Smith JC 2006. Zebrafish promoter microarrays identify actively transcribed embryonic genes. Genome Biol 7: R71 doi: 10.1186/gb-2006-7-8-r71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber CC, Hurst LD 2011. Support for multiple classes of local expression clusters in Drosophila melanogaster, but no evidence for gene order conservation. Genome Biol 12: R23 doi: 1186/gb-2011-12-3-r23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams Nic A, Holland PWH 1996. Old head on young shoulders. Nature 383: 490 doi: 10.1038/383490a0 [Google Scholar]
- Woolfe A, Goodson M, Goode DK, Snell P, McEwen GK, Vavouri T, Smith SF, North P, Callaway H, Kelly K, et al. 2005. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol 3: e7 doi: 10.1371/journal.pbio.0030007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137 doi: 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]