Abstract
One clear hallmark of mammalian promoters is the presence of CpG islands (CGIs) at more than two-thirds of genes, whereas TATA boxes are only present at a minority of promoters. Using genome-wide approaches, we show that GC content and CGIs are major promoter elements in mammalian cells, able to govern open chromatin conformation and support paused transcription. First, we define three classes of promoters with distinct transcriptional directionality and pausing properties that correlate with their GC content. We further analyze the direct influence of GC content on nucleosome positioning and depletion and show that CpG content and CGI width correlate with nucleosome depletion both in vivo and in vitro. We also show that transcription is not essential for nucleosome exclusion but influences both a weak +1 and a well-positioned nucleosome at CGI borders. Altogether our data support the idea that CGIs have become an essential feature of promoter structure defining novel regulatory properties in mammals.
How the transcriptional machinery accesses gene promoters is a central question for understanding gene regulation. Because eukaryotic genomes are highly compacted, most of the DNA is not easily accessible to transcription factors, to the notable exception of promoters that tend to be more open chromatin structures. In recent years, with the development of genome-wide approaches, several studies in various organisms have described that nucleosomes are at least partially dependent on the primary sequence for their positioning (Kaplan et al 2009). In yeast and Drosophila, for example, AT-rich stretches at promoters exclude nucleosomes both in vivo and in vitro, although the correlation of both data sets has been discussed in the literature, as opposed to GC-rich regions, often found in gene bodies, which favor nucleosome occupancy (Kaplan et al. 2009, 2010; Zhang et al. 2009; Pugh 2010). In contrast, most mammalian promoters are enriched for GC-rich areas—also called CpG islands (CGIs)—whereas TATA boxes only appear in a minority of genes (Sandelin et al. 2007). CGIs are defined as large genomic areas of over-enriched CpG dinucleotides estimated to amount to between 20,000 and 30,000 in various mammalian genomes lacking counterparts in cold blooded organisms or other eukaryotes (Illingworth and Bird 2009; Sharif et al. 2010). Although recent works suggest a link between nucleosome depletion at promoters and the presence of CGIs, a direct correlation was never established (Li et al. 2011; Valouev et al. 2011). Furthermore, it remains to be determined whether nucleosome depletion is a cause or a consequence of promoter transcription because in vitro experimental approaches describe apparently contradictory results (Ramirez-Carrozzi et al. 2009; Valouev et al. 2011).
In this study, we report that GC content and CGIs are major promoter elements in mammals able to govern open chromatin conformation and support paused transcription genome-wide. First, we define three classes of promoters with distinct transcriptional directionality and pausing properties that correlate with their GC content. We further analyze the direct influence of GC content on nucleosome positioning and depletion, and show that CpG content and CGI width correlate with nucleosome depletion both in vivo and in vitro. We also show that transcription is not essential for nucleosome exclusion but influences both a weak +1 and a well-positioned nucleosome at CGI borders. Altogether our data support the idea that CGIs arose as an essential feature of promoter structure defining novel regulatory properties in mammals.
Results
Three classes of promoters with distinct pausing and bidirectional transcription properties
We previously showed that mammalian transcription initiation platforms (TIPs), recruiting large arrays of general transcription factors (GTFs) and initiating RNA polymerase (Pol) II, displayed high GC content at promoters (Koch et al. 2011). To further investigate transcription at promoters, we analyzed genome-wide Pol II accumulation by ChIP-seq, nucleosome density by micrococcal nuclease digestion (MNase-seq), and paused bidirectional transcripts associated with transcription start sites (TSS RNAs) (Seila et al. 2008) by strand-specific RNA-seq in mouse primary T-cells. We present two types of analyses of the MNase data that relate either to nucleosome density for quantitative assessment of apparent depletion areas or nucleosome midpoints for nucleosome positioning investigations.
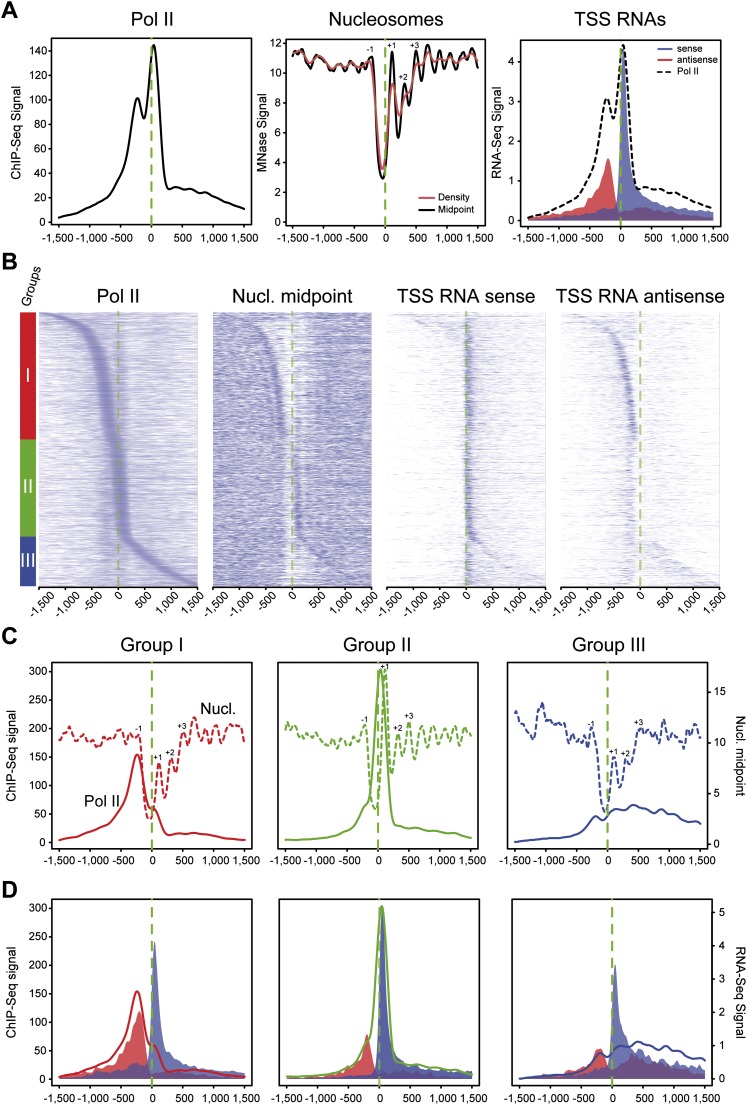
Average profiling on a selection of Pol II–containing promoters (see Supplemental Methods) showed strong overlap between upstream and downstream Pol II peaks around TSSs, borders of the −1/+1 nucleosomes, and antisense/sense TSS RNAs (Fig. 1A), as previously suggested (Core et al. 2008; Seila et al. 2008). Because average profiles may reflect a composite of very heterogeneous populations, we sorted active Pol II–containing promoters based on the position of the main peak/accumulation area from the 5′ to the 3′ of TSSs (Fig. 1B). This analysis revealed three main classes of promoters depending on the main Pol II signals: (I) upstream of, (II) around, and (III) downstream from the TSS. Furthermore, while sense TSS RNA was detected at most genes, antisense (AS) transcription was mainly restricted to class I, overlapping with Pol II and upstream nucleosome density—indicating a block of the transcriptional machinery. In each of the three classes within 3 kb around the TSS, we observed the main Pol II peak overlapping with (class I) or upstream of (class II and III) the center of the nucleosome (Fig. 1B,C). Investigation of corresponding profiles for initiating Pol II phosphorylated on Ser5 of its carboxy-terminal domain (Ser5P), as well as the TBP and TFIIB GTFs, indicated accumulation of the initiation complex in all three classes at the primary Pol II peaks (Supplemental Fig. 1A). It is worth noting that most TSS-proximal TATA boxes were found in class II promoters, whereas the upstream B recognition element (BREu) was enriched in all classes (Supplemental Fig. 1B). Furthermore, the average distance between Pol II and GTFs was relatively constant throughout classes and in agreement with previous structural studies (Kostrewa et al. 2009) and as recently described at various yeast promoters (Rhee and Pugh 2012). As observed in the clusters, all promoters showed paused unidirectional sense transcription around the TSS in average profiling, whereas bidirectional (sense and antisense) transcription was essentially observed in class I (Fig. 1D) and to a lesser extent in class III, which differed from class I, showing more diffuse products of paused transcription and less local accumulation. In class II, a relatively good overlap can be observed between Pol II peaks and TSS RNAs. In class I, however, a larger separation between Pol II and RNAs was measured, in the range of 15–30 nt. This raises the possibility that AS Pol II produces longer paused transcripts at the stage that they reach the nucleosome and suggests that AS transcriptional initiation does not harbor the same constraints as compared with sense transcription.
Figure 1.
Three groups of mammalian promoters are defined by Pol II occupancy and correlate with directionality of paused transcripts and nucleosome occupancy. (A) Average profiling of Pol II (left), nucleosomes (middle), and short directional TSS-RNAs (right) based on ChIP-seq, MNase-seq, and strand-specific RNA-seq, respectively. The analysis was performed on 1727 active promoters in mouse primary CD4+ CD8+ (DP) sorted T-cells (top 20% of Pol II signal distribution on selected promoters). The main aNDR is delimited by −1 and +1 nucleosomes around the TSS (dashed green line), approximately between −200 bp and +100 bp around the TSS. Nucleosomal midpoints (middle, black line) show a more marked periodicity than pure nucleosome occupancy (red) and also reveal strong +2, +3 nucleosomes. (B) Heat map of features described in A on promoters sorted by position of the main Pol II accumulation area (peak) from the most 5′ to the most 3′ around TSSs. Three main groups are defined by Pol II occupancy (left panel): class I most 5′ (red bar), class II TSS-proximal (green bar), and class III most 3′ (blue bar). The corresponding heat maps for nucleosome midpoints and short TSSs RNA heat maps are indicated. (C) Pol II (solid line) and nucleosome midpoint profiling (dashed lines) of the three groups defined in B. (D) Profiling of short sense (blue) and antisense (red) TSS-RNAs from the groups defined in B. Pol II corresponding profiles are indicated as in C.
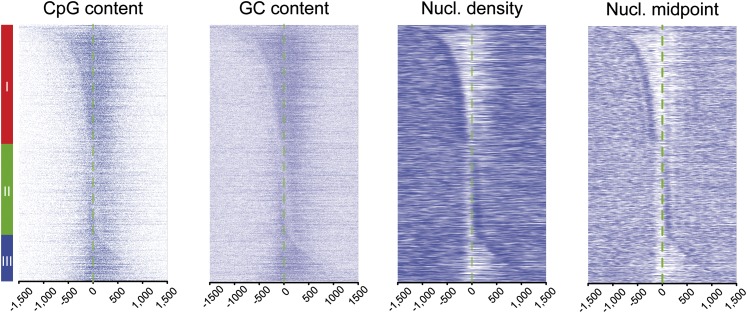
To further characterize the influence of the sequence context in the properties of paused transcription, we next analyzed our promoter groups for their GC and CpG dinucleotide content. We observed classes I and III to display a higher and more spread GC/CpG content as compared with class II (Fig. 2; Supplemental Fig. 1C). Both features as well as the average CGI length were most prominent in class I promoters, possibly accounting for differences observed for paused transcription. Conversely, class II displayed an increased AT content, peaking at approximately −25 bp, the theoretical location of the TATA box. There was also a striking correlation between the nucleosome/Pol II favored position and the boundaries of GC enrichment as well as between apparent nucleosome-depleted regions (aNDR) and the level of GC content. Finally, by assessing a score for transcriptional pausing (Supplemental Fig. 2A–C; Supplemental Methods), we confirmed that classes I and II, unlike class III, showed medium to high pausing levels (Supplemental Fig. 2D). Overall, these data indicate three classes of promoters: (1) paused, GC-rich, and bidirectional with the main Pol II peak upstream of the TSS; (2) paused, GC-poor, and monodirectional with the main Pol II peak at the TSS; and (3) less paused, GC-rich, and diffuse bidirectional with the main Pol II peak downstream from the TSS. In each case, the GC content of the promoters appears to dictate the best nucleosomal position.
Figure 2.
Nucleosome position at the borders of CpG-rich areas in the three classes of Pol II–bound promoters. For the three classes of promoters defined in Figure 1, CpG and GC content are displayed side by side with nucleosome midpoints and densities. A more complete analysis of this cluster is also presented in Supplemental Figure 1.
GC content and CGIs define nucleosome depletion patterns at mammalian promoters
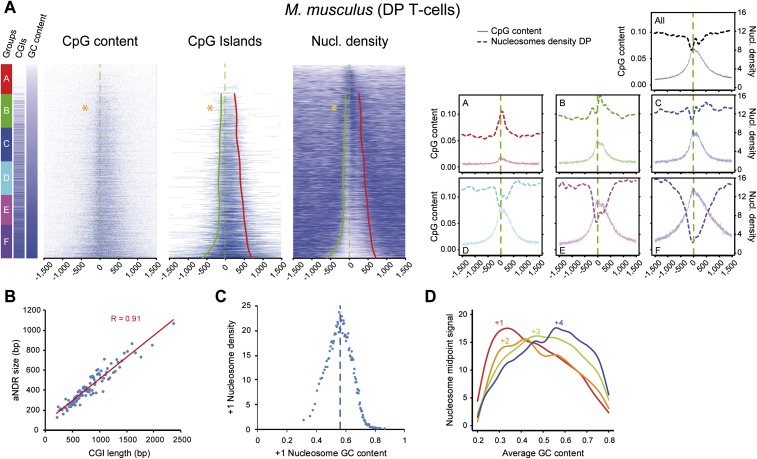
We therefore wondered if there was a direct influence of GC content, including the presence and widths of associated CGIs, on promoter organization and nucleosome positioning. We ranked all promoters according to increasing GC content and established sextiles of genes (Fig. 3A). In the first two AT-rich groups, with a low amount of CGIs (A and B of which 4% and 34% contain CGIs), the +1 and overall nucleosome densities increased with GC/CpG content (defined in Supplemental Methods), as described for promoters of eukaryotes with no CGIs such as yeast (Kaplan et al. 2009). In contrast, within the four remaining groups of promoters (C–F) with high CGI promoter occurrence (55%–93%), increasing GC/CpG +1 nucleosome density decreased, whereas aNDRs increased. Strikingly, the length of the CGIs within the CGI-containing promoters directly correlated with that of aNDRs (Fig. 3B) in these groups, and nucleosome density anticorrelated the GC content around the +1 position beyond a 0.58 threshold (Fig. 3C; indicated by stars in Fig. 3A) that is located between groups B and C. We also validated these trends in mouse embryonic stem (ES) cells and human B-cells and T-cells (Supplemental Fig. 3A,C). To exclude a possible MNase digestion bias or sequencing artifact within CGIs, we performed additional controls such as ChIP-seq with histone H3 in mouse T-cells (Supplemental Fig. 3A) and nucleosome mapping using an alternative method based on DNA digestion by a chemical agent (phenanthroline) in human Raji B-cells (Supplemental Fig. 3C; Tsang et al. 1996). In both cases, we confirmed a correlation between nucleosome depletion and CGI length; we also sequenced sonicated genomic DNA that showed only very little sequencing bias in the promoters containing the largest CpG content (Supplemental Fig. 3A). Finally, the position of the major nucleosome in each promoter varied depending on increasing GC content and its average position was shifted from the +1 to the +2 and +3 (Fig. 3D). A similar analysis was conducted using only those promoters containing CGIs or in orphan intergenic CGIs and control random intergenic regions that confirmed correlation of CpG/GC content with aNDR in all promoters and orphan CGIs (data not shown).
Figure 3.
GC content at promoters conditions +1 nucleosome strength and correlates with aNDRs in GC-rich areas. (A) Heat maps of CpG dinucleotides, CpG islands, and nucleosome density, ordered by increasing GC content, in murine DP T-cells at all 8634 promoters (excluding those in close proximity to other annotations) and centered on the TSSs. On the left of the heat maps, the six groups A–F (red to violet boxes) of increasing GC content are represented along with the presence of CGIs in each group (blue bar represent CGIs, and white bars non-CGI promoter). The percentages of CGI promoters in each group from top to bottom are 4%, 34%, 55%, 65%, 78%, and 93%. Therefore, the first two groups can be classified as CGI-poor. The profiles of CpG content and nucleosome densities for all promoters (top) or the six subgroups A–F are shown on the right. The red and green lines on the CGIs and nucleosome density heat maps show the trend lines of the CGIs borders (see Methods) starting from group B because group A does not contain a significant amount of CGIs. (Orange stars) Transition point from which +1 nucleosome densities switch from a correlation to an anti-correlation pattern with GC content. (B) Correlation of CpG peak length and aNDR size within CGI-rich groups. A linear regression of CGI length versus nucleosome occupancy, divided into 80 classes, shows a high correlation (R = 0.91). (C) GC content correlate (in GC-low areas) and anti-correlate in GC-rich areas with MNase signal at +1 nucleosome proximity. Promoters were divided into 80 equal groups of increasing +1 GC content and plotted against MNase signal. Nucleosome density increases and then decreases after a 0.58 GC content threshold. (D) Transitions in the major nucleosome occupancy from position +1 to +4 with increasing GC content. Occupancy levels for +1 to +4 nucleosomes were measured around the midpoint position calculated on all promoters.
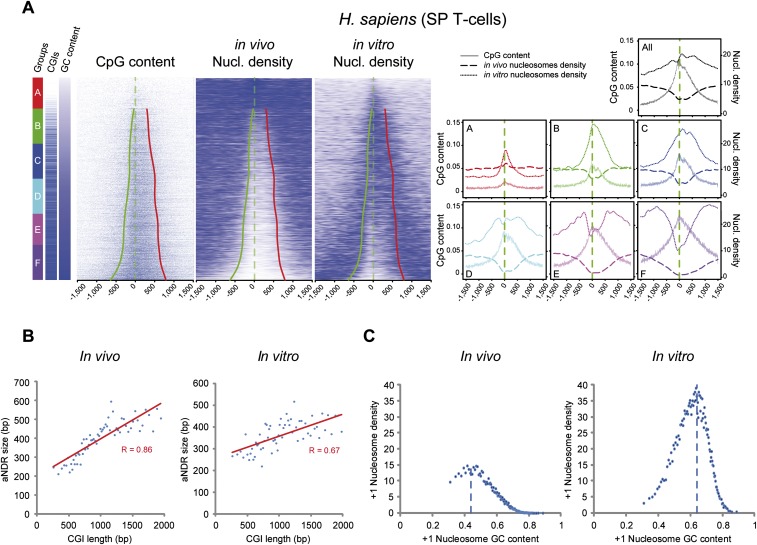
A recent study compared positioning of human nucleosomes in vitro and in vivo. Interestingly, it was suggested that nucleosomes in vivo are overall depleted within CGIs, whereas nucleosomes in vitro tend to prefer these areas for positioning (Valouev et al. 2011). Although the in vitro trend, indeed, shows more nucleosome preference at higher GC content, a more detailed investigation of these data, ranking promoters by increasing GC content, indicates a profile closer to the in vivo situation within the largest CGIs (Fig. 4A). Indeed, nucleosome density shows a similar trend as described above: first increasing nucleosome density with GC content in the three lowest groups (A–C) of GC/CpGs density (Fig. 4A), and then an inverted trend with elevated nucleosome exclusion as CGI length increases (groups D–F, with 65%–93% of CGIs containing promoters, respectively). As before, we observed a strong correlation between CGI and aNDR width (Fig. 4B). Thus, although the inversion of correlation trend is occurring at higher GC content, in vitro and in vivo nucleosomes show similar trends (Fig. 4A,C). The reasons why the trends are shifted might relate to many parameters, including the experimental conditions for nucleosome reconstitution on genomic DNA (average size of fragmented DNA, ratio of nucleosomes to DNA, ionic strength, etc.) or the absence of specific additional factors naturally associated with high GC content or CGI promoters in vivo. Furthermore, investigating an independent in vivo data set in the same CD4 T-cells (Schones et al. 2008), and allowing for a better nucleosome positioning analysis, indicated a much closer correspondence of the in vivo and in vitro data sets (Supplemental Fig. 4).
Figure 4.
In vivo and in vitro human nucleosomes show a similar inversion of correlation trends. (A) Comparison of in vivo– and in vitro–reconstituted nucleosomes in human T-cells (CD4+ or SP) or using T-cell-derived genomic DNA and presented as in Figure 3A at all promoters (7021 regions). Whereas in vivo the aNDR appears in subgroup B, it appears in group D in vitro. (B) Correlation of CpG peak length and aNDR size in vitro and in vivo within CGI-rich groups as in Figure 3B. (C) GC content and MNase signal at +1 nucleosome proximity (as in Fig. 3C) from data presented in A. A similar trend of inversion from correlation to anti-correlation is observed between +1 nucleosome density and GC content but with different GC content thresholds.
Altogether, our analyses show a strong correlation between CGI width and aNDR, on the one hand, and between the major nucleosome position and the boundaries of CGIs at promoters, on the other.
Nucleosome depletion at CGIs promoters is essentially transcription-independent
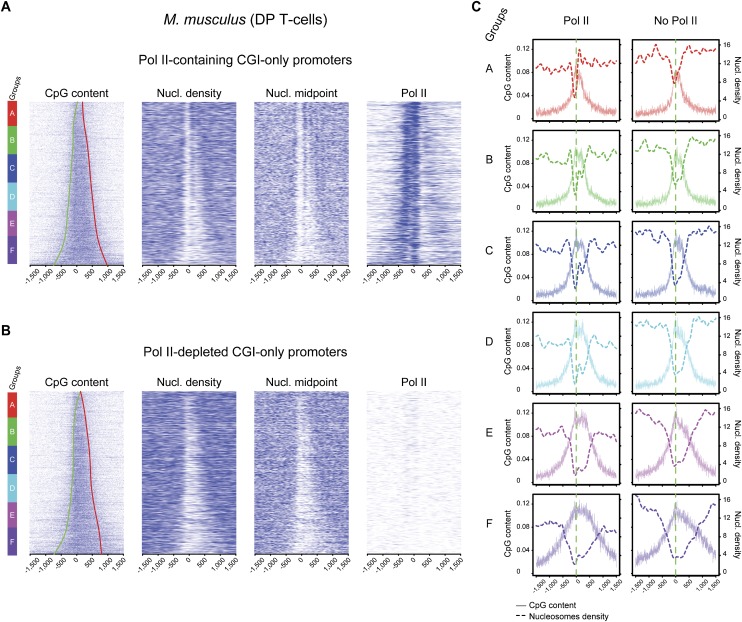
Several studies suggested that Pol II might play a role in the maintenance of aNDRs at promoters in various species, although this point was subject to discussion (Gilchrist et al. 2010; Weiner et al. 2010; Valouev et al. 2011; Zhang et al. 2011). To investigate this phenomenon in the light of CpG content at promoters, we selected two groups of promoters with or without Pol II and ranked them based on GC content (Supplemental Fig. 5). As previously described, we observed that Pol II–containing promoters are over-enriched (>80%) in CGIs (Rozenberg et al. 2008), whereas the Pol II–depleted ones are less enriched (33%). However, when comparing the CGIs promoters directly in both categories, we observed the aNDR lengths to increase similarly with that of CGIs in both cases, suggesting a transcription-independent mechanism. In the Pol II–depleted group of promoters, we observed enrichment for the H3K27me3 epigenetic mark in the CGI area (Supplemental Fig. 5C). This mark depends on the Polycomb group (PcG) complex, previously described to associate with CGIs and hence consistent with our observations (Mikkelsen et al. 2007; Ku et al. 2008; Mendenhall et al. 2010). We also performed a correlation analysis between CGI length or GC content and aNDR length as in Figure 2B, and found similar results for Pol II–containing and Pol II–depleted promoters (Supplemental Fig. 5A,B). To strengthen this observation and perform a more direct comparison, we isolated a similar selection of Pol II–containing and Pol II–depleted only within CGI promoters, based on UCSC annotations (Fig. 5). In both promoter sets, a direct correlation of CGI length with aNDR can be observed. We also note an enhanced positional effect of nucleosomes in Pol II–containing promoters as compared with the overall promoter set from Figure 2 or to the ones with no detectable levels of Pol II (Figs. 5, 6; Supplemental Fig. 5A,B). This observation is supported by nucleosome midpoint analyses that allow a better assessment of nucleosome positions (Fig. 6), whereas nucleosome density is used for aNDR length correlations. Finally, we also analyzed promoters without CGI annotations (AT-rich) that show both an increase of +1 nucleosome and overall promoter nucleosome density when GC content increases, irrespective of the presence of Pol II (Supplemental Fig. 6). This situation reminds us of what was previously described in eukaryotes with AT-rich promoters such as yeast. Taken together, our results suggest that aNDRs are not depending on Pol II at promoters, whereas nucleosome positioning at CGI boundaries is constrained by initiating transcription.
Figure 5.
Pol II occupancy influences nucleosome positioning but not nucleosome depletion at CGI promoters. Heat maps of CpG content, nucleosome density, and nucleosome midpoints ordered by increasing GC content are shown for 2590 Pol II–containing (A) and 2590 Pol II–depleted (B) CGI promoters only. CGI border trend lines are shown on the CpG heat maps as for Figure 3A. (C) Corresponding average profiles of subgroups A–F (gene sextiles from lowest to highest GC content) for nucleosome densities and CpG dinucleotide scores are shown.
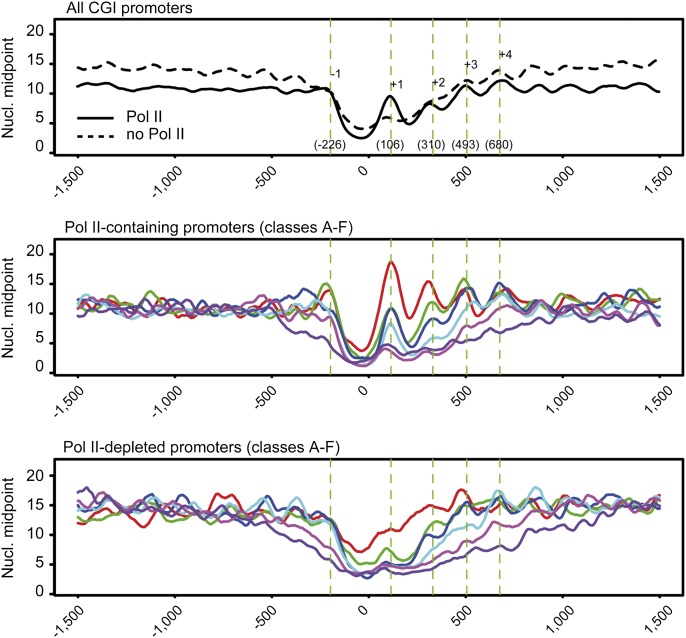
Figure 6.
Enhanced nucleosome positioning in Pol II–containing CGI promoters. Average positions of the −1 to +4 nucleosomes midpoint shown in Figure 5, A and B, are indicated on the top panel (all CGI promoters). Gene group colors A–F in the middle and bottom panels correspond to sextiles of increasing GC content. For Pol II–containing promoters, a clearer nucleosome periodicity as well as positioning is visible when compared with Pol II–depleted promoters.
To further investigate the importance of paused transcription on nucleosome density and organization at promoters, we inhibited transcription in a human B-cell line (Raji) using α-amanitin. This compound binds the active site of the enzyme, blocking transcription, which finally results in Pol II degradation (Nguyen et al. 1996). We performed a kinetic experiment that showed a disappearance of Pol II between 24 h and 36 h after treatment (Fig. 7A). When examining the corresponding genome-wide MNase-seq experiments, we observed that overall nucleosome density slightly increased at 24 h and 36 h at most promoters, and thus chromatin opening became reduced (Fig. 7B). This effect was more pronounced at the +1 nucleosome level as we found a 50% increase of the apparent +1 nucleosome density after stripping most Pol II from promoters. However, we do not observe aNDR clearance even at the latest time point for Pol II–bound promoters (Fig. 7B,C). This effect is neither observed for a selection of highly paused promoters (Supplemental Figs. 7, 8), indicating that paused Pol II transcription is not the cause of nucleosome exclusion in CGIs, although a slight reduction of the depleted area is visible after α-amanitin treatment. Interestingly, however, this reduction is getting more pronounced for groups with high GC content, although the aNDR remain clearly present in the absence of Pol II (Fig. 7D). Overall, our results are consistent with the presence of aNDRs in the absence of Pol II (Fig. 5) as well as with our analysis of in vitro–reconstituted nucleosomes, and plead for CGI-dependent promoter opening being a cause rather than a consequence of paused transcription.
Figure 7.
Pol II disruption in vivo reduces aNDRs only moderately and results in an increase of the +1 nucleosome. (A) To investigate the effect of Pol II presence on aNDRs, we stripped Pol II from the chromatin by treating the human Raji B-cell line with α-amanitin for the indicated time points. Western blot samples were analyzed at 12 h, 18 h, 24 h, 36 h, and 48 h and showed Pol II disappearance in the latter two points. Tubulin (lower panel) was used as an internal control to check equal protein loading. At t0, t24, and t36, Pol II ChIP-seq and MNase-seq experiments were performed as indicated. (B) Genome-wide analysis of nucleosome occupancy and Pol II recruitment following treatment at 1504 Pol II–enriched promoters (top 20%). As expected, Pol II signals around the TSS decreased over time (left). Nucleosome density slightly increased downstream from the TSS, indicating narrowing of the effective aNDR area (right) and an increase of the +1 nucleosome. (C) Examples showing the difference of Pol II occupation and nucleosome density across the TRIM4 and PPIA genes. ChIP-seq and MNase-seq data were scaled such that the represented units correspond to an equivalent amount of sequenced tags. (Arrow) Position of the +1 nucleosome. (D) Effect of GC content on aNDRs after Pol II removal. Three groups of GC content were built for the group of Pol II–enriched promoters shown in B. CpG scores, nucleosome densities (bottom panels), and Pol II ChIP-seq signal (top panels), before and after treatment are displayed. A stronger aNDR reduction, but not disruption, is observed for the group with the highest GC (and CpG) content.
Discussion
Previous studies already suggested that CGIs influence a lower nucleosome density at promoters (Schones et al. 2008; Ramirez-Carrozzi et al. 2009; Choi 2010; Li et al. 2011; Valouev et al. 2011). Our work adds several lines of evidence that these genomic structures directly correlate with the level of nucleosome depletion at promoters and that GC content is the prominent, if not the most essential mark for maintenance of opened chromatin structures. The positioning of the +1 nucleosome in mouse promoters increases with GC content at thresholds lower than 0.5–0.6 (0.4–0.6 in humans in vivo and 0.6–0.7 in vitro). This class resembles yeast or mammalian AT-rich promoters that might depend on SWI/SNIF-related complexes to displace nucleosomes, allowing Pol II to enter elongation (Ramirez-Carrozzi et al. 2009; Zhang et al. 2011). In CGI-containing promoters, Pol II is able to transcribe most likely in a paused conformation within larger areas. Our analyses reveal that these promoters can either be paused (and mostly bidirectional) or not. Pol II pausing within large GC-rich regions should result in high energy requirements, resulting from triple hydrogen bonding and base stacking (Yakovchuk et al. 2006) and explaining why these regions are more prone to paused rather than to elongating transcription. It is also plausible that many GC-rich promoters contain secondary structures such as G-quadruplexes (Lipps and Rhodes 2009) that could disfavor the assembly into nucleosomal structures, resulting in the observed exclusion at these promoter regions. Interestingly, a recent work indicates that certain yeast species harbor promoters with poly(G) tracks that also display anti-nucleosomal properties, although CGIs are not present per se in these genomes (Tsankov et al. 2011).
It has been debated whether paused promoter transcription represents a cause or a consequence of open chromatin structures (Seila et al. 2009). Our experiments support the latter possibility: Open CGI/GC-rich promoters would naturally exclude nucleosomes and allow PIC formation unless specific repressive mechanisms, such as the one mediated by PcG, forbid or restrict Pol II recruitment and transcription. Experiments with the group of promoters containing high GC content in human cells suggest a moderate effect of Pol II for enlargement of the aNDR. Indeed, the widths of aNDRs were reduced 8%, 10%, and 20% in the three GC content classes following α-amanitin treatment and Pol II removal. Thus, we conclude that Pol II is not responsible for aNDR formation but plays a role in both enlargement of this area and precise nucleosome positioning at promoters. We propose that in the absence of transcription, nucleosomes tend to position closer to the border of GC-rich areas, whereas initiating Pol II represents a motor force that will push them further away up to a sequence position where they will stabilize more easily.
Our various analyses with Pol II ChIP-seq profiling suggest that >50% promoters harbor detectable Pol II levels (data not shown). It is conceivable that the remaining promoters are also transcribed at a rate that falls below the experimental detection range. CGIs, however, seem to disfavor naturally nucleosome positioning, even in the absence of Pol II. Several factors were previously described to be recruited in CGIs, resulting in inclusion or exclusion of specific epigenetic marks from promoters (Blackledge et al. 2010; Thomson et al. 2010). It will be of interest to investigate their connection with initiating Pol II and pausing at active TSSs. We believe that our work will open avenues for a better understanding of mammalian promoter elements that govern many aspects of complex gene regulation.
Methods
A detailed description of methods is provided in the Supplemental Material.
Cell culture and sorting
Mouse CD4+ CD8+ DP cells were sorted from thymuses of 5- to 6-wk-old mice. Raji cells were grown in RPMI 1640 medium supplemented with 10% fetal calf serum, 100 units/mL penicillin, 100 mg/mL streptomycin, and 2 mM L-glutamine at 37°C and 5% CO2. For the α-amanitin experiment, cells were treated at 2.5 μg/mL for the indicated times. Murine ES cells (Bl6) were grown as described (Boyer et al. 2006) on a layer of PMEF in the presence of leukemia inhibitory factor on gelatinized plates. Cells were trypsynized and separated from feeders prior to cross-linking for ChIP.
ChIP-seq and MNase-seq
ChIPs were essentially carried out as described (Koch et al. 2011). All antibodies and ChIP conditions are described in Supplemental Table 1. MNase nucleosome digestion of permeabilized cells was optimized to obtain a majority of mononucleosomes (70%–80% as estimated on a Bioanalyzer DNA chip). The corresponding DNA fraction was selected after library preparation. The sequencing procedure was conducted using at least 1 ng of starting material and run on a Genome Analyzer II (Illumina). The in vivo phenanthroline digestion of nucleosomes was adapted from Tsang et al. (1996) and is described in the Supplemental Methods together with the computational processing and analysis pipeline.
Strand-specific TSS RNA-seq
We isolated a 15-nt to 70-nt population from TRIzol-extracted RNA using a denaturing urea polyacrylamide gel. The isolated RNA was processed using the Illumina small RNA kit with modifications (see Supplemental Methods). The resulting complementary DNA (cDNA) was sequenced on an Illumina Genome Analyzer II, using the small RNA sequencing kit (see Supplemental Methods).
Downloaded published data
The in vivo– and in vitro–reconstituted MNase-seq experiments in human CD4+ T-cells (GEO accession no. GSE25133) (Valouev et al. 2011) were downloaded to complement those presented here and our previously published work (GEO accession no. GSE29362) (Koch et al. 2011).
Bioinformatic processing and analyses
The pre-treatment of raw sequence data was essentially carried out as previously described (Koch et al. 2011). For TSS RNA-seq, we used the MIRO pipeline (http://seq.crg.es/main/bin/view/Home/MiroPipeline) to detect and remove adapter sequences before alignment using the Genome Multi-tool (GEM; http://gemlibrary.sourceforge.net).
Data access
ChIP-seq, RNA-seq, and MNase-seq data used in this study have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE38577.
Acknowledgments
Work in the P.F. laboratory is supported by institutional grants from Institut National de la Santé et de la Recherche Médicale (INSERM) and the Centre National de la Recherche Scientifique (CNRS), and by specific grants from the Fondation Princesse Grace de Monaco, the Agence Nationale de la Recherche (ANR), the Institut National du Cancer (INCa), and the Commission of the European Communities. R.F. was supported by Genopole and CNRS, P.C. by grants from INCa and Fondation pour la Recherche Médicale, and F.K. by grants from the Chromatin Plasticity Marie Curie Research Training Network, Association pour la Recherche sur le Cancer (ARC), and ANR. The work was also granted for sequencing costs by an ESGI Consortium grant of the EU. We are grateful to J. Blanc for technical help, as well as M. Ingham and O. Fernando (CNAG) for sequencing and bioinformatics assistance. We also thank G. Almouzni for suggesting the phenanthroline nucleosome mapping as an additional control for our experiments.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.138776.112.
References
- Blackledge NP, Zhou JC, Tolstorukov MY, Farcas AM, Park PJ, Klose RJ 2010. CpG islands recruit a histone H3 lysine 36 demethylase. Mol Cell 38: 179–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441: 349–353 [DOI] [PubMed] [Google Scholar]
- Choi JK 2010. Contrasting chromatin organization of CpG islands and exons in the human genome. Genome Biol 11: R70 doi: 10.1186/gb-2010-11-7-r70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT 2008. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322: 1845–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DA, Dos Santos G, Fargo DC, Xie B, Gao Y, Li L, Adelman K 2010. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell 143: 540–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illingworth RS, Bird AP 2009. CpG islands–‘a rough guide.’ FEBS Lett 583: 1713–1720 [DOI] [PubMed] [Google Scholar]
- Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, et al. 2009. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458: 362–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, Moore I, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, Hughes TR, Lieb JD, Widom J, Segal E 2010. Nucleosome sequence preferences influence in vivo nucleosome organization. Nat Struct Mol Biol 17: 918–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch F, Fenouil R, Gut M, Cauchy P, Albert TK, Zacarias-Cabeza J, Spicuglia S, de la Chapelle AL, Heidemann M, Hintermair C, et al. 2011. Transcription initiation platforms and GTF recruitment at tissue-specific enhancers and promoters. Nat Struct Mol Biol 18: 956–963 [DOI] [PubMed] [Google Scholar]
- Kostrewa D, Zeller ME, Armache KJ, Seizl M, Leike K, Thomm M, Cramer P 2009. RNA polymerase II–TFIIB structure and mechanism of transcription initiation. Nature 462: 323–330 [DOI] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. 2008. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet 4: e1000242 doi: 10.1371/journal.pgen.1000242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Schug J, Tuteja G, White P, Kaestner KH 2011. The nucleosome map of the mammalian liver. Nat Struct Mol Biol 18: 742–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipps HJ, Rhodes D 2009. G-quadruplex structures: In vivo evidence and function. Trends Cell Biol 19: 414–422 [DOI] [PubMed] [Google Scholar]
- Mendenhall EM, Koche RP, Truong T, Zhou VW, Issac B, Chi AS, Ku M, Bernstein BE 2010. GC-rich sequence elements recruit PRC2 in mammalian ES cells. PLoS Genet 6: e1001244 doi: 10.1371/journal.pgen.1001244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen VT, Giannoni F, Dubois MF, Seo SJ, Vigneron M, Kedinger C, Bensaude O 1996. In vivo degradation of RNA polymerase II largest subunit triggered by alpha-amanitin. Nucleic Acids Res 24: 2924–2929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugh BF 2010. A preoccupied position on nucleosomes. Nat Struct Mol Biol 17: 923. [DOI] [PubMed] [Google Scholar]
- Ramirez-Carrozzi VR, Braas D, Bhatt DM, Cheng CS, Hong C, Doty KR, Black JC, Hoffmann A, Carey M, Smale ST 2009. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell 138: 114–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee HS, Pugh BF 2012. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature 483: 295–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenberg JM, Shlyakhtenko A, Glass K, Rishi V, Myakishev MV, FitzGerald PC, Vinson C 2008. All and only CpG containing sequences are enriched in promoters abundantly bound by RNA polymerase II in multiple tissues. BMC Genomics 9: 67 doi: 10.1186/1471-2164-9-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandelin A, Carninci P, Lenhard B, Ponjavic J, Hayashizaki Y, Hume DA 2007. Mammalian RNA polymerase II core promoters: Insights from genome-wide studies. Nat Rev Genet 8: 424–436 [DOI] [PubMed] [Google Scholar]
- Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K 2008. Dynamic regulation of nucleosome positioning in the human genome. Cell 132: 887–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA 2008. Divergent transcription from active promoters. Science 322: 1849–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seila AC, Core LJ, Lis JT, Sharp PA 2009. Divergent transcription: A new feature of active promoters. Cell Cycle 8: 2557–2564 [DOI] [PubMed] [Google Scholar]
- Sharif J, Endo TA, Toyoda T, Koseki H 2010. Divergence of CpG island promoters: A consequence or cause of evolution? Dev Growth Differ 52: 545–554 [DOI] [PubMed] [Google Scholar]
- Thomson JP, Skene PJ, Selfridge J, Clouaire T, Guy J, Webb S, Kerr AR, Deaton A, Andrews R, James KD, et al. 2010. CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature 464: 1082–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang SY, Tam SC, Bremner I, Burkitt MJ 1996. Research communication copper-1,10-phenanthroline induces internucleosomal DNA fragmentation in HepG2 cells, resulting from direct oxidation by the hydroxyl radical. Biochem J 317: 13–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsankov A, Yanagisawa Y, Rhind N, Regev A, Rando OJ 2011. Evolutionary divergence of intrinsic and trans-regulated nucleosome positioning sequences reveals plastic rules for chromatin organization. Genome Res 21: 1851–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valouev A, Johnson SM, Boyd SD, Smith CL, Fire AZ, Sidow A 2011. Determinants of nucleosome organization in primary human cells. Nature 474: 516–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner A, Hughes A, Yassour M, Rando OJ, Friedman N 2010. High-resolution nucleosome mapping reveals transcription-dependent promoter packaging. Genome Res 20: 90–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakovchuk P, Protozanova E, Frank-Kamenetskii MD 2006. Base-stacking and base-pairing contributions into thermal stability of the DNA double helix. Nucleic Acids Res 34: 564–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Moqtaderi Z, Rattner BP, Euskirchen G, Snyder M, Kadonaga JT, Liu XS, Struhl K 2009. Intrinsic histone–DNA interactions are not the major determinant of nucleosome positions in vivo. Nat Struct Mol Biol 16: 847–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Wippo CJ, Wal M, Ward E, Korber P, Pugh BF 2011. A packing mechanism for nucleosome organization reconstituted across a eukaryotic genome. Science 332: 977–980 [DOI] [PMC free article] [PubMed] [Google Scholar]