Abstract
Whole genome duplication (WGD) has made a significant contribution to many eukaryotic genomes including yeast, plants, and vertebrates. Following WGD, some ohnologs (WGD paralogs) remain in the genome arranged in blocks of conserved gene order and content (paralogons). However, the most common outcome is loss of one of the ohnolog pair. It is unclear what factors, if any, govern gene loss from paralogons. Recent studies have reported physical clustering (genetic linkage) of functionally linked (interacting) genes in the human genome and propose a biological significance for the clustering of interacting genes such as coexpression or preservation of epistatic interactions. Here we conduct a novel test of a hypothesis that functionally linked genes in the same paralogon are preferentially retained in cis after WGD. We compare the number of protein–protein interactions (PPIs) between linked singletons within a paralogon (defined as cis-PPIs) with that of PPIs between singletons across paralogon pairs (defined as trans-PPIs). We find that paralogons in which the number of cis-PPIs is greater than that of trans-PPIs are significantly enriched in human and yeast. The trend is similar in plants, but it is difficult to assess statistical significance due to multiple, overlapping WGD events. Interestingly, human singletons participating in cis-PPIs tend to be classified into “response to stimulus.” We uncover strong evidence of biased gene loss after WGD, which further supports the hypothesis of biologically significant gene clusters in eukaryotic genomes. These observations give us new insight for understanding the evolution of genome structure and of protein interaction networks.
Well before genome sequences were available to test the hypothesis, Ohno proposed that two rounds (2R) of whole genome duplication (WGD) occurred in early vertebrate evolution (Ohno 1970). Ultimately, analysis of complete genome sequences verified the 2R hypothesis but did not reveal perfectly symmetric duplicate chromosomes. Instead, several studies uncovered complex fossils of the ancient genome duplication events where only some genes remained duplicated (termed “ohnologs”) (Wolfe 2000), and even these groups of duplicated genes had been broken up into “paralogons” by extensive genome rearrangements (Popovici et al. 2001; McLysaght et al. 2002; Panopoulou et al. 2003; Vandepoele et al. 2004; Dehal and Boore 2005; Nakatani et al. 2007; Putnam et al. 2008). The existence of biased gene loss following WGD due to structural or functional constraints is still considered an open question (Jaillon et al. 2009). Here we consider how functional interactions between genes may influence the patterns of gene loss following WGD.
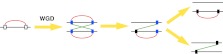
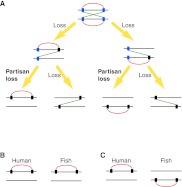
Large-scale linkage conservation between distantly related species has been shown by comparative analyses of vertebrate and invertebrate genomes (Putnam et al. 2007, 2008); however, the biological significance, if any, is unclear. Many functional gene clusters exist in the human genome (Popovici et al. 2001; Hurst et al. 2004; Makino and McLysaght 2008), and some of these, such as the HOX clusters, exist within paralogons (Popovici et al. 2001). We previously showed that interacting gene clusters in the human genome are more numerous than expected and have been conserved in vertebrate genomes more frequently than expected, indicating a functional role for clustering on the chromosome (Makino and McLysaght 2008). If we translate this observation to paralogons, we can consider the patterns of gene loss and test for preferential retention of interacting gene pairs in cis rather than in trans (Fig. 1). Following WGD, all interacting gene clusters will be perfectly duplicated, resulting in exactly equal numbers of cis- and trans-PPIs (protein–protein interactions). For interacting gene pairs that eventually revert to single-copy, the first gene loss is considered to be neutral if all losses are functionally equivalent. However, the second loss will result in either retention of a cis-PPI or of a trans-PPI. If there is no biological significance of the cis positioning of interacting genes, then this is a neutral “choice,” and each scenario should occur with equal frequency. However, if the relative proximity of interacting genes on the genome has biological relevance, then we expect to see non-random gene loss favoring the retention of the cis-PPI.
Figure 1.
Gene losses after whole genome duplication (WGD). Rectangles and horizontal lines represent genes and chromosomes, respectively. Red and green lines indicate cis- and trans-protein-protein interactions (PPIs) between proteins encoded by singletons or extant-paired ohnologs, respectively. (White rectangles) Genes prior to WGD. Blue and black rectangles show extant-paired ohnologs and singletons, respectively. Following WGD, all interacting gene clusters will be perfectly duplicated, resulting in exactly equal numbers of cis- and trans-PPIs. The first gene loss can occur at any locus. Gene loss that reverts the second gene to single copy will result in either retention of a cis- or a trans-PPI. If gene loss is neutral, then both scenarios should occur with equal frequency.
Genome duplication has also been detected in other eukaryotic lineages including yeast (Wolfe and Shields 1997; Dietrich et al. 2004; Dujon et al. 2004; Kellis et al. 2004) and plants (Arabidopsis Genome Initiative 2000; Blanc et al. 2000). Additionally, there is evidence for interacting gene clusters in the yeast genome (Teichmann and Veitia 2004; Poyatos and Hurst 2006).
Here we define protein–protein interactions (PPIs) between genes on the same “side” of a paralogon as cis-PPIs, and PPIs between genes across a paralogon pair as trans-PPIs (Fig. 1, red and green lines, respectively). Although the number of cis-PPIs must have been the same as that of trans-PPIs in a paralogon immediately after WGD, many of these interactions have been removed by gene losses during evolution. We test whether the number of cis-PPIs is greater than that of trans-PPIs in paralogons in human, yeast, and Arabidopsis.
Results and Discussion
Preferential retention of cis-interacting gene pairs in paralogons following WGD and gene loss
We identified 725 paralogon pairs in the human genome based on extant-paired ohnologs, 373 paralogon pairs in yeast based on gene order in the pre-WGD species Kluyveromyces lactis and 253 paralogon pairs in Arabidopsis derived from conserved gene synteny in the plant genome duplication database (http://chibba.agtec.uga.edu/duplication/). We confirmed that the gene content of combined human paralogons is representative of the gene content of at least the ancestral amniote by synteny conservation with chicken (see Methods). We obtained human, yeast, and plant PPI data sets from the Human Protein Reference Database (HPRD), BioGRID, and the Arabidopsis thaliana protein interaction network (AtPIN), respectively. We excluded paralogons in which no genes had any annotated PPIs from this study. We classified interactions between genes in a paralogon into cis- and trans-PPIs (Fig. 1; red and green lines, respectively).
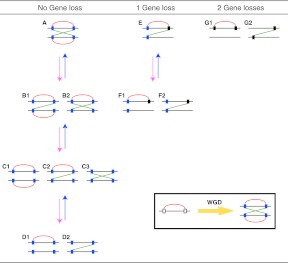
We considered all possible scenarios of retained PPIs in a paralogon after gene loss and/or protein interaction network (PIN) rewiring (Fig. 2). Through whole genome duplication, a single cis-PPI between neighboring genes in a genome would be increased to two cis- and two trans-PPIs (Fig. 2; box insert). After gene and PPI loss events, there are 13 possible PPI scenarios retaining at least one PPI (Fig. 2 A–G). To explore the properties of gene loss, we focused on PPIs among singletons within a paralogon because these have experienced gene loss events (scenario G in Fig. 2). Note that we are using the term “singleton” only to refer to the gene's duplication status within the paralogon, where it once had an ohnolog copy. It is possible that these genes do have other paralogs in the genome and thus are not strictly singletons in the conventional sense of the term.
Figure 2.
Retained network patterns in a paired gene cluster after gene and/or PPI losses. Rectangles and horizontal lines represent genes and chromosomes, respectively. Red and green lines indicate cis- and trans-PPIs, respectively. (White rectangles) Genes before they experienced WGD. Blue and black rectangles show extant-paired ohnologs and singletons, respectively. Pink and blue arrows indicate PPI losses and gains during evolution, respectively. All possible scenarios that retain at least one PPI are shown.
For our analysis, we used 668, 308, and 172 paralogon pairs in human, yeast, and plant, respectively (Table 1). Searching within paralogons rather than within a fixed base-pair distance greatly expanded the physical range for detection of cis-interactions (Fig. 3). The numbers of cis- and trans-PPIs between genes in paralogons were counted (see Methods). The total number of cis-PPIs was larger than that of trans-PPIs for human, yeast, and plant (Table 1). We also found that the number of paralogons in which cis-PPIs outnumber trans-PPIs was larger than that of others in human, yeast, and plant paralogons.
Table 1.
Cis- and trans-interactions between singletons within paralogons
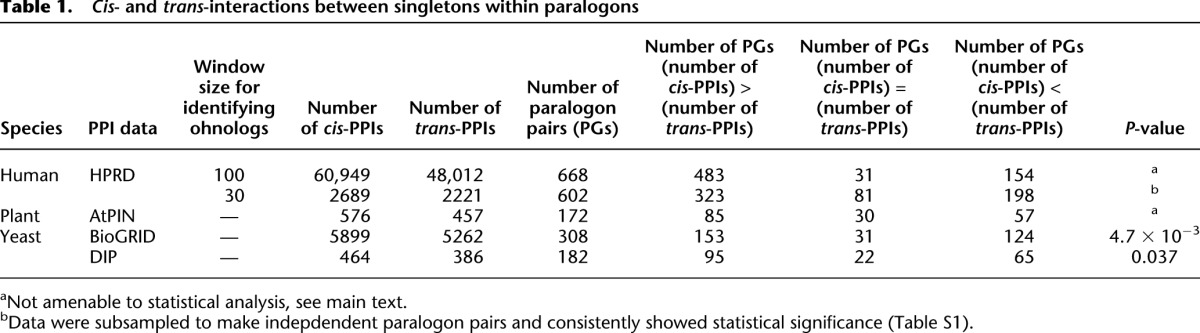
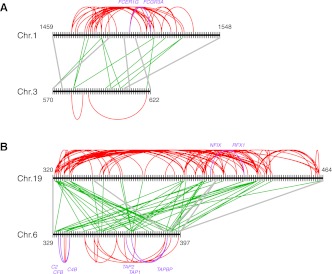
Figure 3.
Cis- and trans-PPIs between genes in human paralogons. Black vertical and horizontal lines represent genes and chromosomes, respectively. The numbers beside chromosomes indicate gene ordinal numbers along the chromosome. (Bold gray lines) Homology relationships between extant-ohnolog pairs. Red and green lines indicate cis- and trans-PPIs between genes in a human paralogon, respectively. Purple lines and gene names denote cis-PPIs previously identified as interacting gene clusters related to “immune response” (Makino and McLysaght 2008). In the case of physical links in paralogons, it is possible to identify cis-PPIs over a wider range compared with searches within a fixed base-pair distance (Makino and McLysaght 2008). There are many more cis-PPIs compared with trans-PPIs in (A) paralogon ID 24 (27 cis and 10 trans) and in (B) paralogon ID 507 (69 cis and 46 trans) (Supplemental File 1). Even after collapsing tandem duplicated genes, the PPIs of singletons are enriched in cis (ID: 24: 18 cis and 8 trans; ID 507: 60 cis and 34 trans).
Both vertebrates and plants have undergone more than one round of WGD. In the case of the vertebrate 2R (two rounds) tetraploidizations, there are potentially four chromosomal regions—nominally A, B, C, and D—that are all paralogons of each other (Supplemental Fig. S1). The ideal situation would be to only have three comparisons; A–B and C–D, which are the two products of the second round of WGD, and another comparison of [A,B]–[C,D], which examines the outcome of the first WGD. All possible comparisons of four paralogons result in six measurements, three of which might be considered redundant. However, not all of the comparisons are the same because of differing gene content, so we preferred to do all comparisons. This does not introduce a bias because after the first genome duplication, any genes that are kept in cis may be resolved to either cis or trans after the subsequent genome duplications. However, any relationships that are resolved to trans after the first genome duplication will only ever be trans, and these may be counted multiple times in the paralogon comparisons. Thus this is more likely to disfavor the hypothesis being tested and does not introduce a favorable bias.
However, tests of statistical significance require independent outcomes, and overlapping paralogons from multiple WGD violate this requirement. Therefore, we clustered paralogons derived from a common ancestral region (see Methods) and considered each cluster as only one occurrence for the purposes of statistical analysis. We randomly sampled one representative paralogon pair from each cluster for statistical analysis and repeated this sampling 1000 times. We performed the Wilcoxon signed rank test with continuity correction on each replicate. We observed that the number of paralogons in which cis-PPIs outnumber trans-PPIs was significantly higher than others for all of the replicates (Supplemental Table S1). Thus, there is strong statistical support for greater retention of cis-PPIs.
A. thaliana has only five chromosomes, and the lineage has experienced WGD at least three times; thus, the number of discriminable subsets of nonoverlapping paralogons with PPIs was very small (only 10 sets including 172 paralogons with PPIs) and not amenable to robust statistical analysis, but we note that the trends are the same as in human.
Similarly, we observed significant differences in the number of cis- and trans-PPIs in yeast paralogons (P = 4.7 × 10−3 Wilcoxon signed rank test with continuity correction) (Table 1). This result was consistent when we used an alternative available yeast PPI data set from the Database of Interacting Proteins (DIP) (P = 0.037) (Table 1). Notably, we observed consistent trends in different species with paralogons created at different times during evolution. These results indicate that there is a general bias in gene losses in eukaryote genomes following WGD.
Tests of independence of gene loss
Our analysis assumes that each gene loss is independent. However, it is possible to imagine a scenario in which two linked and interacting genes are removed, along with all intervening genes on the chromosome, in a single large DNA deletion event. One strategy to exclude the possibility of long deletions is to require the retention in duplicate (i.e., as ohnolog pairs) of at least one of the ancestrally intervening genes. There is insufficient knowledge of the ancestral gene order in vertebrates and plants to conduct this test; however, the ancestral gene order has been carefully reconstructed for yeasts (Gordon et al. 2011). Using this information, we could infer which genes in yeast lay between interacting genes prior to WGD and gene loss. Where at least one of these genes is retained in a present-day ohnolog pair, we can deduce that no single DNA deletion event spanned the entire region and that the return to single copy was an independent event for each of the interacting pair (Supplemental Fig. S2). This requirement reduced the data set of PPIs that we could analyze because only a small fraction of genes remains as ohnolog pairs. Furthermore, the ancestral genomic distance (counted in number of genes) between trans-PPIs tends to be greater than of cis-PPIs (Supplemental Fig. S3), which affords greater opportunity for the retention of an intervening ohnolog for trans-PPIs compared with cis-PPIs; thus, cis-PPIs are disproportionately removed from the data set under this rule. If we correct for differences in the number of intervening genes on the ancestral genome by restricting our search to only genes separated by five genes or fewer, then the number of paralogons with cis-PPIs greater than trans-PPIs is significantly larger than the converse (P = 2.9 × 10−5, Wilcoxon signed rank test with continuity correction; similarly for interacting pairs separated by up to five genes, P = 1.5 × 10−5) (Supplemental Table S2). However, when we only included PPIs between genes separated by at least one retained gene although cis-PPIs still outnumber trans-PPIs, there was no significant difference between the number of paralogons with more cis-PPIs and the number with more trans-PPIs (Supplemental Table S3).
It has been shown that, following WGD, deletion events tend to be no longer than one gene (Woodhouse et al. 2010) and that gene loss events in recent primate evolution are typically by pseudogenization rather than DNA deletion (Schrider et al. 2009). Therefore, although it is difficult to definitively exclude the possibility of single large deletions simultaneously removing interacting genes from the same side of the paralogon, we suggest that such events are unlikely to have contributed a bias to this analysis.
We also assume that these gene loss events are equally likely on each side of the paralogon. If copies in one paralogon are more likely to be lost than those in the other paralogon, we should expect more cis-PPIs than trans-PPIs due to a biased reduction of one paralogon, rather than any functional consequence of the interaction between genes. Biased fractionation after tetraploidization was observed in maize (Woodhouse et al. 2010). To test whether biased loss is occurring irrespective of PPIs, we examined paralogons for which the number of cis-PPIs was larger than that of trans-PPIs and compared the observed number of cis-PPIs with that of expected ones based on biased gene retention (see Methods). We observed that only 0%–23.1% of the paralogons had fewer than expected cis-PPIs (Supplemental Tables S4, S5). This indicates that a biased gene reduction of one paralogon did not cause biased PPI retention.
No preferential PPI retention and/or creation after WGD
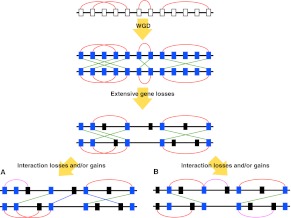
We have already shown that in the case of singletons within a paralogon, we find an excess of cis-PPIs (scenario G1 in Fig. 2). However, PPIs may be lost or gained independently of gene gain and loss, a phenomenon known as “network rewiring” (Wagner 2001; Beltrao and Serrano 2007; Presser et al. 2008), although a recent study reported that the evolutionary rate of PPI rewiring is very slow in yeast (Qian et al. 2011). We examined the possibility that the above results are part of a general phenomenon of biased retention of linked PPIs and/or the creation of interacting clusters occurring independently of gene loss (Fig. 4). If there is no bias in PPI retention or creation, then the number of cis- and trans-PPIs should be equal when summed over all possible scenarios (Fig. 2A–F). We used 532, 192, and 102 paralogon pairs in human, yeast, and plant, respectively (Table 2). We only counted paralogons with at least one PPI between genes of interest, which means that the total number of paralogons analyzed differs slightly between Table 1 (scenario Fig. 2G) and Table 2 (scenarios Figure 2A–F). The numbers of cis- and trans-PPIs between genes in a paralogon were counted as above (see Methods). We found no difference between the number of paralogons with more cis-PPIs and the number with more trans-PPIs in human, yeast, and plant (Wilcoxon signed rank test with continuity correction) (Table 2). This indicates that interaction gain and loss are not biased with respect to relative location and that our observation of more cis-PPIs is a result of biased gene loss rather than biased interaction gain or loss.
Figure 4.
PPIs losses and gains after whole genome duplication (WGD). Rectangles and horizontal lines represent genes and chromosomes, respectively. Red and green lines indicate cis- and trans-PPIs, respectively. (White rectangles) Genes prior to WGD. Blue and black rectangles show extant-paired ohnologs and singletons, respectively. Pink and blue lines indicate newly created cis- and trans-PPIs of ohnologs, respectively. As a matter of convenience, PPI losses and gains are shown after gene losses; however, the events may occur simultaneously in the evolutionary process. (A) Random PPI dynamics model. Some PPIs among ohnologs disappeared and/or appeared randomly after WGD. Trans-PPIs are observed as often as cis-ones. (B) Biased PPI dynamics model. PPIs among ohnologs in the same paralogon are retained preferentially after PPI losses. In addition, new interacting clusters are created by PPI rewiring. The number of cis-PPIs is larger than that of trans-PPIs.
Table 2.
Cis- and trans-interactions of extant-paired ohnologs
Conservation of genes in cis-PPIs across vertebrate genomes
If human cis-PPIs have biological significance, we expect that the cis-PPIs should be observed in other vertebrates. When we examined the relative location of orthologs of genes involved in cis-PPIs across vertebrate genomes, we found that they are likely to be conserved, i.e., they are located within the same paralogon (Table 3).
Table 3.
Cis- and trans-interacting gene pairs between singletons within paralogons of vertebrate genomes
We also considered the possibility that gene loss occurred independently in different vertebrate lineages. Under this scenario, we would expect to observe some cases in which there is no ortholog but only a paralog present due to “independent sorting-out” of the redundancy (Scannell et al. 2006). If independent gene loss also resulted in independent preservation of the cis relationship of the interacting gene pair, that would provide further support for the biological significance of the relative chromosomal location of these genes (Fig. 5). We identified orthologous and paralogous paralogons within 12 vertebrate genomes for human paralogons based on conserved gene order of orthologous ohnologs and surveyed partisan losses. We found several paralogous cis-PPIs in fish genomes (Table 3). However, overall we rarely observed paralogous relationships. This indicates that gene loss occurred quite rapidly before the radiation of most lineages and that cis-PPIs have been conserved since before the fish–tetrapod split. We also examined partisan losses for yeast (see Methods). As we observed in human, there were several paralogous cis-PPIs, but most cis-PPIs had orthologous relationships (Supplemental Table S6).
Figure 5.
PPIs in orthologous/paralogous paralogons in vertebrate genomes. Vertical and horizontal lines represent genes and chromosomes, respectively. Blue and black rectangles show extant-paired ohnologs and singletons, respectively. Red and green lines indicate cis- and trans-PPIs, respectively. (A) Gene loss patterns of cis-linked genes after WGD. Partisan loss can be observed when the second gene loss occurs in a paralogon where the first gene loss occurred. (B) When we observe conserved cis-linked genes in an orthologous paralogon between human and fish, it is difficult to distinguish an independent partisan loss from a cis-PPI derived from a common ancestor. (C) When we observe conserved cis-linked genes in a paralogous paralogon between human and fish, this pattern is strong evidence of independent partisan loss after speciation between human and fish.
Here we had specifically searched for cases in which we could infer independent retention of the cis-PPI by the paralogous rather than orthologous relationship of the extant genes in different vertebrate genomes (Fig. 5; Table 3). The fish-specific genome duplication (FSGD) provides an additional opportunity to test for the retention of the same cis-PPIs. We constructed FSGD paralogons for stickleback, tetraodon, medaka, and zebrafish (see Methods) for examining preservation (independent preservation is no object) of the cis-relationship of the interacting gene pair in fish paralogons after FSGD (Supplemental Fig. S4). We investigated whether fish orthologs (FSGD singletons) of human singletons with cis-PPIs in a paralogon were observed in the FSGD paralogons in a cis-relationship more frequently than in a trans-relationship. We found that the number of FSGD paralogons in which the number of cis-PPIs was larger than that of trans-PPIs was statistically significantly larger than that of others (Wilcoxon signed rank test with continuity correction) (Supplemental Table S7). The result indicates that cis-interacting singletons have been retained in both lineages (human and fishes) independently even after FSGD in which a cis-interacting gene pair would have a chance to change its formation as trans (Supplemental Fig. S4).
Retained cis-PPIs are enriched for function in “response to stimulus”
We attempted to understand the characteristics of singletons participating in cis- and trans-PPIs within paralogons. In a previous study, we showed that interacting genes located within 1 Mb of each other are biased toward a function in “response to stimulus” that includes many genes that operate in adaptive immunity (Makino and McLysaght 2008). Here, we examined the function of cis- and trans-PPIs using GO slim in human (http://www.geneontology.org).
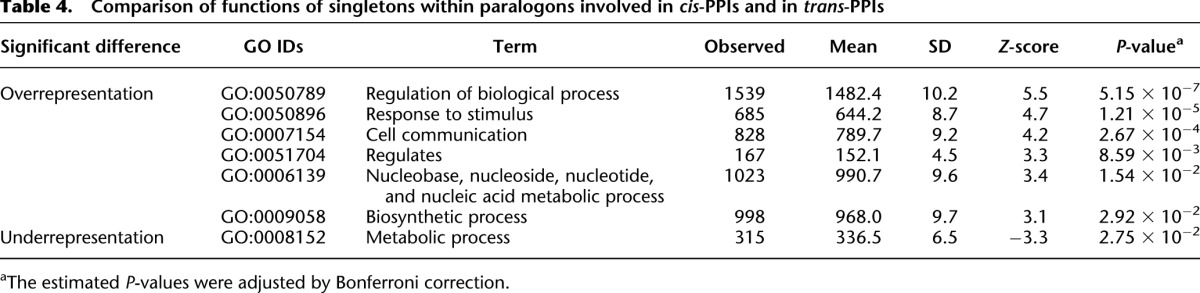
There were two rounds of WGD early in the vertebrate lineage, and therefore it is possible that some genes that are singletons with respect to one paralogon pair are extant-paired ohnologs in another paralogon pair. Furthermore, it has been shown that extant ohnologs are likely to be classified into specific functional classes in vertebrates (Blomme et al. 2006; Brunet et al. 2006; Hufton et al. 2008). Consistent with previous studies, we found that extant-paired ohnologs are often related to developmental processes in human (e.g., multicellular organismal development, cell communication, regulation of biological process, cell motion, cell differentiation, multicellular organismal process) (Supplemental Table S8). The numbers of extant-paired ohnologs classified into functional classes “response to stimulus” or metabolic processes were significantly smaller than expected in human (Supplemental Table S8). On the other hand, functional classes “response to abiotic stimulus” and “response to chemical stimulus” were enriched in ohnologs for plant and yeast, respectively (Supplemental Table S9). Biased functions of ohnologs seem to be different among eukaryotes that experienced WGD (Maere et al. 2005; Wapinski et al. 2007). In particular, “response to stimulus” is likely to be enriched in ohnologs from 2R WGD but not those from 1R/3R WGD in plant (Maere et al. 2005). To minimize the functional bias of extant-paired ohnologs (Supplemental Table S8), we only used singletons that were not included in any extant-paired ohnologs for GO analysis. We compared the number of GO slim terms for singletons in cis-PPIs with that for singletons in trans-PPIs (Table 4). We found “response to stimulus” (GO:0050896) was significantly enriched in cis-interacting singletons in human (Table 4; P = 1.2 × 10−5 after correction for multiple tests). This is consistent with the enrichment for “response to stimulus” in genes in interacting gene clusters in the human genome (Makino and McLysaght 2008). Interestingly, by searching for cis-PPIs within a paralogon rather than within a fixed base-pair distance, we were able to detect conserved linkage of genes over longer regions of chromosome (Fig. 3). These observations show a tendency for genes involved in “response to stimulus” to revert to single copy retaining the cis relationship of interacting genes. Only “RNA metabolic process” was enriched in cis-interacting singletons compared with trans-interacting ones in yeast, although the statistical significance was not high (P = 0.034). There was no observable bias in the function of cis-interacting singletons in plant because only 38 cis-interacting singletons had GO annotation.
Table 4.
Comparison of functions of singletons within paralogons involved in cis-PPIs and in trans-PPIs
Conclusion
We present evidence that functionally linked singletons in the same paralogon were preferentially retained in cis after extensive gene losses in human, yeast, and plant (Table 1). On the other hand, there was no significant enrichment of cis-PPIs between extant-paired ohnologs in the three species (Table 2); i.e., we found no evidence for biased rewiring of PPIs after WGD. Furthermore, the relative location of genes with cis-PPIs tends to be conserved across vertebrate genomes (Table 3).
The analysis of cis-PPI retention reported here assumes independent gene loss rather than large, sweeping DNA deletions that removed one copy of the interacting pair along with all intervening genes. Although we could not conduct a robust test to definitively eliminate the possibility of single large deletion events, we are satisfied that if these occur, they are rare and unlikely to introduce an artifact into this genome-wide analysis. Previous work has shown such large deletion events to be extremely rare following tetraploidization (Woodhouse et al. 2010) and within recent primate evolution gene loss is almost exclusively by means of pseudogenization rather than DNA deletion (Schrider et al. 2009). Although we note that neither of these studies refers specifically to the period following vertebrate WGD, they lend support to the assumption that large deletions were rare.
An alternative and very interesting explanation for the biased retention of cis-PPIs is that rather than reflecting a biological advantage to physical clustering on the chromosome, it instead is a legacy of an allopolyploid rather than autopolyploid event. Allopolyploidy is genome doubling caused by a type of hybridization between related organisms. If this occurred and if the two parent lineages had sufficiently diverged, the interacting proteins in each genome might have coadapted to the extent that retained interacting pairs from the same genome (i.e., in cis) were strongly preferred. This model suggests an explanation for why interacting genes would be initially retained in cis following WGD and fractionation. However, it provides no explanation for the retention of interacting gene clusters that are found in excess in eukaryotic genomes (Teichmann and Veitia 2004; Poyatos and Hurst 2006; Makino and McLysaght 2008).
The observation that functionally linked genes have been preferentially retained in cis following WGD and gene loss and that they have been conserved in cis during vertebrate evolution supports the hypothesis that the physical clustering on the chromosome has biological and functional significance. However, the nature of this biological significance remains unclear and may include co-regulation (Hurst et al. 2004), epistasis (Nei 1967), and epigenetic factors (Thomas et al. 2006). Many of the cis-interacting genes in human are classified as “response to stimulus,” which is consistent with previous studies showing clustering of genes that operate in immunity. We propose that functionally and physically linked genes have influenced the evolution of both genomic structures and protein interaction networks after WGD in fungi, plants, and vertebrates.
Methods
Paralogons
Human paralogons
We used six outgroups, which were amphioxus (Branchiostoma floridae) assembly v1.0 (Putnam et al. 2008) from JGI (http://www.jgi.doe.gov), sea urchin (Strongylocentrotus purpuratus) build 2.1 from NCBI (http://www.ncbi.nlm.nih.gov), two ascidians (Ciona intestinalis and Ciona savignyi), fly (Drosophila melanogaster), and worm (Caenorhabditis elegans) from Ensembl release 52 (Hubbard et al. 2007) for identification of ohnologs and combined them as shown in Makino and McLysaght (2010). Full details of the identification of human ohnologs are given in Makino and McLysaght (2010). We constructed human paralogons using the combined ohnolog data sets. Two genomic regions having two or more ohnologous pairs (within 100 genes) in which an ohnolog of the pairs was located in different genomic region from its ohnologous partner were defined as “paralogons.” We obtained 725 paralogon pairs in human (Supplemental File 1). We also used an alternative, stricter “paralogon” definition in which the maximum distance between ohnologs in a single paralogon was 30 genes; these data give consistent results.
Out of 70,624 human singleton pairs having a cis-PPI as shown in Figure 2G before collapsing tandem duplicates, 21,352 pairs were unique. Out of 21,352 gene pairs, we found 9669 pairs in which both genes of a pair had one-to-one orthologs in the chicken genome (Ensembl v52). Both genes in 6057 out of 9669 pairs were on the same chromosome of the chicken genome. When we use the tighter, alternative definition of paralogons (window size = 30), most of the chicken orthologs for singletons with a cis-PPI were in the same chromosome (95.9%, 792/826). This result indicates that the locations of genes involved in cis-PPIs in human paralogons have been conserved during at least land vertebrate evolution.
Yeast paralogons
We used ohnologs in Saccharomyces cerevisiae and their orthologs in pre-WGD species K. lactis to detect yeast paralogons in the Yeast Gene Order Browser (http://wolfe.gen.tcd.ie/ygob/). We removed nonsyntenic genes from our data set, because they have been possibly relocated from other chromosomal regions. We also removed ohnologs in a paralogon where the genomic location of the paired paralogon was unknown. We obtained 373 yeast paralogon pairs (Supplemental File 2).
We observed that 97.6% (5876/6021) of cis-interacting gene pairs in yeast (PPIs in BioGRID database) were in the same chromosome of pre-WGD ancestor (Gordon et al. 2011). The similar result was observed using PPIs in the DIP database (96.4%, 449/466). This indicates that the locations of genes involved in cis-PPIs in yeast paralogons have been conserved after WGD.
Plant paralogons
We downloaded conserved gene synteny blocks in the A. thaliana Genome from the Plant Genome Duplication Database (PGDD; http://chibba.agtec.uga.edu/duplication) and used them as plant paralogons (253 paralogon pairs).
Cis- and trans-interactions of genes in paralogons
We downloaded human protein interaction network (PIN) data from Human Protein Reference Database release 7 (Peri et al. 2003), yeast PIN data from the Database of Interacting Proteins (http://dip.doe-mbi.ucla.edu/) and BioGRID (only physical interactions; http://www.thebiogrid.org/), and plant PIN data from the A. thaliana protein interaction network (only experimentally determined interactions; http://bioinfo.esalq.usp.br/atpin/atpin.pl) to identify cis- and trans-interactions of genes in paralogons. We used protein–protein interactions (PPIs) between genes with distance <3 in the protein interaction networks as shown in Poyatos and Hurst (2006). To minimize tandem duplication effects, we removed PPIs among duplicated genes (BLAST, E-value < 0.2) (Lercher et al. 2003) using protein sequences of human and yeast genes from Ensembl and plant genes from PGDD, and we furthermore collapsed tandemly duplicated genes in the same paralogon as genes in the same family (BLAST, E-value < 0.2). Note that we removed self-interactions because they do not represent a relationship between different loci. If collapsing paralogs into a gene family generated self-interactions, we removed them. Finally, we counted PPIs between genes in the same side of a paralogon as cis-PPIs, and PPIs across a paralogon as trans-PPIs.
Preparation of independent (nonoverlapping) paralogons for statistical analysis
We clustered sets of human paralogons derived from the same ancestral region (such as nominal paralogons A, B, C, and D shown in Supplemental Fig. S1). We grouped paralogons into clusters when paralogons shared at least one ohnolog. A paralogon pair was not clustered when the paralogon pair was from the same chromosome, because they possibly belonged to the same ancestral region, but the paralogon that has become segmented. There were not many sets of nonoverlapping paralogons for human with window size = 100 (only 11 sets including 668 paralogons with PPIs). Therefore, we prepared a set of overlapping human paralogons using window size = 30 (169 sets including 380 paralogons with PPIs). Some paralogons are paired with many other paralogons, and these tend to cause large paralogon clusters by gathering subsets of clusters and thus reducing the number of clusters to a tiny number. Therefore, we deleted paralogons with many relationships (three to seven) (Supplemental Table S1) to maximize the number of paralogon clusters. Note that even when we did not delete them, our conclusions were the same (Supplemental Table S1). We prepared independent paralogons by choosing one paralogon pair randomly from each set and examined the enrichment of paralogons in which the number of cis-PPIs was significantly larger than that of trans-PPIs (Wilcoxon signed rank test with continuity correction). We repeated this 1000 times and performed the Wilcoxon signed rank test with continuity correction on each replicate.
Theoretical calculations of expected numbers of cis-PPIs
If gene loss is biased based on location in the genome rather than PPIs, then genes may be disproportionately removed from one side of the paralogon, and a greater number of cis-PPIs may simply reflect a greater number of cis gene relationships. We constructed a hypothetical scenario with differing rates of gene deletion in the two sides of a paralogon, where the probabilities of deletion of a gene from each side of the paralogon are p and 1 − p, respectively. We only consider genes that return to single copy. We estimate p as the number of singletons on one side of the paralogon (n1) expressed as a fraction of the total singletons over the two sides of the paralogon (n1 + n2), that is, p = n1/(n1 + n2). A pre-WGD cis-PPI is retained in cis after WGD with probability p2 + (1 − p)2 and retained in trans with probability 2p(1 − p). If there are c cis-PPIs between singletons and t trans-PPIs between singletons, the expected number of cis-PPIs, E1, is given by E1 = (c + t)[((n1/(n1 + n2))2 + (n2/(n1 + n2))2)]. This calculation gives an expected number of cis-PPIs proportional to the sizes of the two sides of the paralogon. If both sides have equal numbers of singletons, then p = 1/2 and E1 = 1/2(c + t).
Orthologous and paralogous paralogons
To identify orthologous and paralogous paralogons within 12 vertebrate genomes (chimpanzee, macaque, mouse, rat, dog, cow, opossum, chicken, medaka, zebrafish, tetraodon, and stickleback), we downloaded vertebrate orthologs for human genes and their genomic locations from Ensembl release 52. We identified orthologous paralogons of the vertebrates based on conserved gene synteny of orthologous ohnolog pairs by using the same algorithm (window size: 100) reported by Makino and McLysaght (2010). Note that, for human genes in a paralogon, it is traceable whether the homologs in vertebrate paralogons are in an orthologous paralogon or a paralogous one.
We obtained orthologous and paralogous paralogons within 10 yeast genomes (Vanderwaltozyma polyspora, Tetrapisispora phaffii, Tetrapisispora blattae, Naumovozyma dairenensis, Naumovozyma castellii, Saccharomyces castellii, Kazachstania naganishii, Kazachstania africana, Candida glabrata, and Saccharomyces bayanus) based on syntenic orthologs from YGOB (http://wolfe.gen.tcd.ie/ygob).
FSGD paralogons
We identified paralogons generated by FSGD for stickleback, medaka, tetraodon, and zebrafish using the same algorithm (window size: 100) reported by Makino and McLysaght (2010). Note that we used protein sequences for five teleost fishes (stickleback, medaka, tetraodon, zebrafish, and Fugu) and human (outgroup) from Ensembl release 52 to find FSGD candidate ohnolog pairs generated by duplication between speciation of teleost fishes and the fish–tetrapod split.
Gene Ontology
Gene Ontology (GO) “slim” annotations for biological processes of human, plant and yeast were downloaded from ftp://ftp.geneontology.org/pub/go/GO_slims. We excluded the GO ID GO:0008150 (biological process unknown). We calculated the P-value for each GO ID by comparison of the observed frequency in extant-paired ohnologs with expectations based on hypergeometric distribution using whole genes with at least one GO ID (Supplemental Tables S8, S9). We also calculated the P-value for each GO ID by comparison of the observed frequency in singletons with cis-PPIs with expectations based on hypergeometric distribution using singletons with trans-PPIs (Table 4). The estimated P-values were adjusted by Bonferroni correction.
Acknowledgments
This work is supported by Science Foundation Ireland.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.131953.111.
References
- The Arabidopsis Genome Initiative 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Beltrao P, Serrano L 2007. Specificity and evolvability in eukaryotic protein interaction networks. PLoS Comput Biol 3: e25 doi: 10.1371/journal.pcbi.0030025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Barakat A, Guyot R, Cooke R, Delseny M 2000. Extensive duplication and reshuffling in the Arabidopsis genome. Plant Cell 12: 1093–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomme T, Vandepoele K, De Bodt S, Simillion C, Maere S, Van de Peer Y 2006. The gain and loss of genes during 600 million years of vertebrate evolution. Genome Biol 7: R43 doi: 10.1186/gb-2006-7-5-r43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet FG, Crollius HR, Paris M, Aury JM, Gibert P, Jaillon O, Laudet V, Robinson-Rechavi M 2006. Gene loss and evolutionary rates following whole-genome duplication in teleost fishes. Mol Biol Evol 23: 1808–1816 [DOI] [PubMed] [Google Scholar]
- Dehal P, Boore JL 2005. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol 3: e314 doi: 10.1371/journal.pbio.0030314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich FS, Voegeli S, Brachat S, Lerch A, Gates K, Steiner S, Mohr C, Pohlmann R, Luedi P, Choi S, et al. 2004. The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science 304: 304–307 [DOI] [PubMed] [Google Scholar]
- Dujon B, Sherman D, Fischer G, Durrens P, Casaregola S, Lafontaine I, De Montigny J, Marck C, Neuveglise C, Talla E, et al. 2004. Genome evolution in yeasts. Nature 430: 35–44 [DOI] [PubMed] [Google Scholar]
- Gordon JL, Armisén D, Proux-Wéra E, ÓhÉigeartaigh SS, Byrne KP, Wolfe KH 2011. Evolutionary erosion of yeast sex chromosomes by mating-type switching accidents. Proc Natl Acad Sci 108: 20024–20029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard TJ, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, et al. 2007. Ensembl 2007. Nucleic Acids Res 35: D610–D617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hufton AL, Groth D, Vingron M, Lehrach H, Poustka AJ, Panopoulou G 2008. Early vertebrate whole genome duplications were predated by a period of intense genome rearrangement. Genome Res 18: 1582–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst LD, Pal C, Lercher MJ 2004. The evolutionary dynamics of eukaryotic gene order. Nat Rev Genet 5: 299–310 [DOI] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Wincker P 2009. “Changing by doubling,” the impact of Whole Genome Duplications in the evolution of eukaryotes. C R Biol 332: 241–253 [DOI] [PubMed] [Google Scholar]
- Kellis M, Birren BW, Lander ES 2004. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428: 617–624 [DOI] [PubMed] [Google Scholar]
- Lercher MJ, Blumenthal T, Hurst LD 2003. Coexpression of neighboring genes in Caenorhabditis elegans is mostly due to operons and duplicate genes. Genome Res 13: 238–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S, De Bodt S, Raes J, Casneuf T, Van Montagu M, Kuiper M, Van de Peer Y 2005. Modeling gene and genome duplications in eukaryotes. Proc Natl Acad Sci 102: 5454–5459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino T, McLysaght A 2008. Interacting gene clusters and the evolution of the vertebrate immune system. Mol Biol Evol 25: 1855–1862 [DOI] [PubMed] [Google Scholar]
- Makino T, McLysaght A 2010. Ohnologs in the human genome are dosage balanced and frequently associated with disease. Proc Natl Acad Sci 107: 9270–9274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLysaght A, Hokamp K, Wolfe KH 2002. Extensive genomic duplication during early chordate evolution. Nat Genet 31: 200–204 [DOI] [PubMed] [Google Scholar]
- Nakatani Y, Takeda H, Kohara Y, Morishita S 2007. Reconstruction of the vertebrate ancestral genome reveals dynamic genome reorganization in early vertebrates. Genome Res 17: 1254–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M 1967. Modification of linkage intensity by natural selection. Genetics 57: 625–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. 1970 Evolution by gene duplication. Springer, Berlin. [Google Scholar]
- Panopoulou G, Hennig S, Groth D, Krause A, Poustka AJ, Herwig R, Vingron M, Lehrach H 2003. New evidence for genome-wide duplications at the origin of vertebrates using an amphioxus gene set and completed animal genomes. Genome Res 13: 1056–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peri S, Navarro JD, Amanchy R, Kristiansen TZ, Jonnalagadda CK, Surendranath V, Niranjan V, Muthusamy B, Gandhi TK, Gronborg M, et al. 2003. Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Res 13: 2363–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovici C, Leveugle M, Birnbaum D, Coulier F 2001. Coparalogy: Physical and functional clusterings in the human genome. Biochem Biophys Res Commun 288: 362–370 [DOI] [PubMed] [Google Scholar]
- Poyatos JF, Hurst LD 2006. Is optimal gene order impossible? Trends Genet 22: 420–423 [DOI] [PubMed] [Google Scholar]
- Presser A, Elowitz MB, Kellis M, Kishony R 2008. The evolutionary dynamics of the Saccharomyces cerevisiae protein interaction network after duplication. Proc Natl Acad Sci 105: 950–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam NH, Srivastava M, Hellsten U, Dirks B, Chapman J, Salamov A, Terry A, Shapiro H, Lindquist E, Kapitonov VV, et al. 2007. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 317: 86–94 [DOI] [PubMed] [Google Scholar]
- Putnam NH, Butts T, Ferrier DE, Furlong RF, Hellsten U, Kawashima T, Robinson-Rechavi M, Shoguchi E, Terry A, Yu JK, et al. 2008. The amphioxus genome and the evolution of the chordate karyotype. Nature 453: 1064–1071 [DOI] [PubMed] [Google Scholar]
- Qian W, He X, Chan E, Xu H, Zhang J 2011. Measuring the evolutionary rate of protein–protein interaction. Proc Natl Acad Sci 108: 8725–8730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannell DR, Byrne KP, Gordon JL, Wong S, Wolfe KH 2006. Multiple rounds of speciation associated with reciprocal gene loss in polyploid yeasts. Nature 440: 341–345 [DOI] [PubMed] [Google Scholar]
- Schrider DR, Costello JC, Hahn MW 2009. All human-specific gene losses are present in the genome as pseudogenes. J Comput Biol 16: 1419–1427 [DOI] [PubMed] [Google Scholar]
- Teichmann SA, Veitia RA 2004. Genes encoding subunits of stable complexes are clustered on the yeast chromosomes: An interpretation from a dosage balance perspective. Genetics 167: 2121–2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas BC, Pedersen B, Freeling M 2006. Following tetraploidy in an Arabidopsis ancestor, genes were removed preferentially from one homeolog leaving clusters enriched in dose-sensitive genes. Genome Res 16: 934–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K, De Vos W, Taylor JS, Meyer A, Van de Peer Y 2004. Major events in the genome evolution of vertebrates: Paranome age and size differ considerably between ray-finned fishes and land vertebrates. Proc Natl Acad Sci 101: 1638–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A 2001. The yeast protein interaction network evolves rapidly and contains few redundant duplicate genes. Mol Biol Evol 18: 1283–1292 [DOI] [PubMed] [Google Scholar]
- Wapinski I, Pfeffer A, Friedman N, Regev A 2007. Natural history and evolutionary principles of gene duplication in fungi. Nature 449: 54–61 [DOI] [PubMed] [Google Scholar]
- Wolfe K 2000. Robustness—it's not where you think it is. Nat Genet 25: 3–4 [DOI] [PubMed] [Google Scholar]
- Wolfe KH, Shields DC 1997. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387: 708–7139192896 [Google Scholar]
- Woodhouse MR, Schnable JC, Pedersen BS, Lyons E, Lisch D, Subramaniam S, Freeling M 2010. Following tetraploidy in maize, a short deletion mechanism removed genes preferentially from one of the two homologs. PLoS Biol 8: e1000409 doi: 10.1371/journal.pbio.1000409 [DOI] [PMC free article] [PubMed] [Google Scholar]