Abstract
DNA methylation and nucleosome positioning work together to generate chromatin structures that regulate gene expression. Nucleosomes are typically mapped using nuclease digestion requiring significant amounts of material and varying enzyme concentrations. We have developed a method (NOMe-seq) that uses a GpC methyltransferase (M.CviPI) and next generation sequencing to generate a high resolution footprint of nucleosome positioning genome-wide using less than 1 million cells while retaining endogenous DNA methylation information from the same DNA strand. Using a novel bioinformatics pipeline, we show a striking anti-correlation between nucleosome occupancy and DNA methylation at CTCF regions that is not present at promoters. We further show that the extent of nucleosome depletion at promoters is directly correlated to expression level and can accommodate multiple nucleosomes and provide genome-wide evidence that expressed non-CpG island promoters are nucleosome-depleted. Importantly, NOMe-seq obtains DNA methylation and nucleosome positioning information from the same DNA molecule, giving the first genome-wide DNA methylation and nucleosome positioning correlation at the single molecule, and thus, single cell level, that can be used to monitor disease progression and response to therapy.
Epigenetic mechanisms including DNA methylation and nucleosome positioning work together to generate specific chromatin states which facilitate, inhibit, or allow for the potential of gene expression. Active promoters have unmethylated DNA and lack nucleosomes just prior to the gene's transcriptional start site (TSS), while inactive promoters have densely packed nucleosomes and can be unmethylated (poised or repressed) or methylated (silent). Due to this variety of chromatin structures, gene activation potential cannot be predicted by looking at nucleosome occupancy or DNA methylation alone.
Pioneering work by Michael Kladde and colleagues has demonstrated the ability of methyltransferase-based footprinting to determine nucleosome positioning in yeast and mammalian cells (Xu et al. 1998; Jessen et al 2004, 2006; Kilgore et al. 2007; Pardo et al. 2010). Using next generation sequencing, we describe a genome-wide nucleosome footprinting method termed NOMe-seq (nucleosome occupancy and methylome sequencing), which uses a GpC methyltransferase (M.CviPI) (Xu et al. 1998) to obtain nucleosome positioning information based on enzyme accessibility to GpC sites, while obtaining endogenous DNA methylation information at the same time from CpG sites. Importantly, both pieces of epigenetic information are obtained from the same individual DNA molecule, revealing the relationship between these two chromatin features on a single chromosome. Thus, using a single methodology, one can generate genome-wide maps of multiple epigenetic modifications at the single molecule level.
Using NOMe-seq with whole-genome bisulfite sequencing, we generated an integrated map and show distinct nucleosome/methylation configurations associated with specific genomic features and that the strength of the NDR upstream of the TSS is indicative of expression level and can accommodate several nucleosomes. By examining promoters with reads from two distinct chromatin states, as defined by nucleosome occupancy and methylation, we identified genes likely to be in two divergent allelic states, which are strongly enriched on the X chromosome and at known imprinted loci. Simultaneously measuring nucleosome occupancy and DNA methylation within individual DNA strands is an important tool for examining how chromatin structure across the genome is altered in disease states.
Results
Identifying optimal treatment conditions for accurate footprinting of a variety of genomic loci
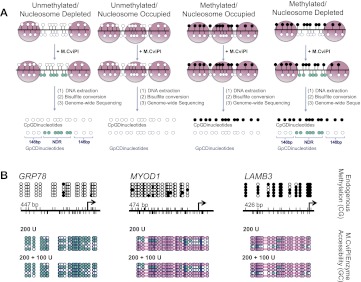
To generate integrated DNA methylation and nucleosome occupancy information, nuclei are treated with M.CviPI, which methylates GpC dinucleotides not protected by nucleosomes or tight binding proteins. Following bisulfite conversion to differentiate between methylated and unmethylated cytosine residues, cytosines contained within a CpG dinucleotide context provide endogenous methylation information, while nucleosome positioning is derived from cytosines within GpC dinucleotides. Nucleosome occupancy and endogenous DNA methylation information is obtained as the methylation of each individual cytosine is calculated as the fraction of methylated reads divided by all reads covering that position. Combining CpG and GpC methylation profiles, four distinct chromatin structures can be visualized (Fig. 1A). We first identified a set of reaction conditions, which allowed for accurate footprinting (i.e., accessibility of nucleosome depleted regions, while not aberrantly accessing nucleosome-occupied regions, defined as 146 bp or larger that are inaccessible to M.CviPI) of a variety of chromatin configurations (Fig. 1B; Supplemental Fig. S1A).
Figure 1.
NOMe-seq can footprint a variety of chromatin structures. (A) After IMR90 cell nuclei are treated with M.CviPI, DNA is extracted, bisulfite-converted, and sequencing is performed. DNA methylation status is obtained from CpG dinucleotides, and nucleosome occupancy information is gained from the inaccessibility of the M.CviPI methyltransferase to GpC dinucleotides. The combination of DNA methylation and nucleosome occupancy data can reveal four distinct chromatin signatures: unmethylated and nucleosome-depleted, unmethylated and nucleosome-occupied, methylated and nucleosome-occupied, and methylated and nucleosome-depleted. (Black circles) Methylated CpG sites; (teal circles) accessible (methylated) GpC sites. (B) We found that 200 units of M.CviPI for 7.5 min followed by a boost of 100 units accurately revealed an NDR upstream of the TSS of HSPA5 (also known as GRP78), an active CGI promoter, while also showing that the polycomb repressed MYOD1 CGI promoter and methylation-silenced CpG-poor LAMB3 promoter were occupied by nucleosomes and inaccessible to M.CviPI, as expected. M.CviPI-inaccessible regions greater than 146 bp are covered by a pink rectangle indicating nucleosome occupancy. PCR amplicon sizes: HSPA5–447 bp, MYOD–474 bp, and LAMB3–426 bp.
NOMe-seq reveals expected nucleosome occupancy patterns at CTCF and transcription start sites
We generated whole-genome NOMe-seq libraries and adapted our whole-genome bisulfite-processing pipeline (Berman et al. 2012; Liu et al. 2012; Supplemental Material) to segregate cytosines based on the trinucleotide containing the cytosine in the central position. GCH cytosines were generally used to plot enzyme accessibility (nucleosome protection or occupancy), while HCGs (where H = C, T, or A) were used for endogenous methylation. GCGs were excluded due to ambiguity between endogenous and enzymatic methylation. Exclusion of GCGs is not likely to dramatically hurt the ability of M.CviPI to footprint nucleosomes since GCGs represent less than 0.24% of the genome and make up only 5.6% of all GC dinucleotides (Supplemental Table S1). Furthermore, 93.4% of GCG trinucleotides have a GCH within 20 bp (half of which are within 5 bp) from which nucleosome occupancy information can be derived (Supplemental Fig. S1B).
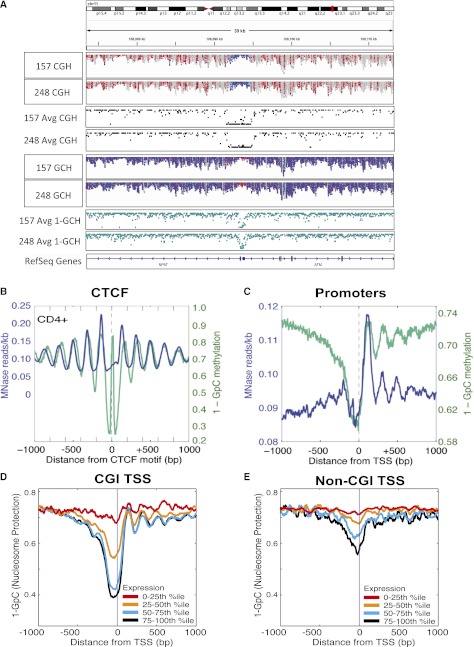
Due to the availability of genome-wide data (Lister et al. 2009; Bernstein et al. 2010), we performed whole-genome NOMe-seq in IMR90 cells and obtained 156 million uniquely alignable reads which can be displayed from raw BAM alignment files using a newly developed module of the IGV viewer (Fig. 2A; Supplemental Fig. S2; Thorvaldsdottir et al. 2012). We compared the ability of NOMe-seq to accurately map the well-positioned nucleosomes flanking CTCF binding sites. We aligned reads to conserved CTCF binding motifs (Xie et al. 2007) located more than 2 kb away from TSSs that have been experimentally validated as bound in vivo by CTCF (Supplemental Table S2; Kim et al. 2007; Cuddapah et al. 2009) and found that NOMe-seq mapped nucleosomes similar to MNase-seq data (Fig. 2B; Schones et al. 2008). Nucleosome occupancy is plotted as inaccessibility to M.CviPI (1-GpC methylation) (Fig. 2B, teal line); thus, regions of protection appear as peaks in the graph. The first and second nucleosomes to the right of the CTCF binding site appear to be slightly out of phase using NOMe-seq compared to MNase; however, here we are comparing MNase-seq data from the benchmark CD4+ T-cell data set (Schones et al. 2008) with NOMe-seq data collected in IMR90 cells. This phase shifting is not apparent when we compare NOMe-seq and MNase-seq data both generated from IMR90 cells (Supplemental Fig. S3). The high resolution generated by NOMe-seq also reveals a region of protection coinciding with the CTCF binding site, likely reflective of CTCF binding. To investigate this further, we examined a CTCF binding region at high resolution (Supplemental Fig. S4) and show a discreet protection pattern overlapping the CTCF motif with a size (<40 bp) consistent with a nonnucleosomal protein. This protected region is surrounded by clear nucleosome depletion (accessibility), which is, in turn, surrounded by larger protected regions whose size is consistent with nucleosome occupancy. We next aligned NOMe-seq reads to all TSSs and again found that NOMe-seq was comparable to MNase-seq and able to identify a nucleosome-depleted region (NDR) upstream of TSSs and well-positioned nucleosomes downstream from TSSs (Fig. 2C).
Figure 2.
NOMe-seq diplays nucleosome occupancy profiles at specific loci and globally. (A) Broad view of the ATM promoter using a newly developed module of the IGV viewer (Thorvaldsdottir et al. 2012) to visualize NOMe-seq BAM alignment files. The top two tracks indicate endogenous DNA methylation (at HCG sites) in each of two GBM samples, while tracks 5 and 6 indicate GCH accessibility of the same GBM samples. (Red) Methylated sites (for both HCG and GCH); (blue) unmethylated sites (for both HCG and GCH). The promoters of ATM and NFAT are unmethylated (blue in top two tracks) and nucleosome-depleted (i.e., accessible and therefore methylated, and thus red in tracks 5 and 6). The same methylation and nucleosome occupancy pattern is seen for both GBM samples. Tracks 3 and 4 show average methylation levels derived from these tracks—at each individual HCG, the number of reads methylated at that HCG is divided by the total number of reads methylated and unmethylated. Average GCH methylation in tracks 7 and 8 is calculated as before but inverted (1-GCH) to indicate nucleosome protection as used throughout the main figures. The tool and source code are publicly available for download at the IGV project website: http://www.broadinstitute.org/igv/. (B,C) NOMe-seq reads were aligned to CTCF (B) and TSSs (C). Nucleosome positioning in IMR90 cells is indicated on the y-axis by inaccessibility to M.CviPI (1-GpC methylation; teal line) and the number of MNase sequencing reads (blue line). For MNase-seq, reads were aligned to 8709 CTCF sites, while 8687 CTCF sites had at least one GpC site that was covered by a minimum of three reads (B). For TSS, 42,103 promoters were used for MNase-seq, and 41,292 promoters had at least one GpC site that was covered by a minimum of three reads. (D,E) Gene promoters were divided into quartiles based on transcription level (Hawkins et al. 2010), and the corresponding M.CviPI inaccessibility (1-GCH, teal line) is plotted on the y-axis. (D) CpG island promoters. (E) Non-CpG island promoters. The NDR is stronger in more highly expressed genes and, in some cases, can be several hundred bp long to accommodate multiple nucleosomes.
We next examined the relationship between nucleosome depletion and expression by dividing promoters into quartiles based on expression level (Hawkins et al. 2010) and found that promoters in the lowest bin (0%–25%) were nucleosome-occupied regardless of whether they were CGI or non-CGI promoters (Fig. 2D,E). With increasing expression quartiles, the NDR upstream of the TSSs and the positioning of the nucleosomes after the TSS became more apparent for both CGI and non-CGI promoters. These results suggest that an NDR upstream of the TSS and positioned nucleosomes downstream from the TSS are strongly predictive of expression level and indicate similar epigenetic regulation of CGI and non-CGI promoters.
NOMe-seq reveals distinct chromatin configurations at specific promoter types
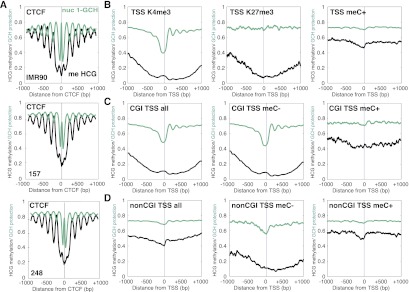
We examined the combined nucleosome occupancy and methylation patterns at CTCF sites, specific promoter classifications (Fig. 3; Hawkins et al. 2010), and other genomic regions including enhancers and intron/exon boundaries (Supplemental Figs. S6, S7). Interestingly, DNA methylation and nucleosome occupancy were strongly anti-correlated surrounding CTCF sites such that DNA methylation peaked in the linker regions between nucleosomes (Fig. 3A; Supplemental Fig. S5). To examine whether this correlation was cell type-specific, we performed NOMe-seq in two primary cultures from glioblastoma tumors (157 and 248) and found that DNA methylation and nucleosome positioning were also anti-correlated at CTCF sites in these cells (Fig. 3A). At promoters, nucleosome occupancy and DNA methylation were consistent with transcription potential (Fig. 3B): H3K4me3-marked (active) promoters were unmethylated with a distinct NDR upstream of TSSs and at least four nucleosomes downstream from TSSs, while H3K27me3-marked (repressed) promoters were unmethylated but nucleosome occupied as indicated by inaccessibility to M.CviPI. DNA methylated (silent) promoters were completely nucleosome-occupied. NOMe-seq is able to distinguish these three important and distinct promoter architectures in a single experiment. Surprisingly, there was a “bump” in DNA methylation just upstream of the TSS of promoters marked by H3K4me3, which we found to be due to “off-target” activity of M.CviPI and only affected the endogenous methylation information obtained from cytosines that were preceded by another cytosine at regions of peak M.CviPI accessibility. This artifact could be removed completely by eliminating CCGs (Supplemental Fig. S8; Supplemental Material), and future analysis methods can better adjust for this known off-target activity rate.
Figure 3.
NOMe-seq reveals distinct chromatin configurations at CTCF sites and associated with specific histone modifications and promoter types. (A) NOMe-seq demonstrates unmethylated NDRs at CTCF sites in IMR90 and GBM cells, which are marked by a peak in inaccessibility at the CTCF site itself. Well-positioned nucleosomes flank CTCF sites, with DNA methylation peaking in between nucleosomes. 0 indicates the middle of the CTCF binding motif. CTCF binding sites were obtained from GSM935404. (B) NOMe-seq distinguishes the three major promoter states at promoters in IMR90 cells—active H3K4me3-marked promoters are unmethylated and contain a NDR upstream and well-positioned nucleosomes after the TSS. TSSs are indicated on the x-axes as 0. Repressed/poised H3K27me3-marked promoters are unmethylated and nucleosome-occupied. Methylated promoters are nucleosome-occupied. The y-axis indicates M.CviPI inaccessibility (1-CpG; teal) and CpG methylation level. (C) In IMR90 cells, CpG island promoters are characterized by a lack of CpG methylation, an upstream NDR, and well-positioned nucleosomes after the TSS. The majority of CpG island promoters are unmethylated (11,165) and display the same pattern, while methylated CpG island promoters (781) are nucleosome-occupied and inaccessible to M.CviPI. (D) Non-CpG island promoters are generally characterized by CpG methylation and inaccessibility to M.CviPI, indicating nucleosome occupancy. The few unmethylated non-CpG island promoters (1397) are depleted of nucleosomes upstream of the TSS, while the majority of non-CpG island promoters (4668) are nucleosome-occupied and inaccessible to M.CviPI. M.CviPI inaccessibility is plotted (1-GCH) in teal and CpG methylation (CGH) in black.
We next investigated chromatin configurations of CGI and non-CGI promoters (Fig. 3C,D; Supplemental Fig. S7A,B). In general, CGI promoters had low levels of cytosine methylation near the TSS (relative to 1 kb away from the TSS), a distinct NDR upstream of the TSS, and well-positioned nucleosomes downstream from the TSS. Separating CGI promoters into those that are methylated and unmethylated reveals that the CGI promoter pattern is largely driven by unmethylated CGI promoters and the few CGI promoters that were methylated were nucleosome-occupied. Separating non-CGI promoters into those that were methylated and unmethylated revealed that the relatively few non-CGI promoters that were unmethylated also had an NDR upstream of the TSS and a nucleosome immediately downstream from the TSS, while the more commonly methylated non-CGI promoters were nucleosome-occupied.
To demonstrate NOMe-seq's reproducibility, we sequenced two glioblastoma (GBM) primary cell cultures and found similar nucleosome positioning patterns at promoters and enhancers in the GBM cells as we did in IMR90 cells (Supplemental Fig. S7). Using a statistical test to identify NDRs near TSSs (see Methods), we found high concordance among all samples at CGI promoters; the two GBMs had NDRs that were 90% overlapping with each other and 88% and 91% overlapping with IMR90, respectively (Supplemental Fig. S7C). Many genes which are essential for cellular function (i.e., housekeeping genes) have CGI promoters; thus, it was not surprising to have such significant overlap between the GBM and IMR90 cells. Nevertheless, the probability of getting such a 90% overlap of NDRs in the two GBMs by chance is 10−518 using a hypergeometric test. We found significantly less overlap at non-CGI promoters between cell types, consistent with the greater cell-type specificity of non-CGI genes. In these gene promoters, the two GBM samples overlapped by 58%, while they overlapped IMR90 by 43% and 47%, respectively.
Combinatorial epigenomic signatures reveal functional chromatin
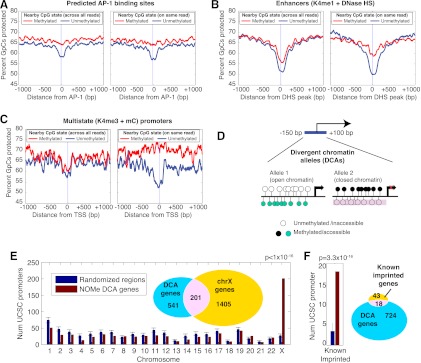
Unlike any other method used to assess nucleosome occupancy or DNA methylation, NOMe-seq includes both nucleosome positioning and DNA methylation data for individual DNA strands, enabling a correlation between the two features at the single molecule level. Because different chromatin states can exist on the two alleles in a single cell or in different subpopulations of cells within a sample, we expected the combination of two marks on a single molecule to yield more information than average levels taken across a population of cells. To investigate this, we calculated nucleosome protection patterns around genomic elements as a function of DNA methylation state, first using methylation information from population averages from any read covering the same position in the genome (Fig. 4A–C, left panels) and then using only the methylation state from the same read (Fig. 4A–C, right panels). Some regulatory elements we investigated, such as sequences with predicted AP-1 binding motifs, had a visible NDR but showed almost no difference between population averages (Fig. 4A, left) and within-read averages (Fig. 4A, right). These elements suggest uniformity of a specific chromatin state across the entire population of cells. Other elements we investigated, such as those annotated as DNase hypersensitive enhancers in IMR90 cells (Hawkins et al. 2010), had a much stronger correlation between DNA methylation and accessibility within individual reads than across the population of reads, suggesting that a combinatorial chromatin signature exists within a subset of cells or alleles within the sample (Fig. 4B).
Figure 4.
Combinatorial epigenomic signatures reveal functional chromatin. (A–C) Nucleosome occupancy levels (“percent of GpCs protected”) are shown stratified by the methylation status of nearby CpGs (within 20 bp). For each element type, this analysis was performed twice—once sampling randomly across all reads covering the same genomic position as the GpC (left plots, labeled “across all reads”) and a second time using only the methylation status from the same read (right plots, labeled “on same read”) (see Methods). All three examples show nucleosome depletion associated primarily with the unmethylated state, but while predicted AP-1 binding motifs (A) display this in both population and within-read profiles, enhancers and promoters marked by the opposing K4me3 and meC (B,C) show this association only in the within-read analysis. 0 refers to the center of the AP-1 binding motif (A), the peak of DNase HS within K4me1-marked regions (B) and TSSs (C). (D) Search strategy for finding divergent chromatin alleles (DCA) by searching TSS regions for at least two reads with opposing chromatin profiles in IMR90 cells. (E) Promoters that exist in both nucleosome-depleted and unmethylated and nucleosome-occupied and methylated are enriched on the X chromosome. Seven hundred and forty-two DCA genes were compared to randomized sets of 742 genes—1000 trials were performed and the standard deviation is shown for the number on each chromosome. A P-value was determined from the X chromosome using a binomial test with the probability determined by the random trials. (F) DCA genes were compared to 1000 randomized gene sets for the number within 50 kb of known imprinted genes.
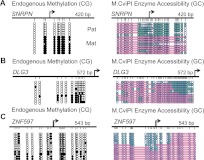
To investigate whether we could detect combinatorial chromatin signatures within regions likely to be monoallelic, we applied this same approach to gene promoters identified as having both DNA methylation and H3K4me3 marks in IMR90 cells (Fig. 4C; Hawkins et al. 2010). These two states are generally antagonistic at promoters, suggesting that they might exist on two different alleles in the same cell, especially in a genetically female cell line like IMR90. The across-read vs. within-read comparison shows that any correlation between methylation and nucleosome occupancy is lost when averaging across all reads (Fig. 4C, left) but clear when looking at within-read correlations (Fig. 4C, right). To test whether we could exploit this within-read correlation to identify allele-specific regions, we searched all promoters (−150 to +100 bp from TSS) for a combination of the two opposing chromatin conformations (Fig. 4D), one containing unmethylated CpGs and no nucleosome protection (Allele 1) and the other, methylated CpGs and nucleosome protection (Allele 2). We found that 742 promoter regions met this “divergent chromatin alleles” (DCA) criteria, of which 201 mapped to the X chromosome (27% of DCA promoters compared to 2.7% of promoters genome-wide) (Fig. 4E). Eighteen DCA promoters were associated with one of 58 known imprinted genes (http://www.geneimprint.org/), compared to an average of four in matched sets of randomly selected promoters (Fig. 4F). To validate our genome-wide findings, we performed locus-specific NOMe-seq analysis on one X-linked (DLG3), one imprinted gene (SRNPN), and one novelly identified DCA promoter (ZNF597), which was recently suggested to be imprinted (Fig. 5; Choufani et al. 2011; Nakabayashi et al. 2011). Our results clearly show the presence of two distinct chromatin structures. We further showed more overlap in DCA alleles between the two GBM samples compared to the number of DCA alleles shared amongst the GBM and IMR90 samples (Supplemental Fig. S9). The incorporation of both DNA methylation and nucleosome positioning information from individual DNA strands enabled the identification of several monoallelic genes that have not been previously described, and we expect that increased sequencing depth will greatly increase our sensitivity for these regions.
Figure 5.
Validation of DCA promoters. PCR amplicons were cloned, and several colonies were sequenced to visualize two distinct chromatin configurations of an imprinted gene, SNRPN (A), and an X-linked locus, DLG3 (B), and a novelly identified DCA gene, ZNF597 (C). (Black) DNA methylation of CpG sites; (teal) GpC accessibility. Pink bars indicate nucleosome positioning.
Discussion
Using a novel approach to examine nucleosome occupancy and endogenous DNA methylation genome-wide, we footprinted chromatin architecture at a variety of promoter and nonpromoter regions. We showed that the NDR upstream of the TSS can accommodate multiple nucleosomes and is indicative of expression level for both CGI and non-CGI promoters. We further show that the relationship between nucleosome occupancy and DNA methylation is context-specific—depending on genomic location—and that incorporation of DNA methylation and nucleosome information within the same DNA strand can facilitate the identification of monoallellic chromatin patterns. Importantly, we show that NOMe-seq alone can distinguish among the three major chromatin states known to be found at promoters—active (H3K4me3, meC−, NDR+), repressed/poised (H3K27me3, meC−, NDR−), and silenced (mec+, NDR−). The ability to distinguish these three promoter architectures within a single experiment, let alone a single molecule, holds great promise for epigenomic mapping.
Traditionally, genome-wide mapping of nucleosome positioning has been done using MNase-seq or H3 ChIP-seq, which rely on DNA breakage. FAIRE-seq relies on enhanced sensitivity to DNA breakage of nucleosome-depleted regions (Giresi et al. 2007; Nagy and Price 2009). Rather than using a nuclease, methyltransferase-based footprinting, with the CpG methyltransferase M.SssI (Gal-Yam et al. 2006; Lin et al. 2007; Bouazoune et al. 2009; Kelly et al. 2010) or GpC methyltransferase M.CviPI (Kelly et al. 2010; Wolff et al. 2010; Andreu-Vieyra et al. 2011; Taberlay et al. 2011; You et al. 2011), uses the placement of a biochemical mark (i.e., methylation) on DNA to assess nucleosome occupancy. Since GpC dinucleotides are not endogenously methylated, NOMe-seq provides both nucleosome positioning and DNA methylation within the same individual DNA strand. In addition, since NOMe-seq signal is interpreted as a percentage of sequencing reads at a given position, it provides a normalized and unskewed measurement that does not rely upon the number of reads that map to a particular genomic locus and, thus, is an independent method of assessing nucleosome occupancy that can complement and validate results from the established enrichment methods.
Using MNase-seq combined with DNA methylation information from bisulfite sequencing libraries, previous work has found that nucleosomal DNA is preferentially methylated (Chodavarapu et al. 2010). While this is true for the majority of the genome, specific types of elements did not adhere to this genome-wide trend. For example, nucleosomes surrounding CTCF sites were unmethylated while the linker regions between nucleosomes were methylated, demonstrating a novel relationship at a functionally important class of chromatin.
One hallmark of an active gene is the presence of a NDR immediately upstream of the TSS. Previous work found the levels of active histone marks correlated with expression level (Barski et al. 2007); however, nucleosome occupancy itself was not measured, and the regions upstream of the TSS appeared equally depleted of nucleosomes regardless of transcript level. Here, we show that NDRs are more prominent at more highly expressed genes. Importantly, this correlation between expression and nucleosome depletion was similar for CGI and non-CGI promoters, suggesting that at least some key aspects of epigenetic gene regulation are shared between CGI and non-CGI promoters. In addition, our results show that NDRs are large enough to accommodate multiple nucleosomes. The inability to detect subtle nucleosome depletion differences based on expression and the underestimation of NDR size by previous studies is potentially reflective of the variability in fragment sizes generated by sonication, and highlights the subtleties of chromatin organization that can be identified using NOMe-seq and that have been overlooked in previous studies.
Whole genome NOMe-seq is a novel approach that footprints nucleosome occupancy while retaining DNA methylation information to identify chromatin structures of a variety of genomic regions including promoters, enhancers, and insulators. The combination of these two epigenetic marks on the same molecule can identify combinatorial profiles within a mixed population of cells or alleles with greater sensitivity than the two marks in isolation. The epigenetic landscape generated by these combinatorial epigenomic profiles has several important implications for biology, especially in the context of profiling complex tissues containing multiple cell types. Furthermore, as mutations in chromatin remodeling complexes are becoming increasingly associated with cancer (Wilson and Roberts 2011), whole-genome NOMe-seq is an ideal approach to address the effects that these mutations have, both on nucleosome positions and DNA methylation, and can further investigate whether chromatin remodeling defects are dependent on DNA methylation state.
Methods
Cell culture
IMR90 cells were cultured according to ATCC recommendations. Primary GBM cells were cultured as previously described (Laks et al. 2009). Briefly, neurosphere media contained DMEM/F12 supplemented with B27 (GIBCO), bFGF (20 ng/mL, R&D Systems Inc.), epidermal growth factor (EGF; 50 ng/mL, Peprotech), penicillin/streptomycin (1%, Invitrogen), and heparin (5 μg/mL, Sigma-Aldrich). Heparin, bFGF, and EGF were added to the media every 3 or 4 d. Spheres were passaged every 7 to 14 d following dissociation with TrypLE Express (Invitrogen).
Nucleosome footprinting
NOMe-seq is a modified version of our methylation-dependent single promoter assay (Miranda et al. 2010). Nuclei from IMR90 cells (ATCC) were isolated as previously described (Miranda et al. 2010). Previous publications using locus-specific NOMe-seq have used the minimal amount of M.CviPI that resulted in optimal footprinting of the specific region of interest: 100 units (Wolff et al. 2010), 200 units (Taberlay et al. 2011; You et al. 2011), or 200 + 100 units (Andreu-Vieyra et al. 2011). Since whole-genome NOMe-seq required accurate footprinting of a variety of genomic regions, we performed a dose response curve (Fig. 1; Supplemental Fig. S1); nuclei were incubated with 100 or 200 units of GpC methyltransferase (M.CviPI) and S-Adenosyl methionine (SAM) for 15 min at 37°C or 200 units of GpC methyltransferase (M.CviPI) and SAM for 7.5 min at 37°C followed by a boost with an additional 100 units M.CviPI and SAM for 7.5 min. For whole-genome NOMe-seq, libraries were generated from nuclei that were incubated with 200 units of GpC methyltransferase (M.CviPI) and SAM for 7.5 min at 37°C followed by a boost with an additional 100 units M.CviPI and SAM for 7.5 min. The reaction was stopped, DNA extracted and bisulfite-converted to distinguish methylated from unmethylated Cs. For individual regions of interest, PCR was performed, using PCR primers that do not contain any CpG or GpC dinucleotides, followed by TA cloning and sequencing. Sequences of PCR primers are available upon request.
Library construction and sequencing
For NOMe-seq, libraries were prepared from 5 ug of DNA as previously described (Lister et al. 2009; Kelly et al. 2010; Berman et al. 2012). Briefly, M.CviPI-treated DNA was fragmented into ∼200-bp pieces, END-repaired (Epicenter), methylated adaptors ligated (Illumina), bisulfite-converted (Zymo EZ DNA methylation), and subjected to PCR. Clusters were generated following Illumina protocols, and the resulting library was sequenced on Illumina Hi-seq 2000 using the 76-bp single-end configuration. Each glioblastoma sample was sequenced using the same approach, except that they were sequenced using the Illumina Hi-seq 2000 Paired-End protocol. Base calling was performed by Illumina Real Time Analysis (RTA) software, yielding a total of 1.180 million reads that passed the Illumina quality filter (IMR90). GBM culture #157 was sequenced with one lane of 50-bp paired-end (310 million reads) and one lane of 100-bp paired-end (291 million reads), while culture #248 was sequenced with one lane of 50-bp paired-end (313 million reads) and one lane of 100-bp paired-end (301 million reads) (Supplemental Table S4).
Sequence alignment and extraction of CG and GC methylation levels
Genomic alignment and bisulfite sequence analysis was performed largely as previously described (Berman et al. 2012), with some adjustments for paired-end sequencing. For single-end IMR90 libraries, MAQ (Li et al. 2008) was used with the “-c” bisulfite mode (as in Berman et al. 2012), and for paired-end GBM libraries, BSMAP (Xi and Li 2009) was used. IMR90 reads were aligned to NCBI reference genome hg18 and GBM sequences to hg19. Genomic alignments with a mapping quality of less than 30 were filtered out, resulting in 678 million reads (IMR90), 587 million (GBM #157), and 691 million (GBM #248). For IMR90 and GBM cells, we removed reads starting at exactly the same genomic position as another read (PCR “duplicate” reads), yielding a total of 156 million analyzable reads for IMR90 (11.8 gigabases). For GBM paired-end, we additionally removed reads not “properly paired” (mapping to opposing strands within 500 bp of each other), yielding a total of 462 million analyzable reads (34.0 gigabases) for GBM #157 and 492 million analyzable reads (36.4 gigabases) for GBM #248.
It is difficult or impossible to distinguish C to T SNPs in bisulfite sequencing data, but our Illumina protocol only recovers bisulfite data from one of the two strands (G residues complementary to cytosines are read as G whether or not the complementary cytosine is methylated). For this “directional” bisulfite library protocol, cytosine positions appear on the sequence reads as C or T depending on bisulfite conversion, whereas the complementary G on the strand opposite the C will only be read as G (Krueger et al. 2012). We, therefore, refer to two strands relative to a given cytosine position—the “bisulfite-C strand” (BCS) and the “genotype G strand” (GGS). The genotype G strand is thus named because it reveals the true genotype of the position, unaffected by bisulfite conversion. Because of the specifics of Hi-Seq paired-end sequencing, the second end of a paired-end run is always the reverse complement of the BCS sequence and must be reverse-complemented before analysis to obtain the true BCS sequence.
We only included cytosines present in the reference genome if at least 90% of reads mapping to the BSC strand were C or T, and this included at least three reads. Additionally, we only included cytosines where 90% of the reads mapped to the GGS were G (any other base indicates a genetic variant; importantly, only the GGS strand can reveal the C > T transitions that can lead to false methylation calling). A cytosine was determined to be in a particular XCX trinucleotide context using the same criteria, e.g., GCH positions were only included if 90% of reads were G for the preceding base and 90% of the reads were A, C, or T (IUPAC “H” symbol includes A,C,T) for the following base. Reads on the BCS strand were treated as described above, i.e., either a C or T could match a C in either of the “X” context positions. This approach was used to determine the following trinucleotides discussed in this study: HCG (H includes A, C, or T), GCG, WCG (W includes A or T), and GCH.
As in Berman et al. (2012), we filter out the 5′ ends of reads that have apparent bisulfite nonconversion, which is common in the Illumina protocol presumably due to reannealing of base pairs adjacent to the adapter sequences which are methylated and thus have 100% base complementarity (Hansen et al. 2011; Berman et al. 2012). We accomplish this by walking inward from the 5′ of the sequencing read and disregarding any unconverted cytosine (in any sequence context) until the first converted cytosine is encountered. From that point and all 3′ positions within the read, we include all converted and unconverted cytosines in methylation counts.
For all downstream analyses, we included CCG trinucleotides, despite the slight off-target M.CviPI activity described that only affects CG methylation information. Thus, methylation averages include all HCG trinucleotides The single exception was the within-read combination plots (Fig. 4), where the very large number of data points being averaged allowed us to exclude CCGs and use only WCG trinucleotides (W: A,T).
Genomic element average profile plots
Methylation values were extracted from regions surrounding genomic landmarks of interest (promoters, CTCF sites, etc.), and all methylation values were averaged within moving windows of 20 bp for all plots (genomic positions without cytosines of the correct type were not included in averages). Twenty bp was chosen because it is smaller than the average distance between adjacent GCs in the genome and clearly able to resolve nucleosome phasing/positioning (as evidenced in CTCF alignments).
Promoter positions, chromatin marks, and expression values were taken from Supplemental Table 7 (mmc6.xls) of a previous reference (Hawkins et al. 2010; GEO ID GSE16256). Enhancers with a H3K4me1+/H3K4me3− profile were taken from Supplemental Table 12 (mmc11.xls) of the same reference (Hawkins et al. 2010; GEO ID GSE16256). IMR90 DNase hypersensitivity data is from GEO ID GSM468792. Histone and EP300 (also known as p300) locations from Neural Progenitor Cells were taken from a second reference (Rada-Iglesias et al. 2011). H3K27me3-enriched regions are those elements beginning with “R” (for region) in GEO record GSM602301, while EP300 calls are from GEO record GSM602299. H3K4me3 marks were not included in GEO and were provided by Alvaro Rada-Iglesias (available upon request). For CpG island and non-CpG island promoters, we used the Takai-Jones definition (Takai and Jones 2002). For CTCF annotations, we used evolutionarily conserved CTCF binding motifs (Xie et al. 2007) that were bound in vivo in either HeLa cells (Kim et al. 2007) or CD4+ T-cells (Figs. 2B, 3A, GBM; Cuddapah et al. 2009) and those obtained using ChIP-seq in IMR90 cells from GEO record GSM935404 (Fig. 3A, IMR90). We removed ∼10% of these sites that fell within 2 kb of a known TSS. Our final set contained 8722 nonpromoter CTCF sites (Supplemental Table S2).
Promoter nucleosome-depleted region detection
Identification of promoter nucleosome-depleted regions (Supplemental Fig. S7) was performed as follows: each unique TSS from the UCSC KnownGenes track was considered independently. All sequencing reads overlapping the candidate NDR region (−100 to +50 bp) were collected, and GCHs were analyzed on each read. Every GCH on each of the overlapping reads was counted as an independent nucleosome protection measurement, and only those with base quality phred scores of greater than 10 were included. Those TSSs with 10 or less such data points were removed from the analysis as regions of inadequate sequence coverage. This coverage filter removed 27,312 of 41,054 (66%) hg18 TSSs for IMR90, and 6225 (15%) and 4009 (10%) of 41,017 hg19 TSSs for GBM cultures #157 and #248, respectively. For each sample, the frequency of methylation among these independent GCH measurements within the candidate NDR region was compared to the frequency within the surrounding 8 kb—the 4 kb directly upstream of the candidate −100- to +50-bp region and the 4 kb directly downstream. We used a one-tailed binomial test to test whether the frequency of GCH methylation within the candidate NDR region was higher (i.e., less nucleosome protection) than the surrounding 8 kb. The binomial test resulted in raw P-values, which were corrected for multiple hypotheses (Benjamini-Hochberg) in each sample independently, using the number of TSSs passing the initial coverage filter in that particular sample as the number of hypotheses. Lists of all TSSs, methylation frequencies in candidate NDR and surrounding regions, and raw and corrected P-values for each sample are available as Supplemental Tables S5–S7.
Intersections between NDR calls from the three samples and histone marks (Venn diagrams in Supplemental Fig. S7) were generated as follows: The “universe” of TSSs considered for a given Venn diagram included only those that passed the coverage filter for all the samples included in the intersection, i.e., for Supplemental Figure S7C, only the 12,424 TSSs covered by all three cell types were included, while in Supplemental Figure S7D, only the 33,425 TSSs covered by both GBM samples were included; all histone-marked TSSs within this given subset were considered.
Combinatorial epigenomic signatures
Nucleosome protection comparisons (from GCH) stratified by DNA methylation state (Fig. 4) were performed as follows: Each GCH in the reference genome within 1 kb of a TSS (or other element, as listed) was evaluated independently. For each such genomic position, each read mapping to the bisulfite-C strand was analyzed for within-read associations with “nearby” CGs. Each WCG within 20 bp upstream or downstream was considered “nearby” (chosen as a distance that could resolve nucleosome positioning). If the nearby WCG was methylated, the GCH methylation value for the read was stratified into the “methylated” bin (red lines in Fig. 4); likewise, those reads where the nearby CG was unmethylated went into the unmethylated bin (Fig. 4, blue lines). If a single GCH was within 20 bp of multiple CGs, the methylation value of each of the multiple CGs in each read went into the appropriate (methylated or unmethylated) bin as an independent observation.
To generate the plots in the right-hand plots of Figure 4A (labeled “on same read”), these methylated and unmethylated GCH bins were averaged across all genomic elements to yield two average GC profiles for the methylated (red) and unmethylated (blue) bins. For the left-hand plots (labeled “across all reads”), the entire analysis was performed identically, except that “nearby” CG methylation values were taken from a randomly selected read mapping to the same location, rather than the same read as the GC. Generally, multiple reads overlapped the same position, but we only selected one read at random to keep the number of observations identical to the “on same read” condition, eliminating any possible effects from differences in variance between the two conditions.
Divergent chromatin allele promoter detection
Identification of promoters with “divergent chromatin alleles” (Fig. 4; Supplemental Fig. S9) was performed as follows: we only counted reads that had two or more GCHs and two or more HCGs, with 90% of cytosines in each category being in agreement. For each TSS from the UCSC KnownGenes track, we selected those reads where more than half of the read fell within (−150 to +100 bp). Any gene with at least one read in the “active” chromatin combination state (CG unmethylated and GC nucleosome-accessible) and another read in the “silenced” state (CG methylated and GC nucleosome-protected) was counted as a DCA gene. The fraction of these falling onto chromosome X or associated with imprinted genes was compared to size-matched sets picked randomly from the genome, as described in the Figure 4 legend.
Data access
NOMe-seq tracks for genomic viewers (Fig. 2; Supplemental Fig. S2) are available as a supplemental document and at http://epigenome.usc.edu and the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE21823. All source code tools are available at http://sourceforge.net/projects/uecgatk/. See Supplemental Material for instructions on using these tools. The tool and source code for the new module of the IGV viewer to display NOMe-seq data from raw BAM alignment files are publicly available for download at the IGV project website, http://www.broadinstitute.org/igv/.
Acknowledgments
We thank Charlie Nicolet, Selene Tyndale, and Helen Truong for support in generating sequencing data and members of the Jones lab and USC Epigenome Center for helpful discussions. GBM cells were a generous gift from Dr. Harley Kornblum, of the UCLA Intellectual and Developmental Disabilities Research Center, Human Cell Core. We also thank Alvaro Rada-Iglesias and Joanna Wysocka for providing NEC ChIP-sequencing data. This work was funded by NCI 5R37CA082422-13 to P.A.J., T32-CA009320-77 and 1K99CA160349-01 to T.K.K., T32 CBM Training Grant to F.D.L., and the generous support of the Kenneth T. and Eileen L. Norris Foundation to Y.L. and B.P.B. Computation was performed at the USC High Performance Computing and Communications Center (http://www.usc.edu/hpcc/).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.143008.112.
References
- Andreu-Vieyra C, Lai J, Berman BP, Frenkel B, Jia L, Jones PA, Coetzee GA 2011. Dynamic nucleosome depleted regions at androgen receptor enhancers in the absence of ligand in prostate cancer cells. Mol Cell Biol 31: 4648–4662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Berman BP, Weisenberger DJ, Aman JF, Hinoue T, Ramjan Z, Liu Y, Noushmehr H, Lange CP, van Dijk CM, Tollenaar RA, et al. 2012. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nat Genet 44: 40–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, Kellis M, Marra MA, Beaudet AL, Ecker JR, et al. 2010. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol 28: 1045–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouazoune K, Miranda TB, Jones PA, Kingston RE 2009. Analysis of individual remodeled nucleosomes reveals decreased histone-DNA contacts created by hSWI/SNF. Nucleic Acids Res 37: 5279–5294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodavarapu RK, Feng S, Bernatavichute YV, Chen PY, Stroud H, Yu Y, Hetzel JA, Kuo F, Kim J, Cokus SJ, et al. 2010. Relationship between nucleosome positioning and DNA methylation. Nature 466: 388–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choufani S, Shapiro JS, Susiarjo M, Butcher DT, Grafodatskaya D, Lou Y, Ferreira JC, Pinto D, Scherer SW, Shaffer LG, et al. 2011. A novel approach identifies new differentially methylated regions (DMRs) associated with imprinted genes. Genome Res 21: 465–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, Zhao K 2009. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res 19: 24–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal-Yam EN, Jeong S, Tanay A, Egger G, Lee AS, Jones PA 2006. Constitutive nucleosome depletion and ordered factor assembly at the GRP78 promoter revealed by single molecule footprinting. PLoS Genet 2: e160 doi: 10.1371/journal.pgen.0020160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD 2007. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res 17: 877–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KD, Timp W, Bravo HC, Sabunciyan S, Langmead B, McDonald OG, Wen B, Wu H, Liu Y, Diep D, et al. 2011. Increased methylation variation in epigenetic domains across cancer types. Nat Genet 43: 768–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, Pelizzola M, Edsall LE, Kuan S, Luu Y, Klugman S, et al. 2010. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell 6: 479–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessen WJ, Dhasarathy A, Hoose SA, Carvin CD, Risinger AL, Kladde MP 2004. Mapping chromatin structure in vivo using DNA methyltransferases. Methods 33: 68–80 [DOI] [PubMed] [Google Scholar]
- Jessen WJ, Hoose SA, Kilgore JA, Kladde MP 2006. Active PHO5 chromatin encompasses variable numbers of nucleosomes at individual promoters. Nat Struct Mol Biol 13: 256–263 [DOI] [PubMed] [Google Scholar]
- Kelly TK, Miranda TB, Liang G, Berman BP, Lin JC, Tanay A, Jones PA 2010. H2A.Z maintenance during mitosis reveals nucleosome shifting on mitotically silenced genes. Mol Cell 39: 901–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilgore JA, Hoose SA, Gustafson TL, Porter W, Kladde MP 2007. Single-molecule and population probing of chromatin structure using DNA methyltransferases. Methods 41: 320–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B 2007. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell 128: 1231–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F, Kreck B, Franke A, Andrews SR 2012. DNA methylome analysis using short bisulfite sequencing data. Nat Methods 9: 145–151 [DOI] [PubMed] [Google Scholar]
- Laks DR, Masterman-Smith M, Visnyei K, Angenieux B, Orozco NM, Foran I, Yong WH, Vinters HV, Liau LM, Lazareff JA, et al. 2009. Neurosphere formation is an independent predictor of clinical outcome in malignant glioma. Stem Cells 27: 980–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ruan J, Durbin R 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res 18: 1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JC, Jeong S, Liang G, Takai D, Fatemi M, Tsai YC, Egger G, Gal-Yam EN, Jones PA 2007. Role of nucleosomal occupancy in the epigenetic silencing of the MLH1 CpG island. Cancer Cell 12: 432–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. 2009. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462: 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Siegmund KD, Laird PW, Berman BP 2012. Bis-SNP: Combined DNA methylation and SNP calling for Bisulfite-seq data. Genome Biol 13: R61 doi: 10.1186/gb-2012-13-7-r61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda TB, Kelly TK, Bouazoune K, Jones PA 2010. Methylation-sensitive single-molecule analysis of chromatin structure. Curr Protoc Mol Biol 89: 21.17.1–21.17.16 [DOI] [PubMed] [Google Scholar]
- Nagy PL, Price DH 2009. Formaldehyde-assisted isolation of regulatory elements. Wiley Interdiscip Rev Syst Biol Med 1: 400–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabayashi K, Trujillo AM, Tayama C, Camprubi C, Yoshida W, Lapunzina P, Sanchez A, Soejima H, Aburatani H, Nagae G, et al. 2011. Methylation screening of reciprocal genome-wide UPDs identifies novel human-specific imprinted genes. Hum Mol Genet 20: 3188–3197 [DOI] [PubMed] [Google Scholar]
- Pardo CE, Carr IM, Hoffman CJ, Darst RP, Markham AF, Bonthron DT, Kladde MP 2010. MethylViewer: Computational analysis and editing for bisulfite sequencing and methyltransferase accessibility protocol for individual templates (MAPit) projects. Nucleic Acids Res 39: e5 doi: 10.1093/nar/gkq716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J 2011. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470: 279–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K 2008. Dynamic regulation of nucleosome positioning in the human genome. Cell 132: 887–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taberlay PC, Kelly TK, Liu CC, You JS, De Carvalho DD, Miranda TB, Zhou XJ, Liang G, Jones PA 2011. Polycomb-repressed genes have permissive enhancers that initiate reprogramming. Cell 147: 1283–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai D, Jones PA 2002. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc Natl Acad Sci 99: 3740–3745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdottir H, Robinson JT, Mesirov JP 2012. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief Bioinform doi: 10.1093/bib/bbs017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson BG, Roberts CW 2011. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer 11: 481–492 [DOI] [PubMed] [Google Scholar]
- Wolff EM, Byun HM, Han HF, Sharma S, Nichols PW, Siegmund KD, Yang AS, Jones PA, Liang G 2010. Hypomethylation of a LINE-1 promoter activates an alternate transcript of the MET oncogene in bladders with cancer. PLoS Genet 6: e1000917 doi: 10.1371/journal.pgen.1000917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Y, Li W 2009. BSMAP: Whole genome bisulfite sequence MAPping program. BMC Bioinformatics 10: 232 doi: 10.1186/1471-2105-10-232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Mikkelsen TS, Gnirke A, Lindblad-Toh K, Kellis M, Lander ES 2007. Systematic discovery of regulatory motifs in conserved regions of the human genome, including thousands of CTCF insulator sites. Proc Natl Acad Sci 104: 7145–7150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Kladde MP, Van Etten JL, Simpson RT 1998. Cloning, characterization and expression of the gene coding for a cytosine-5-DNA methyltransferase recognizing GpC. Nucleic Acids Res 26: 3961–3966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You JS, Kelly TK, De Carvalho DD, Taberlay PC, Liang G, Jones PA 2011. OCT4 establishes and maintains nucleosome-depleted regions that provide additional layers of epigenetic regulation of its target genes. Proc Natl Acad Sci 108: 14497–14502 [DOI] [PMC free article] [PubMed] [Google Scholar]