Abstract
Chromosome rearrangements in small apes are up to 20 times more frequent than in most mammals. Because of their complexity, the full extent of chromosome evolution in these hominoids is not yet fully documented. However, previous work with array painting, BAC-FISH, and selective sequencing in two of the four karyomorphs has shown that high-resolution methods can precisely define chromosome breakpoints and map the complex flow of evolutionary chromosome rearrangements. Here we use these tools to precisely define the rearrangements that have occurred in the remaining two karyomorphs, genera Symphalangus (2n = 50) and Hoolock (2n = 38). This research provides the most comprehensive insight into the evolutionary origins of chromosome rearrangements involved in transforming small apes genome. Bioinformatics analyses of the human–gibbon synteny breakpoints revealed association with transposable elements and segmental duplications, providing some insight into the mechanisms that might have promoted rearrangements in small apes. In the near future, the comparison of gibbon genome sequences will provide novel insights to test hypotheses concerning the mechanisms of chromosome evolution. The precise definition of synteny block boundaries and orientation, chromosomal fusions, and centromere repositioning events presented here will facilitate genome sequence assembly for these close relatives of humans.
The small apes (gibbons) are one of the most dramatic examples of extremely rapid karyotype evolution, and it is intriguing in this respect to note that among the apes (superfamily Hominoidea) the hylobatids have the highest number of species. There is little agreement on the exact numbers, but there are from 14 to 19 living gibbon species. Chromosomal changes in gibbons are up to 20 times that of the average mammalian rate and are only surpassed by some muroid rodents. The importance of understanding their rapid genome evolution is also provided by their phylogenetic affinity to humans. Gibbons and humans diverged about 17–23 million years ago (Matsudaira and Ishida 2010) and are classified in the same superfamily Hominoidea.
Yet, a satisfactory explanation of why gibbons experienced such an accelerated rate of evolution has escaped our understanding. One reason is that the full extent of their chromosomal changes is not yet well-documented, even at the molecular cytogenetic level. Because the rearrangements are so complex, chromosome painting, the mostly widely applied molecular cytogenetic technique (Muller et al. 2003), did not allow final conclusions about the number of rearrangements and about the steps that led to the four existing karyomorphs that typify each genus of small apes: Hoolock (2n = 38), Hylobates (2n = 44), Symphalangus (2n = 50), and Nomascus (2n = 52). Molecular cytogenetic techniques now allow a significantly higher resolution than chromosome painting through the use of hybridization of precisely mapped BAC clones, microarrays, and selective sequencing. Using these approaches we recently defined the chromosomal changes and synteny block organization of Hylobates lar (HLA, lar gibbon) and Nomascus leucogenys (NLE, white-cheeked gibbon) (Carbone et al. 2006; Roberto et al. 2007; Misceo et al. 2008; Girirajan et al. 2009). Yet, the reconstruction of the evolutionary history of rearrangements is only as complete as the taxonomic array of relevant species. Here, we report a comparable detailed analysis of the chromosomes of the two remaining karyomorphs, Hoolock leuconedys (HLE, eastern hoolock gibbon) and Symphalangus syndactylus (SSY, siamang gibbon). For the first time, we provide an analysis involving the complete taxon set of the four small ape karyomorphs. These data permit a more complete and accurate reconstruction of their ancestral genome and provides the basis to understand the steps that led to the amazing chromosomal diversity found today among small apes. This study provides the most comprehensive insight into the evolutionary origins of chromosome rearrangements involved in transforming the genome in small apes. In addition, the close evolutionary relationship between small apes, Hominidae, and Old World monkeys also means that the results are set against the detailed evolutionary history of these species, the human genome in particular. The comparison provides exquisite resolution to understand the flow of chromosome rearrangements and to place each rearrangement on a phylogenetic tree.
Results
The arrangement of conserved syntenic blocks in Hoolock and Symphalangus were defined by cohybridization FISH experiments of 450 and 550 selected human BAC clones on HLE and SSY chromosomes, respectively. In Hoolock, array-painting experiments were also performed.
Cross-species BAC array-paintings in Hoolock
Cross-species array-painting is an effective strategy for mapping breakpoints of evolutionary interchromosomal rearrangements and obtaining preliminary physical maps of complex karyotypes such as that of HLE. Chromosomes of HLE were flow-sorted, the chromosomal DNAs labeled with fluorescent dyes (Cy3 and Cy5) and hybridized onto human BAC-arrays, as previously described (Carbone et al. 2006). In order to reduce the number of experiments, flow-sorted HLE chromosomes were grouped into five pools (Supplemental Table ST1). Each pool was hybridized onto a microarray slide containing >32,000 BACs, covering the whole human genome (Krzywinski et al. 2004), and the results were analyzed as described by Carbone et al. (2006) (Methods; Supplemental Fig. SF1). This high-throughput approach enabled us to quickly identify HLE/human synteny breakpoints with an average resolution of 340 Kb.
FISH experiments in Hoolock and Symphalangus
Microarray results (for the Hoolock) and all published data on Hylobatidae provided criteria for the selection of BACs. In particular, we took into consideration the Hylobatidae ancestral karyotype proposed by Misceo et al. (2008) and the FISH data already published on other gibbon species (Muller et al. 2003; Roberto et al. 2007; Misceo et al. 2008; Girirajan et al. 2009).
FISH was first used to test whether the rearrangements hypothesized to have occurred in the Hylobates ancestor (HyA) (Misceo et al. 2008) were also present in H. leuconedys and S. syndactylus. We then set off to characterize species-specific rearrangements. Every breakpoint was identified by a splitting human BAC or by two overlapping BACs mapping to opposite sides of the break. These experiments allowed a fine definition of the synteny organization of the HLE and SSY with respect to the human genome. The results are graphically summarized in Figures 1 and 2. The results of all the FISH experiments are reported in the Supplemental Tables ST2 and ST3. In order to facilitate the comparison with previous data on HLA and NLE, the BAC positions in these Supplemental Tables refer to the UCSC May 2006 release (hg18). The relevant data are also displayed in a user friendly, figurative way, at the website www.biologia.uniba.it/hoolock for Hoolock and www.biologia.uniba.it/siamang for siamang.
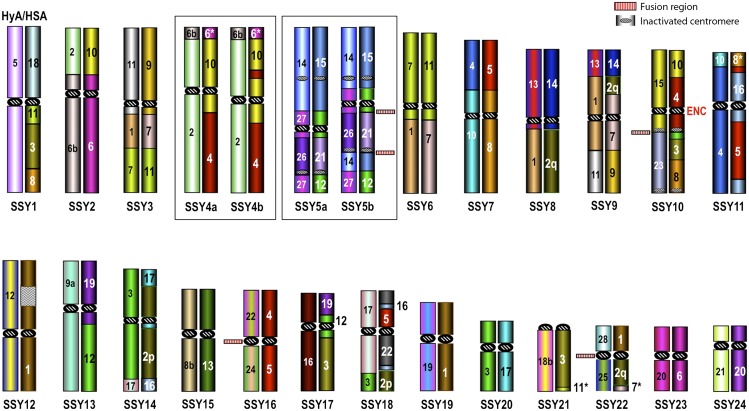
Figure 1.
Two distinct ideograms for each of the 18 HLE autosomes. The ideogram on the left reports the synteny block arrangement with respect to the Hylobatidae ancestral karyotype (HyA) reported in Figure 3, to which the colors also refer. The one on the right reports the homologous human blocks. For details, see Supplemental Table ST2 or the website http://www.biologia.uniba.it/hoolock. This figure also reports the fusion points, the evolutionary new centromeres (ENC), and inactivated centromeres.
Figure 2.
Two distinct ideograms for each of the 24 SSY autosomes. The ideogram on the left reports the synteny block arrangement with respect to the Hylobatidae ancestral karyotype (HyA), reported in Figure 3, to which the colors also refer. The one on the right reports the homologous human blocks. For details see Supplemental Table ST3 or the website http://www.biologia.uniba.it/siamang. This figure also reports the fusion points, the evolutionary new centromeres (ENC), and inactivated centromeres.
Comparison of the karyotypes of each of the four Hylobatidae karyomorphs, genera Hoolock, Hylobates, Symphalangus, and Nomascus, allowed a precise reconstruction of the karyotype of their common ancestor (HyA). We compared the four gibbon species to see how many of them shared a specific breakpoint, assuming the hypothesized Hominoidea ancestor as the starting point (Stanyon et al. 2008). By applying cladistics principles we were able to discriminate the gibbon-specific rearrangements from those that occurred in the lineage leading to humans. The breakpoints shared by all of the four genera and not present in the Hominoidea ancestor were considered to have occurred in the common ancestor of all living small apes species (HyA) (see Fig. 3). Those not present in all four species were considered to have occurred after gibbon divergence from the common ancestor. The results for HLE and SSY are summarized in Figures 1 and 2 and below. Data on HLA and NLE have been already reported (Carbone et al. 2006; Roberto et al. 2007; Misceo et al. 2008; Girirajan et al. 2009).
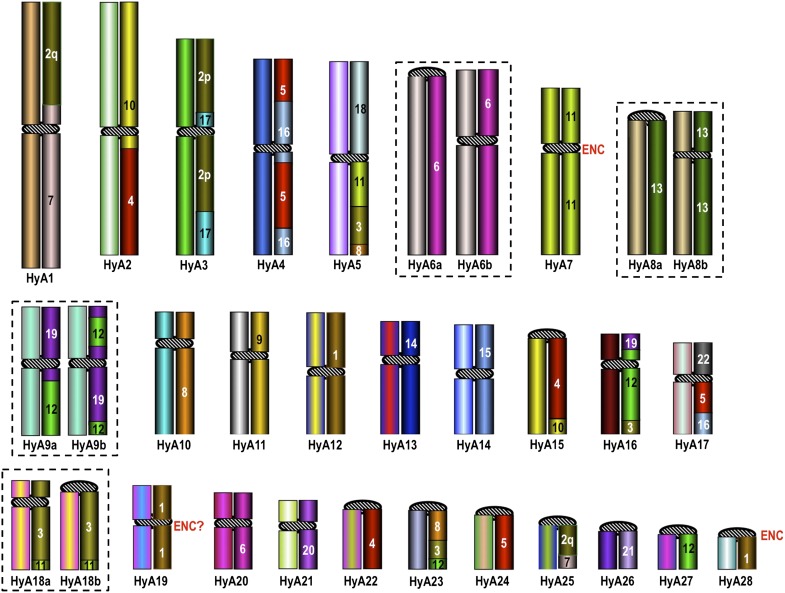
Figure 3.
Hylobatidae ancestral karyotype (HyA). The chromosomes on the right report the human chromosomes contributing to each HyA chromosome. In most cases, HyA chromosomes are a mosaic of various fragments of human chromosomes. These details, not annotated in the Figure, are reported in Supplemental Table ST4.
Novel chromosomal rearrangements in Hoolock and Symphalangus
The previously available synteny arrangements in HLE and SSY were based on chromosome painting (Koehler et al. 1995a,b; Nie et al. 2001; Muller et al. 2003). It is generally appreciated that BAC-FISH has a considerably higher resolution than chromosome painting. It was no surprise, therefore, to discover small chromosomal segments from various human chromosomes, which had previously gone undetected with chromosome paints. They are indicated by an asterisk in Figures 1 and 2. In detail:
HLE: HSA4p16.3 (∼2 Mb) in HLE1; HSA7p22.3 (∼2.5 Mb) in HLE4; HSA11p15.5 (∼1 Mb) in HLE6; HSA7p22.2 (∼2 Mb) in HLE7; HSA15q21.1 (∼1.2 Mb) and HSA11p15.5 (∼2 Mb) in HLE11; HSA16p12.3 (∼3.3 Mb) and HSA22q11 (∼0.5 Mb) in HLE13;
SSY: HSA6q14.1-14.3 (∼10 Mb) in SSY4a/b; HSA8q13.2-13.3 (∼5 Mb) in SSY11; HSA11p15.5 (∼1 Mb) in SSY21; HSA7p22.3 (∼2.5 Mb) in SSY22. Note that the last two segments were also discovered by BAC-FISH in HLE.
Of note, chromosomes 4 and 5 in the siamang, as reported in Figure 2, exist in two forms. However, while a single inversion can reconcile 4b (derivative) with 4a, both 5a and 5b are, very likely, the results of two independent inversions in a chromosome that was formed in the siamang from the fusion of the three distinct small chromosomes HyA14, HyA26, and HyA27 of the Hylobatidae ancestor (HyA) (see below). We cannot exclude, however, that the initial chromosome 5 form is still present in the siamang population, but has not yet been sampled.
Genomic features associated with chromosomal breakpoints
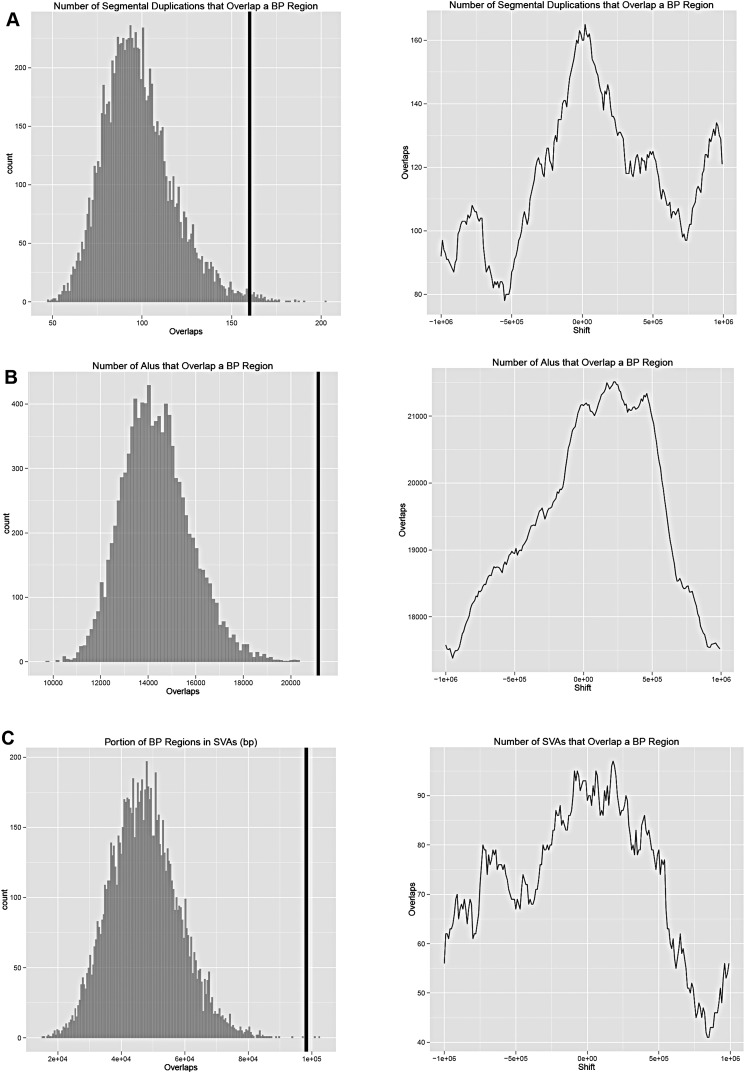
To search for genomic features potentially associated with gibbon chromosomal breakpoints we computed the significance of the overlap between the breakpoint regions and a set of genomic features of interest using permutation tests. The analysis was performed on the human hg19 assembly. The features examined were genes, human segmental duplications, and some repeat families (Alu, LINE, ERV, and SVA). Moreover, we investigated the associations between breakpoint regions and chromatin structure by testing the overlap with open chromatin regions in human embryonic stem cells reported by the ENCODE consortium (The Encode Project Consortium 2011). We found a significant enrichment for genes (Bonferroni adjusted P-value = 0.0287), human segmental duplications (P = 0.0366), Alu (P < 0.0001), and SVA (P = 0.0008) (Fig. 4A). We did not find significant enrichment for LINE and ERV repeats, nor for the ENCODE open chromatin regions. Systematically shifting the location of breakpoint regions by increments of 10 kb up- and downstream of their actual location, up to a maximum of 1 MB, shows that the locations of the breakpoint regions gives the greatest or close to the greatest number of overlaps with the four significantly overlapping features (genes, segmental duplications, Alu, and SVA) in the local genomic neighborhood (Fig. 4B).
Figure 4.
Enrichment of genomic features in breakpoint regions. Permutation tests were used to assess the overlap between the gibbon breakpoints and genomic features. (A) Segmental duplications; (B) Alu elements; and (C) SVA elements. The black vertical line indicates the observed value for the breakpoints identified in the study. In all three cases it is evident that the genomic features have a higher overlap with the breakpoints than one could expect by chance.
SVA elements are infrequent in gibbons and greatly expanded in human and chimpanzee (Wang et al. 2005). It is therefore very unlikely that these elements had an impact on the gibbon rearrangements. However, SVAs are known to correlate with Alu elements due to their preference for G+C and gene-rich regions of the genome (Wang et al. 2005) and the fact that they use the same L1-based insertion site and mechanism (Raiz et al. 2012). It seems likely that the association between human SVA locations and gibbon breakpoint regions is therefore an indirect one, dependent on the presence of additional genomic features present in both humans and gibbons. We therefore tested the association between SVAs and Alus in the breakpoint regions. First, we adjusted for the varying lengths of the breakpoint regions by finding the rate of occurrence of Alu and SVA per 10 kb in each breakpoint. These rates are correlated (Kendall's tau = 0.214, P = 0.002), indicating that Alus tend to colocalize with SVAs in these regions.
Discussion
Using cross-species array painting and FISH mapping of large panels of human BAC clones we were able to generate a detailed synteny map of the H. leuconedys and S. syndactylus genomes. Figures 1 and 2 report the synteny block organization of these two species with respect to the reconstructed Hylobates ancestral karyotype (see below) and to humans. Moreover, we precisely narrowed down the boundaries of each synteny block to an on average of ∼200 Kb.
Chromosomal rearrangements are one of the tools that have helped clarify the phylogenetic tree of numerous taxa. We compared the four gibbon genera in search of a coherent temporal order of genus divergence. In this context and according to cladistics procedures, we considered only shared derived rearrangements as illustrated in Figure 5. Chromosome rearrangements did not provide a consistent, simple dichotomic phylogeny. This result is typical of species that diverged in a relatively short period of time and can be attributed to incomplete lineage sorting. In agreement with this scenario we hypothesized that the HyA was heterozygous for variant forms of chromosomes 6, 8, 9, and 18, as reported in Figure 3. Our results are in agreement with the pattern found by Matsudaira and Ishida (2010) who sequenced the full mtDNA of the genera Hylobates, Nomascus, and Syndactylus and with Kim et al. (2011), who based their conclusions on 60 kb of sequence data from a panel of 19 gibbons representing nine species from all four genera.
Figure 5.
The ellipse in the middle shows the four chromosomes (6, 8, 9, and 18) hypothesized to be heterozygous for variant forms in the HyA. (Arrows) Segregations of each variant chromosome in the four genera. The table at the bottom makes clear the conflicting results generated by the grouping of the four genera when the variant forms are separately considered.
Refinement of the karyotype of the last common ancestor of the Hylobatidae (HyA)
In order to better understand the organization of the karyotype of the last common ancestor of gibbons we compared the karyotypes of HLE, SSY, HLA, and NLE (Roberto et al. 2007; Misceo et al. 2008; present data) with the karyotype of Hominoidea ancestor (Stanyon et al. 2008). The analysis showed that 33 rearrangements probably occurred in the gibbon ancestor after its divergence from Hominidae and before gibbon radiation. The results of the comparison allowed us to make modifications of the ancestral karyotype of all small apes (HyA) previously hypothesized by Misceo et al. (2008), which are reported in detail in Supplemental Table ST4 and summarized in Figure 3. The marker order of HyA27 (former HyA28), corresponding to a portion of HSA1, not resolved in Misceo et al. (2008), was clarified, and the former HyA24 (portions of HSA5 and HSA16) and HyA27 (HSA22) now form a single chromosome (now HyA17) in HyA. As a consequence, the diploid number of HyA was reduced from 60 to 58, and the chromosome numbering was revised accordingly. Finally, the presence of a small portion of HSA11 at the telomeric region of HyA18 (former HyA17) was definitely established as part of the HyA
HLE- and SSY-specific chromosomal changes
Once the HyA was reconstructed we could demonstrate that 22 rearrangements (11 translocations, one inversion, and 10 fusions) were Hoolock-specific (Supplemental Table ST2). The most notable difference between HLE and the Hylobatidae ancestor is the dramatic evolutionary reduction in chromosomal number from 58 to 38. The reduction was due to 10 chromosomal fusions: two fusions formed HLE1, HLE2, and HLE4, and one fusion each formed HLE3, HLE6, HLE8, HLE9, and HLE12. The ancestral chromosomes involved in the fusions are indicated in Figure 1. One fusion was Robertsonian (HLE4). In four fusions, one of the two chromosomes involved was an acrocentric (for details see Supplemental Table ST2). However, no interstitial telomeric signals were detected in the fusion chromosomes after FISH with telomeric specific probes (data not shown). The number of fusions found in the HLE genome is perhaps only exceeded by those found in the evolution of muntjac chromosomes (2n = 6/7) (Chi et al. 2005).
The many chromosomal changes that occurred in the H. leuconedys genome, fusions in particular, resulted in the inactivation of 13 centromeres. Their location is reported in Figure 1 and Supplemental Table ST2.
The siamang showed, with respect to the HyA, 14 specific rearrangements. Chromosomes 2, 3, 4a/b, 6, 7, 8, 9, 11, 14, and 18 were the result of seven translocations. Two inversions originated chromosomes SSY15 and SSY18. Four chromosomes (SSY5a/b, SSY10, SSY16, SSY22) were formed by chromosomal fusions. A fission of HyA3 gave origin to SSY14 and SSY20. As a result, the siamang karyotype dropped from 58 to 50 chromosomes (see Fig. 2). Three additional inversions generated chromosomal forms that were not fixed in the population (see below). All of these changes are reported in detail in Supplemental Table ST3.
The genomic analyses of the chromosomal breakpoints showed a significant association with segmental duplications, genes, Alu, and SVA elements. Such association had already been reported for NLE (Carbone et al. 2009b) and it seems to extend to the breakpoints shared by the four genera. The association between breakpoint regions and genes is surprising since breakage of coding and/or regulatory regions can be supposed to negatively affect the fitness. This effect seems to be somehow mitigated in gibbons in which the rearranged chromosomes were selected despite their falling in gene-dense regions (Carbone et al. 2009b). In evaluating these results, however, it is worth noting that they were obtained using the human sequence as a reference. They have to be considered, therefore, as very preliminary results to be tested on the gibbon reference genomes when they become available.
Evolutionary new centromeres (ENC)
ENC emergence consists in the appearance of a new centromere along a chromosome during evolution. We compared the mapping position of HLE and SSY centromeres with respect to NLE, HLA, and Hominoidea ancestor (Roberto et al. 2007; Misceo et al. 2008; Stanyon et al. 2008) in search of potential ENCs. The comparison showed that six hoolock and three siamang chromosomes harbor an ENC. Five were Hoolock- (HLE1, HLE5, HLE11, and HLE17) or Symphalangus-specific (SSY10) (Figs. 1, 2), while two (HLE9/SSY22 and HLE12/SSY6) were shared with all of the other gibbons and presumably occurred in their common ancestor (Fig. 3). The position of neocentromeres with respect to the human sequence is reported in Table 1. The ENC of HyA19 (Fig. 3) is flanked by two noncontiguous regions, with homology to human chromosome 1 (see Supplemental Table 4). Therefore, we could not precisely determine the time of seeding, indicated by the question mark on the figure.
Table 1.
Evolutionary new centromeres in HLE and SSY

The HLE11 neocentromere (HSA7q11.23) was seeded in a region that in humans is dense with multiple segmental duplications. In addition, it is noteworthy that a 250-kb gap is present in the middle of this domain. The more recent hg19 assembly (February 2009) did not resolve this sequence gap. Recently, Carbone et al. (2009a) have reported that the ENC of macaque chromosome 18 (HSA18q21.2) exactly corresponded to a gap in the human genome assembly (chr18:50,313,129–50,360,135). This gap was almost fully sequenced and it was shown to be ∼9 kb in size and composed of a satellite-like DNA that was hypothesized to have played a role in triggering neocentromere emergence. Similarly, it could be hypothesized that the gap corresponding to the seeding point of the HLE11 contained a satellite-like DNA that could have triggered the neocentromere formation.
Variant chromosomes and the gibbon radiation
As mentioned, variant forms of chromosomes 4 and 5 exist in the siamang. Examples of gibbon chromosomal heterozygosity have been reported: in Nomascus: an inversion of NLE7 generated chromosome NLE7b, and a translocation between chromosomes NLE1 and NLE22 generated variants 1b and 22b (Couturier and Lernould 1991; Koehler et al. 1995b). There is a polymorphism of chromosome 8, with three distinct forms, in the genera Hylobates (Stanyon et al. 1987; Van Tuinen et al. 1999; Hirai et al. 2003). These findings are not unexpected considering the very high rate of chromosomal reshuffling in gibbons. It would not be a surprise if additional chromosomal forms are present in natural populations, because cytogenetic studies in gibbons are scarce. Our knowledge about chromosomal variability in small ape species is derived from the analysis of very few individuals, most of them from animals maintained in captivity.
Concluding remarks
The high-resolution analysis of the Hoolock and Symphalangus karyotypes and the comparison with those of Hylobates and Nomascus enabled us to draw an exceptionally precise and more comprehensive picture of chromosome evolution in gibbons.
Our results also remove the discrepancy between chromosome phylogenies for small apes and those from other biomolecular studies. Indeed, the uncertainty in drawing a reliable phylogenetic tree of gibbons either with cytogenetic or molecular data is most likely due to the lineage sorting, a phenomena that has apparently occurred at both levels during the rapid evolutionary radiation of gibbons
There is no doubt that the small ape lineage has experienced an incredible burst of chromosomal rearrangements. A host of evolutionary processes and mechanisms may have operated to fix these changes including meiotic drive, recombination reduction, molecular divergence in rearranged chromosomes methylation and epigenetic architecture, as well as the demographic parameter (Misceo et al. 2008; Brown and Xu 2009; Carbone et al. 2009b; Israfil et al. 2011). A comprehensive picture of small ape evolution should also probably include consideration of demographic dynamics and the structure of reproductive units.
The future will very likely witness a flourish of projects based on next-generation sequencing technology, including a good number of small apes species. This methodology, however, poses serious problems in the correct sequence assembly, especially if applied to genomes whose synteny has been deeply reshuffled. In the panda genome assembly, for instance, even the position of the centromeres was not annotated (Li et al. 2010). The present data, therefore, constitute a significant contribution to future gibbon sequencing efforts.
Methods
Cross-species array-painting
The same set of flow-sorted chromosomes from H. leuconedys reported previously in Ferguson-Smith et al. (2005) were used in this study. DOP-amplified HLE sorted chromosomes were used as templates for a second round of DOP-PCR. The product resulting from this secondary amplification was purified and quantified and used for hybridization. Labeling and hybridization were performed as described in Veltman et al. (2002). The following changes were applied: 1 μg of each purified DOP-PCR product derived from flow-sorted chromosomes was labeled with Cy3-dUTP dye. Anonymous human reference DNA was obtained from the Children's Hospital Oakland Research Institute, amplified by DOP-PCR, and labeled with Cy5-dUTP. Labeling reactions were performed using the BioPrime total genomic labeling system (Invitrogen). Hybridization was carried overnight. Hybridization images were obtained by scanning the slides on a 4000B scanner (Axon Instruments, http://www.moleculardevices.com) and images were processed with the software GenePix Pro 5.1 (Axon Instruments). The GenePix Results files were analyzed using standard procedures (flagged spots removal, background subtraction, and loess normalization) using BASE (BioArray Software) (Saal et al. 2002). The final output was human chromosome-specific plots of Log2 ratio values/chromosome location. The subtraction method described by Carbone et al. (2006) was applied in order to reduce the background noise and enhance the resolution of breakpoint mapping.
FISH experiments
Metaphase preparations were obtained from a female lymphoblastoid, cell lines obtained from Hoolock leuconedys, and Symphalangus Syndactylus from the Gibbon Conservation Center. Most BAC clones were from the RP11 human library. Some BACs were from the CHORI-271 Nomascus leucogenys library (for both libraries, see http://www.chori.org/bacpac/). Extraction of total DNA from BACs was performed according to standard methods. Chromosome preparations were hybridized in situ with probes directly labeled with Cy3-dCTP, FluorX-dCTP, DEAC, and Cy5-dCTP by nick-translation, essentially as described by Lichter et al. (1990), with minor modifications. Briefly, 300 ng of labeled probe (total BAC DNA) was used for the FISH experiments; hybridization was performed at 37°C in 10 μL of hybridization buffer containing 2×SSC, 50% (v/v) formamide, 10% (w/v) dextran sulphate, 5 mg of COT1 DNA (Roche), and 3 mg of sonicated salmon sperm DNA. Post-hybridization washes of FISH experiments were performed at lower stringency: 37°C in 2 SSC-50% formamide (three times), followed by three washes at 42°C in 2 SSC (three times).
Chromosome identification was obtained by simultaneous DAPI staining, producing a Q-banding pattern. Some chromosomes are difficult to distinguish on the basis of DAPI banding. In these cases an appropriate BAC clone was always cohybridized, as a reference, to unambiguously identify the chromosome and/or the chromosome arm under study.
The use of human arrays and human BACs imply that all of the rearrangements are referred to the human genome. Consequently, chromosomal changes that occurred in the human lineage appear as “apparent” breaks in HLE and SSY. However, the human karyotype is relatively well conserved with respect to the common ancestor of Hominoidea and the 14 rearrangements that occurred in the human lineage are very well characterized (Stanyon et al. 2008). Therefore, we could easily discriminate the apparent breaks from the actual HLE and SSY breaks. The karyotype of the Hominoidea ancestor, as reconstructed by Stanyon et al. (2008), was used as a reference genome and compared to the Hoolock karyotype.
Digital images were obtained using a Leica DMRXA2 epifluorescence microscope equipped with a cooled CCD camera (Princeton Instruments). Cy3 (red), FluorX (green), DEAC, Cy5, and DAPI (blue) fluorescence signals, detected with specific bandpass filters, were recorded separately as grayscale images. Pseudocoloring and merging of images were performed using Adobe Photoshop software.
Karyotype reconstruction
The reconstruction of the karyotype of HyA was accomplished by comparing the synteny arrangement of the four gibbon species (HLA, HLE, NLE, and SSY) with respect to humans, obtained by BAC-FISH experiments, and with respect to the reconstructed karyotype of the Hominoidea ancestor (Stanyon et al. 2008). The rearrangements shared by all of the four gibbons and not present in the Hominoidea ancestor were assumed to have occurred in HyA. The karyotype of the Hominoidea ancestor was crucial to discriminate the apparent rearrangement that, in fact, had occurred in the lineage leading to humans. Then we defined the rearrangements that were specific to a single genus or shared by two or three genera. This information is reported in Figures 1 and 2.
Permutation analysis
To determine the significance of the overlap of features with the breakpoint regions we used a permutation approach. While maintaining the chromosomal assignment and length of breakpoint regions, we permuted their start coordinates 10,000 times using BEDTools version 2.16.2 (Quinlan and Hall 2010). Genomic regions annotated as centromeres and telomeres in the “Gaps” track of the hg19 build were excluded from possible random placements of the regions. Locations of the features were held constant. We then compared the number of features that overlapped a breakpoint region to the observed distribution of results among the randomly permuted regions, and used the quantile of the real observed value in that distribution as an estimate of the P-value of observing a value equal to or greater than the real observation. Estimated P-values were computed in the R programming language and plotted using the ggplot2 library (Wickham 2009).
Repeat elements were extracted from the rmsk table downloaded from the UCSC Genome Browser for human build hg19. OCRs were identified using the ENCODE track wgEncodeOpenChromSynthH1hescPk for hg19.
Data access
Microarray data have been deposited in the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE40024.
Acknowledgments
This publication is dedicated to the late Alan Mootnick. His passion for gibbons, scientific curiosity, and generosity over almost four decades led to a better understanding of these fascinating small apes. This work was supported by PRIN (Programmi di Ricerca di Interesse Nazionale) to M.R., N.A., and R.R.S.; CEGBA (Centro di Eccellenza Geni in campo Biosanitario e Agroalimentare) to M.R. and N.A.; and NIH (OD011092) to L.C.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.138651.112.
References
- Brown WR, Xu ZY 2009. The ‘kinetochore maintenance loop': The mark of regulation? Bioessays 31: 228–236 [DOI] [PubMed] [Google Scholar]
- Carbone L, Vessere GM, ten Hallers BFH, Zhu B, Osoegawa K, Mootnick A, Kofler A, Wienberg J, Rogers J, Humphray S et al. 2006. A high-resolution map of synteny disruptions in gibbon and human genomes. PLoS Genet 2: e223 doi: 10.1371/journal.pgen.0020223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone L, D'Addabbo P, Cardone MF, Teti MG, Misceo D, Vessere GM, de Jong PJ, Rocchi M 2009a. A satellite-like sequence, representing a “clone gap” in the human genome, was likely involved in the seeding of a novel centromere in macaque. Chromosoma 118: 269–277 [DOI] [PubMed] [Google Scholar]
- Carbone L, Harris RA, Vessere GM, Mootnick AR, Humphray S, Rogers J, Kim SK, Wall JD, Martin D, Jurka J, et al. 2009b. Evolutionary breakpoints in the gibbon suggest association between cytosine methylation and karyotype evolution. PLoS Genet 5: e1000538 doi: 10.1371/journal.pgen.1000538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi JX, Huang L, Nie W, Wang J, Su B, Yang F 2005. Defining the orientation of the tandem fusions that occurred during the evolution of Indian muntjac hromosomes by BAC mapping. Chromosoma 114: 167–172 [DOI] [PubMed] [Google Scholar]
- Couturier J, Lernould JM 1991. Karyotypic study of four gibbon forms provisionally considered as subspecies of Hylobates (Nomascus) concolor (Primates, Hylobatidae). Folia Primatol (Basel) 56: 95–104 [DOI] [PubMed] [Google Scholar]
- The Encode Project Consortium 2011. A user's guide to the Encyclopedia of DNA elements (ENCODE). PLoS Biol 9: e1001046 doi: 10.1371/journal.pbio.1001046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson-Smith MA, Yang F, Rens W, O'Brien PC 2005. The impact of chromosome sorting and painting on the comparative analysis of primate genomes. Cytogenet Genome Res 108: 112–121 [DOI] [PubMed] [Google Scholar]
- Girirajan S, Chen L, Graves T, Marques-Bonet T, Ventura M, Fronick C, Fulton L, Rocchi M, Fulton RS, Wilson RK, et al. 2009. Sequencing human-gibbon breakpoints of synteny reveals mosaic new insertions at rearrangement sites. Genome Res 19: 178–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai H, Mootnick AR, Takenaka O, Suryobroto B, Mouri T, Kamanaka Y, Katoh A, Kimura N, Maeda N 2003. Genetic mechanism and property of a whole-arm translocation (WAT) between chromosomes 8 and 9 of agile gibbons (Hylobates agilis). Chromosome Res 11: 37–50 [DOI] [PubMed] [Google Scholar]
- Israfil H, Zehr SM, Mootnick AR, Ruvolo M, Steiper ME 2011. Unresolved molecular phylogenies of gibbons and siamangs (Family: Hylobatidae) based on mitochondrial, Y-linked, and X-linked loci indicate a rapid Miocene radiation or sudden vicariance event. Mol Phylogenet Evol 58: 447–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SK, Carbone L, Becquet C, Mootnick AR, Li DJ, de Jong PJ, Wall JD 2011. Patterns of genetic variation within and between Gibbon species. Mol Biol Evol 28: 2211–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler U, Arnold N, Wienberg J, Tofanelli S, Stanyon R 1995a. Genomic reorganization and disrupted chromosomal synteny in the siamang (Hylobates syndactylus) revealed by fluorescence in situ hybridization. Am J Phys Anthropol 97: 37–47 [DOI] [PubMed] [Google Scholar]
- Koehler U, Bigoni F, Wienberg J, Stanyon R 1995b. Genomic reorganization in the concolor gibbon (Hylobates concolor) revealed by chromosome painting. Genomics 30: 287–292 [DOI] [PubMed] [Google Scholar]
- Krzywinski M, Bosdet I, Smailus D, Chiu R, Mathewson C, Wye N, Barber S, Brown-John M, Chan S, Chand S, et al. 2004. A set of BAC clones spanning the human genome. Nucleic Acids Res 32: 3651–3660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Fan W, Tian G, Zhu H, He L, Cai J, Huang Q, Cai Q, Li B, Bai Y, et al. 2010. The sequence and de novo assembly of the giant panda genome. Nature 463: 311–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichter P, Tang Chang C-J, Call K, Hermanson G, Evans GA, Housman D, Ward DC 1990. High resolution mapping of human chromosomes 11 by in situ hybridization with cosmid clones. Science 247: 64–69 [DOI] [PubMed] [Google Scholar]
- Matsudaira K, Ishida T 2010. Phylogenetic relationships and divergence dates of the whole mitochondrial genome sequences among three gibbon genera. Mol Phylogenet Evol 55: 454–459 [DOI] [PubMed] [Google Scholar]
- Misceo D, Capozzi O, Roberto R, Dell'Oglio MP, Rocchi M, Stanyon R, Archidiacono N 2008. Tracking the complex flow of chromosome rearrangements from the Hominoidea ancestor to extant Hylobates and Nomascus gibbons by high-resolution synteny mapping. Genome Res 18: 1530–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S, Hollatz M, Wienberg J 2003. Chromosomal phylogeny and evolution of gibbons (Hylobatidae). Hum Genet 113: 493–501 [DOI] [PubMed] [Google Scholar]
- Nie W, Rens W, Wang J, Yang F 2001. Conserved chromosome segments in Hylobates hoolock revealed by human and H. leucogenys paint probes. Cytogenet Cell Genet 92: 248–253 [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM 2010. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiz J, Damert A, Chira S, Held U, Klawitter S, Hamdorf M, Lower J, Stratling WH, Lower R, Schumann GG 2012. The non-autonomous retrotransposon SVA is trans-mobilized by the human LINE-1 protein machinery. Nucleic Acids Res 40: 1666–1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberto R, Capozzi O, Wilson RK, Mardis ER, Lomiento M, Tuzun E, Cheng Z, Mootnick AR, Archidiacono N, Rocchi M, et al. 2007. Molecular refinement of gibbon genome rearrangement. Genome Res 17: 249–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saal LH, Troein C, Vallon-Christersson J, Gruvberger S, Borg A, Peterson C 2002. BioArray Software Environment (BASE): A platform for comprehensive management and analysis of microarray data. Genome Biol 3: software0003.1–software0003.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanyon R, Sineo L, Chiarelli B, Camperio-Ciani A, Haimoff AR, Mootnick EH, Sutarman D 1987. Banded karyotypes of the 44-chromosome Gibbons. Folia Primatol (Basel) 48: 56–64 [Google Scholar]
- Stanyon R, Rocchi M, Capozzi O, Roberto R, Misceo D, Ventura M, Cardone M, Bigoni F, Archidiacono N 2008. Primate chromosome evolution: Ancestral karyotypes, marker order and neocentromeres. Chromosome Res 16: 17–39 [DOI] [PubMed] [Google Scholar]
- Van Tuinen P, Mootnick AR, Kingswood SC, Hale DW, Kumamoto AT 1999. Complex, compound inversion/translocation polymorphism in an ape: Presumptive intermediate stage in the karyotypic evolution of the agile gibbon Hylobates agilis. Am J Phys Anthropol 110: 129–142 [DOI] [PubMed] [Google Scholar]
- Veltman JA, Schoenmakers EF, Eussen BH, Janssen I, Merkx G, van Cleef B, van Ravenswaaij CM, Brunner HG, Smeets D, van Kessel AG 2002. High-throughput analysis of subtelomeric chromosome rearrangements by use of array-based comparative genomic hybridization. Am J Hum Genet 70: 1269–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Xing J, Grover D, Hedges DJ, Han K, Walker JA, Batzer MA 2005. SVA elements: A hominid-specific retroposon family. J Mol Biol 354: 994–1007 [DOI] [PubMed] [Google Scholar]
- Wickham H. 2009 ggplot2: elegant graphics for data analysis. Springer, New York. [Google Scholar]