Abstract
Cell fusion plays a well-recognized physiological role during development, while its function during progression is still unclear. Here, we show that acute myeloid leukemia (AML) cells spontaneously fused with murine host cells in vivo. AML cells fused in most cases with mouse macrophages. Other targets of AML cell fusion were dendritic and endothelial cells. Cytogenetic and molecular analysis revealed that successive recipients conserved detectable amounts of parental DNA. Moreover, in a mouse AML1-ETO model where female AML1-ETO-leukemic cells, expressing CD45.2, were injected in congenic CD45.1 male mice AML cells, we found hybrid cells expressing both allelic types of CD45 and XXY set of sexual chromosomes. More importantly, the fusion protein AML1-ETO was transferred in the hybrid cells. When sorted hybrid cells were reinjected in a secondary recipient, they gave rise to leukemia with 100% penetrance and similar time of onset of leukemic cells. Our data indicate that in vivo fusion of cancer cells with host cells may be a mechanism of gene transfer for cancer dissemination and suggest that fused cells may be used to identify still unrecognized leukemogenic genes that are conserved in hybrid cells and able to perpetuate leukemia in vivo.
Introduction
The theory that cell fusion contributes to cancer progression was introduced almost 100 years ago providing a non-mutational mechanism that could explain the aberrant gene expression pattern associated with malignant cells (reviewed in [1–4]). It has been proposed that cancer cells may fuse with different cell types including macrophages [5], stromal [6], epithelial [7], and endothelial cells [8,9]. In solid tumors such as melanomas [10] and breast cancer [6,11], fusion events have been proposed to play a possible role in cancer progression. The resulting hybrid cells were reported to be more aggressive than their parental cells because of faster growth, acquisition of drug resistance properties [12], higher propensity to metastasize, and different organotropism than the parental cells [13]. In addition, hybrids of myeloid and tumor cells expressed genes and differentiated traits from both parental cells. This has been postulated to contribute to immune escape [5]. Moreover, it has been recently reported that hybrid cells derived from spontaneous fusion events exhibited stem cell-like characteristics [14] and resulted in nuclear reprogramming [15]. However, most of these studies reported spontaneous cell fusion in vitro or the presence of hybrid cells in tumors in vivo, and the lack of a model where hybrid cells can be detected, subsequently isolated, and eventually implanted in vivo has not allowed further studies on gene transfer through cell fusion and its implications for tumor progression. In addition, these studies regarded essentially solid tumors and there is no data suggesting a similar mechanism for leukemia. While the manuscript was under review, Skinner et al. reported that intra-hematopoietic cell fusion acts as a source of somatic variation in the hematopoietic system [16], suggesting a putative role for increasing tumor heterogeneity in leukemia.
Several cell fusion-associated molecules have been characterized. Most of them are expressed on cell types that undergo cell fusion in physiological processes, whereby some of them might also play a role in tumor cell fusion such as the macrophage fusion receptor (also known as SIRPα) and its ligands CD44 and CD47. Interestingly, CD44 has been reported to play a role in leukemia initiation and progression and targeting this receptor eradicates acute myeloid leukemia (AML) in mouse models [17]. Moreover, it has been reported that expression of CD44 variant exons in AML is more common and more complex than that observed in normal blood, bone marrow (BM), or CD34+ cells and that a strong expression of CD44-6v correlates with shorter survival of patients with AML [18,19]. Expression of CD47 has been suggested to be an adverse prognostic factor for patients with AML and the use of a CD47 antibody targeting AML stem cells has been proposed for a possible therapeutic use [20]. More recently, Theocharides et al. showed that disruption of SIRPα signaling in macrophages eliminates human AML stem cells in xenografts [21]. We speculate a putative role for SIRPα and its ligands as a fusion mechanism.
We designed this study to investigate the role of cell fusion in leukemia. Transplantation of human leukemias in NOD/Scid mice successfully engrafts and faithfully recapitulates the pathology of the original human leukemia. Weeks after injection, leukemia onset is usually evaluated by expression of specific leukocyte markers, such as human CD45 [21,22] on flow cytometry. This type of single specie analysis and the low frequency of hybrid cell events have probably contributed to the misestimation of the cell fusion during leukemia progression in the past. Moreover, the induction of mouse leukemia by transplantation of transduced AML1-ETO leukemic cells in congenic mice allowed us to determine the fusion protein transfer from the leukemic to the hybrid cell lending its leukemic potential.
We now report evidence for the malignant potential of hybrid cells resulting from cell fusion of human primary and mouse leukemia cells with host macrophages.
Materials and Methods
Collection of Patient Samples and Cell Lines
Peripheral blood (PB) and BM blood cells were collected from patients with newly diagnosed AML and acute lymphoblastic leukemia (ALL) after obtaining informed consent. Individuals were diagnosed with AML according to the standards of the World Health Organization classification. Patients' samples were selected on the basis of availability of materials and cells from 14 different samples representing five AML subtypes, and five ALL cases were investigated for in vivo studies. Detailed characteristics of the patients included in this study are shown in Table W1. Cells were separated using Biocoll Separating Solution (Biochrom AG, Berlin, Germany) to obtain a mononuclear cell population, washed in RPMI 1640 (EuroClone, Milano, Italy) supplemented with 10% FBS (Gibco-Invitrogen, Life Technologies, Carlsbad, CA), and counted. Cells were then washed and freshly inoculated into mice or otherwise frozen in FBS plus 10% CryoSure-DMSO (WAK-Chemie Medical GmbH, Steinbach, Germany) and stored in liquid nitrogen. As controls, umbilical cord blood CD34+ cells were immunomagnetically purified with a CD34 microbead kit (Miltenyi Biotec, Bergisch Gladbach, Germany) according to the manufacturer's instructions. HL60, KG-1 AML lines, MOLT-16, and 697 ALL lines were used in this study, cultured according to the bank's protocols, and purchased at DSMZ Bank (Braunschweig, Germany).
Mice and Human Leukemia Transplants
NOD/LtSz-Prkdcscid (NS), NOD.Cg-PrkdcscidB2mtm1Unc (NSB), and NOD.cg-PrkdcscidIl2rgtm1Wjll (NSG) were kindly donated by Dr L. Shultz, bred, and housed at Charles River Laboratories (Calco, Italy). The following mice strains were obtained from Charles River Laboratories: C57B6/J (C57-CD45.1) and B6.SJL-Ptprca Pep3b/BoyJ (C57-CD45.2). All animals used were in a range of 6 to 8 weeks old. Experiments involving animals were done in the animal facilities at Istituto FIRC di Oncologia Molecolare (IFOM)-Istituto Europeo di Oncologia (IEO) campus (Milan, Italy) and all procedures were carried out in accordance with national and international laws and policies. For induction of AML-like leukemia, 1 x 107 low-density mononuclear cells from the BM of newly diagnosed AML patients were injected intraperitoneally (ip) in non-irradiated mice. Human engraftment was defined by means of percentage human CD45+ cells in PB from tail vein of the recipient animals. Mice were checked periodically for signs of disease (ruffled fur coat and hunched back), for complete blood cell count, and for the presence of blast cells by May-Grünwald-Giemsa staining of blood smears. Flow cytometry analyses were performed for the first time a month after transplant and repeated every month. When engraftment rose 50% hCD45+ cells, animals were monitored daily and blood sampling and subsequent fluorescence-activated cell sorting (FACS) analysis was performed every week until mice were sacrificed. Secondary transplants were performed by injecting spleen or BM cells of primary transplanted mice and successively in secondary and tertiary recipients.
Generation of Mouse Models of AML1-ETO Leukemias
Lin- cells were purified with a lineage cell depletion kit (StemSep Mouse Hematopoietic Progenitor Cell Enrichment Kit; Stem Cell Technologies, Vancouver, Canada) according to the manufacturer's instructions from the BM of C57-CD45.2 female mice. Briefly, after centrifugation through a Ficoll gradient, mononucleate cells were enriched for progenitors by negative selection of cells presenting myeloid, erythroid, and lymphoid differentiation markers. Unwanted cells are targeted for removal with biotinylated antibodies directed against non-hematopoietic progenitor cells [CD5 (Ly-1), CD11b (Mac-1), CD45R/B220, Ly-6G/C (Gr-1), neutrophils (7-4), TER119] and tetrameric antibody complexes recognizing biotin and dextran-coated magnetic particles. Labeled cells are then separated and StemSep Columns and Magnets. Flow-through containing Lin- cells was collected, centrifugated, and resuspended in medium containing interleukin-3, interleukin-6, and stem cell factor for 36 hours in a cell incubator. Cells were then plated onto retronectin-coated (Takara-Shuzo, Shiga, Japan), non-tissue culture-treated plates and exposed to the supernatant of packaging, ecotropic Phoenix cells transiently transfected with the PINCO-AML1-ETO retroviral vector as previously described [23]. Transduced cells were sorted using a BD FACS Vantage instrument and iv injected into lethally irradiated C57-CD45.2 female mice. Four weeks later, mice were treated with a single dose of N-ethyl-nitrosourea (Sigma-Aldrich, St Louis, MO; 50 mg/kg, ip). The animals develop myeloblastic leukemia with a latency of 20 weeks as previously described [23]. Leukemic spleens were then dissociated and reinjected ip at different doses into lethally irradiated C57-CD45.1 male mice. Recipient mice were checked periodically for signs of disease, for complete blood cell count, and for the presence of blast cells by May-Grunwald-Giemsa staining of blood smears. Leukemia burden was detected in the PB by the presence of CD45.2+ cells by cytometry analyses repeated every 2 weeks. Secondary transplants were performed injecting spleen or BM cells of primary transplanted mice and successively in secondary recipients.
Antibody Blocking Experiments
Whole IgG or F(ab′)2 fragments of clone IM7.8.1 (ATCC, Rockville, MD), a rat IgG2B monoclonal antibody recognizing all human and mouse isoforms of the CD44 expressed on hematopoietic cells, were ip injected at a dosage of 750 or 500 µg, respectively, per injection three times a week, for 4 weeks, into mice transplanted with AML cells 10 days earlier. As isotype control (IgG2b), an antibody to CD8, clone 2.43 (ATCC), was injected with the same schedule.
Anti-human CD47 antibody, B6H12.2 (ATCC), was given daily at a dosage of 100 or 50 µg by ip injection for 4 weeks into mice transplanted with AML cells 3 days earlier. An IgG1 antibody against Myc (9E10; Santa Cruz Inc, Capitola, CA) was injected at the same dose in control mice.
Flow Cytometry
Leukocytes from PB, spleen, and BM were isolated by mechanical dissociation of the organs, filtered on a 40-µm cell strainer, and analyzed by four- or six-color analysis as previously described [22]. For human AML engraftment evaluation, cells were previously incubated with CD16/CD32 (2.4G2) and then incubated with anti-human CD45 (clone 2D1), anti-mouse CD45 (30-F11), or with the following antibodies: for human antigens—anti-CD34 (8G12), anti-CD33 (P67.6), anti-CD13 (WM15), anti-CD31 (WM59), anti-CD14 (M5E2); for murine antigens—anti-CD45 (30-F11), anti-VEGFR2 (AVAS12a1), anti-CD11b (M1/70), anti-CD11c (HL3), anti-Gr-1 (RB6-805), anti-Sca-1 (D7), anti-Ter-119, anti-CD117 (2B8). These antibodies were purchased from BD Bioscience Pharmingen Inc (San Diego, CA). Anti-mouse CD11b (5C6) from Abcam (Cambridge, United Kingdom) was also used. Mouse AML engraftment evaluation was performed by flow cytometric analysis of CD45.1 and CD45.2 mouse isotypes; anti-CD45.1 (clone A20) and CD45.2 (clone 104) antibodies were from eBioscience (San Diego, CA). Apoptotic cells were depicted and excluded from the analysis by 7AAD staining (Sigma-Aldrich). DNA content was measured by propidium iodide staining as described elsewhere and analyzed by flow cytometry in a BD-FACScan flow cytometer.
Cell sorting experiments were performed in a BD-FACSAria cell sorter platform. Dead cells were excluded by a combination of scatter gates, and doublets were eliminated using the pulse-width parameter.
Immunohistochemistry
Immunostaining for human and murine antigens was performed on serial sections of formalin-fixed and paraffin-embedded biopsies with an automated immunostainer (Autostainer; Dako, Glostrup, Denmark), using a commercial detection kit (Dako EnVision Plus-HRP; Dako), according to the manufacturer's instructions. Antigen retrieval was obtained by placing tissue sections in 0.01 M EDTA buffer at pH 8.0 and underwent three cycles at 90°C in a microwave oven operating at 780 W as previously described [24]. Negative control sections were incubated with non-immune mouse serum in place of the specific primary antibodies and consistently lacked any staining. Myeloperoxidase cytochemical staining in blood smears was performed with a commercial kit (Sigma Aldrich) following manufacturer's instructions.
Fluorescence In Situ Hybridization
To better characterize the hybrid phenotype of fused cells, we performed fluorescence in situ hybridization (FISH) on BM and spleen cells as previously described [25]. For BM experiments, animals were treated with 200 µl of KaryoMAX Colcemid (Gibco, Invitrogen Corporation) ip injected for 1 hour before sacrifice. For FISH experiments, cell suspensions obtained from spleen dissociation were treated with KaryoMAX Colcemid for 45 minutes. Cell suspension were treated with hypotonic solution (KCl, 0.075 M) and fixed in methanol/acetic acid. Chromosome spreads and interphase nuclei were hybridized in situ with mouse Cot-1 DNA and BAC RP23-403C21 (for murine X chromosome) labeled with Cy3 (Amersham Biosciences, GE Healthcare, Buckinghamshire, United Kingdom) and with human cot-1 DNA and BAC RP4-567G7 (for murine Y chromosome) labeled with FluorX (Amersham Biosciences) by nick translation as described previously [25]. The commercial probe LSI ETO SpectrumOrange/AML1 SpectrumGreen dual color (Abbott Molecular Inc, Des Plaines, IL) was used for the presence of the fusion gene. Chromosomes and interphase nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). Digital images were obtained using a SkyVision workstation (Applied Spectral Imaging, Migdal Ha'Emek, Israel).
DNA Extraction and Polymerase Chain Reaction Analysis
High-molecular weight DNA was extracted from the PB, the spleen, and the BM of each mouse by the Dneasy Blood and Tissue Kit (Qiagen GmbH, Hilden, Germany). The presence of human-specific DNA within the BM or the spleen of transplanted mice was confirmed by polymerase chain reaction (PCR) of the human specific Alu sequence [26]. The amplification fragments were detected using the forward primer 5′CACCTGTAATCCCAGCACTTT and the reverse primer 5′CCCAGGCTGGAGTGCAGT. The reaction was performed in a final volume of 50 µl using 0.5 U of AmpliTaq Gold (Applied Biosystems, Carlsbad, CA), 250 nM of each primer, 0.4 mM of each nucleotide, and 25 mM MgCl2. Following an initial DNA denaturation at 94°C for 2 minutes, thirty-five 1-minute cycles of denaturation at 94°C, 1-minute annealing at 55°C, and 1-minute extension at 72°C were performed before a final elongation step at 72°C for 7 minutes. For the PCR of chromosome 17, amplification of an 850-bp fragment was detected using the forward primer 5′GGGATAATTTCAGCTGACTAAA and the reverse primer 5′TTCCGTTTAGTTAGGTGCAGTTATC. The reaction was performed using a Taq PCR Master Mix (Qiagen GmbH) with 250 nM primer concentration. Following an initial DNA denaturation at 94°C for 2 minutes, thirty-five 45-second cycles of denaturation at 94°C, annealing at 60°C, and extension at 72°C were performed before a final elongation step at 72°C for 7 minutes. All amplified DNA fragments were stained with SybrSafe DNA gel stain (Invitrogen Corp), electrophoresed through 1% agarose gel, and visualized in UV light. As an internal control, the presence of the glyceraldehyde-phosphate dehydrogenase (GAPDH) housekeeping gene was analyzed using the human/mouse GADPH primer set (Millipore, Billerica, MA).
Results
Human AML Cells Form Hybrid Cells with Murine Cells In Vivo
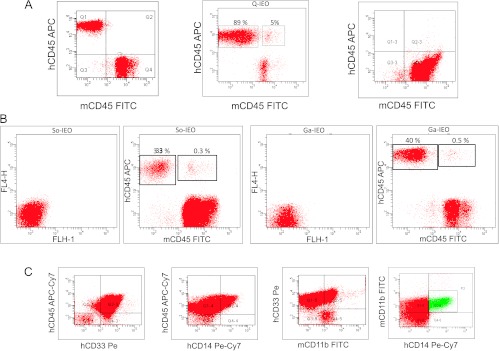
We injected non-irradiated NSB mice ip with 1 x 107 AML cells from 14 AML patients (for details, see Table W1). Human leukemia engraftment was first evaluated as the presence of human CD45+ cells by flow cytometry in BM, PB, and spleen. Data regarding the species specificity of anti-human and anti-murine monoclonal antibodies used are provided as supporting information (Figure W1). In addition, non-specific binding in cell mixing experiments in which human leukemia and mouse PB cells were mixed and then stained with human and mouse antibodies are shown as supporting information (Figure W2). Human leukemia was developed in 75.8% of mice with a latency range of 4 to 16 weeks. In most cases, human engraftment was detected by the presence of human CD45+ cells (Figure 1A, left panel), and in some cases, leukemia do not engraft (Figure 1A, right panel). However, in mice injected with cells derived from three different AML cases (Q-IEO, So-IEO, and Ga-IEO), we observed that a well-defined cell population of about 0.3% to 5% of the human leukemic cells also co-expressed the murine CD45 antigen (Figure 1A, central panel, Q-IEO; Figure 1B, So-IEO and Ga-IEO). At sacrifice, fused cells were observed in three cellular compartments (PB, BM, and spleen; Figure W3). After fusion, most of the cells sharing human and murine CD45 antigens showed a murine myeloid (CD11b/CD33/CD14-positive) phenotype, suggesting that hybrids were from human leukemic cells and murine macrophages (Figure 1B). Human leukemic cells of these three patients did not express CD11b (Table W1), and the absence of non-specific binding in cell mixing experiments of human leukemia and mouse PB cells is shown as supporting information (Figure W2).
Figure 1.
Human AML cells fuse with mouse CD11b-positive macrophages in vivo. (A) Flow cytometry analysis of PB from a representative leukemic mouse without cell fusion events (left panel), a leukemic mouse showing human engraftment (Q-IEO) and hybrid cells (0.5%, center), and from mouse without human engraftment (right panel). (B) Flow cytometry analysis of different AML showing distinct population of hybrid cells. (C) Flow cytometry analysis ofmouse PB showing a myeloid leukemic population expressing human CD45, CD33, and CD14, as well as murine CD11b.
Human and Mouse DNA Are Detectable in Hybrid Cells and Hybrid Cells Are Leukemogenic
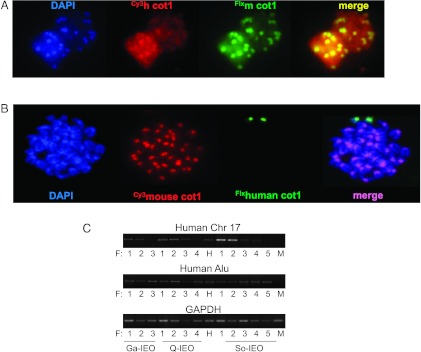
Human engraftment was evaluated by PCR analysis for the α-satellite region of human chromosome 17. PCR products performed in different groups of serial transplanted mice showed human engraftment in several of the primary and secondary recipients but not in all the tertiary and quaternary recipients nor in the DNA of a healthy mouse used as a control (Figure 2C, upper panel). To detect small amounts of genomic DNA transferred, we took advantage of the species-related specificity and of the highly repetitive Alu sequences to perform a sensitive and specific quantification of human genomic DNA by PCR analysis. A PCR product was detected in all recipient leukemic mice but not in the DNA of a healthy mouse used as a control (Figure 2C, middle panel). PCR detection of the GAPDH human/mouse housekeeping gene confirmed that a comparable amount of DNA was amplified in the above-described PCR experiments (Figure 2C, lower panel).
Figure 2.
Human DNA content is detectable in hybrid cells. (A) Dual-color FISH of interphase nuclei showing co-localization of mouse (FluorX-mcot-1 DNA) and human (Cy3-hcot-1 DNA) genomic DNA content in cells from spleen of a secondary recipient. (B) Dual-color FISH of a metaphase spread from a spleen cell of a tertiary recipient containing mouse (Cy3-mcot-1 DNA) and human chromosomes (FluorX-hcot-1 DNA) and counterstained with DAPI. Arrows, human chromosomes. (C) PCR analysis of human engraftment of spleen cells of three representative series of transplanted mice. PCR analysis of DNA of α-satellite region of chromosome 17 (upper row) and human Alu-specific sequences (medium row) detecting human DNA content in serial mouse recipients. Lower row, PCR detection of the GAPDH housekeeping gene confirmed that the same amount of DNA was amplified in PCR experiments. F, recipient (F1, primary; F2, secondary; F3, tertiary; and F4, quaternary recipient); H, human DNA of a healthy donor; M, mouse DNA of a healthy mouse.
FISH analysis labeling human and murine Cot-1 DNA with the same efficacy-specific hybridization probes showed the concomitant presence of both human and murine genomic content in cell interphases of cells from recipient mice (Figure 2A). Although spontaneous metaphases were rare in these cytogenetic preparations and small fragments of genomic DNA were difficult to visualize with the cot-1 labeling, Figure 2B shows a complete mouse karyotype with two additional human chromosomes observed in cells from a spleen of patient Q-IEO recipient mouse. Taken together, these results suggest that human DNA was transferred to the hybrid human/murine AML cells.
To investigate the leukemic potential of hybrid cells, we sorted human CD45/mouse CD45 double-positive cells from five BMs of leukemic xenotransplanted mice and injected them in secondary recipient. The sorting strategy was design to first discard dead cells and deplete cell doublets. Human CD45+ cells and hybrid human CD45+/mouse CD45+ cells were then separated by cell sorting with a purity of 93.1% and 90.2%, respectively (Figure W4). Hybrid cells promoted leukemia with a time of onset similar to human AML cells (40 vs 46 days for animals injected iv with 2 x 105 cells hCD45/mCD45 or hCD45, respectively). At sacrifice, the percent of human engraftment of animals that received human leukemic cells was similar to that of animals that received sorted fused cells (82–85% in the PB, 93–95% in the BM, and 95–98% in the spleen of human CD45+ leukemic cells). The presence of fused cells was detected in all the recipients in a variable ratio of 0.1% to 7% human CD45/mouse CD45 positive cells.
The Cell Fusion Occurs in a Murine Model of AML and the Gene AML1-ETO Can Be Transferred to the Hybrid Cells Conferring Leukemogenic Potential
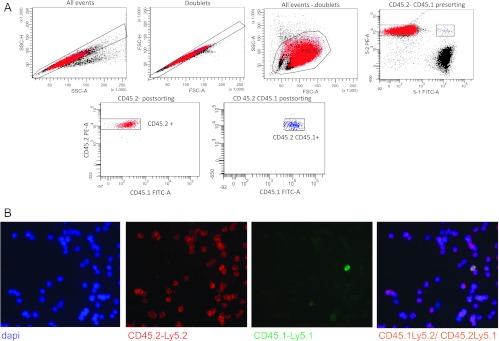
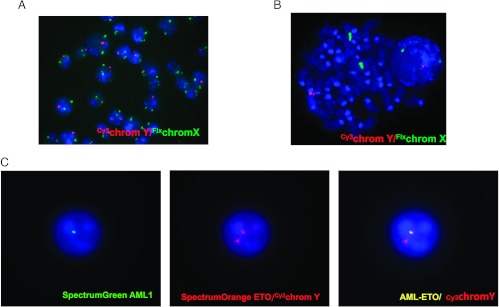
The absence of chromosomal aberrations or known fusion proteins in the three human AML cases of this study made difficult the determination of putative leukemic gene transferred from human to hybrid cells. To further study the mechanism of DNA transfer from cancer to normal cells, we decided to analyze whether the AML1-ETO leukemic gene, which can be followed as a cytogenetic alteration, should be transfer to hybrid cells in vivo in an AML murine leukemia model. C57-CD45.1 male mice were injected with leukemic murine cells from C57-CD45.2 donor female mice. Immunophenotype of AML1-ETO CD45.2 leukemia was analyzed. As reported in Figure W5, 1.8% of the Lin. population expressed Sca1 and cKit. When mice were overtly leukemic, their BM, PB, and spleen were investigated by flow cytometry for the expression of both allelic isoforms CD45.1 and CD45.2. About 4% to 6% of BM cells expressed both CD45.1 and CD45.2 antigens, suggesting that also in this AML mouse model fusion events might occur (Figure 3A). Non-specific staining of CD45.1 and CD45.2 antibodies on both single staining and mixing cell staining experiments of PB from C57-CD45.1 and C57-CD45.2 male mice was excluded as shown in Figure W6. Immunofluorescence staining and image analysis confirmed the presence of cells that co-express CD45.1 and CD45.2 antigens (Figure 3B). CD45.1/CD45.2 double-positive cells were then isolated by cell sorting from the spleen of leukemic mice. Sorting strategy involves discarding dead cells and doublets as shown in the upper panels of Figure 3A. Sorted CD45.1/CD45.2 double-positive cells were then investigated by FISH analysis. As shown in Figure 4, A and B, a cell population carried two X and one Y chromosome set providing evidence of their hybrid origin, from the donor and the recipient mice. Furthermore, in sorted CD45.1/CD45.2 double-positive cells, the presence of AML1-ETO fusion protein was detected in cells with the Y chromosome (Figure 4C), suggesting that fusion can be a mechanism for the transfer of leukemic genes to donor normal cells. Interestingly, sorted CD45.1/CD45.2 double-positive cells (purity 91.07%) reinjected at different doses in recipient animals promoted leukemia with a kinetics similar to CD45.2 AML1-ETO leukemic cells (purity, 98.8%), 21 versus 24 days of latency for three animals injected with 106 cells and 23.8 versus 21.6 for six animals injected with 105 cells (CD45.1/CD45.2 or CD45.2, respectively).
Figure 3.
AML1-ETO AML cells expressing CD45.2 fuse with CD45.1 cells from host mice. (A) Flow cytometry analysis of a leukemic spleen for the expression of both allelic isoforms CD45.1 and CD45.2 showing three different populations: CD45.2+ cells were AML1-ETO-engrafted cells, CD45.1 were host cells, and cells with both allelic isoforms CD45.1 and CD45.2 were hybrid cells. Left upper panels show discarding of dead cells and doublets. Lower panels show sorted CD45.2 cells at 98.8% purity and CD45.2-CD45.1 double-positive cells at 91.1%purity. (B) CD45.2-PE-Cy5 (red) and CD45.1-fluorescein isothiocyanate (green) double immunofluorescence staining on BM cells confirmed the presence of cells that co-express both antigens (merge). Immunostaining was performed on cells analyzed in Figure 3A, and cells were counterstained with DAPI.
Figure 4.
FISH analysis of sorted CD45.1/CD45.2 double-positive cells. (A, B) Dual-color staining of X (FluorX; green) and Y (Cy3; red) chromosomes counterstained with DAPI. (C) Dual-color staining of AML1-ETO fusion protein (ETO SpectrumOrange/AML1 Spectrum-Green; AML1-ETO fusion protein; yellow) and Y (Cy3; red) counterstained with DAPI.
AML Cells Can Fuse also with Endothelial and Dendritic Cells
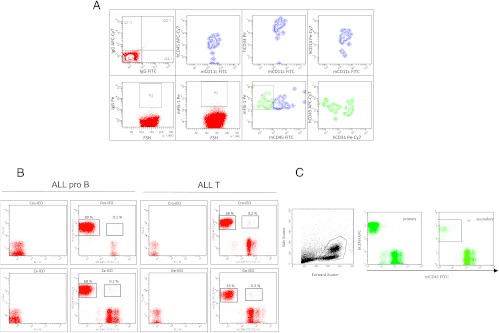
To further investigate what type of mouse cells fuse with the human leukemic cells, we analyzed BM of leukemic mice by flow cytometry. In addition to macrophages, some human AML cells formed hybrids also with dendritic (i.e., CD11c-positive; Figure 5A, upper panels) or endothelial (CD45-negative, Flk-1-positive; Figure 5A, lower panels) murine cells.
Figure 5.
Human AML cells fuse also with dendritic and endothelial cells. (A) Upper panels show flow cytometry analysis of cells coexpressing human CD45, CD33, and CD14 that express also murine CD11c, a dendritic cell-specific marker. Lower panels show flow cytometry analysis of mouse endothelial cells (Flk-1-positive) expressing human CD45 and human CD31. (B) In addition to AML, ALL cell transplantation induces cell fusion in vivo; 1 x 107 primary human ALL cells were injected ip, and 47 α 6 (ALL-B) or 72 ± 13 (ALL-T) days later, animals were sacrificed and spleen cells were analyzed by flow cytometry for the co-expression of human and mouse CD45. (C) Human CD45 and mouse CD45 flow cytometry expression patterns of BM cells from a primary and a secondary recipient of human cord blood CD34+ cells showing absence of hybrid cells.
AML and ALL but Not CD34+ Cord Blood Cells Promoted Cell Fusion
Acute leukemia cell fusion was not AML-type restricted, because it was observed after the ip injection of different AML FAB types (M1, M4, M5) or AML cell lines (HL60, KG1; Figure W6A) and also after the ip injection of ALL-B and ALL-T (Figure 5B) primary cells or cell lines (MOLT-16, 697; Figure W7A). In contrast, from the injection of CD34+ cells isolated from cord blood, we did not obtain fused cells in neither primary nor secondary recipients (Figure 5C), suggesting that fusion ability might be a characteristic of tumor cells.
Human leukemia cell fusion was not restricted to a specific NS-related strain. In fact, fusion events were observed by flow cytometry in BM samples of NS, NSB, and NSG mice, albeit at different frequencies (ranging from 0.2% to 11% of human AML cells in primary recipients; Figure W7B).
Monoclonal Antibodies Directed against CD44 or CD47 Do Not Inhibit Specifically AML Cell Fusion
It has been reported that targeting of CD44 eradicates human AML leukemic stem cells [17]. IM7 is a biologically active anti-CD44 antibody and it has been shown that treatment with this antibody inhibits BCR-ABL blast engraftment and subsequent chronic myeloid leukemia (CML)-like myeloproliferative disease (MPD) development in mice. In our hands, administration of IM7 induced systemic shock and death (data not shown), but its F(ab′)2 fragments did not induce a reaction in mice. However, treatment of AML-transplanted mice with these F(ab′)2 fragments did not have any effect on human engraftment in terms of time of onset of the leukemia symptoms and percent of human cells in PB, spleen, and BM at mice sacrifice. Blocking CD44 neither modified significantly the percent of fusion events obtained in PB and BM at sacrifice of 10 mice in two different experiments (Figure W8A).
In contrast, administration of the anti-CD47 monoclonal antibody B6H12.2, used either at 100 or 50 µg/mouse daily 3 days after AML transplant, completely prevented AML engraftment in spleen (91% of human cells in isotype Ig control-treated mice vs 0% of anti-CD47-treated animals), BM (53% vs 0%), and PB (59% vs 0%), thus hampering the detection of any fusion event. In some experiments, the administration of anti-CD47 monoclonal antibody (mAb) was interrupted after 4 weeks of treatment, and 4 months later, human engraftment was measured in the PB. Complete absence of human CD45+ cells (Figure W8B) suggested that the inhibition of CD47 targeted also cancer stem cells in these AML cases. Importantly, a very recent work while this paper was under review, reported that disruption of SIRPα signaling in macrophages eliminates AML stem cells in xenografts [21], as we suggest from our secondary transplants of anti-CD47-treated mice.
Discussion
Our data provide novel evidence that human and murine leukemic cells may spontaneously fuse in vivo with mouse normal cells resulting in hybrid cells containing DNA and protein markers from both fusion partners. None of the human cases examined presented abnormal cytogenetic characteristics and it was not possible to follow the putative “leukemic” gene transferred from human to mouse cells. However, the mouse model of AML1-ETO leukemia allowed us to confirm that hybrid cells contained the leukemia-generating AML1-ETO fusion protein. When transplanted in a secondary recipient, these hybrids were capable to transfer leukemia in vivo. Our data indicate that tumor cell fusion might be a mechanism of gene transfer for leukemia dissemination and suggest that further studies on the screening of human genes in hybrid cells can be a useful tool in the identification of new human genes involved in leukemia.
Most of the hybrid leukemic cells were CD11b-positive. One of the main functions of macrophages is internalization of apoptotic cells, pathogens, and foreign bodies and their routing toward lysosomes for degradation. However, macrophages have alternative internalization pathways toward homotypic fusion with other macrophages, leading to multinucleation. Thus, abortive digestion of tumor cells might be a potential mechanism for hybrid formation [2,3]. Macrophage cell fusion can be mediated by CD44 and its occupancy prevents multi-nucleation [27]. It has also been reported that targeting CD44 with the activating antibody H90 inhibits engraftment of human AML in vivo [17]. Here, we presented the data obtained with another CD44 antibody, IM7 (reported to be able to inhibit BCR-ABL engraftment [28]), showing no effect on AML engraftment. These data suggest that different epitopes of CD44 might play different roles on leukemia cell homing and engraftment and do not exclude a CD44 involvement in AML fusion events and successive engraftment. CD47 is another ligand for macrophage fusion receptor (MFR)/SIRPα that participates in macrophage multinucleation, and its expression has been reported to be an adverse prognostic factor in patients with AML [20]. Weissman's group showed that targeting CD47 with the monoclonal antibody B6H12.2 inhibits AML engraftment by increasing phagocytosis of cancer cells [20]. The same group recently reported that CD47 is overexpressed and acts as an anti-phagocytic signal in tumor cells, contrasting and so regulating the function of calreticulin [29]. Calreticulin is only highly expressed in tumor cells and the antibody anti-CD47 is able to elicit phagocytosis only in tumor cells that express calreticulin [29]. The reason why tumors cells should express a prophagocytic signal in the cell surface is not fully understood and we speculate a possible involvement of the CD47-calreticulin balance on cell fusion. However, when we treated AML-transplanted mice with the CD47 antibody at different doses, the complete inhibition of AML engraftment prevented any fusion event. Further experiments to investigate the role of calcericulin-CD47 in cell fusion should be addressed in future studies.
Cancer cell fusion with macrophages or stroma cells might induce immune escape [2,6]. Now, we present evidence that some human AML cells fuse also with endothelial (CD45-negative, Flk-1-positive) murine cells. It has been suggested that through fusions with endothelial cells, cancer cells may temporarily or permanently acquire endothelial cell characteristics that enhance their capability to traverse the endothelial barrier and metastasize [8].
Past studies about individual X-chromosome inactivation pattern (XCIP)-related clonality in AML have failed to identify multiple clones of the disease. However, there is recent evidence of XCIP skewing (>75% expression of one allele) occurring in a large proportion of hematologically normal females, probably because of the stem pool size at the time of lyonization. Moreover, limits in the sensitivity of the procedures used to assess clonality through XCIP may hamper the detection of small subclones [30], particularly considering that our data show a very small window of contemporary presence of the full genome of cancer and healthy host cells. In addition, a recent study has found the presence of apparently non-clonal AML foci in the spleen of some patients [31].
Fusion between cancer and normal cells can generate genomic and epigenetic variability at a rate exceeding that achievable by random mutagenesis [32]. This suggests a new possible use of our in vivo model. Thus, one can speculate that human cancer-inducing genes are “enriched” in hybrids between human leukemia and host murine cells (Figure W9). According to this working hypothesis, hybrid cells able to transfer leukemia to host mice are a particularly interesting target for screening of human cancer-driving genes.
Our findings offer new insight into a number of still unresolved clinical questions. Fusion of cancer cells with stroma (macrophages and endothelial cells) may explain cancer recurrence after therapy in patients who achieved a remission and had no morphologic or molecular evidence of residual disease. In fact, the expression of CD11b (a human macrophage marker), in the presence of progenitor cell-related markers and in the absence of markers of differentiation along the monocytic lineage, has been found to be a negative prognostic factor in human AML [33]. Likewise, cell fusion can explain enigmas such as BM cell-derived gastric cancer [34], the observation of donor cell-derived cancer [4] or leukemia [35] in recipients of allogenic BM transplants, the presence at leukemia relapse of clones different from those observed at diagnosis [36], the presence of cancer-specific genetic aberrations in the endothelial cells of some cancer vessels [9], the presence of a stem cell-like population of ALL cells co-expressing EC markers [37], and on a more general basis, the observed correlation between chronic inflammation (where macrophages play a crucial role) and tumorigenesis [3,4]. Overall, our data indicate, for the first time to our knowledge, that leukemia cells fuse with stromal cells in vivo and the malignant potential of the leukemia cell is maintained in the resulting hybrid cells.
Supplementary Material
Acknowledgments
The authors thank John Dick and Pier Giuseppe Pelicci for critical data review and for suggestions. We apologize to the numerous investigators whose papers could not be cited because of space limitations.
Abbreviations
- AML
acute myeloid leukemia
- ALL
acute lymphoblastic leukemia
- NS
NOD/LtSz-Prkdcscid
- NSB
NOD.Cg-PrkdcscidB2mtm1Unc
- NSG
NOD.cg-PrkdcscidIl2rgtm1Wjll
- MFR
macrophage fusion receptor
- BM
bone marrow
- PB
peripheral blood
- FISH
fluorescence in situ hybridization
- ip
intraperitoneally
- iv
intravenously
- IL
interleukin
Footnotes
This investigation was supported in part by Associazione Italiana per la Ricerca sul Cancro (AIRC), Istituto Superiore di Sanita (ISS), Italian Ministry of Health, and Fondazione Umberto Veronesi.
This article refers to supplementary materials, which are designated by Table W1 and Figures W1 to W9 and are available online at www.neoplasia.com.
References
- 1.Duelli D, Lazebnik Y. Cell fusion: a hidden enemy? Cancer Cell. 2003;3:445–448. doi: 10.1016/s1535-6108(03)00114-4. [DOI] [PubMed] [Google Scholar]
- 2.Vignery A. Macrophage fusion: are somatic and cancer cells possible partners? Trends Cell Biol. 2005;15:188–193. doi: 10.1016/j.tcb.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 3.Pawelek JM. Tumour-cell fusion as a source of myeloid traits in cancer. Lancet Oncol. 2006;6:988–993. doi: 10.1016/S1470-2045(05)70466-6. [DOI] [PubMed] [Google Scholar]
- 4.Pawelek JM, Chakraborty AK. Fusion of tumour cells with bone marrow-derived cells: a unifying explanation for metastasis. Nat Rev Cancer. 2008;8:377–384. doi: 10.1038/nrc2371. [DOI] [PubMed] [Google Scholar]
- 5.Pawelek JM, Chakraborty AK. The cancer cell-leukocyte fusion theory of metastasis. Adv Cancer Res. 2008;101:397–444. doi: 10.1016/S0065-230X(08)00410-7. [DOI] [PubMed] [Google Scholar]
- 6.Jacobsen BM, Harrell JC, Jedlicka P, Borges VF, Varella-Garcia M, Horwitz KB. Spontaneous fusion with, and transformation of mouse stroma by, malignant human breast cancer epithelium. Cancer Res. 2006;66:8274–8279. doi: 10.1158/0008-5472.CAN-06-1456. [DOI] [PubMed] [Google Scholar]
- 7.Rizvi AZ, Swain JR, Davies PS, Bailey AS, Decker AD, Rizvi AZ, Swain JR, Davies PS, Bailey AS, Decker AD. Bone marrow-derived cells fuse with normal and transformed intestinal stem cells. Proc Natl Acad Sci USA. 2006;103:6321–6325. doi: 10.1073/pnas.0508593103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mortensen K, Lichtenberg J, Thomsen PD, Larsson LI. Spontaneous fusion between cancer cells and endothelial cells. Cell Mol Life Sci. 2004;61:2125–2131. doi: 10.1007/s00018-004-4200-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Streubel B, Chott A, Huber D, Exner M, Jäger U, Wagner O, Schwarzinger I. Lymphoma-specific genetic aberrations in microvascular endothelial cells in B-cell lymphomas. N Engl J Med. 2004;351:250–259. doi: 10.1056/NEJMoa033153. [DOI] [PubMed] [Google Scholar]
- 10.Chakraborty AK, Pawelek J. β1,6-Branched oligosaccharides regulate melanin content and motility in macrophage-melanoma fusion hybrids. Melanoma Res. 2007;17:9–16. doi: 10.1097/CMR.0b013e3280114f34. [DOI] [PubMed] [Google Scholar]
- 11.Shabo I, Stal O, Olsson H, Doré S, Svanvik J. Breast cancer expression of CD163, a macrophage scavenger receptor, is related to early distant recurrence and reduced patient survival. Int J Cancer. 2010;123:780–786. doi: 10.1002/ijc.23527. [DOI] [PubMed] [Google Scholar]
- 12.Carloni V, Mazzocca A, Mello T, Galli A, Capaccioli S. Cell fusion promotes chemoresistance in metastatic colon carcinoma. Oncogene. 2012;31:1–12. doi: 10.1038/onc.2012.268. [DOI] [PubMed] [Google Scholar]
- 13.Lu X, Kang Y. Efficient acquisition of dual metastasis organotropism to bone and lung through stable spontaneous fusion between MDA-MB-231 variants. Proc Natl Acad Sci USA. 2009;106:9385–9390. doi: 10.1073/pnas.0900108106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dittmar T, Schwitalla S, Seidel J, Haverkampf S, Reith G, Meyer-Staeckling S, Brandt BH, Niggemann B, Kanker KS. Characterization of hybrid cells derived from spontaneous fusion events between breast epithelial cells exhibiting stem-like characteristics and breast cancer cells. Clin Exp Metastasis. 2011;28:75–90. doi: 10.1007/s10585-010-9359-3. [DOI] [PubMed] [Google Scholar]
- 15.Powell A, Anderson E, Davies P, Silk A, Pelz C, Impey S, Wong MH. Fusion between intestinal epithelial cells and macrophages in a cancer context results in nuclear reprogramming. Cancer Res. 2011;71:1497–1505. doi: 10.1158/0008-5472.CAN-10-3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skinner AM, Grompe M, Kurre P. Intra-hematopoietic cell fusion as a source of somatic variation in the hematopoietic system. J Cell Sci. 2012;15:2837–2843. doi: 10.1242/jcs.100123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin L, Hope KJ, Zhai Q, Smadja-Joffe F, Dick JE. Targeting of CD44 eradicates human acute myeloid leukemic stem cells. Nat Med. 2006;12:1167–1174. doi: 10.1038/nm1483. [DOI] [PubMed] [Google Scholar]
- 18.Bendall LJ, Bradstock KF, Gottlieb DJ. Expression of CD44 variant exons in acute myeloid leukemia is more common and more complex than that observed in normal blood, bone marrow orCD34+ cells. Leukemia. 2000;14:1239–1246. doi: 10.1038/sj.leu.2401830. [DOI] [PubMed] [Google Scholar]
- 19.Legras S, Günthert U, Stauder R, Curt F, Oliferenko S, Kluin-Nelemans HC, Marie JP, Proctor S, Jasmin C, Smadja-Joffe F. A strong expression of CD44-6v correlates with shorter survival of patients with acute myeloid leukemia. Blood. 1998;91:3401–3413. [PubMed] [Google Scholar]
- 20.Majeti R, Chao MP, Alizadeh AA, Pang WW, Jaiswal S, Gibbs KD, van Rooijen N, Weissman IL. CD47 is an adverse prognostic factor and therapeutic antibody target on human acute myeloid leukemia stem cells. Cell. 2009;138:286–299. doi: 10.1016/j.cell.2009.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Theocharides AP, Jin L, Cheng PY, Prasolava TK, Malko AV, Ho JM, Poeppl AG, van Rooijen N, Minden MD, Danska JS, et al. Disruption of SIRPα signaling in macrophages eliminates human acute myeloid leukemia stem cells in xenografts. J Exp Med. 2012;209:1883–1899. doi: 10.1084/jem.20120502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Agliano A, Martin-Padura I, Mancuso P, Marighetti P, Rabascio C, Pruneri G, Shultz LD, Bertolini F. Human acute leukemia cells injected in NOD/LtSz-scid/IL-2Rgamma null mice generate a faster and more efficient disease compared to other NOD/scid-related strains. Int J Cancer. 2008;123:2222–2227. doi: 10.1002/ijc.23772. [DOI] [PubMed] [Google Scholar]
- 23.Viale A, De Franco F, Orleth A, Cambiaghi V, Giuliani V, Bossi D, Ronchini C, Ronzoni S, Muradone I, Monestiroli S, et al. Cell-cycle restriction limits DNA damage and maintains self-renewal of leukaemia stem cells. Nature. 2009;457:51–56. doi: 10.1038/nature07618. [DOI] [PubMed] [Google Scholar]
- 24.Pruneri G, Fabris S, Baldini L, Carboni N, Zagano S, Colombiu MA, Ciceri G, Lombardi L, Ronchi M, Buffa R, et al. Immunohistochemical analysis of cyclin D1 shows deregulated expression in multiple myeloma with the t(11;14) Am J Pathol. 2000;156:1505–1513. doi: 10.1016/S0002-9440(10)65022-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Belloni E, Trubia M, Mancini M, Derme V, Nanni M, Lahortiga I, Riccioni R, Confalonieri S, Lo-Coco F, Di Fiore PP, et al. A new complex rearrangement involving the ETV6, LOC115548, and MN1 genes in a case of acute myeloid leukemia. Genes Chromosomes Cancer. 2004;41:272–277. doi: 10.1002/gcc.20081. [DOI] [PubMed] [Google Scholar]
- 26.Schneider T, Osl F, Friess T, Stockinger H, Scheuer WV. Quantification of human Alu sequences by real-time PCR—an improved method to measure therapeutic efficacy of anti-metastatic drugs in human xenotransplants. Clin Exp Metastasis. 2002;19:571–582. doi: 10.1023/a:1020992411420. [DOI] [PubMed] [Google Scholar]
- 27.Cui W, Ke JZ, Zhang Q, Ke HZ, Chalouni C, Vignery A. The intracellular domain of CD44 promotes the fusion of macrophages. Blood. 2006;107:796–805. doi: 10.1182/blood-2005-05-1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krause DS, Lazarides K, von Andrian UH, van Etten RA. Requirement for CD44 in homing and engraftment of BCR-ABL-expressing leukemic stem cells. Nat Med. 2006;12:1175–1180. doi: 10.1038/nm1489. [DOI] [PubMed] [Google Scholar]
- 29.Chao MP, Jaiswal S, Weissman-Tsukamoto R, Alizadeh AA, Gentles AJ, Volkmer J, Weiskopf K, Willingham SB, Raveh T, Park CY, et al. Calreticulin is the dominant pro-phagocytic signal on multiple human cancers and is counterbalanced by CD47. Sci Transl Med. 2010;2:63ra94. doi: 10.1126/scitranslmed.3001375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gale RE, Linch DC. Clonality studies in acute myeloid leukemia. Leukemia. 1998;12:117–120. doi: 10.1038/sj.leu.2400935. [DOI] [PubMed] [Google Scholar]
- 31.O'Malley DP, Orazi O, Wang M, Cheng L. Analysis of loss of heterozygosity and X chromosome inactivation in spleens with myeloproliferative disorders and acute myeloid leukemia. Mod Pathol. 2005;18:1562–1568. doi: 10.1038/modpathol.3800481. [DOI] [PubMed] [Google Scholar]
- 32.Duelli DM, Padilla-Nash HM, Berman D, Murphy KM, Ried T, Lazebnik Y. A virus causes cancer by inducing massive chromosomal instability through cell fusion. Curr Biol. 2007;17:431–437. doi: 10.1016/j.cub.2007.01.049. [DOI] [PubMed] [Google Scholar]
- 33.Paietta E, Andersen J, Yunis J, Rowe JM, Cassileth PA, Tallman MS, Bennett JM, Wiernik PH. Acute myeloid leukaemia expressing the leucocyte integrin CD11b—a new leukaemic syndrome with poor prognosis: result of an ECOG database analysis. Eastern Cooperative Oncology Group. Br J Haematol. 1998;100:265–272. doi: 10.1046/j.1365-2141.1998.00561.x. [DOI] [PubMed] [Google Scholar]
- 34.Houghton J, Stoicov C, Nomura S, Rogers AB, Carlson J, Li H, Cai X, Fox JG, Goldenring JR, Wang TC. Gastric cancer originating from bone marrow-derived cells. Science. 2004;306:1568–1571. doi: 10.1126/science.1099513. [DOI] [PubMed] [Google Scholar]
- 35.Fraser CJ, Hirsch BA, Dayton V, Creer MH, Neglia JP, Wagner JE, Baker KS. First report of donor cell-derived acute leukemia as a complication of umbilical cord blood transplantation. Blood. 2005;106:4377–4380. doi: 10.1182/blood-2005-06-2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi S, Henderson MJ, Kwan E, Beesley AH, Sutton R, Bahar AY, Giles J, Venn NC, Pozza LD, Baker DL, et al. Relapse in children with acute lymphoblastic leukemia involving selection of a preexisting drug-resistant subclone. Blood. 2007;110:632–639. doi: 10.1182/blood-2007-01-067785. [DOI] [PubMed] [Google Scholar]
- 37.Wang L, O'Leary H, Fortney J, Gibson LF. Ph+/VE-cadherin+ identifies a stem cell-like population of acute lymphoblastic leukemia sustained by bone marrow niche cells. Blood. 2007;110:3334–3344. doi: 10.1182/blood-2007-01-068122. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.