A cortical myosin-dependent mechanism induces cell elongation to ensure clearance of trailing chromatids from the cleavage plane at the correct time during cytokinesis.
Abstract
Chromosome segregation must be coordinated with cell cleavage to ensure correct transmission of the genome to daughter cells. Here we identify a novel mechanism by which Drosophila melanogaster neuronal stem cells coordinate sister chromatid segregation with cleavage furrow ingression. Cells adapted to a dramatic increase in chromatid arm length by transiently elongating during anaphase/telophase. The degree of cell elongation correlated with the length of the trailing chromatid arms and was concomitant with a slight increase in spindle length and an enlargement of the zone of cortical myosin distribution. Rho guanine-nucleotide exchange factor (Pebble)–depleted cells failed to elongate during segregation of long chromatids. As a result, Pebble-depleted adult flies exhibited morphological defects likely caused by cell death during development. These studies reveal a novel pathway linking trailing chromatid arms and cortical myosin that ensures the clearance of chromatids from the cleavage plane at the appropriate time during cytokinesis, thus preserving genome integrity.
Introduction
Coordination of chromatid segregation with cell cleavage is crucial for equal partitioning of the genetic material and maintaining genome stability. However, it remains unclear how the coordination of these two mitotic events is achieved. Recently, studies in budding yeast and mammalian cells revealed an Aurora B–dependent pathway (NoCut pathway) that inhibits abscission, the final step of cytokinesis, when chromatin is trapped in the contractile ring (Norden et al., 2006; Mendoza et al., 2009; Steigemann et al., 2009). In metazoa, this mechanism involves the stabilization of the contractile ring via Aurora B–mediated phosphorylation of endosomal sorting complex required for transport and mitotic kinesin-like protein 1 (Steigemann et al., 2009; Capalbo et al., 2012; Carlton et al., 2012).
To probe the mechanism by which chromosome segregation is coordinated with cleavage furrow ingression, other studies have monitored the effect of abnormally long chromatids on cell division. Plant cells can divide properly with long chromatids, provided that the chromatid length does not exceed half of the spindle length at telophase (Schubert and Oud, 1997). Budding yeast can adapt to a 45% increase in chromosome size by enhancing the compaction of the long chromatid during anaphase. This depends on Aurora B activity and serine 10 of Histone H3 (Neurohr et al., 2011). In Drosophila melanogaster, the ability to correctly segregate long chromatids varies with cell type: errors in segregation occur frequently during syncytial embryonic divisions, whereas larval neuroblasts divide properly via an unknown mechanism (Sullivan et al., 1993). Our study focuses on understanding how Drosophila neuroblasts coordinate chromatid segregation with cell division by monitoring cells transiting through mitosis with abnormally long chromatids.
Results and discussion
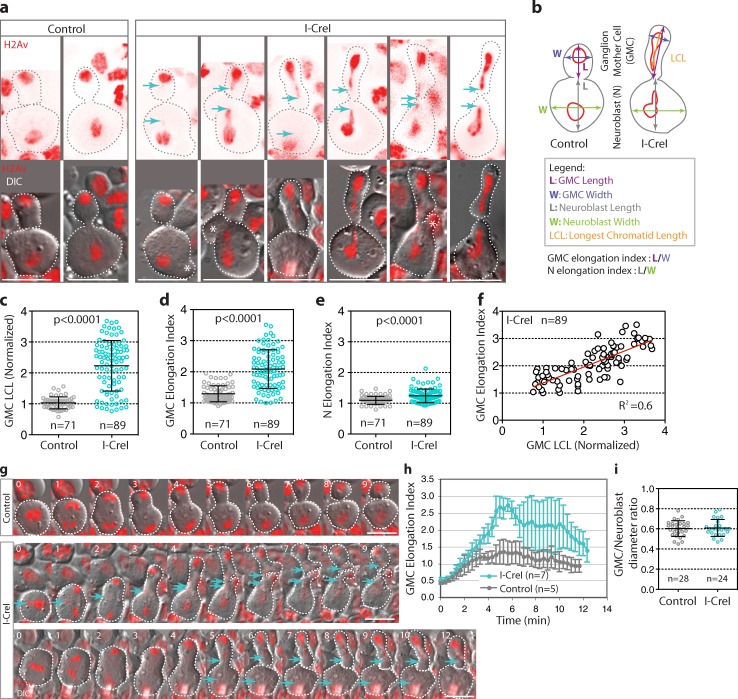
To generate long chromatids, we used the induction of I-CreI endonuclease, which creates DNA double-strand breaks in the ribosomal DNA repeats located on the Drosophila X and Y chromosomes (Rong et al., 2002). The resulting acentric and centric chromosome fragments remain attached through a DNA “tether,” which transiently increases the length of the chromatid arms during anaphase (Royou et al., 2010). By monitoring Drosophila larval neuroblasts labeled with the histone variant H2Av fused to red fluorescent protein (H2Av::RFP), we found that, after I-CreI expression, the transient increase in chromatid arm length varied considerably between cells, ranging from 1 to nearly 4 times the length of the chromatid mass (Fig. 1 a, cyan arrows; and Fig. 1 c).
Figure 1.
Increase in chromatid length is associated with cell elongation during anaphase/telophase. (a) Still images from time-lapse movies of individual neuroblasts expressing the H2Av::RFP (red) during late cytokinesis. The top row shows the histone variant H2Av fused with RFP (H2Av::RFP) images (red), and the bottom row shows differential interference contrast (DIC) images merged with H2Av::RFP signal (red). All images are oriented with the GMC at the top. The cyan arrows indicate the position of the tip of the longest chromatids lagging behind the mass of chromatids. The gray and white broken lines outline the cells. Asterisks designate cell blebs. Bars, 8 µm. (b) Schemes indicating how the various lengths, widths, and distances were measured for graphs c–f in live cells (see Materials and methods). (c) Scatter dot plot with mean ± SD showing the range of the longest chromatid length in the GMC (GMC LCL) for control and I-CreI cells. (d) Scatter dot plot with mean ± SD showing the GMC elongation index for control and I-CreI cells. (e) Scatter dot plot with mean ± SD showing the neuroblast (N) elongation index for control and I-CreI cells. (f) Graph showing the linear correlation (R2 = 0.6) of the GMC elongation index with the longest chromatid length for I-CreI cells. (g) Time-lapse images of control and I-CreI neuroblasts expressing H2Av::RFP (red). Each image is one sagittal DIC image merged with the H2Av::RFP signal. The cyan arrows indicate the positions of the tip of the longest chromatids. The white broken lines outline the cells. Time is given in minutes. 0 = anaphase onset. The top row shows a control cell (see Video 1). The middle row shows a cell with I-CreI–induced long chromatids that elongates and starts rounding up 7 min after anaphase onset in parallel with the decrease in chromatid arm length (see Video 2). The third row shows a cell with long chromatids that takes longer to round up. The cell stays elongated until 12 min after anaphase onset before starting to round up (see Video 3). Bars, 8 µm. (h) Graph showing the GMC elongation index (mean ± SD) over time for control and I-CreI cells exhibiting long chromatids. Time starts at anaphase onset. (i) Scatter dot plot with mean ± SD showing the diameter of the GMC at the end of mitosis relative to the neuroblast diameter for control and I-CreI cells. n = number of cells.
Neuroblasts divide asymmetrically to generate a large neuroblast and a small ganglion mother cell (GMC). Next, we examined the effect of the variation in chromatid arm length on neuroblast and GMC cell shape. While in control cells, the neuroblast and GMC appeared spherical during late cytokinesis; in cells expressing I-CreI, the neuroblast and GMC daughter cells adopted various shapes that were dependent on the length of the trailing chromatid arm. The neuroblast shape varied from a nearly spherical shape to a pear-like shape (Fig. 1 a, compare the first I-CreI cell with the last I-CreI cell). The change in shape of the GMC was more striking, as it varied from a sphere to a tubular shape (Fig. 1 a, compare the first two I-CreI cells with the other I-CreI cells). In addition, the cells with long chromatids blebbed frequently near the site of the cleavage furrow (Fig. 1 a, asterisks). We quantified the change in cell shape by measuring the elongation index (length-to-width ratio) of the daughter cells during late anaphase in control and I-CreI cells (Fig. 1 b). GMCs with long chromatids were more elongated than control cells (Fig. 1, a and d). This difference in cell elongation between control and I-CreI cells was less severe in the neuroblast (Fig. 1, a and e). In addition, we found that the longer the chromatid arm the more elongated the GMC (Fig. 1 f).
To control that the elongation of the cell was dependent on the length of the trailing chromatid arm and not on the DNA “tether” present in the I-CreI–induced long chromosome, we studied mitotic neuroblasts carrying an abnormally long compound chromosome known as C(2)EN (Novitski et al., 1981). In contrast to I-CreI–induced long chromosomes, neither BubR1 nor γ-H2Av signals were detected on C(2)EN chromosome arms, confirming the absence of a “tether” on the C(2)EN (Fig. S1, a and b). In live neuroblasts, C(2)EN chromatids displayed some variations in length, ranging from 1 to 3 times the length of the chromatid mass during anaphase (Fig. S1, c and d). We found that an increase in C(2)EN length was correlated with an increase in GMC index of elongation (Fig. S1, c and e). These observations indicate that cell elongation is not triggered by DNA breaks or BubR1-coated DNA tethers present in the I-CreI–induced long chromatid. Therefore, we conclude that trailing long chromatid arms promote cell elongation.
We next monitored the rate of GMC elongation from anaphase onset to the end of cytokinesis for control and I-CreI cells. No change in the rate of cell elongation was observed between control and I-CreI neuroblasts during anaphase A, when the sister chromatids move poleward (Fig. 1, g and h). However, differences between control and I-CreI cells were detected during anaphase B and telophase. Control cells generated two spherical daughter cells within 8 min after anaphase onset (Fig. 1, g and h; and Video 1). In contrast, neuroblasts with long chromatids produced daughter cells whose shapes evolved during cytokinesis. The index of elongation of the nascent GMC cells increased constantly throughout cytokinesis, reaching a peak of elongation at around 5 min after anaphase onset (Fig. 1 h). Afterward, the behavior of elongated cells varied greatly. Some cells rounded up rapidly, 8–9 min after anaphase onset (Fig. 1 g, second row; and Video 2), whereas other cells remained elongated for a longer time (>10 min after anaphase onset) and then abruptly rounded up (Fig. 1 g, third row; and Video 3). The extended elongated state of the GMC was concomitant with a delay in the regression of long chromatid arms back into the main chromatid mass (Fig. 1 g, cyan arrows, compare the second and third row). At the end of mitosis, two unequally sized daughter cells of similar diameter ratio were generated in control and I-CreI cells (Fig. 1 i). This indicated that the presence of long chromatids and the transient elongation of the nascent cells during cytokinesis did not impair the inherent asymmetry of GMC-neuroblast divisions.
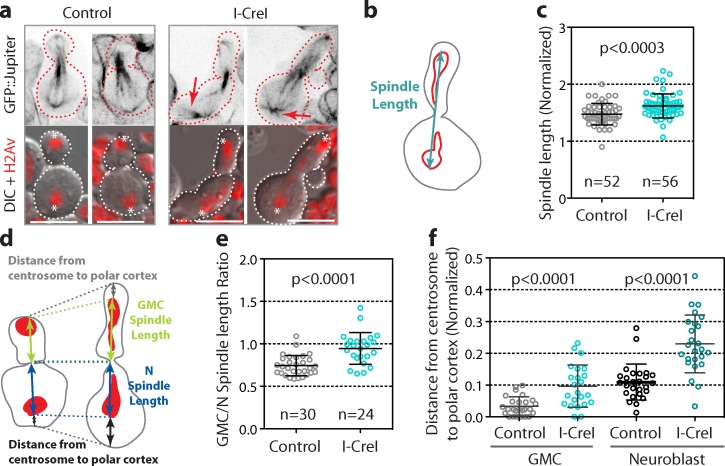
To determine if the segregation of long chromatids affected the spindle, we monitored mitotic cells expressing a microtubule marker (GFP::Jupiter). No noticeable spindle defect was observed in I-CreI elongated cells (Fig. 2 a). Next, we measured the spindle length from one centrosome to the other during late anaphase (Fig. 2 b). We found that the spindle was slightly, yet significantly longer in I-CreI cells than in control cells (Fig. 2 c). Similar to embryonic neuroblasts, the spindle was asymmetrically positioned toward the neuroblast in control cells (Fig. 2, d and e; Kaltschmidt et al., 2000). In contrast, the spindle was nearly symmetrically positioned in I-CreI cells, revealing a transient shift of the spindle toward the GMC during cytokinesis (Fig. 2, d and e). In addition, we noticed that although in control cells the centrosomes were apposed to the polar cortex, in I-CreI elongated cells the centrosomes were frequently detached from the neuroblast and GMC polar cortex (Fig. 2, d and f). This was associated with the formation of prominent astral microtubules (Fig. 2 a, red arrows). To determine whether asters play a role in this pathway, we monitored long chromosome segregation in the centrosomin (cnn) mutant in which asters are depleted (Li and Kaufman, 1996; Megraw et al., 1999). In the presence of long chromatids, the GMC cnn mutant adopted a tubular shape comparable to that in wild-type cells (Fig. S2 a). The elongation index of the GMC was substantially increased in cnn cells expressing I-CreI compared with cnn control cells (Fig. S2 b). Moreover, the index of elongation correlated with the length of the chromatid arm (Fig. S2 c). These results indicate that astral microtubules are not required for cell elongation.
Figure 2.
Segregation of long chromatids is associated with a slight increase in spindle length and a shift of the spindle toward the GMC. (a) Still images from time-lapse movies of control and I-CreI neuroblasts expressing H2Av::RFP and GFP::Jupiter during cytokinesis. Top panels are inverted images of GFP::Jupiter signal. Bottom panels are DIC images merged with H2Av::RFP signal (red). Asterisks designate the position of the centrosomes. The arrows point to the prominent asters in elongated cells. Broken lines outline the cells. Bar, 8 µm. (b) Scheme of an elongated cell showing how the spindle length is measured (see Materials and methods). (c) Scatter dot plot with mean ± SD showing the normalized spindle length for control and elongated I-CreI cells. (d) Scheme of control and I-CreI cells indicating how the GMC, neuroblast spindle length, and centrosome–to–polar cortex distances were measured for the graphs in e and f. (e) Scatter dot plot with mean ± SD showing the ratio of the GMC spindle length to the neuroblast spindle length. This ratio reflects the displacement of the spindle toward the GMC in I-CreI cells. (f) Scatter dot plot with mean ± SD showing the detachment of the centrosome from the polar cortex in both neuroblast and GMC I-CreI cells compared with control cells. n = number of cells.
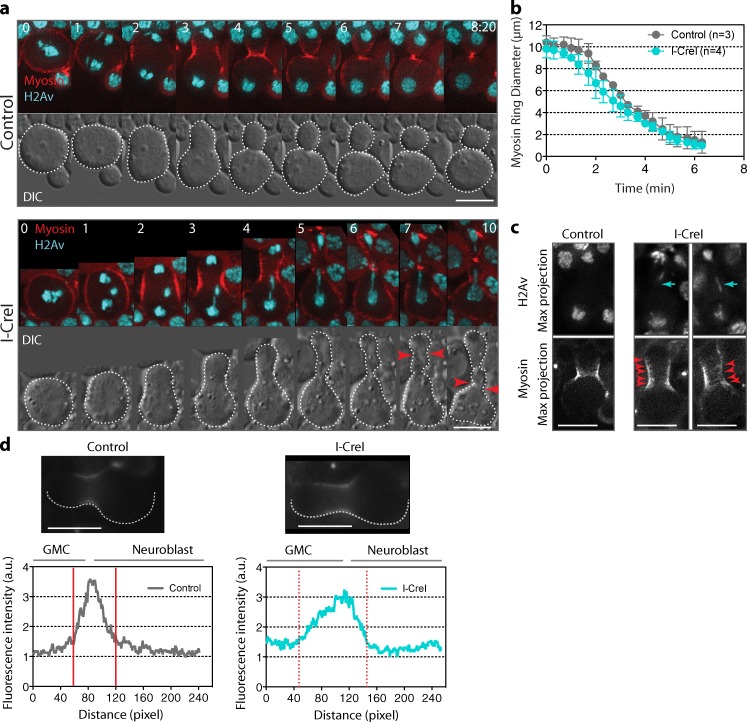
Cell cleavage requires the assembly of an actomyosin-based contractile ring between the two sets of chromatids. The instructive signal promoting the assembly of the ring is thought to depend on the spindle microtubules (Somers and Saint, 2003; D’Avino et al., 2005; Piekny et al., 2005; Yüce et al., 2005). Therefore, we next determined if cell elongation associated with the displacement of the spindle toward the GMC affected the localization of cortical myosin, the assembly of the contractile ring, and the rate of the cleavage furrow ingression. To do so, we monitored neuroblasts expressing the nonmuscle myosin II regulatory light chain (encoded by the spaghetti squash gene) fused to GFP (referred to as myosin), which marks the contractile ring (Royou et al., 2004). In both control and I-CreI cells, myosin was uniformly distributed around the cortex at metaphase and relocalized to the future cleavage site during anaphase (Fig. 3 a, Video 4, and Video 5). The rate of furrow ingression in cells bearing long chromatids was similar to that of control cells (Fig. 3, a and b), which indicates that the segregation of long chromatids does not delay the initiation and progression of cytokinesis. However, the cortical myosin localization at the cleavage site was broader in cells with long chromatids than in control cells (Fig. 3, a, c and d; and Video 5). Furthermore, we frequently observed discrete myosin rings at the cortex (Fig. 3 c, red arrowheads) and transient formation of extra furrows on both sides of the main cleavage furrow (Fig. 3 a, red arrows). We quantified the fluorescence intensity of myosin in one half of the cortex from the GMC cortical pole to the neuroblast pole, 4 min after anaphase onset for control and I-CreI cells. We found that cortical myosin distribution was wider in cells expressing I-CreI (Fig. 3 d). Moreover, the distribution of the cortical myosin was more spread toward the GMC (Fig. 3 d, broken red lines). Together, our results indicate that the presence of trailing chromatid arms induces a transient cortical myosin spreading associated with cell elongation without affecting the position or rate of cleavage furrow ingression.
Figure 3.
Cell elongation during segregation of long chromatids does not affect cleavage furrow ingression but is associated with a broader cortical myosin distribution. (a) Time-lapse images of control (Video 4) or I-CreI (Video 5) neuroblasts expressing H2Av::RFP (H2Av, cyan) and RLC::GFP (myosin, red). The top panels are merges of maximum projections of deconvolved H2Av and myosin signals. The bottom panels are DIC images. The outline of the cell is shown as a white broken line. In I-CreI cells, cortical myosin starts spreading beyond the cytokinetic ring 4 min after anaphase onset. In addition, pseudocleavage furrows form later during cytokinesis, bracketing the main cytokinetic furrow (red arrowheads). Time is given in minutes:seconds. 0 = anaphase onset. (b) Graph showing the rate of constriction of the myosin ring for control and I-CreI cells. The diameter of the ring is measured from anaphase onset (time = 0 min) to the end of cytokinesis. (c) Localization of myosin during cytokinesis, 4 min after anaphase onset for control and I-CreI cells. The top panels show maximum projections of deconvolved H2Av::RFP signal. The cyan arrows show the positions of the long chromatids. The bottom panels show maximum projections of deconvolved myosin signal. Red arrowheads point to the multiple myosin rings. Bars, 8 µm. (d) The panels show one sagittal plane of myosin nondeconvolved images from control and I-CreI cells. Bars, 8 µm. The graphs show cortical pixel intensity of myosin around one half of the cell cortex, from the GMC cortical pole to the neuroblast cortical pole as delineated by the broken white line in the picture. Note that the distribution curve of the cortical myosin signal is wider and shifted toward the GMC in the I-CreI cell (broken red lines) compared with the control cell (solid red lines).
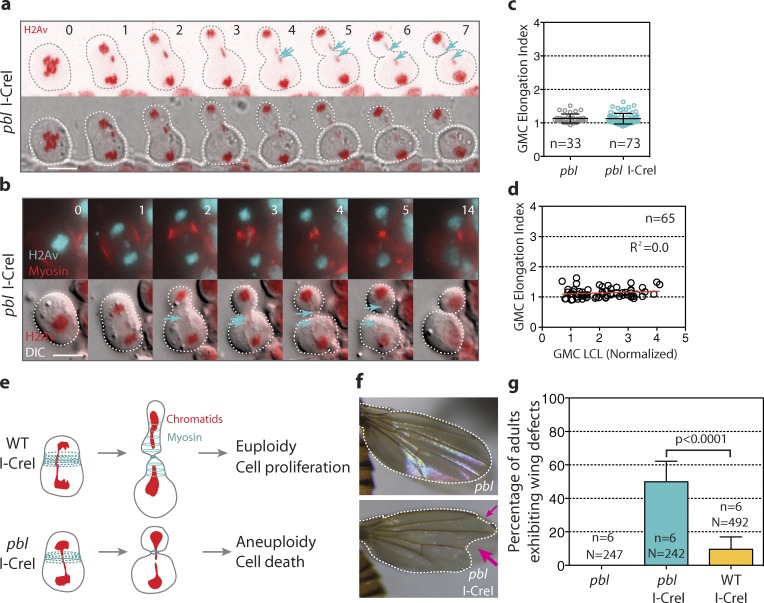
Our observation that GMCs with long chromatids adopt a transient tubular shape concomitant with broader myosin rings resembles the mechanism for polar body extrusion resulting from an extreme asymmetric division during female meiosis in Caenorhabditis elegans (Dorn et al., 2010). The authors reported that the polar body protrusion from the cortex requires the formation of an actomyosin tube. Furthermore, in Drosophila S2 cells, the elongation of the cell during anaphase, before cytokinesis, is attributed to the relaxation of the polar cortex and the broad equatorial contraction via myosin activity (Hickson et al., 2006). Therefore, we reason that the additional myosin rings formed during the segregation of long chromatids partially constrict the cortex, promoting the temporary deformation of the nascent daughter cells. Rho-kinase is one of the kinases that activate myosin during cytokinesis (Amano et al., 1996). Rho-kinase is activated by the GTPase Rho1 (Matsumura, 2005). In Drosophila, Rho1 is activated by the Rho guanine nucleotide exchange factor (RhoGEF) Pebble (Pbl; Prokopenko et al., 1999). To test the idea that myosin contractile activity is required for cell elongation during the segregation of long chromatids, we monitored I-CreI mitotic neuroblasts with reduced Pbl activity (see Materials and methods). pbl mutant neuroblasts failed to elongate during the segregation of abnormally long chromatids (Fig. 4, a and b; and Video 6). No difference in the index of elongation was observed between control and I-CreI pbl mutant cells (Fig. 4 c). Consequently, we found no correlation between the length of the trailing chromatid arms and the GMC elongation index (Fig. 4 d). Furthermore, cortical myosin formed a focused contractile ring throughout cytokinesis in the pbl mutant (Fig. 4 b and Video 6). These results are consistent with a model in which the formation and transient constriction of additional myosin rings induces cell elongation, allowing the clearance of long chromatids from the cleavage plane at the appropriate time before completion of cytokinesis. We hypothesized that the absence of this mechanism in pbl mutants may result in cell death because of the long chromatid arm being trapped by the contractile ring (Fig. 4 e). To test this model, we monitored the survival rate and the morphology of the adult wings after I-CreI expression in wild type and pbl mutants. We found no difference in the survival rate between wild-type and pbl mutant cells expressing I-CreI (Fig. S3). However, nearly half of adult pbl mutants exhibited wing morphological defects likely caused by cell death during development compared with 8.7% in the wild type (Fig. 4, f and g). These wing morphological defects in pbl mutants resulted from I-CreI induction, as no morphological defects were observed in pbl mutants without I-CreI expression (Fig. 4 g). We propose that cell elongation serves as a backup mechanism to prevent defects in chromosome segregation.
Figure 4.
pbl mutant cells fail to elongate during the segregation of abnormally long chromatids. (a) Time-lapse images of a pbl mutant cell expressing H2Av::RFP and I-CreI. The top panels are images of H2Av::RFP and the bottom panels are DIC images merged with H2Av signal (red). (b) Time-lapse images of a pbl mutant cell expressing H2Av::RFP (cyan) and myosin (red; see Video 6). The top panels are merges of H2Av and myosin signals. The bottom panels show one sagittal DIC image merged with H2Av signal (red). The broken lines show the contour of the cell. The cyan arrows point to the tip of the long chromatids. Time is given in minutes. Bars, 8 µm. (c) Scatter dot plot with mean ± SD showing the GMC elongation index for pbl mutant control cells or cells expressing I-CreI. n = number of cells. (d) Graph showing no linear correlation of the GMC elongation index with the length of the longest chromatid for pbl I-CreI cells. n = number of cells. (e) Scheme showing the scenarios that may arise in wild-type and pbl mutant cells with long chromatids. The wild-type cell adjusts its length to clear the long chromatid from the cleavage plane, hence producing euploid daughter cells, which will proliferate and differentiate properly. The pbl mutant cell fails to elongate in the presence of long chromatids. This produces aneuploid daughter cells, which will undergo apoptosis. (f) Images of wings from pbl mutant adult flies with our without I-CreI, heat shocked for 1 h at 37°C during the third instar larval stage. The pink arrows point to the wing morphological defects. (g) Histogram showing the frequency of adult flies exhibiting wings with morphological defects. n = number of experiments. N = total number of adult flies scored.
Our study reveals that chromosomes are capable of exerting an effect on cortical myosin recruitment during cytokinesis. Previous studies in mouse oocytes have shown that chromosomes promote the formation of an actomyosin-rich cortical domain in a manner dependent on their distance from the cortex (Maro et al., 1986; Van Blerkom and Bell, 1986; Brunet and Maro, 2005; Deng et al., 2007). Similarly, we found that during the segregation of long chromatids, the small daughter cell (GMC) is systematically more elongated than the large cell (neuroblast), and myosin spreads preferentially toward the GMC. This suggests that the effect of chromosomes on myosin depends on their distance from the cortex. Thus, we speculate that the sensor that signals to cortical myosin via Pbl activity is associated with the trailing chromatid arm and forms a gradient of activity that diffuses to the cortex. Interestingly, studies in mouse oocytes have shown that chromosomes generate a gradient of active Rac1 and Ran, which in turn affect cortical polarity (Deng et al., 2007; Halet and Carroll, 2007).
We found that spindle length is increased in elongated cells. Since chromosomes promote microtubule nucleation, this raises the possibility that the presence of trailing chromatid arms induces elongation of the central spindle. Consequently, the area of centralspindlin accumulation, the complex that recruits Pbl at the cleavage site, may broaden, which enlarges the zone of cortical myosin localization. Consistently, studies in human cells have reported that abnormally elongated central spindles are associated with the formation of a broader cleavage furrow (Hu et al., 2011). However, if trailing chromatids were to enlarge the zone of centralspindlin localization, this would have a negative impact on cytokinesis because it might impede the formation of a tight ring. This is likely to have an effect on the rate of cleavage furrow ingression. One argument against this model is that cell elongation does not affect the rate of cleavage furrow invagination. An alternative hypothesis is that cell elongation and cytokinesis are triggered through Pbl acting on different pathways. Our finding that subtle reduction of Pbl activity completely abolished cell elongation and myosin broadening regardless of the length of the chromatid arms, while only mildly affecting cytokinesis, supports this possibility. It suggests that cell elongation is more sensitive to the level of Pbl activity than cleavage furrow ingression. When Pbl activity is decreased, cytokinesis is triggered by the central spindle–dependent pool of Pbl acting on Rho1 at the expense of the chromatid-dependent pool of Pbl acting on another GTPase. Interestingly, recent studies have demonstrated that Pbl activates Rac1 (van Impel et al., 2009). A third explanation arises from the finding that myosin cytokinetic ring positioning is mediated by two pathways in Drosophila larval neuroblasts: a central spindle pathway and a parallel polarity cue–dependent pathway (Giansanti et al., 2001; Cabernard et al., 2010). Reduced Pbl activity might affect the formation of extra myosin rings required for cell elongation without affecting the polarity-mediated pathway involved in cytokinesis.
Budding yeast has evolved a mechanism for segregating long chromatids, which involves the supercompaction of the trailing chromatid arm via Aurora B activity (Neurohr et al., 2011). We found that Drosophila neuroblasts use a different mechanism to adapt to the presence of chromatid arms in the cleavage plane. The cell adjusts its cortical shape proportionally to the length of the longest chromatid during cytokinesis. This difference between Drosophila and yeast cells may be explained by the presence of a cell wall in yeast that restricts cortical plasticity, hence constraining cell elongation. Recent studies have revealed a novel Aurora B–dependent checkpoint that inhibits abscission in the presence of chromatin at the cleavage site (Norden et al., 2006; Mendoza et al., 2009; Steigemann et al., 2009). We do not yet know if this checkpoint exists in Drosophila neuroblasts. Nonetheless, it is possible that the Aurora B–mediated and Pbl-mediated mechanisms exist in all cell types, optimizing the robust nature of faithful chromosome transmission throughout evolution. Alternatively, the process of cell elongation may be specific to asymmetric stem cell divisions. Either way, it will be important to determine if the mechanism that we report in Drosophila neuroblasts exists in human cells. The fact that key proteins involved in these mechanisms (Pbl/RhoGEF and myosin) are highly conserved in humans supports this idea.
Materials and methods
Fly stocks
The stocks were raised on maize and yeast media with or without molasses at 25°C. We used a GFP::Jupiter construct to mark the spindle (a gift from A. Debec, Institut Jacques Monod, Paris, France). The GFP is fused to the N terminal of Jupiter, a microtubule-binding protein (Karpova et al., 2006). Myosin was labeled with the RLC::GFP construct, where the GFP is fused to the C terminal of the nonmuscle myosin II regulatory light chain (Royou et al., 2004). Since Pbl is required for cytokinesis (Prokopenko et al., 1999), a lethal allele (pbl5, a mismatched mutation in the highly conserved DH domain; Somers and Saint, 2003) combined with a viable allele (pblmale sterile, also annotated as pblZ4836, a point mutation within the coding sequence; Giansanti et al., 2004) of pbl was used to create a viable transheterozygote (referred as pbl), in which cytokinesis was affected in <10% of the cells with or without I-CreI expression. The rate of furrow ingression was dramatically reduced or the furrow partially invaginated and then regressed in 3 out of 36 and 8 out of 81 pbl mutant control and I-CreI cells, respectively. Fly stocks were crossed to generate the following genotype: w; p[w+, RLC::GFP]/+;p[w+, H2Av::RFP], pblmale sterile/pbl5 and w; p[w+, RLC::GFP]/+; p[w+, H2Av::RFP], pblmale sterile/pbl5, p[v+, hs70-I-CreI], Sb. The H2Av::RFP stock and the pbl5 stock were obtained from the Bloomington Drosophila Stock Center. The hs70-I-CreI stock was a gift from K. Golic (University of Utah, Salt Lake City, UT). The pblmale sterile allele was provided by M. Gatti (La Sapienza, Rome, Italy). The stock carrying the compound 2 Entire, 2R2L-2R2R, abbreviated C(2)EN stock was described by Novitski et al. (1981) and was obtained from the Bloomington Drosophila Stock Center (no. 2974). The cnnHK21 mutant allele has a point mutation forming a stop codon within the amino terminal of the coding region (Li and Kaufman, 1996; Megraw et al., 1999; Vaizel-Ohayon and Schejter, 1999).
Microscope image acquisition
To induce I-CreI expression, late third instar larvae were heat shocked for 1 h at 37°C and left to recover at room temperature for 1–2 h. The dividing neuroblasts were observed within 1 h after recovery. Third instar larval brains were dissected in PBS and slightly squashed between a slide and a coverslip by capillary forces as described previously (Buffin et al., 2005). The preparation was visualized immediately under the microscope and for a maximum period of 30 min. Images from Figs. 1 a (except the last panel), 1 g, 2 a, 3 (a, c, and d), and S1 (b and c) were acquired with a wide-field inverted microscope (DMI6000B; Leica) equipped with an EM charge-coupled device (CCD) camera (ORCA C9100-02; Hamamatsu) with a binning of 1. The last images of Fig. 1 a were acquired with a wide-field inverted DMI6000B microscope (Leica) equipped with a CoolSnap HQ2 camera (Photometrics, Roper Scientific SAS) with a binning 2. Images from Fig. 4 a were acquired with a confocal microscope (SP2; Leica). Images from Figs. 4 b, S1 a, and S2 a were acquired with a wide-field inverted microscope (Axio-observer; Carl Zeiss) equipped with an EMCCD camera (Evolve; Photometrics and Roper Scientific). All images were observed with a 100× oil Plan-Apochromat objective lens (NA 1.4). A z series of 0.5 µm steps was acquired every 20 s. LAS AF6000 (Leica) or MetaMorph software (Roper Scientific) were used for the acquisition. 10 iterations of a blind deconvolution algorithm were run, using a refractive index of 1.5. Images from Figs. 1 (a and g), 2 a, and 3 (a, c, and d) are deconvolved maximum intensity projections. Images from Figs. 4 (a and b), S1 c, and S2 a are maximum intensity projections.
Quantification
ImageJ and Prism software (GraphPad) were used for image quantification and statistical analysis, respectively. Measurement described in Figs. 1 (c–f), 1 (h and i), 2 (c, e, and f), 4 (c and d), S1 (d and e), and S2 (b and c) were done at one time point during cytokinesis, when the cell is the most elongated. The variation in length of the longest chromatid arm in the GMC was calculated by measuring the length of the long chromatid from the telomere to the centromere located at the tip of the chromosome mass at the pole (Fig. 1 b), and dividing this length by the mean length of normal chromatid arms for each genotype at a similar time point. The elongation index of the neuroblast and GMC was measured by calculating the length-to-width ratio of the ellipsoid (Fig. 1 b). The spindle length was measured from the tip of the segregated chromatids on the neuroblast pole side to the tip of the segregated chromatids on the GMC pole side, where the centrosomes are located and normalized by dividing this length to the diameter of the cell at metaphase (Fig. 2 b). The distance from centrosome to polar cortex was normalized with the diameter of the cell at metaphase (Fig. 2 f). For Fig. 3 b, the diameter of the contractile ring was measured from one tip of the invaginating furrow to the other at the position of the ring marked with RLC::GFP over time. From time 0–2 min, when the ring is not yet formed, the diameter of the cell was calculated by drawing a line across the cell at an equal distance from the segregating sister chromatids. An unpaired t test with a 95% confidence was used to calculate the p-value for all statistical analysis.
Survival assay and wing morphological scoring
5–6 d after egg deposition, larvae of each genotype were transferred to separate vials (16–25 larvae per vials) with standard food and heat shocked in a 37°C water bath for 1 h. The larvae were kept at 25°C and the adults were scored after 5 d for 3 d. Adults with distinct “notched” wings were scored for each genotype. This experiment was repeated six times.
Immunostaining
For BubR1 immunostaining, the brains were dissected in PBS and fixed in PBS + 3.7% formaldehyde for 30 min to 1 h, then fixed for 30 s in 45% acetic acid and transferred in a 60% acetic acid drop on a coverslip for 3 min. The samples were squashed and frozen in liquid nitrogen, then blocked for 30 min in PBT (PBS + 0.1% Triton X-100; Williams and Goldberg, 1994). The samples were incubated overnight at 4°C with rabbit anti-BubR1 antibodies (1:1,000; a gift from C. Sunkel, Universidade do Porto, Porto, Portugal). The preparations were washed four times for 10 min in PBT, rinsed once in PBS, and mounted with Slow Fade containing DAPI (Invitrogen). For γ-H2Av immunostaining, the brains were dissected in 0.7% NaCl and then incubated in 0.5% sodium citrate for 5 min. The samples were fixed in 4% paraformaldehyde in PBST (PBS + 0.2% Triton X-100), squashed, and blocked for 1 h in PBST + 5% milk. The samples were incubated with rabbit anti–γ-H2Av antibodies (1:250; a gift from G. Karpen, Lawrence Berkeley National Lab, Berkley, CA) overnight at 4°C. The samples were washed three times for 5 min in PBST, then incubated for 1 h at room temperature with secondary goat 488 anti–rabbit antibodies (1:300; Invitrogen). The samples were washed three times for 5 min in PBST, rinsed with PBS, and mounted with Vectashield containing DAPI (Vector laboratories).
Online supplemental material
Fig. S1 shows that cell elongation occurs during the segregation of the long compound chromosome 2 (C(2)EN). Fig. S2 shows that astral microtubules are not required for cell elongation. Fig. S3 illustrates the survival rate to adulthood of wild-type and pbl mutant larvae expressing I-CreI. Videos 1, 2, and 3 correspond to Fig. 1 g (top, middle, and bottom rows, respectively). Videos 4 and 5 correspond to Fig. 3 a (top and bottom rows, respectively). Video 6 correspond to Fig. 4 b. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201208041/DC1.
Supplementary Material
Acknowledgments
We thank Roger Karess (Institut Jacques Monod, Paris, France), Derek McCusker (University of Bordeaux, Institut Européen de Chimie et Biologie, France), and Denis Dupuy (University of Bordeaux, Institut Européen de Chimie et Biologie, France) for sharing equipment and reagents. We thank Maurizio Gatti (La Sapienza, Rome, Italy), Kent Golic, Gary Karpen, and Claudio Sunkel (Universidade do Porto, Porto, Portugal) for reagents. We thank Marie-Charlotte Claverie (University of Bordeaux, Institut Européen de Chimie et Biologie, France) and the plateform ImagoSeine (Institut Jacques Monod, Paris, France) for technical assistance.
A. Royou and E. Montembault were supported by Atip-Avenir/Institut National du Cancer 2010-291 grant. A. Royou was also supported by FP7 Marie Curie PIRG08GA-2010-277117/BroChroMito, Conseil Régional d’Aquitaine (20111301010), and Centre National de la Recherche Scientifique (CNRS). W. Sullivan was supported by National Institutes of Health (GM046409-19). S. Kotadia was supported by the California Institute for Regenerative Medicine (TG2-01157, FA1-00617-1).
Footnotes
Abbreviations used in this paper:
- cnn
- centrosomin
- DIC
- differential interference contrast
- GMC
- ganglion mother cell
- Pbl
- Pebble
References
- Amano M., Ito M., Kimura K., Fukata Y., Chihara K., Nakano T., Matsuura Y., Kaibuchi K. 1996. Phosphorylation and activation of myosin by Rho-associated kinase (Rho-kinase). J. Biol. Chem. 271:20246–20249 10.1074/jbc.271.34.20246 [DOI] [PubMed] [Google Scholar]
- Brunet S., Maro B. 2005. Cytoskeleton and cell cycle control during meiotic maturation of the mouse oocyte: integrating time and space. Reproduction. 130:801–811 10.1530/rep.1.00364 [DOI] [PubMed] [Google Scholar]
- Buffin E., Lefebvre C., Huang J., Gagou M.E., Karess R.E. 2005. Recruitment of Mad2 to the kinetochore requires the Rod/Zw10 complex. Curr. Biol. 15:856–861 10.1016/j.cub.2005.03.052 [DOI] [PubMed] [Google Scholar]
- Cabernard C., Prehoda K.E., Doe C.Q. 2010. A spindle-independent cleavage furrow positioning pathway. Nature. 467:91–94 10.1038/nature09334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capalbo L., Montembault E., Takeda T., Bassi Z.I., Glover D.M., D’Avino P.P. 2012. The chromosomal passenger complex controls the function of endosomal sorting complex required for transport-III Snf7 proteins during cytokinesis. Open Biol. 2:120070 10.1098/rsob.120070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton J.G., Caballe A., Agromayor M., Kloc M., Martin-Serrano J. 2012. ESCRT-III governs the Aurora B-mediated abscission checkpoint through CHMP4C. Science. 336:220–225 10.1126/science.1217180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Avino P.P., Savoian M.S., Glover D.M. 2005. Cleavage furrow formation and ingression during animal cytokinesis: a microtubule legacy. J. Cell Sci. 118:1549–1558 10.1242/jcs.02335 [DOI] [PubMed] [Google Scholar]
- Deng M., Suraneni P., Schultz R.M., Li R. 2007. The Ran GTPase mediates chromatin signaling to control cortical polarity during polar body extrusion in mouse oocytes. Dev. Cell. 12:301–308 10.1016/j.devcel.2006.11.008 [DOI] [PubMed] [Google Scholar]
- Dorn J.F., Zhang L., Paradis V., Edoh-Bedi D., Jusu S., Maddox P.S., Maddox A.S. 2010. Actomyosin tube formation in polar body cytokinesis requires Anillin in C. elegans. Curr. Biol. 20:2046–2051 10.1016/j.cub.2010.10.030 [DOI] [PubMed] [Google Scholar]
- Giansanti M.G., Gatti M., Bonaccorsi S. 2001. The role of centrosomes and astral microtubules during asymmetric division of Drosophila neuroblasts. Development. 128:1137–1145 [DOI] [PubMed] [Google Scholar]
- Giansanti M.G., Farkas R.M., Bonaccorsi S., Lindsley D.L., Wakimoto B.T., Fuller M.T., Gatti M. 2004. Genetic dissection of meiotic cytokinesis in Drosophila males. Mol. Biol. Cell. 15:2509–2522 10.1091/mbc.E03-08-0603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halet G., Carroll J. 2007. Rac activity is polarized and regulates meiotic spindle stability and anchoring in mammalian oocytes. Dev. Cell. 12:309–317 10.1016/j.devcel.2006.12.010 [DOI] [PubMed] [Google Scholar]
- Hickson G.R., Echard A., O’Farrell P.H. 2006. Rho-kinase controls cell shape changes during cytokinesis. Curr. Biol. 16:359–370 10.1016/j.cub.2005.12.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C.K., Coughlin M., Field C.M., Mitchison T.J. 2011. KIF4 regulates midzone length during cytokinesis. Curr. Biol. 21:815–824 10.1016/j.cub.2011.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltschmidt J.A., Davidson C.M., Brown N.H., Brand A.H. 2000. Rotation and asymmetry of the mitotic spindle direct asymmetric cell division in the developing central nervous system. Nat. Cell Biol. 2:7–12 10.1038/71323 [DOI] [PubMed] [Google Scholar]
- Karpova N., Bobinnec Y., Fouix S., Huitorel P., Debec A. 2006. Jupiter, a new Drosophila protein associated with microtubules. Cell Motil. Cytoskeleton. 63:301–312 10.1002/cm.20124 [DOI] [PubMed] [Google Scholar]
- Li K., Kaufman T.C. 1996. The homeotic target gene centrosomin encodes an essential centrosomal component. Cell. 85:585–596 10.1016/S0092-8674(00)81258-1 [DOI] [PubMed] [Google Scholar]
- Maro B., Johnson M.H., Webb M., Flach G. 1986. Mechanism of polar body formation in the mouse oocyte: an interaction between the chromosomes, the cytoskeleton and the plasma membrane. J. Embryol. Exp. Morphol. 92:11–32 [PubMed] [Google Scholar]
- Matsumura F. 2005. Regulation of myosin II during cytokinesis in higher eukaryotes. Trends Cell Biol. 15:371–377 10.1016/j.tcb.2005.05.004 [DOI] [PubMed] [Google Scholar]
- Megraw T.L., Li K., Kao L.R., Kaufman T.C. 1999. The centrosomin protein is required for centrosome assembly and function during cleavage in Drosophila. Development. 126:2829–2839 [DOI] [PubMed] [Google Scholar]
- Mendoza M., Norden C., Durrer K., Rauter H., Uhlmann F., Barral Y. 2009. A mechanism for chromosome segregation sensing by the NoCut checkpoint. Nat. Cell Biol. 11:477–483 10.1038/ncb1855 [DOI] [PubMed] [Google Scholar]
- Neurohr G., Naegeli A., Titos I., Theler D., Greber B., Díez J., Gabaldón T., Mendoza M., Barral Y. 2011. A midzone-based ruler adjusts chromosome compaction to anaphase spindle length. Science. 332:465–468 10.1126/science.1201578 [DOI] [PubMed] [Google Scholar]
- Norden C., Mendoza M., Dobbelaere J., Kotwaliwale C.V., Biggins S., Barral Y. 2006. The NoCut pathway links completion of cytokinesis to spindle midzone function to prevent chromosome breakage. Cell. 125:85–98 10.1016/j.cell.2006.01.045 [DOI] [PubMed] [Google Scholar]
- Novitski E., Grace D., Strommen C. 1981. The entire compound autosomes of Drosophila melanogaster. Genetics. 98:257–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piekny A., Werner M., Glotzer M. 2005. Cytokinesis: welcome to the Rho zone. Trends Cell Biol. 15:651–658 10.1016/j.tcb.2005.10.006 [DOI] [PubMed] [Google Scholar]
- Prokopenko S.N., Brumby A., O’Keefe L., Prior L., He Y., Saint R., Bellen H.J. 1999. A putative exchange factor for Rho1 GTPase is required for initiation of cytokinesis in Drosophila. Genes Dev. 13:2301–2314 10.1101/gad.13.17.2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong Y.S., Titen S.W., Xie H.B., Golic M.M., Bastiani M., Bandyopadhyay P., Olivera B.M., Brodsky M., Rubin G.M., Golic K.G. 2002. Targeted mutagenesis by homologous recombination in D. melanogaster. Genes Dev. 16:1568–1581 10.1101/gad.986602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royou A., Field C., Sisson J.C., Sullivan W., Karess R. 2004. Reassessing the role and dynamics of nonmuscle myosin II during furrow formation in early Drosophila embryos. Mol. Biol. Cell. 15:838–850 10.1091/mbc.E03-06-0440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royou A., Gagou M.E., Karess R., Sullivan W. 2010. BubR1- and Polo-coated DNA tethers facilitate poleward segregation of acentric chromatids. Cell. 140:235–245 10.1016/j.cell.2009.12.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert I., Oud J.L. 1997. There is an upper limit of chromosome size for normal development of an organism. Cell. 88:515–520 10.1016/S0092-8674(00)81891-7 [DOI] [PubMed] [Google Scholar]
- Somers W.G., Saint R. 2003. A RhoGEF and Rho family GTPase-activating protein complex links the contractile ring to cortical microtubules at the onset of cytokinesis. Dev. Cell. 4:29–39 10.1016/S1534-5807(02)00402-1 [DOI] [PubMed] [Google Scholar]
- Steigemann P., Wurzenberger C., Schmitz M.H., Held M., Guizetti J., Maar S., Gerlich D.W. 2009. Aurora B-mediated abscission checkpoint protects against tetraploidization. Cell. 136:473–484 10.1016/j.cell.2008.12.020 [DOI] [PubMed] [Google Scholar]
- Sullivan W., Daily D.R., Fogarty P., Yook K.J., Pimpinelli S. 1993. Delays in anaphase initiation occur in individual nuclei of the syncytial Drosophila embryo. Mol. Biol. Cell. 4:885–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaizel-Ohayon D., Schejter E.D. 1999. Mutations in centrosomin reveal requirements for centrosomal function during early Drosophila embryogenesis. Curr. Biol. 9:889–898 10.1016/S0960-9822(99)80393-5 [DOI] [PubMed] [Google Scholar]
- Van Blerkom J., Bell H. 1986. Regulation of development in the fully grown mouse oocyte: chromosome-mediated temporal and spatial differentiation of the cytoplasm and plasma membrane. J. Embryol. Exp. Morphol. 93:213–238 [PubMed] [Google Scholar]
- van Impel A., Schumacher S., Draga M., Herz H.M., Grosshans J., Müller H.A. 2009. Regulation of the Rac GTPase pathway by the multifunctional Rho GEF Pebble is essential for mesoderm migration in the Drosophila gastrula. Development. 136:813–822 10.1242/dev.026203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams B.C., Goldberg M.L. 1994. Determinants of Drosophila zw10 protein localization and function. J. Cell Sci. 107:785–798 [DOI] [PubMed] [Google Scholar]
- Yüce O., Piekny A., Glotzer M. 2005. An ECT2-centralspindlin complex regulates the localization and function of RhoA. J. Cell Biol. 170:571–582 10.1083/jcb.200501097 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.