Rlp24 recruits Drg1 to pre-60S particles and stimulates its ATP hydrolysis to promote downstream maturation through specific extraction of Rlp24.
Abstract
Formation of eukaryotic ribosomes is driven by energy-consuming enzymes. The AAA-ATPase Drg1 is essential for the release of several shuttling proteins from cytoplasmic pre-60S particles and the loading of late joining proteins. However, its exact role in ribosome biogenesis has been unknown. Here we show that the shuttling protein Rlp24 recruited Drg1 to pre-60S particles and stimulated its ATPase activity. ATP hydrolysis in the second AAA domain of Drg1 was required to release shuttling proteins. In vitro, Drg1 specifically and exclusively extracted Rlp24 from purified pre-60S particles. Rlp24 release required ATP and was promoted by the interaction of Drg1 with the nucleoporin Nup116. Subsequent ATP hydrolysis in the first AAA domain dissociated Drg1 from Rlp24, liberating both proteins for consecutive cycles of activity. Our results show that release of Rlp24 by Drg1 defines a key event in large subunit formation that is a prerequisite for progression of cytoplasmic pre-60S maturation.
Introduction
Ribosomes are essential for all living cells by translating the genetic information into the amino acid sequence of proteins. Eukaryotic ribosomes are composed of one 40S and one 60S subunit containing different ribosomal RNA species and ribosomal proteins. The formation of the subunits starts in the nucleolus with the assembly of a long precursor (pre) ribosomal RNA with ribosomal and nonribosomal proteins. This precursor particle gives birth to progenitors of the large and small subunit. Further maturation steps lead to export-competent pre-60S and pre-40S particles, which are transported through the nuclear pore complex (NPC) into the cytoplasm, where the final maturation steps take place (for ribosome biogenesis see Henras et al. [2008] and Kressler et al. [2010]). For the pre-60S particle, these cytoplasmic maturation steps involve formation of the characteristic ribosomal stalk structure, incorporation of the last ribosomal proteins, and release and recycling of shuttling proteins and export factors (Panse and Johnson, 2010).
A key player in cytoplasmic pre-60S maturation in Saccharomyces cerevisiae is the AAA (ATPases associated with diverse cellular activities) protein Drg1 (Pertschy et al., 2007; Lo et al., 2010). Drg1 is a cytoplasmic protein that forms hexamers and exhibits high homology to Cdc48 from yeast (47% sequence identity of the AAA domains) and its mammalian orthologue p97. Similar to these proteins, Drg1 contains an N-terminal domain and two AAA domains, D1 and D2 (see Fig. 3 A; Thorsness et al., 1993; Zakalskiy et al., 2002). The AAA domains consist of a conserved stretch of ∼230 amino acid residues with Walker A and B motifs. AAA proteins use the energy of ATP hydrolysis to generate mechanical force that acts on specific substrates and results in ATP-dependent remodeling of proteins or macromolecular complexes (for short overviews on general aspects of AAA proteins see Lupas and Martin [2002], Hanson and Whiteheart [2005], and White and Lauring [2007]). The D1 domain of p97 is required for oligomerization, whereas D2 is regarded as the main catalytic site, which generates tension by means of ATP hydrolysis that is finally transmitted to the substrate proteins (DeLaBarre and Brunger, 2005; Pye et al., 2006; Briggs et al., 2008). The N domains of p97 and Cdc48 serve as interaction platforms for adaptor proteins that target the proteins into different cellular pathways (Dreveny et al., 2004; Yeung et al., 2008). Based on the sequence homology, a similar structural and functional organization was proposed for the N, D1, and D2 domains of Drg1 (Kressler et al., 2012).
Figure 3.
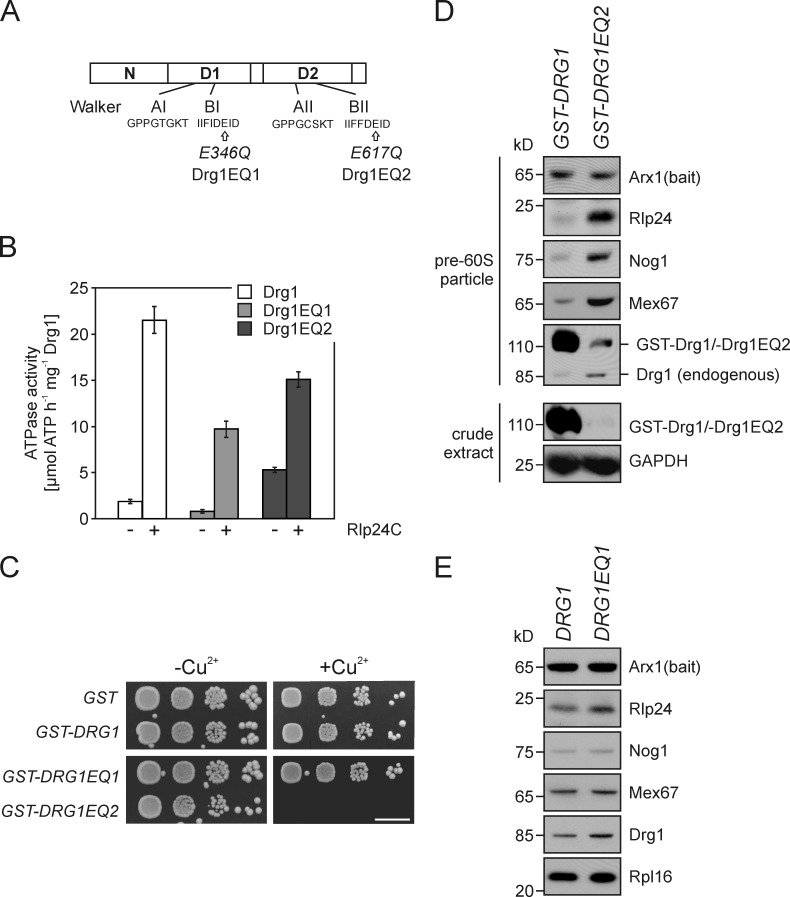
Inactivation of ATP hydrolysis in the D2 domains of Drg1 blocks the release of shuttling proteins. (A) Schematic representation of the domain structure of Drg1. The positions of the respective E to Q amino acid exchanges in the Walker B motifs of the first (Drg1EQ1) and second (Drg1EQ2) AAA domain of the two mutants are indicated. (B) The ATPase activity of both AAA domains is stimulated by Rlp24. The ATPase activity of Drg1, Drg1EQ1, or Drg1EQ2 in the presence (+) or absence (−) of 800 nM Rlp24C was determined. Error bars: SD of at least two biological replicates. (C) Overexpression of the Drg1EQ2 but not of the Drg1EQ1 protein results in a dominant-negative growth phenotype. Spot assay to monitor growth under uninduced (−Cu2+) or induced (+Cu2+) conditions. Serial dilutions of strains expressing GST fusions of Drg1, Drg1EQ1, and Drg1EQ2 under the control of the CUP1 promoter were spotted on SDC–ura plates containing 0.5 mM CuSO4. Growth was monitored after incubation at 30°C for 3 d. Bar, 10 mm. (D) Overexpression of the Drg1EQ2 protein causes accumulation of shuttling proteins and export factors on late pre-60S particles in a wild-type background. The Drg1EQ2 protein or wild-type Drg1 were expressed as GST fusions under the control of the Cu2+-inducible CUP1 promoter in an Arx1-TAP strain. Cells were grown to early log phase and the CUP1 promoter was induced with 0.5 mM CuSO4 for 3 h. Afterward, pre-60S particles were isolated by protein A affinity purification and TEV elution. Purified particles were analyzed for the presence of pre-60S factors by Western blotting. Note that despite the low expression level of Drg1EQ2 (see protein levels in the crude extract), binding of the mutant protein and an accumulation of shuttling proteins occurred. (E) The composition of late pre-60S particles does not change significantly when ATP hydrolysis is blocked in the D1 AAA domain. Pre-60S particles were affinity purified with Arx1-TAP as bait protein from the drg1Δ strain ectopically expressing Drg1 or Drg1EQ1 from centromeric plasmids under the control of their native promoters. The TEV eluates were analyzed for the presence of shuttling proteins by Western blotting.
Functional inactivation of Drg1 is lethal. It leads to a failure to release shuttling proteins from pre-60S particles in the cytoplasm and prevents association of late joining maturation factors and ribosomal proteins. This results in a block in the transition to mature 60S subunits (Pertschy et al., 2007). Consequently, aberrant pre-60S particles accumulate in the cytoplasm of temperature-sensitive drg1 mutants that contain accumulated shuttling proteins and lack late joining cytoplasmic maturation factors. The accumulation of several shuttling proteins in the drg1-ts mutant raises the question of whether the release of these proteins occurs in a concerted action or in a strictly ordered process with Drg1 catalyzing the first reaction. However, the direct release substrate and the exact function of Drg1 in this process were hitherto unknown.
We show here that Drg1 is recruited to pre-60S particles by the shuttling factor Rlp24. Binding to Rlp24 causes enhanced ATP hydrolysis of Drg1, which is used to specifically extract Rlp24 from pre-60S particles but none of the other shuttling proteins. Besides the requirement for ATP, we show that the nucleoporin Nup116 is necessary for the release reaction in vitro, suggesting a close cooperation between nuclear export and cytoplasmic maturation of pre-60S particles in vivo.
Results
Rlp24 interacts with Drg1 and stimulates its ATPase activity
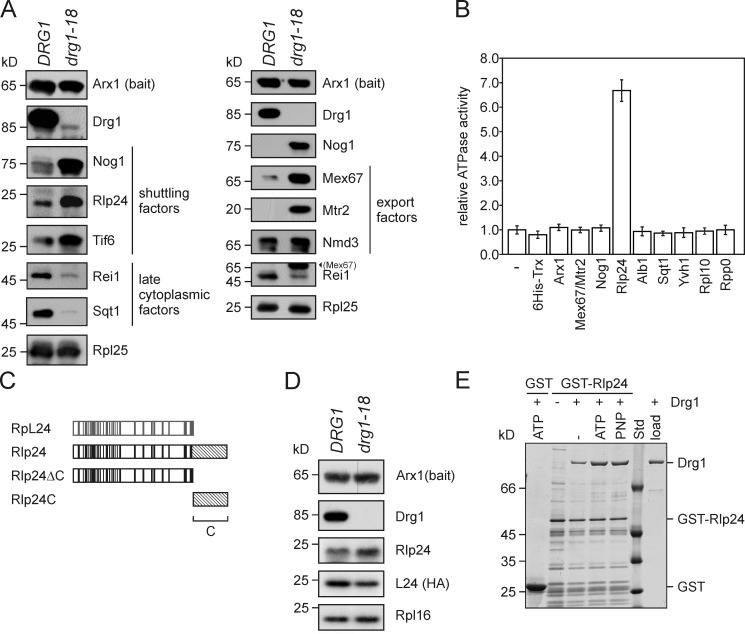
Drg1 binds to cytoplasmic pre-60S particles and is required for the release of Nog1, Rlp24, Arx1, and Tif6 (Pertschy et al., 2007). To identify additional factors that require Drg1 for their release we performed a detailed analysis of the composition of pre-60S particles in the temperature-sensitive drg1-18 mutant. As shown in Figs. 1 A and S1, functional Drg1 is necessary for the release of the shuttling proteins Nog1, Rlp24, Tif6, and Mrt4 and the export factors Mex67/Mtr2 as well as for joining of the late cytoplasmic factors Rei1, Sqt1, and Yvh1. As joining of Yvh1 and release of Mrt4 have been shown to be required for assembly of the characteristic ribosomal stalk of 60S subunits, we conclude that in drg1-18 mutants this structure cannot be formed. This suggestion is further supported by cytoplasmic accumulation of Mrt4-YFP in the drg1-18 mutant (Lo et al., 2010).
Figure 1.
Rlp24 is the direct interaction partner of Drg1 on the pre-60S particle. (A) Drg1 inactivation results in accumulation of shuttling proteins and export factors on pre-60S particles. Late logarithmic phase wild-type and temperature-sensitive drg1-18 cells were incubated at 37°C for 1 h. Afterward, pre-60S particles were isolated by TAP using Arx1-TAP as bait protein. Purified particles were analyzed for the presence of shuttling proteins (left) or export factors (right) by Western blotting. Nog1 and Rei1 were detected in both preparations, confirming comparability of the two preparations. (Mex67) in the Rei1 blot denotes residual Mex67 signal after stripping. (B) The ATPase activity of Drg1 is stimulated by Rlp24. 10 µg Drg1 were incubated with 5 µg of the indicated purified proteins in the presence of 1 mM ATP at 30°C for 30 min. The ATPase activity was determined as described in Materials and methods. Mex67/Mtr2 indicates that purified Mex67/Mtr2 heterodimer was added. The relative ATPase activity compared with the activity of Drg1 alone (−) is plotted. Error bars: SD of at least two biological replicates. (C) Schematic representation of the homology regions of the large subunit ribosomal protein L24 (gray) and the preribosomal protein Rlp24 (black). Residues identical in both proteins are indicated as vertical lines. The nonhomologous 53–amino acid extension at the C terminus, which is unique to Rlp24, is indicated as a hatched area. The C-terminally truncated Rlp24ΔC variant and the Rlp24C domain, which were used in subsequent experiments are also indicated. (D) Inactivation of Drg1 results in decreased levels of L24 on pre-60S particles. Late logarithmic phase wild-type and temperature-sensitive drg1-18 cells expressing chromosomally HA-tagged L24A (YGL031C) were incubated at 37°C for 1 h. Afterward, pre-60S particles were isolated by protein A affinity purification and TEV elution using Arx1-TAP as bait protein. Purified particles were analyzed for the presence of L24 using an HRP-conjugated rat anti-HA antibody (Roche). Western blots with polyclonal antibodies directed against Rlp24, Arx1, or the ribosomal protein L16 using a secondary HRP-conjugated goat anti–rabbit antibody served as controls. (E) In vitro binding of Drg1 to Rlp24 is enhanced in the presence of nucleotide. GST-Rlp24 immobilized on glutathione beads was incubated with Drg1 in the presence of 1 mM ATP or 1 mM of the nonhydrolyzable analogue AMP-PNP (PNP) at room temperature for 2 h. As a control for nonspecific binding, the GST tag bound to the beads was incubated with Drg1. After extensive washing, GST-tagged Rlp24 was eluted using free glutathione and eluates were investigated by SDS-PAGE and Coomassie staining. Std, protein standard; load, an aliquot of purified Drg1 used for the binding assay was loaded.
Up to now, no direct substrate of Drg1 has been identified, although it is likely that Drg1 uses ATP hydrolysis to actively strip one or more nonribosomal proteins from the pre-60S particle or to load a late-acting factor. In vivo, ATP hydrolysis in the D2 AAA domain of Drg1 is strictly required for the release of shuttling proteins and export factors (Fig. 3 D; Pertschy et al., 2007). However, the in vitro ATP hydrolysis rate of Drg1 is low (Zakalskiy et al., 2002). We therefore reasoned that interaction with a substrate protein or cofactor might stimulate the ATPase activity of the AAA protein. To test this hypothesis, we purified proteins that we considered potential binding partners of Drg1 and investigated their influence on its ATPase activity. Particularly, proteins were chosen that showed altered levels on late pre-60S particles from the drg1-18 mutant. This set of proteins included the export factors Mex67/Mtr2 and Arx1, the shuttling proteins Rlp24, Nog1, and Alb1, the late cytoplasmic factors Sqt1 and Yvh1, as well as the late binding ribosomal proteins Rpl10 and Rpp0. Factors that are known to be released or to bind at a later stage of maturation (e.g., Tif6 and Lsg1) were not taken into consideration in this experiment. Drg1 was incubated with the purified proteins and the ATPase activity was determined (see Materials and methods for details). As shown in Fig. 1 B, Rlp24 stimulated the ATPase activity of Drg1, whereas none of the other proteins had an influence on ATP hydrolysis by Drg1. ATP was not hydrolyzed when Drg1 was omitted and only Rlp24 was present in the assay (unpublished data). Rlp24 is a shuttling pre-60S maturation factor that accompanies the preribosomal particle into the cytoplasm (Saveanu et al., 2003), where it is released in a Drg1-dependent manner (Fig. 1 A; Pertschy et al., 2007). Rlp24 exhibits significant sequence identity with the ribosomal protein L24 (Saveanu et al., 2003). The most evident difference between Rlp24 and L24 is the presence of a 53-residues-long C-terminal extension rich in amino acids with acidic side chains that is specific to the nonribosomal protein Rlp24 (Fig. 1 C). Both proteins are thought to recognize the same binding site on (pre-)60S particles (Saveanu et al., 2003). Consistent with this view, inactivation of Drg1 results in decreased levels of L24 on pre-60S particles, whereas Rlp24 increases (Fig. 1 D).
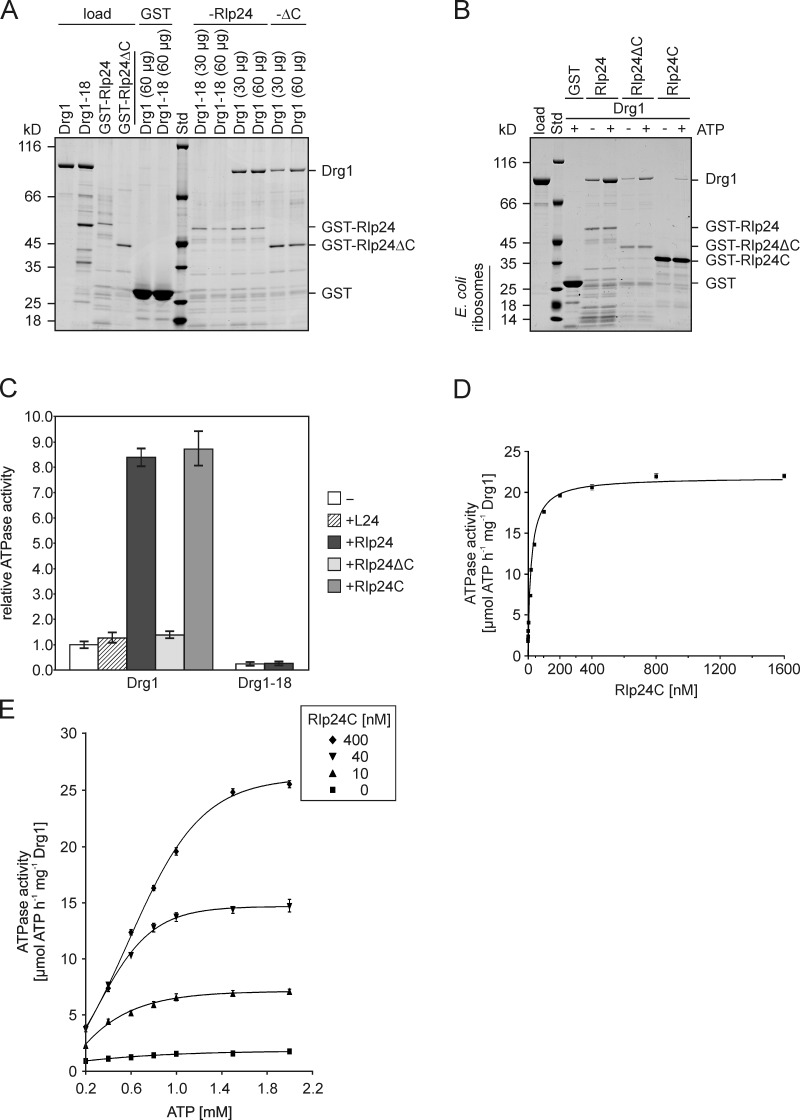
Stimulation of the ATPase activity of Drg1 by Rlp24 necessitates a direct interaction of the two proteins. Indeed, pull-down experiments demonstrated in vitro interaction of Drg1 with purified GST-Rlp24. The amount of copurified Drg1 was increased by the presence of ATP or the nonhydrolyzable analogue AMP-PNP (Fig. 1 E). Thus, ATP binding, but not hydrolysis, is important for the interaction of Drg1 with Rlp24. Because nucleotide binding is known to trigger oligomerization of Drg1 (Zakalskiy et al., 2002), we propose that only hexameric Drg1 binds to Rlp24. The binding detected in the absence of nucleotide is likely caused by the presence of ∼30% hexameric Drg1 in the protein preparations (Fig. S2). Indeed, the protein from the drg1-18 mutant, which is unable to form hexamers (Zakalskiy et al., 2002), does not bind GST-Rlp24 in vitro (Fig. 2 A) and fails to be activated by Rlp24 (Fig. 2 C). Notably, Drg1-18 does not associate with pre-60S particles (Fig. 1 A; Pertschy et al., 2007), suggesting that the interaction with Rlp24 is necessary to recruit Drg1 to the pre-60S particle. Consistent with a role of Rlp24 in recruitment of Drg1, deletion of the last 53 amino acid residues of Rlp24 prevents interaction of Drg1 with preribosomal particles in vivo (Lo et al., 2010). To examine the contribution of this C-terminal domain to in vitro interaction with Drg1, a C-terminally truncated Rlp24 variant was used for GST pull-down experiments. As shown in Fig. 2 A, GST-Rlp24ΔC still bound Drg1, albeit binding was reduced compared with full-length Rlp24. However, Rlp24ΔC did not activate ATP hydrolysis of Drg1 (Fig. 2 C). Furthermore, L24, which lacks this C-terminal extension, did not stimulate the ATPase activity of Drg1. In contrast, the C-terminal 53 amino acid residues of Rlp24 (designated Rlp24C) are sufficient to bind Drg1 and to stimulate its ATPase activity (Fig. 2, B and C). These results suggest that Drg1 interacts with two domains of Rlp24, the C-terminal domain that stimulates the ATPase activity of Drg1 and a second region that does not have any stimulatory effect on ATP hydrolysis.
Figure 2.
The C-terminal domain of Rlp24 interacts with Drg1 and stimulates its ATPase activity. (A and B) GST pull-down assays to test the interaction between Rlp24 fragments and Drg1 or Drg1-18. The GST-Rlp24 fragments were immobilized on glutathione-agarose and incubated for 2 h with the respective Drg1 variants. After washing and elution with glutathione, eluates were analyzed by SDS-PAGE and Coomassie staining. Std, protein standard. GST served as control. (A) Binding of the temperature-sensitive Drg1-18 protein (30 or 60 µg) to full-length Rlp24 and binding of wild-type Drg1 to full-length Rlp24 and C-terminally truncated Rlp24 (Rlp24ΔC) in the presence of 1 mM ATP. Note that, despite the higher amount of bait protein used, significantly less Drg1 bound to C-terminally truncated Rlp24 compared with the full-length protein. (B) Comparison of the binding of Drg1 to full-length Rlp24, Rlp24ΔC, and Rlp24C in the presence and absence of ATP. (C) The ATPase activity of Drg1 in the presence of 6His-tagged versions of L24, Rlp24, Rlp24C, or Rlp24ΔC as well as of Drg1-18 in the presence of 6HisRlp24 was measured. Relative values compared with the ATPase activity of the unstimulated protein (−) were calculated. Error bars: SD of two biological replicates. (D) ATPase activity of Drg1 in the presence of different 6HisRlp24C concentrations (0–1600 nM). The ATPase activity in micromoles of ATP h−1 mg−1 Drg1 was plotted as a function of the Rlp24C concentration. Error bars: SEM of three biological replicates. (E) Dependency of the ATPase activity of Drg1 on the ATP concentration. The ATPase activities (micromoles of ATP h−1 mg−1 Drg1) in the presence of different ATP (0.2 to 2 mM) and Rlp24C (0, 10, 40, and 400 nM) concentrations were determined and plotted as a function of the ATP concentration. Error bars: SEM of three biological replicates.
Although heterologously expressed full-length GST-Rlp24 and 6HisRlp24 were integrated into Escherichia coli 50S ribosomal subunits, the fusion protein containing only the last 53 amino acid residues of Rlp24 did not copurify E. coli ribosomes (Fig. 2 B) and was therefore used for further enzymatic characterization. As shown in Fig. 2 D, in the presence of 1 mM ATP, Rlp24C stimulated the ATPase activity of Drg1 to a Vmax of 22.4 (±0.5) µmol ATP h−1 mg−1. The curve could be fitted best to a sigmoid function with a Hill factor of 0.96 (±0.05). The required concentration for half-maximal activity was calculated to be 28.3 (±0.6) nM Rlp24C (mean and SD of three biological replicates). Full saturation in terms of maximal ATPase activity was achieved at an Rlp24 concentration of ∼200 nM, corresponding to a molar ratio of one Rlp24 molecule to one hexamer of Drg1 in the reaction mixture.
To gain further mechanistic insights into the activation of Drg1 by Rlp24, we measured the ATPase activity in the presence of different ATP concentrations. In the absence of Rlp24, a kMapp for ATP of 0.27 mM (±0.07) and a Vmax of 1.99 (±0.29) µmol ATP h−1 mg−1 were measured (mean of two biological replicates). However, when Rlp24 was present in the assay, sigmoid kinetics were observed (Fig. 2 E). With saturating concentrations of 400 nM Rlp24 a Hill coefficient of 2.4 (±0.17) was determined, indicating pronounced positive cooperativity of the Drg1 subunits in response to the nucleotide. The half-maximal activation (EC50) was achieved with 0.8 mM ATP and Vmax was calculated to 28.7 (±0.7) µmol ATP h−1 mg−1. These data show that binding of Rlp24 to Drg1 induces increased cooperativity of the individual subunits of Drg1 and increases both Vmax and EC50 to fuel ATP hydrolysis.
The D2 AAA domain is required for shuttling protein release
To investigate whether the increased ATPase activity arises from the first or second ATPase domain, we tested two mutant variants of Drg1. These variants contain exchanges of the conserved glutamate residues in Walker motif B of the first (Drg1EQ1) and second ATPase domain (Drg1EQ2) to glutamine (Fig. 3 A). As the carboxylic group of the glutamate activates a catalytic water molecule for nucleophilic attack on the γ phosphate, these exchanges render the respective domains nonfunctional in ATP hydrolysis (Hanson and Whiteheart, 2005). As shown in Fig. 3 B, in the absence of Rlp24, Drg1EQ1 showed lower ATPase activity, whereas Drg1EQ2 showed higher ATPase activity than the wild-type protein. In this respect Drg1 resembles the AAA-ATPases Hsp104 and ClpB, which also show higher ATPase activity of one AAA domain upon inactivation of the other AAA domain (Watanabe et al., 2002; Mogk et al., 2003; Doyle et al., 2007; Schaupp et al., 2007). The higher ATPase activity of the EQ2 mutant suggests a tight coordination of ATP hydrolysis in the two ATPase domains of Drg1. In the presence of Rlp24C, increased activity was measured for wild-type, Drg1EQ1, and Drg1EQ2, demonstrating that the interaction with Rlp24 activates ATP hydrolysis in both AAA domains.
To investigate the contribution of ATP hydrolysis in the D1 and D2 domains to the in vivo function of Drg1, we analyzed the growth behavior of strains expressing only Drg1EQ1 or Drg1EQ2. The strain expressing Drg1EQ1 showed no obvious growth phenotype, whereas the strain expressing Drg1EQ2 was nonviable (unpublished data). Furthermore, overexpression of Drg1EQ2 from the strong CUP1 promoter resulted in a dominant-negative growth phenotype, whereas Drg1EQ1 overexpression did not affect growth (Fig. 3 C). To correlate these results with the effect of Drg1 on the composition of pre-60S particles, we analyzed Arx1-TAP containing particles from strains expressing Drg1EQ1 or Drg1EQ2 variants by Western blotting. As the EQ2 exchange is nonfunctional, this variant was overexpressed in the wild-type background. To allow distinguishing Drg1EQ2 from endogenous wild-type protein, the mutant variant was fused to GST. Although very little GST-Drg1EQ2 fusion protein was present in the crude extract of overexpressing cells compared with overexpressed wild-type protein, it was detected in preribosomal particles, suggesting a failure to release Drg1EQ2 (Fig. 3 D). The overexpression of the Drg1EQ2 protein resulted in accumulation of shuttling proteins and export factors similar as observed for the temperature-sensitive drg1-18 mutant (Fig. 1 A). Therefore, the ATP hydrolysis deficiency in D2, although supporting binding to pre-60S particles, renders the protein nonfunctional for its physiological activity. In contrast, strains expressing Drg1EQ1 did not show an altered composition of pre-60S particles (Fig. 3 E).
ATP hydrolysis in D1 dissociates Drg1 from Rlp24
To further characterize the interaction of Drg1 with Rlp24 we used the surface plasmon resonance (SPR) technology. SPR allows real-time monitoring of the interaction between two proteins after immobilization of one binding partner on a solid surface. GST-Rlp24C fusion protein was covalently linked to sensor chips and probed for binding with purified Drg1. As shown in Fig. S3 A, the presence of nucleotide during the association phase was required for interaction of Drg1 or its variants with immobilized Rlp24C. However, during the dissociation phase a rapid detachment of all Drg1 proteins from the chip was observed. This dissociation is caused by washing out the nucleotide from Drg1 and could be prevented by addition of 1 mM ATP or AMP-PNP (Fig. S3 B). Therefore, the corresponding nucleotides were added to the running buffer in all subsequent experiments.
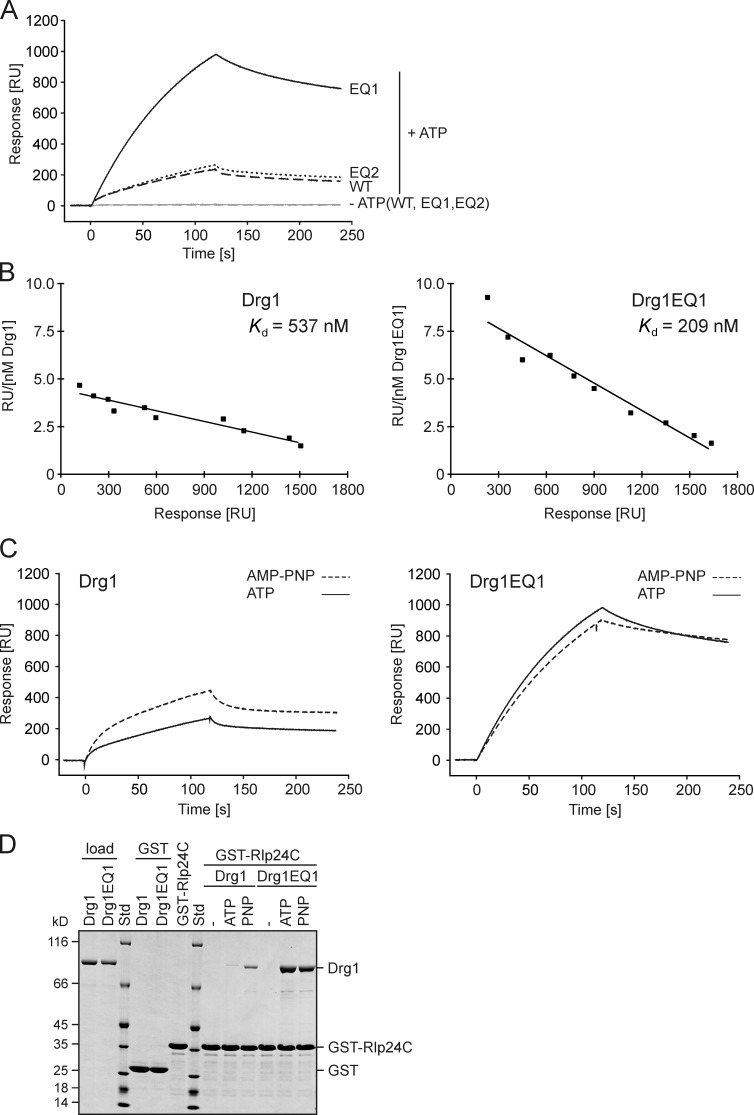
The contribution of the two AAA domains to the interaction with Rlp24 was addressed by including the ATP hydrolysis-deficient variants in this analysis. The Drg1EQ2 variant reached only slightly higher response units (RU) than the wild type, suggesting that a failure to hydrolyze ATP in the second ATPase domain has little influence on binding to Rlp24. In contrast, the Drg1EQ1 variant gained about fourfold higher RU values than the wild-type and EQ2 proteins (Fig. 4 A). Thus, inhibition of ATP hydrolysis in D1 enhances the interaction between Rlp24 and Drg1. Scatchard blot analysis showed that the equilibrium dissociation constant Kd was significantly lower for the EQ1 mutant compared with the wild-type protein (Fig. 4 B). To further dissect the role of ATP hydrolysis for the Drg1–Rlp24 interaction, we monitored binding of the wild-type protein and the EQ1 variant in the presence of AMP-PNP. As shown in Fig. 4 C, the wild-type protein exhibited a longer initial phase and a steeper binding curve in the presence of the nonhydrolyzable analogue, resulting in higher RU values compared with ATP. This result suggests that nucleotide hydrolysis during the experiment significantly reduces the amount of bound Drg1. In contrast, the measured RU values for the Drg1EQ1 variant were lower in the presence of AMP-PNP compared with those in the presence of ATP (Fig. 4 C). Therefore, the ability to hydrolyze ATP in the D1 domain is a major determinant for interaction with Rlp24.
Figure 4.
ATP hydrolysis in D1 dissociates Drg1 from Rlp24C. (A–C) SPR analyses with GST-Rlp24C immobilized on a sensor chip. The data shown are from a representative experiment out of two (B) or three (A and C) biological replicates. (A) The exchange in D1 but not D2 results in decreased release from the GST-Rlp24C fusion. 100 nM of Drg1 (WT), Drg1EQ1, or Drg1EQ2 proteins were injected over the sensor chip in the presence (+) or absence (−) of ATP. RU were recorded and plotted over time. (B) For Scatchard analysis, different concentrations of Drg1 or Drg1EQ1 were injected in the presence of AMP-PNP. The response at the end of the association phase was determined for each concentration from the sensorgrams. The RU per nanomole concentration of Drg1 (left) or Drg1EQ1 (right) were plotted as a function of RU. (C) Comparison of the interaction between Rlp24C and Drg1 (left) or Drg1EQ1 (right) in the presence of ATP or AMP-PNP. (D) ATP hydrolysis in D1 is important for the release of Drg1 from Rlp24C in vitro. GST pull-down experiments using GST-Rlp24C as bait. GST-Rlp24C was immobilized on glutathione agarose and incubated with 60 µg Drg1 or Drg1EQ1 in the presence of the indicated nucleotide. PNP, AMP-PNP; −, incubation in the absence of nucleotide. The eluates were analyzed by SDS-PAGE and Coomassie staining.
The stronger interaction under conditions where nucleotide hydrolysis is blocked could indicate a higher affinity of Drg1 to Rlp24 when ATP is bound in D1. Alternatively, ATP hydrolysis in D1 could trigger dissociation of Drg1 from Rlp24. To distinguish between these possibilities, kinetic SPR analyses of the Drg1–Rlp24 interaction were performed. See Fig. S3 C for a representative experiment. The increase of dRU/dt for the wild-type protein and the mutant variant could not be fitted to a simple 1:1 Langmuir relationship, but follows the two-state reaction formula (Fig. S3 D), which introduces a conformational change term upon binding of Drg1 to Rlp24. This formula postulates the formation of the additional complex [Drg1.Rlp24]*, which is described by the constants kon2 and koff2. Quantitative assessment of the kinetic constants showed that Drg1 and Drg1EQ1 exhibit similar association rates (kon1 and kon2; Table 1). However, the dissociation rates were much lower for the mutant. In particular koff2 was several orders of magnitude lower for Drg1EQ1 than for the wild-type protein. Thus, Drg1EQ1 binds to Rlp24 with similar kinetics as the wild-type protein, but dissociates with a very low rate. In addition, the wild-type protein showed a lower koff2 in the presence of nonhydrolyzable nucleotide compared with ATP. We conclude that ATP hydrolysis in the D1 domain results in dissociation of Drg1 from Rlp24.
Table 1.
Kinetic constants of the interaction of Drg1 and Drg1EQ1 with immobilized Rlp24C (two-state reaction)
| Protein | Nucleotide | kon1 | koff1 | kon2 | koff2 |
| M−1 s−1 | s−1 | s−1 | s−1 | ||
| Drg1 | ATP | 5.4 ± 0.9 × 104 | 1.3 ± 0.1 × 10−1 | 2.8 ± 0.5 × 10−2 | 2.5 ± 0.5 × 10−3 |
| Drg1 | AMP-PNP | 1.1 ± 0.1 × 105 | 6.0 ± 0.9 × 10−2 | 1.5 ± 0.2 × 10−2 | 6.9 ± 0.5 × 10−4 |
| Drg1EQ1 | ATP | 5.8 ± 0.4 × 104 | 5.1 ± 1.5 × 10−3 | 7.2 ± 1.2 × 10−3 | 9.8 ± 0.2 × 10−14 |
| Drg1EQ1 | AMP-PNP | 5.4 ± 0.7 × 104 | 3.5 ± 1.1 × 10−3 | 1.1 ± 0.2 × 10−2 | 8.4 ± 7.4 × 10−14 |
Mean values determined from five independent injections over at least three different CM5-chips.
To confirm these results using an independent approach, in vitro binding studies of wild-type Drg1 protein and Drg1EQ1 with GST-Rlp24C were performed. As shown in Fig. 4 D, the presence of ATP stimulated binding of the wild-type protein to GST-Rlp24C. However, significantly increased Drg1 levels were observed when AMP-PNP was used instead of ATP. In this respect, the C-terminal domain behaves differently from the full-length Rlp24, which shows similar binding in the presence of either nucleotide (compare with Fig. 1 D). The Drg1EQ1 variant showed even stronger binding to GST-Rlp24C than the wild-type protein in the presence of nucleotide and AMP-PNP did not further enhance binding, confirming the results from the SPR analysis.
Drg1-mediated release of Rlp24 from pre-60S particles is promoted by nucleoporins
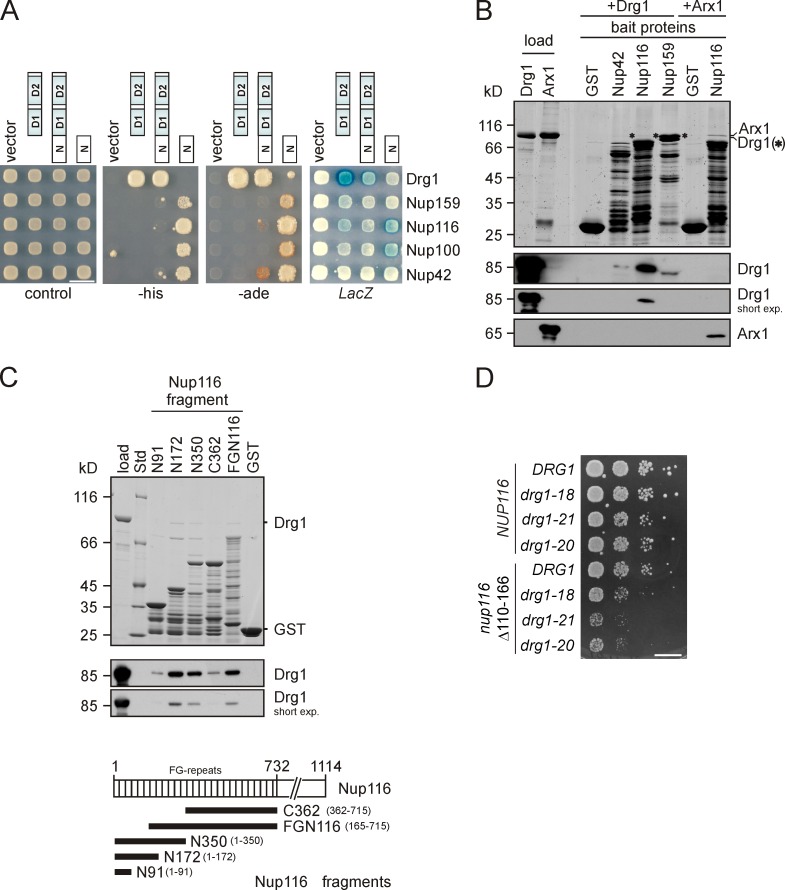
The increased ATPase activity of Drg1 upon binding to Rlp24 raised the question of whether ATP hydrolysis triggers the direct release of Rlp24 or is used to liberate one or more other shuttling proteins from pre-60S particles. To address this issue, we aimed to develop an in vitro release assay. For this purpose, pre-60S particles were purified from the drg1-18 mutant. These particles lack Drg1 but contain increased levels of shuttling proteins (Fig. 1 A). The purified particles were incubated with purified Drg1 and ATP. However, no release of shuttling proteins was observed (see Fig. 6 A, third lane). We therefore speculated that an additional factor might be required for the release reaction that was missing in our in vitro reaction mixture. To identify such a cofactor of Drg1, we performed a genomic two-hybrid screen (James et al., 1996). Because the N-terminal domain is the main interaction surface of AAA-ATPases, we specifically used this region of the protein for screening. The two-hybrid screen resulted in the isolation of 13 interacting clones from 8 × 106 transformants. DNA sequence analysis of interacting clones identified fragments of the genes NUP42 (seven isolates representing three different clones), NUP100 (three isolates representing three different clones), NUP116 (two clones), and NUP159 (one clone). These genes encode FG repeat–containing nucleoporins present in the Nup82 subcomplex and are preferentially or exclusively located at the cytosolic surface of the NPC (Alber et al., 2007). Interaction occurred between the N-terminal domain of Drg1 and the FG repeat portion of the nucleoporins and was strongest for Nup116 (Fig. 5 A). Direct interaction between Drg1 and nucleoporins was confirmed by GST pull-down assays (Fig. 5 B). Further mapping of the Drg1 binding sites of Nup116 showed strongest binding for a fragment ranging from the N terminus of Nup116 to residue 172, with only minor contributions of the first 91 amino acid residues to this interaction (Fig. 5 C). To investigate whether this interaction is important in vivo, a mutant of Nup116 containing a deletion of amino acid residues 110–166 was tested for genetic interaction with temperature-sensitive drg1-ts alleles. Indeed, a synthetic enhancement of growth defects of all drg1-ts alleles was observed, implicating that the interaction between Drg1 and Nup116 is of physiological relevance (Fig. 5 D).
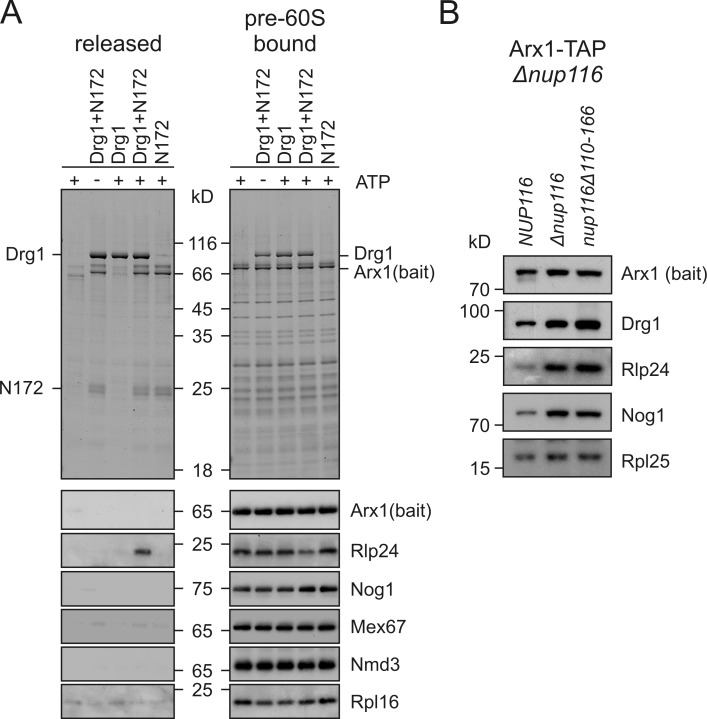
Figure 6.
Drg1 specifically releases Rlp24 from pre-60S particles. (A) Pre-60S particles from the temperature-sensitive drg1-18 mutant incubated at 37°C were purified with Arx1-TAP as bait and immobilized on calmodulin beads. After incubation with Drg1 in the presence (+) or absence (−) of ATP and the Nup116 fragment N172, supernatants (containing released and unbound proteins) were collected and TCA precipitated. pre-60S particles were eluted from the calmodulin beads and concentrated by TCA precipitation. Supernatants (released) and eluates (pre-60S bound) were analyzed by SDS-PAGE and Coomassie staining, as well as Western blotting. (B) Pre-60S particles purified from the Δnup116 as well as from the nup116Δ110-166 mutant show increased amounts of Drg1 and Rlp24. The nup116Δ strain carrying centromeric plasmids with wild-type NUP116 or the nup116 variant lacking codons 110–166 were grown to late log phase and pre-60S particles were isolated via Arx1-TAP. Purified particles were separated by SDS-PAGE and analyzed for the levels of pre-60S maturation factors by Western blotting.
Figure 5.
Drg1 interacts with Nup116. (A) Two-hybrid analysis showing the interaction of Drg1 with FG repeat–containing nucleoporins Nup159, Nup116, Nup100, and Nup42 isolated in a genomic library screen. Interaction was tested by monitoring growth on SD medium lacking histidine (−his) or adenine (−ade) or by detection of blue color formation caused by the LacZ reporter gene activity using overlay assays (LacZ). Bar, 10 mm. The tested Drg1 fragments are indicated by schematic drawings. (B) In vitro interaction of Drg1 with the FG repeat portions of nucleoporins. GST pull-down experiments were performed with purified Drg1 incubated with GST fusions of the FG repeat portions of Nup42, Nup116, or Nup159 for 2 h at 4°C. Purified Arx1, which was shown previously to interact with Nup116 (Bradatsch et al., 2007; Hung et al., 2008), was used as a positive control. After extensive washing, the GST fusion proteins were eluted. Eluates were analyzed by SDS-PAGE and Coomassie blue staining as well as Western blotting to detect bound Drg1 and Arx1. (C) The main Drg1 interaction domain of Nup116 resides between amino acid residues 91 and 172. 100 µg of purified Drg1 was incubated with 100 µg of various GST-Nup116 fragments for 1 h at 4°C. Afterward, glutathione beads were added and incubation continued for another 45 min. After extensive washing, bound proteins were eluted with glutathione-containing buffer. Eluates were analyzed by SDS-PAGE and Coomassie staining, as well as Western blotting using Drg1 antibody. Below the Western blots, a schematic representation of the FG repeat regions of Nup116 (vertical lines) and fragments generated for the characterization of the interaction domain with Drg1 is shown. (D) Deletion of the residues 110–166 in Nup116 leads to an enhanced synthetic growth defect in drg1 temperature-sensitive strains. Spot assay of a nup116Δ drg1Δ strain carrying plasmids with a nup116 variant lacking codons 110–166 and different temperature-sensitive drg1 alleles (18, 21, and 20). Wild-type DRG1 and NUP116 served as controls. The strains carrying the indicated wild-type and mutant alleles were spotted in serial 10-fold dilutions onto SDC–leu-trp and incubated at 28°C for 3 d. Bar, 10 mm.
The Nup116 fragment exhibiting the strongest in vitro interaction was tested for an influence on the ATPase activity of Drg1. No change in the ATPase activity of Drg1 was observed when Nup116 was added in addition to Rlp24 (unpublished data). Such a behavior is characteristic for adaptor proteins of related AAA proteins, which have little effect on ATPase activity (Wang et al., 2004). Next we tested the effect of addition of the Nup116 fragment to our in vitro release assay. As shown in Fig. 6, in the presence of Nup116 and ATP, Drg1 specifically released Rlp24 from pre-60S particles purified from the drg1-ts mutant. No release was observed in the absence of ATP, corroborating the importance of ATP hydrolysis in the release reaction. In contrast to Rlp24, other tested shuttling proteins like Nog1, Mex67, and Nmd3 remained bound to the pre-60S particles. The influence of Nup116 on the release of Rlp24 was also tested in vivo. As shown in Fig. 6 B, deletion of the Drg1 binding site on Nup116 as well as full deletion of the nucleoporin resulted in increased levels of Rlp24 on purified pre-60S particles. As a consequence, other shuttling factors like Nog1 are also accumulating (Fig. 6 B and not depicted). The increased levels of Drg1 on the particles from the mutants suggest that Nup116 is required for full activity and subsequent dissociation of Drg1 in vivo. We conclude that by means of ATP hydrolysis Drg1 specifically releases Rlp24 from pre-60S particles and that this release reaction is assisted by nucleoporins.
Discussion
The AAA-ATPase Drg1 functions in the initial steps of cytoplasmic pre-60S maturation where it is required for the release of several shuttling proteins including Mex67, Tif6, Nog1, and Rlp24 (Fig. 1; Pertschy et al., 2007). Here we show that the direct binding target and release substrate of Drg1 on preribosomal particles is the shuttling protein Rlp24. This protein joins the pre-60S particle in the nucleolus and accompanies it into the cytoplasm. After its release from the particle, it is substituted by the ribosomal protein L24 (Saveanu et al., 2003). In vitro, Drg1 interacts with two independent binding sites within Rlp24. One lies in the part of Rlp24 that is related to the ribosomal protein L24, whereas the second site comprises the 53-residues-long Rlp24-specific C-terminal domain. Because this domain is unique for the shuttling protein, it could provide some degree of specificity to target Drg1 to the immature 60S subunit. Although each individual domain is sufficient for interaction with Drg1, the strongest binding was achieved with full-length Rlp24 (Fig. 2 B). Direct interaction between Drg1 and Rlp24 suggests that Rlp24 is the attachment site for Drg1 on pre-60S particles. Consistently, Rlp24, and particularly its C-terminal domain, is required for association of Drg1 with pre-60S particles in vivo (Lo et al., 2010).
Binding of Rlp24 stimulates the ATPase activity of Drg1. Intriguingly, this stimulatory effect is exclusively conferred by the C-terminal domain of Rlp24. Hence, Rlp24 not only acts in recruitment of Drg1 but also functions as an activator that stimulates ATP hydrolysis by the AAA-ATPase. Full activation of Drg1 is reached at a stoichiometry of one molecule of Rlp24 per hexamer Drg1, which presumably reflects the cellular situation where hexameric Drg1 encounters pre-60S particles containing one Rlp24 molecule. The interaction with Rlp24 triggers a structural change in Drg1 that leads to positive cooperativity of the subunits and an increased ATP hydrolysis rate. In the presence of saturating concentrations of Rlp24 this rate showed a strong dependency on the nucleotide concentration. In contrast, in the absence of Rlp24, ATP hydrolysis showed only little increase when the nucleotide concentration was raised. We therefore propose that the ATPase domains of Drg1 adopt a repressed conformation, which is converted into an active form by binding of Rlp24. The transition into the active conformation could, for example, increase accessibility of the nucleotide binding pocket and thus facilitate exchange of the spent nucleotide.
The results obtained with the ATP hydrolysis–deficient mutants show that the interaction of Rlp24 with Drg1 stimulates ATP hydrolysis in both the D1 and D2 AAA domains. What is the contribution of the two ATPase domains of Drg1 to Rlp24 release? Only ATP hydrolysis in D2 is essential for viability and is required for the release of shuttling proteins including Rlp24 in vivo (Fig. 3 D). Consequently, D2 provides the energy for extracting Rlp24 from the preribosome. In contrast, inactivation of ATP hydrolysis in D1 does not affect growth and has only minor impact on the release of shuttling proteins from the pre-60S particle (Fig. 3 E). Notably, however, Drg1EQ1 forms more stable hexamers than the wild-type protein and shows very slow dissociation rates from Rlp24. We therefore speculate that ATP hydrolysis in D1 is necessary for disassembly of Drg1 into monomers and, possibly as a consequence, dissociation of Rlp24 from Drg1. Considering that Rlp24 does not interact with monomeric Drg1 variants (i.e., Drg1-18), it is tempting to speculate that one Rlp24 molecule requires at least two Drg1 protomers for efficient binding. Hence, disassembly into monomers might be used as a mechanism to liberate Drg1 from Rlp24.
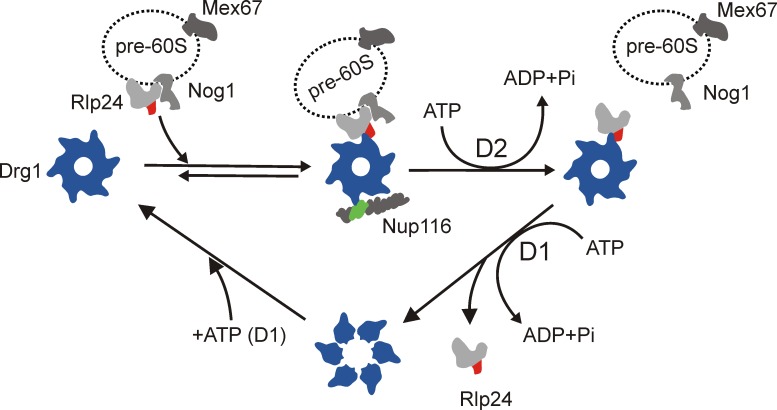
In summary, binding of Rlp24 to Drg1 not only triggers the extraction of Rlp24 from the pre-60S particle by ATP hydrolysis in D2 but also the disassembly of Drg1 into monomers and dissociation of Drg1 from Rlp24 by ATP hydrolysis in D1 (Fig. 7). The switch between monomeric and oligomeric states within its ATPase cycle is a remarkable feature that distinguishes Drg1 from its closest relatives, Cdc48, and its mammalian orthologue, p97. The latter forms stable oligomers throughout its whole ATPase cycle (Wang et al., 2003). Thus, despite the structural similarities between these proteins, the ATP hydrolysis cycles and the consequences for the oligomeric structure are different.
Figure 7.
Schematic model of the ATPase cycle of Drg1 and its function in stripping Rlp24 from pre-60S particles. Binding of ATP to D1 of Drg1 leads to hexamerization of the protein and subsequent association with pre-60S-bound Rlp24. This binding results in stimulation of ATP hydrolysis in D2, which catalyzes extraction of Rlp24 from preribosomal particles. Nup116 is further required in this step to release Rlp24 from the pre-60S particle in vitro. Finally, ATP hydrolysis in D1 triggers dissociation from Rlp24 and likely disassembly of the Drg1 hexamer. The domains interacting with Drg1 are indicated in red for Rlp24 and green for Nup116.
Although Rlp24 binds Drg1 and stimulates its ATPase activity, the dual interaction between Rlp24 and Drg1 is not sufficient for in vitro release of Rlp24 from preribosomal particles. Intriguingly, another direct interaction partner of Drg1, the nucleoporin Nup116, is needed for Drg1 to release Rlp24. Apart from Nup116 we also found interactions with other cytoplasmically exposed FG repeat nucleoporins, suggesting a certain redundancy of the interaction with Drg1. As the interaction with Nup116 was significantly stronger than with any of the other tested nucleoporins, Nup116 is most likely the main nucleoporin binding partner of Drg1. Because Nup116 does not affect the ATPase activity of Drg1, the nucleoporin has to serve a different function. The interaction of Nup116 with the N domain of Drg1 is reminiscent of binding of adaptor proteins to the N domain of p97. These adaptor proteins are thought to target the ATPase into distinct cellular pathways (Wang et al., 2004). However, the strict requirement for Nup116 in the in vitro release assay suggests a more active role of the nucleoporin in the release reaction and not a sole targeting function. For example, Nup116 could allow conversion of an otherwise nonproductive futile ATP hydrolysis cycle into successful extraction of Rlp24. Nevertheless, the interaction of Drg1 with nuclear pore proteins has to be rather transient, as GFP-Drg1 fusions do not exhibit nuclear rim staining (Pertschy et al., 2007; unpublished data). Regardless of the exact function of Nup116 in the release of Rlp24 by Drg1, its requirement suggests that export of the preribosome and the initiation of cytoplasmic pre-60S maturation are tightly coupled events. This coordination could ensure that the cytoplasmic maturation cascade starts as soon as the particle appears at the cytoplasmic side of the NPC complex.
In vivo, Drg1 is necessary for the release of several shuttling proteins from the pre-60S particle; however, in our in vitro assay, Drg1 specifically extracted Rlp24 but none of the other tested nonribosomal proteins (like Nog1 or Mex67). This suggests that Rlp24 is released before other shuttling proteins and export factors can leave the particle. Moreover, these factors do not dissociate spontaneously once Rlp24 has been released, but require additional, active stripping processes. The identification of factors catalyzing these processes will be a major task for future studies. By gradually supplying the assay with additional factors, the in vitro system we developed will provide an excellent tool to reconstitute cytoplasmic maturation steps in vitro and to finally obtain a detailed lineup of all cytoplasmic pre-60S maturation steps.
Materials and methods
Yeast strains and growth conditions
The yeast and bacterial strains used in the present study are listed in Table S1. All plasmids used are listed in Table S2. Chromosomal deletions or gene fusions were generated by homologous recombination using PCR products to transform the respective yeast strain as described previously (Longtine et al., 1998). For generation of deletion strains, PCR products were generated with plasmids pFA6a-hphNT1 as templates and 47- to 50-nucleotide-long gene-specific primers hybridizing to the regions immediately upstream and downstream of the open reading frames. For generating gene fusions, the plasmids pFA6a-TAP(Tcyc1), pFA6a 3HA-kanMX, and pDH5 (Yeast Resource Center, University of Washington) were used as templates. Correct integration was confirmed by colony PCR using one gene-specific primer and one primer specific to the selection cassette. Strains were grown at different temperatures (25, 30, or 37°C) either in YPD complex medium or for plasmid maintenance, in synthetic dextrose (SDC) or galactose medium supplemented with the appropriate amino acids.
Two-hybrid screen
For the yeast two-hybrid screen, the N domain (codon 1–255) of Drg1 was cloned in frame with the Gal4 DNA-binding domain in pGBDU-C2 (URA3 selection marker) after introduction of EcoRI (forward primer) and BamHI (reverse primer) sites. The plasmid was transformed into the reporter strain PJ69-4A (James et al., 1996). The resulting strain was transformed with a two-hybrid library derived from a mixture of pGAD-C1, -C2, and -C3 libraries (LEU2 selection marker) containing short yeast genomic DNA fragments (James et al., 1996). The library plasmids were isolated from transformants positive for all three reporter genes (HIS3, ADE2, and LacZ) after growth in 5-FOA media lacking leucine, and then amplified in E. coli and characterized by DNA sequencing. For further characterization, plasmids were retransformed into PJ69-4A carrying pGAD-C1 with full-length DRG1, the N domain, or the two AAA domains (codon 235 to termination codon).
Tandem affinity purification (TAP)
Complexes were purified according to the standard TAP protocol (Rigaut et al., 1999; Puig et al., 2001) starting from 4 liters of yeast culture. Cells were grown to an OD600 of ∼2, harvested by centrifugation, and resuspended in buffer A (20 mM Hepes-NaOH, pH 7.5, 10 mM KCl, 2.5 mM MgCl2, 1 mM EGTA, 1 mM DTT, 0.5 mM PMSF, and Complete protease inhibitor cocktail [Roche]). Cells were broken by vigorous shaking in the presence of glass beads four times for 30 s in a bead mill (Merkenschlager) with CO2 cooling. After removal of cell debris by centrifugation, the supernatant was incubated with 300 µl of settled IgG beads (GE Healthcare). After 60 min of incubation, IgG beads with bound complexes were transferred to Mobicol columns (MoBiTec) and washed with 8 ml buffer A and with 2 ml TEV cleavage buffer (20 mM Hepes-NaOH, pH 7.5, 100 mM NaCl, 10 mM KCl, 2.5 mM MgCl2, 1 mM EGTA, and 0.5 mM DTT). Complexes were eluted by incubation in 300 µl of cleavage buffer containing 150 units of AcTEV protease (Invitrogen) for 60 min at 22°C. Eluates were collected and mixed 1:1 with calmodulin binding buffer (20 mM Hepes-NaOH, pH 7.5, 100 mM NaCl, 0.075% NP-40, and 0.5 mM DTT) containing 4 mM CaCl2 and incubated with 300 µl of settled calmodulin beads (GE Healthcare) in Mobicol columns at 4°C for 60 min. After washing with 5 ml calmodulin binding buffer containing 2 mM CaCl2, protein complexes were eluted with 500 µl of elution buffer (20 mM Hepes-NaOH, pH 7.5, 5 mM EGTA, 50 mM NaCl, 0.1% Triton X-100, and 1 mM DTT) and concentrated by TCA precipitation.
Protein expression and purification
DRG1 and mutant variants thereof were expressed in yeast cells under the control of the Cu2+-inducible CUP1 promoter as GST fusions (Zakalskiy et al., 2002). Strains were inoculated into SDC–ura medium to an OD600 of 0.01 and expression was induced by the addition of 0.025 mM CuSO4. After 24 h of incubation at 25°C, cells were harvested by centrifugation. Cells were suspended in lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM DTT, 0.5 mM PMSF, and Complete protease inhibitor cocktail) and disrupted by vigorous shaking in the presence of glass beads four times for 30 s in a bead mill with CO2 cooling. After removal of cell debris by centrifugation, the supernatant was incubated with glutathione agarose beads (Sigma-Aldrich) at 4°C for 90 min. After washing three times with lysis buffer and once with binding buffer (20 mM Hepes-NaOH, pH 6.8, 150 mM KOAc, 5 mM Mg(OAc)2, and 0.1% Tween 20), Drg1 variants were eluted by cleaving off the GST tag using PreScission protease (GE Healthcare) and incubating overnight at 4°C.
For purification of heterologously expressed Rlp24, Arx1, Alb1, Nog1, Sqt1, Rpl10, Rpp0, Yvh1, and Mtr2, the genes were cloned into pET32a. For Mex67/Mtr2 coexpression, MEX67, provided with an artificial Shine-Dalgarno sequence, was cloned into the SacI–HindIII site of pET32-Mtr2. The 6His-tagged fusion proteins were expressed in E. coli BL21 codon plus (Table S2) and purified by Ni2+ chelating chromatography (Schmitt et al., 1993) from a 500-ml culture grown in LB medium. Cells were grown to early log phase and expression was then induced by addition of IPTG (Thermo Fisher Scientific) to a final concentration of 0.4 mM and incubation for 2 h at 22°C. Cells were harvested by centrifugation, resuspended in lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 20 mM imidazole, 1 mM DTT, 0.5 mM PMSF, and HP protease inhibitor cocktail [Serva]), and broken by sonification. After removal of cell debris by centrifugation, the supernatant was incubated with Ni-NTA beads (QIAGEN) at 4°C for 60 min. After washing of the beads three times with lysis buffer and once with binding buffer, proteins were eluted with binding buffer containing 300 mM imidazole. To remove imidazole, buffer exchange was performed using Zeba spin desalting columns 7K (Thermo Fisher scientific) according to the manual. GST fusions and truncated versions of Rlp24 were generated by cloning into pGEX6-P1. Expression and crude extract preparation for purification of GST-Rlp24 variants and FG repeat containing fragments of nuclear pore proteins Nup42, Nup116, and Nup159 (Allen et al., 2001) was performed as described for the 6His-tagged proteins. Crude extracts were incubated with glutathione agarose beads (Sigma-Aldrich) at 4°C for 90 min and after extensive washing with lysis buffer bead-bound proteins were used for GST pull-down assays. Fragments of NUP116 were PCR amplified with primers containing EcoRI (forward primer) and SalI (reverse primer) sites and cloned into pGEX6-P1 and pET28a and purified as described earlier in this paragraph. All expression plasmids are listed in Table S2.
Protein–protein interaction
Interaction assays of purified Drg1 and Arx1 proteins with FG repeat containing nuclear pore proteins were performed in binding buffer using 30 µg of purified Drg1 or Arx1. Interaction assays of Rlp24 with Drg1 variants were performed similarly with 30 or 60 µg of purified Drg1. The GST-tagged bait proteins (Nups and Rlp24) were bound to GSH beads and incubated with Drg1 variants and Arx1. ATP or the nonhydrolyzable analogue AMP-PNP (Sigma-Aldrich) was added to some samples to a final concentration of 1 mM. After incubation for 2 h at 4 or 22°C and extensive washing steps the bait and interacting proteins were eluted from GSH beads by treatment with 20 mM of reduced glutathione in 100 mM Hepes-NaOH, pH 7.5. The eluates were analyzed by SDS-PAGE followed by detection of the proteins by Western blotting.
Western blotting
Unless otherwise stated, all antisera used in this study were raised in rabbits with GST- or 6His-tagged fusion proteins purified from E. coli as antigene. Antisera directed against Nog1, Rlp24, Arx1, and Rei1 were provided by M. Fromont-Racine (Institute Pasteur, Paris, France). The anti-Rpl16 antiserum was a gift from S. Rospert (University of Freiburg, Freiburg, Germany). The Sqt1 and Rpl10, Nmd3, and Mex67/Mtr2 antisera were provided by B.L. Trumpower (Dartmouth Medical School, Hanover, NH), A.W. Johnson (University of Texas at Austin, Austin, TX), and E. Hurt (Biochemie-Zentrum der Universität Heidelberg, Heidelberg, Germany), respectively. The anti-Drg1 antiserum was described previously (Pertschy et al., 2007). Antisera directed against Rpl25 and Rpp0 were a gift from J.P. Ballesta (Universidad Autónoma de Madrid, Madrid, Spain). Anti-GFP antibody was obtained from Roche and anti-GST antibody was purchased from Sigma-Aldrich. Antisera from goat against E. coli ribosomal S3 and L5 proteins were provided by O. Vesper (Max F. Perutz Laboratories, Vienna, Austria). Secondary antibodies goat anti–rabbit HRP (Roche) and donkey anti–goat HRP (Santa Cruz Biotechnology, Inc.) were used at a 1:15,000 dilution. The peroxidase activity was visualized with the ECL Western chemiluminescence kit (GE Healthcare).
SPR
SPR measurements were performed using a BiacoreX system. GST-Rlp24C and GST (in the reference flow cell) were immobilized on a CM5 sensor chip (GE Healthcare) using amine-coupling chemistry. The sensor surface was activated by injection of a 1:1 mixture of 0.1 M N-hydroxysuccinimide and 0.4 M 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (Sigma-Aldrich). GST-Rlp24C and GST were diluted to 20 µg/ml in 10 mM sodium acetate, pH 5.0, and injected over the activated surface. The remaining nonreacted ester groups were blocked with 1 M ethanolamine (Sigma-Aldrich). The amounts of immobilized GST-Rlp24C and GST were ∼1,200 and 1,400 RU, respectively. All measurements were performed at 22°C using binding buffer containing 1 mM ATP or AMP-PNP at a flow rate of 40 µl/min (2-min contact time). For Scatchard blot analysis the flow rate was reduced to 20 µl/min to achieve a contact time of 4 min. The response observed in the GST-immobilized flow cell served as an in-line control and was subtracted for each sensorgram. Surface regeneration was accomplished by injection of 1 M NaCl after each binding cycle to remove bound analyte. The sensorgrams and kinetic parameters were analyzed by the BIAevaluation software (version 3.2) using the two-state (conformational change) model.
ATPase assay
ATPase activity was determined using the Malachite green phosphate assay (Lanzetta et al., 1979) through measurement of the formation of free inorganic phosphate from BioAssaySystems according to the manual. For determination of the ATPase activity of Drg1, 10 µg of purified Drg1 or its variants were adjusted to 100 µl with ice cold binding buffer. Some samples were prepared with an additional 0.0005–10 µg of purified recombinant 6His-tagged proteins after buffer exchange using Zeba spin desalting columns 7K to remove imidazole. ATP was added to a final concentration of 1 mM. Samples were incubated 30 min at 30°C in a water bath and after addition of the Malachite Green reagent absorption was measured in a plate reader (GENios Pro; Tecan). All samples were measured in triplicates for at least two independent experiments. The steady-state constants Vmax, the EC50, and Hill slope were obtained by fitting semi-logarithmic progress curves (obtained from plotting ATPase activity against the log of Rlp24 concentration) using GraphPad Prism (GraphPad Software). Error estimates associated with the fitted values were determined by the nonlinear fitting procedure from a mean of at least two biological replicates with at least six measurements each.
In vitro release assay
Pre-60S particles stalled at an early cytoplasmic maturation step were purified from 12 liters of late log phase cultures of the Arx1-TAP drg1-18 strain after incubation at 37°C for 1 h using the TAP protocol (see Tandem affinity purification section). After the particles were bound to the calmodulin beads and washing was performed, the buffer was exchanged for binding buffer containing 2 mM CaCl2. Beads were split into five to six aliquots. 10 µg Drg1 preincubated with 1 mM ATP were added per aliquot in a reaction volume of 150 µl to the binding buffer. 8 µg 6HisNup116 fragment (N172; codon 1 to 172) purified from E. coli after expression from pET28a and pretreated with RNAsin (Promega) were added to some samples. After incubation for 45 min at room temperature, the supernatants containing released proteins were collected after centrifugation. The remaining pre-60S particles were eluted from the calmodulin beads with 150 µl of elution buffer containing 20 mM Hepes-NaOH, pH 7.5, 50 mM NaCl, 5 mM EGTA, 0.075% Nonidet P-40, and 1 mM DTT. Supernatants and eluates were analyzed after TCA precipitation by SDS-PAGE and immunoblotting using polyclonal antibodies.
Online supplemental material
Fig. S1 shows accumulation of Mrt4 and decrease of Yvh1 on pre-60S particles upon inactivation of Drg1. Fig. S2 shows the distribution of hexameric and monomeric forms in preparations of Drg1 and Drg1EQ1. Fig. S3 shows the influence of nucleotide on the interaction between Drg1 and Rlp24, the kinetic analysis of binding, and the two-state reaction model applied for mathematical description of the interaction. Table S1 lists the yeast and bacterial strains and Table S2 lists the plasmids used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201205021/DC1.
Supplementary Material
Acknowledgments
This work is dedicated to the memory of Dr. Gregor Högenauer, who died on 2 May 2012. His passion for science will always be exemplary to us.
We thank Daniel Luschnig for construction of drg1-ts mutants. We thank Micheline Fromont-Racine, Bernard L. Trumpower, Juan P. Ballesta, Arlen W. Johnson, Ed Hurt, Sabine Rospert, and Oliver Vesper for supplying reagents. We thank Arlen W. Johnson for communicating results before publication and Helmut Schwab for the use of the Biacore facility. In addition, we thank Mauro Acchione for helpful discussion on SPR analysis and Tamsyn Stanborough for critically reading the manuscript.
This work was supported by the Graz advanced school of science grant and Austrian Science Foundation FWF P21991 to H. Bergler and FWF T404-B12 to B. Pertschy.
Footnotes
Abbreviations used in this paper:
- NPC
- nuclear pore complex
- RU
- response unit
- SDC
- synthetic dextrose
- SPR
- surface plasmon resonance
- TAP
- tandem affinity purification
References
- Alber F., Dokudovskaya S., Veenhoff L.M., Zhang W., Kipper J., Devos D., Suprapto A., Karni-Schmidt O., Williams R., Chait B.T., et al. 2007. The molecular architecture of the nuclear pore complex. Nature. 450:695–701 10.1038/nature06405 [DOI] [PubMed] [Google Scholar]
- Allen N.P., Huang L., Burlingame A., Rexach M. 2001. Proteomic analysis of nucleoporin interacting proteins. J. Biol. Chem. 276:29268–29274 10.1074/jbc.M102629200 [DOI] [PubMed] [Google Scholar]
- Bradatsch B., Katahira J., Kowalinski E., Bange G., Yao W., Sekimoto T., Baumgärtel V., Boese G., Bassler J., Wild K., et al. 2007. Arx1 functions as an unorthodox nuclear export receptor for the 60S preribosomal subunit. Mol. Cell. 27:767–779 10.1016/j.molcel.2007.06.034 [DOI] [PubMed] [Google Scholar]
- Briggs L.C., Baldwin G.S., Miyata N., Kondo H., Zhang X., Freemont P.S. 2008. Analysis of nucleotide binding to P97 reveals the properties of a tandem AAA hexameric ATPase. J. Biol. Chem. 283:13745–13752 10.1074/jbc.M709632200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLaBarre B., Brunger A.T. 2005. Nucleotide dependent motion and mechanism of action of p97/VCP. J. Mol. Biol. 347:437–452 10.1016/j.jmb.2005.01.060 [DOI] [PubMed] [Google Scholar]
- Doyle S.M., Shorter J., Zolkiewski M., Hoskins J.R., Lindquist S., Wickner S. 2007. Asymmetric deceleration of ClpB or Hsp104 ATPase activity unleashes protein-remodeling activity. Nat. Struct. Mol. Biol. 14:114–122 10.1038/nsmb1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreveny I., Pye V.E., Beuron F., Briggs L.C., Isaacson R.L., Matthews S.J., McKeown C., Yuan X., Zhang X., Freemont P.S. 2004. p97 and close encounters of every kind: a brief review. Biochem. Soc. Trans. 32:715–720 10.1042/BST0320715 [DOI] [PubMed] [Google Scholar]
- Hanson P.I., Whiteheart S.W. 2005. AAA+ proteins: have engine, will work. Nat. Rev. Mol. Cell Biol. 6:519–529 10.1038/nrm1684 [DOI] [PubMed] [Google Scholar]
- Henras A.K., Soudet J., Gérus M., Lebaron S., Caizergues-Ferrer M., Mougin A., Henry Y. 2008. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell. Mol. Life Sci. 65:2334–2359 10.1007/s00018-008-8027-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung N.J., Lo K.Y., Patel S.S., Helmke K., Johnson A.W. 2008. Arx1 is a nuclear export receptor for the 60S ribosomal subunit in yeast. Mol. Biol. Cell. 19:735–744 10.1091/mbc.E07-09-0968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P., Halladay J., Craig E.A. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 144:1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressler D., Hurt E., Bassler J. 2010. Driving ribosome assembly. Biochim. Biophys. Acta. 1803:673–683 10.1016/j.bbamcr.2009.10.009 [DOI] [PubMed] [Google Scholar]
- Kressler D., Hurt E., Bergler H., Bassler J. 2012. The power of AAA-ATPases on the road of pre-60S ribosome maturation—molecular machines that strip pre-ribosomal particles. Biochim. Biophys. Acta. 1823:92–100 10.1016/j.bbamcr.2011.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzetta P.A., Alvarez L.J., Reinach P.S., Candia O.A. 1979. An improved assay for nanomole amounts of inorganic phosphate. Anal. Biochem. 100:95–97 10.1016/0003-2697(79)90115-5 [DOI] [PubMed] [Google Scholar]
- Lo K.-Y., Li Z., Bussiere C., Bresson S., Marcotte E.M., Johnson A.W. 2010. Defining the pathway of cytoplasmic maturation of the 60S ribosomal subunit. Mol. Cell. 39:196–208 10.1016/j.molcel.2010.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A., III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961 [DOI] [PubMed] [Google Scholar]
- Lupas A.N., Martin J. 2002. AAA proteins. Curr. Opin. Struct. Biol. 12:746–753 10.1016/S0959-440X(02)00388-3 [DOI] [PubMed] [Google Scholar]
- Mogk A., Schlieker C., Strub C., Rist W., Weibezahn J., Bukau B. 2003. Roles of individual domains and conserved motifs of the AAA+ chaperone ClpB in oligomerization, ATP hydrolysis, and chaperone activity. J. Biol. Chem. 278:17615–17624 10.1074/jbc.M209686200 [DOI] [PubMed] [Google Scholar]
- Panse V.G., Johnson A.W. 2010. Maturation of eukaryotic ribosomes: acquisition of functionality. Trends Biochem. Sci. 35:260–266 10.1016/j.tibs.2010.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertschy B., Saveanu C., Zisser G., Lebreton A., Tengg M., Jacquier A., Liebminger E., Nobis B., Kappel L., van der Klei I., et al. 2007. Cytoplasmic recycling of 60S preribosomal factors depends on the AAA protein Drg1. Mol. Cell. Biol. 27:6581–6592 10.1128/MCB.00668-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig O., Caspary F., Rigaut G., Rutz B., Bouveret E., Bragado-Nilsson E., Wilm M., Séraphin B. 2001. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 24:218–229 10.1006/meth.2001.1183 [DOI] [PubMed] [Google Scholar]
- Pye V.E., Dreveny I., Briggs L.C., Sands C., Beuron F., Zhang X., Freemont P.S. 2006. Going through the motions: the ATPase cycle of p97. J. Struct. Biol. 156:12–28 10.1016/j.jsb.2006.03.003 [DOI] [PubMed] [Google Scholar]
- Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Séraphin B. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030–1032 10.1038/13732 [DOI] [PubMed] [Google Scholar]
- Saveanu C., Namane A., Gleizes P.-E., Lebreton A., Rousselle J.-C., Noaillac-Depeyre J., Gas N., Jacquier A., Fromont-Racine M. 2003. Sequential protein association with nascent 60S ribosomal particles. Mol. Cell. Biol. 23:4449–4460 10.1128/MCB.23.13.4449-4460.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaupp A., Marcinowski M., Grimminger V., Bösl B., Walter S. 2007. Processing of proteins by the molecular chaperone Hsp104. J. Mol. Biol. 370:674–686 10.1016/j.jmb.2007.04.070 [DOI] [PubMed] [Google Scholar]
- Schmitt J., Hess H., Stunnenberg H.G. 1993. Affinity purification of histidine-tagged proteins. Mol. Biol. Rep. 18:223–230 10.1007/BF01674434 [DOI] [PubMed] [Google Scholar]
- Thorsness P.E., White K.H., Ong W.C. 1993. AFG2, an essential gene in yeast, encodes a new member of the Sec18p, Pas1p, Cdc48p, TBP-1 family of putative ATPases. Yeast. 9:1267–1271 10.1002/yea.320091114 [DOI] [PubMed] [Google Scholar]
- Wang Q., Song C., Li C.-C.H. 2003. Hexamerization of p97-VCP is promoted by ATP binding to the D1 domain and required for ATPase and biological activities. Biochem. Biophys. Res. Commun. 300:253–260 10.1016/S0006-291X(02)02840-1 [DOI] [PubMed] [Google Scholar]
- Wang Q., Song C., Li C.-C.H. 2004. Molecular perspectives on p97-VCP: progress in understanding its structure and diverse biological functions. J. Struct. Biol. 146:44–57 10.1016/j.jsb.2003.11.014 [DOI] [PubMed] [Google Scholar]
- Watanabe Y.H., Motohashi K., Yoshida M. 2002. Roles of the two ATP binding sites of ClpB from Thermus thermophilus. J. Biol. Chem. 277:5804–5809 10.1074/jbc.M109349200 [DOI] [PubMed] [Google Scholar]
- White S.R., Lauring B. 2007. AAA+ ATPases: achieving diversity of function with conserved machinery. Traffic. 8:1657–1667 10.1111/j.1600-0854.2007.00642.x [DOI] [PubMed] [Google Scholar]
- Yeung H.O., Kloppsteck P., Niwa H., Isaacson R.L., Matthews S., Zhang X., Freemont P.S. 2008. Insights into adaptor binding to the AAA protein p97. Biochem. Soc. Trans. 36:62–67 10.1042/BST0360062 [DOI] [PubMed] [Google Scholar]
- Zakalskiy A., Högenauer G., Ishikawa T., Wehrschütz-Sigl E., Wendler F., Teis D., Zisser G., Steven A.C., Bergler H. 2002. Structural and enzymatic properties of the AAA protein Drg1p from Saccharomyces cerevisiae. Decoupling of intracellular function from ATPase activity and hexamerization. J. Biol. Chem. 277:26788–26795 10.1074/jbc.M201515200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.