Abstract
Single-nucleotide polymorphisms (SNPs, >2000) were discovered by using RNA-seq and allele-specific sequencing approaches in pigeonpea (Cajanus cajan). For making the SNP genotyping cost-effective, successful competitive allele-specific polymerase chain reaction (KASPar) assays were developed for 1616 SNPs and referred to as PKAMs (pigeonpea KASPar assay markers). Screening of PKAMs on 24 genotypes [23 from cultivated species and 1 wild species (Cajanus scarabaeoides)] defined a set of 1154 polymorphic markers (77.4%) with a polymorphism information content (PIC) value from 0.04 to 0.38. One thousand and ninety-four PKAMs showed polymorphisms between parental lines of the reference mapping population (C. cajan ICP 28 × C. scarabaeoides ICPW 94). By using high-quality marker genotyping data on 167 F2 lines from the population, a comprehensive genetic map comprising 875 PKAMs with an average inter-marker distance of 1.11 cM was developed. Previously mapped 35 simple sequence repeat markers were integrated into the PKAM map and an integrated genetic map of 996.21 cM was constructed. Mapped PKAMs showed a higher degree of synteny with the genome of Glycine max followed by Medicago truncatula and Lotus japonicus and least with Vigna unguiculata. These PKAMs will be useful for genetics research and breeding applications in pigeonpea and for utilizing genome information from other legume species.
Keywords: pigeonpea, SNP, linkage map, comparative genomics, molecular breeding
1. Introduction
Recent advances in genomics have provided various opportunities to a number of crop species of significant agronomical importance for enhancing crop productivity. One of the foremost applications of genomics in breeding is the prediction of a phenotype from the genotype and the process is called genomics-assisted breeding (GAB).1 Several success stories of GAB are available in many temperate cereal crops2 and some legume species3 also. However, legume crops like pigeonpea (Cajanus cajan L. Millspaugh), which is grown in ∼5 million hectares in the developing countries of Asia and Africa have remained untouched by GAB. This may be attributed mainly to two reasons: (i) paucity of genomic tools and (ii) narrow genetic diversity in the gene pool of the pigeonpea. Following 5 years of intensive research efforts and investment in genomics, however, a significant amount of genomic resources such as a large collection of expressed sequence tags (ESTs) or transcript reads,4–7 large-scale molecular markers including simple sequence repeats (SSRs),8,9 single-feature polymorphisms,10 single-nucleotide polymorphisms (SNPs)5–7 and diversity array technology (DArT)11,12 markers have been developed. Very recently, the draft genome sequence has also become available.13
Identification of molecular markers for applying in a breeding programme requires the development of a genetic map and quantitative trait locus (QTL) analysis. In the case of pigeonpea, although a few genetic maps have been developed,9,12,14,15 the marker density in those maps is very low. A major challenge, therefore, before the pigeonpea community is the development of a saturated genetic map. In this context, SNP markers have attracted significant attention as these markers represent the most abundant class of polymorphisms in genomes and are amenable for high-throughput genotyping.3,16 In general, the implementation of SNPs for genetic studies involves a three-step process: (i) SNP discovery after aligning the ESTs, sequence reads generated by Sanger or next generation sequencing (NGS) technologies for different genotypes in a given species; (ii) validation of SNPs to distinguish DNA polymorphisms of actual allelic variants from those of other biological phenomenon, such as gene duplication events; i.e. paralogous or homeologous genes, as well as those of technical errors, primarily sequencing errors, in case SNPs have been identified using in silico approaches; (iii) SNP genotyping of germplasm collection or genetic/breeding populations. A wide range of molecular techniques suitable for pursuing the mentioned three steps have been available,17 each characterized by a distinct cost scale and throughput capacity, and utilizing different technology platforms. NGS technologies are bringing us the capacity to identify, at affordable cost, large numbers of SNPs for even non-model species. Similarly, the availability of a number of SNP genotyping platforms, in a high-throughput manner, is making SNP genotyping cost-effective.
Depending on the sample size and number of SNPs to be analysed, medium- to high-throughput assay platforms such as BeadXpress and GoldenGate assays from Illumina Inc. with a varying set of multiplexes (96, 384, 768 or 1536 SNPs per assay) are available. Such platforms have been developed and used in several crop species such as barley,18 wheat,19 maize20 and oil seed rape,21 and legumes such as soybean,22 cowpea23 and pea.24 Furthermore, in some crops like maize, Infinium assays with the capacity of genotyping ∼50 000 SNPs have become available.25 Such platforms, however, are cost-effective only when a minimum of 96, 384, 762, 1536 or thousands of SNPs are used with a large number of samples. In cases like marker-assisted selection where only a few markers are required for genotyping large-scale segregating populations, Illumina-based genotyping assays do not seem to be cost-effective. In such cases, the competitive allele-specific polymerase chain reaction (PCR) (KASPar) assay from KBiosciences (www.kbioscience.co.uk) seems to be an effective marker assay. The KASPar genotyping assay is a competitive allele-specific PCR-based fluorescent SNP genotyping system. It is a user-friendly system that provides flexibility in the numbers of SNPs and genotypes to be used for assays. Details about this technology are available at http://www.kbioscience.co.uk/reagents/KASP.html. Because of the importance of KASPar assays in genotyping more samples with a few SNPs, they have been developed in wheat,26 common bean27 and chickpea.28
With an objective to develop a flexible and cost-effective SNP genotyping platform in pigeonpea, this study reports the compilation of informative SNP data sets, development and characterization of KASPar assays, and development of an SNP-based genetic linkage map of pigeonpea and its use for comparative genomics with closely related legume species like soybean (Glycine max), cowpea (Vigna ungiculata), Medicago truncatula and Lotus japonicus.
2. Materials and methods
2.1. Mapping population and DNA isolation
Two Cajanus spp., one from cultivated pigeonpea (C. cajan) ICP 28 and the another accession from the wild relative of pigeonpea (C. scarabaeoides) ICPW 94, were used as crossing parents for the development of an F2 population of 167 individuals. Accordingly, a single plant of ICP 28 accession was used as a female parent and crossed with the pollen parent ICPW 94 plant and F1s were produced. All F1s were selfed under nylon bags and grown at Patancheru in southern India (17°N). A single F1 plant having the highest number of F2 seeds was selected to develop a mapping population of 167 F2 individuals. To characterize developed SNP markers, a set of 24 genotypes was utilized for screening the polymorphism (Supplementary Table S1). These genotypes represent parents of 14 mapping populations which are segregating for various agronomically important traits. Total DNA from the parents of different mapping populations and F2 lines derived from ICP 28 × ICPW 94 were isolated from two to three young leaves following the standard DNA isolation protocol as mentioned in the study of Cuc et al.29 The DNA quantity for each sample was assessed on 0.8% agarose gel.
2.2. Identification of SNPs
The complete Illumina GA IIx data set was comprised 128.9 million, 36 bp short single-end reads from 12 genotypes (ICPL 87119, ICPL 87091, BSMR 736, TAT 10, ICP 7035, TTB 7, ICPL 332, ICPL 20096, ICPB 2049, ICPL 99050, ICP 28 and ICPW 94; Table 1). Identification of SNPs from assembled data was carried out using the Alpheus software system.30 SNPs were identified on the basis of alignment of sequence reads generated from each of the counter genotypes against the reference assembly, i.e. pigeonpea transcriptome assembly which was developed by assembling four different sequence data sets7 and allowing not more than two mismatches. Based on the alignment results, variants at a particular nucleotide position were identified. Significant variants were selected based on two criteria: (i) the allele frequency between two genotypes >0.8 (the number of a specific allele divided by the number of all alleles for the specific SNP between two genotypes should be higher than 80%) and (ii) the number of tags aligned to the reference >5.
Table 1.
Summary on the identification of SNPs in Cajanus spp. accessions
| Genotypes | Illumina GA IIx reads | Total number of SNPs | Number of unique genes with identified SNPs |
|---|---|---|---|
| ICPL 87119 | 7 182 619 | 7488 | 3116 |
| ICPL 87091 | 8 977 567 | ||
| BSMR 736 | 11 065 219 | 2115 | 1454 |
| TAT 10 | 7 932 691 | ||
| ICP 7035 | 1 3 223 516 | 1256 | 983 |
| TTB 7 | 4 122 216 | ||
| ICPL 332 | 16 361 115 | 2452 | 1819 |
| ICPL 20096 | 9 507 797 | ||
| ICPB 2049 | 11 494 670 | 1892 | 1435 |
| ICPL 99050 | 13 498 156 | ||
| ICP 28 | 9 721 562 | 1910 | 1352 |
| ICPW 94 | 15 828 791 | ||
| Total | 128 915 919 | 17 113 | 10 159 |
| TOGs | |||
| ICP 28 | 752 | 670 | |
| ICPW 94a | |||
aICPW 94 is an accession of C. scarabaeoides, a wild relative of pigeonpea (C. cajan).
2.3. KASPar genotyping assay
For each putative SNP, the criteria used for the selection of high-quality SNPs for KBioscience competitive allele-specific PCR (KASPar) assay31 included: (i) an SNP flanked by at least 50 bases on either side; (ii) frequency difference between the two genotypes ≥5; (iii) read depth ≥5. For each SNP, two allele-specific forward primers and one common reverse primer were designed. By using these primers, KASPar assays were performed in a final reaction volume of 5 μL containing 1× KASP reaction mix (KBioscience, Hoddesdon, UK), 0.07 μL of assay mix (12 μM each allele-specific forward primer and 30 μM reverse primer) and 10–20 ng of genomic DNA. The Gene Pro Thermal cycler (Bioer Technology, Hangzhou, China) was used for the following cycling conditions: 15 min at 94°C; 10 touchdown cycles of 20 s at 94°C and 60 s at 65–57°C (the annealing temperature for each cycle being reduced by 0.8°C per cycle); and 26–35 cycles of 20 s at 94°C and 60 s at 57°C. Fluorescence detection of the reactions was performed using an Omega Fluorostar scanner (BMG LABTECH GmbH, Offenburg, Germany) and the data were analysed using the KlusterCaller 1.1 software (KBioscience). Details on the KASPar principle, amplification of targeted region, fluorescence detection and allele calling are available at http://www.kbioscience.co.uk/reagents/KASP_manual.pdf. The polymorphism information content (PIC) values for developed markers across 24 genotypes (Supplementary Table S1) were calculated by using the PowerMarker software (http://statgen.ncsu.edu/powermarker/).
2.4. Linkage mapping
Genotyping data generated using KASPar assays on 167 F2 individuals of an ICP 28 × ICPW 94 population were analysed for linkage analysis using JoinMap version 4.0 with ‘regression mapping algorithm’.32 Prior to linkage analysis, marker segregation data were subjected to the goodness-of-fit test (χ2) to assess deviations from the expected Mendelian segregation ratio of 1:2:1 at a 5% level of significance. Map calculations were performed at a logarithm (base 10) of odds (LOD) value of 5.0, recombination frequency ≤0.40 and χ2 jump threshold for removal of loci = 5. A Kosambi map function was used to convert the recombination frequency into cM distances33 and the third round was set to allow the mapping of an optimum number of loci in the genetic map. Mean χ2 contributions or average contributions to the goodness of fit of each locus were also checked to determine the best fitting position for markers in genetic maps. The markers showing negative map distances or a large jump in mean χ2 values were subsequently discarded. The graphical maps of the linkage groups (LGs) were constructed by using MapChart version 2.2.34
2.5. Comparative genome analysis
DNA sequences for corresponding mapped SNP markers were used for comparative analysis with the genetic map of cowpea23 and the genome assemblies of soybean (ftp://ftp.jgi-psf.org/pub/JGI_data/phytozome/v7.0/Gmax/assembly), M. truncatula (http://www.medicagohapmap.org/downloads.php) and L. japonicus (ftp://ftp.kazusa.or.jp/pub/lotus/lotus_r2.5/pseudomolecule). Vmatch35 was used to identify reciprocal best matches between the pigeonpea sequences and other legume sequences. Hits matching a minimum of 70% sequence identity were retained for the comparative study. Identification of homologous blocks was performed using i-ADHoRe v2.1.36 For the purpose of developing Circos images, cM distances on the pigeonpea LGs were scaled up by a factor of 250 000 to match similar bp lengths of the chromosomes of other legumes' genomes. Synteny blocks were visualized by using Circos26.37 Scales along the outer edges of the pigeonpea and cowpea LGs show actual cM distances, whereas those along the outer edges of the soybean, Medicago and Lotus chromosomes are in Mb.
3. Results
3.1. Development of a mapping population
Although a set of 72 F2 plants were available from an earlier cross (C. cajan ICP 28 × C. scarabaeoides ICPW 94) that were used to develop an SSR-based genetic map,9 a new cross with the same accessions was made to develop a bigger population (167 F2 lines) for developing a high-resolution genetic map.
3.2. Assembly of informative SNPs
With a goal of increasing the cost-effective and high-throughput genetic marker repertoire in pigeonpea, the following two different sequence resources were surveyed for the presence of SNPs: (i) Illumina GA IIx transcript sequence data and (ii) tentative orthologous genes (TOGs) of closely related legumes.
3.2.1. SNPs from Illumina GA IIx transcript sequence data
For the identification of SNPs, 128.9 million Illumina reads of 12 different genotypes (ICPL 87119, ICPL 87091, BSMR 736, TAT 10, ICP 7035, TTB 7, ICPL 332, ICPL 20096, ICPB 2049, ICPL 99050, ICP 28 and ICPW 94) were aligned against the transcriptome assembly (CcTA v2).7 The CcTA v2 comprised 21 434 transcriptome assembly contigs (TACs) developed from transcriptome data sets from 21 pigeonpea genotypes (128.9 million Illumina GA IIx reads from 12 genotypes, 2.19 million FLX/454 reads from 3 genotypes and 18 353 Sanger ESTs from 6 genotypes).7 Variants were identified using the ‘Alpheus’ program30 by comparing the sequence tags from two genotypes of a given mapping population combination. In total, a set of 17 113 SNPs in 10 159 unique sequences were identified between six crosses (Table 1). The number of SNPs in an individual cross ranged from 1256 (TTB 7 × ICPL 7035) to 7488 (ICPL 87119 × ICPL 87091) (Table 1). However, only six SNPs were found common across three populations (ICPL 20096 × ICPL 332, TTB 7 × ICPL 7035 and BSMR 736 × TAT 10). For the ICP 28 and ICPW 94 combination, a total of 1910 SNPs were identified in 1352 TACs. By considering only one SNP per TAC (gene) and primer designing criteria of KASPar assays, 1167 SNPs were further selected.
3.2.2. SNPs from TOGs
After sequencing ICP 28 and ICPW 94 accessions with 670 TOGs, a set of 752 SNPs were identified and used in designing the GoldenGate assay in a separate study.38 From these 752 SNPs, a total of 660 SNPs satisfied the required primer designing criteria for KASPar assays.
After combining the above-mentioned data sets, a non-redundant set of 1827 SNPs (1167 from Illumina GA IIx transcript data, referred as GAIIx-SNPs and 660 TOG-SNPs) was complied.
3.3. Development and validation of KASPar assays
A total of 1827 non-redundant SNPs were used for the development of KASPar assays (Supplementary Table S2). However, successful assays could be developed for 1616 SNPs (88.4%) with scorable allele calls. These marker assays have been referred to as PKAMs (pigeonpea KASPar assay markers). All 1616 PKAMs were screened on 24 pigeonpea genotypes, including 23 cultivated and one wild-type ICPW 94, representing parents of 14 mapping populations (Supplementary Table S1), further defined a subset of 1154 polymorphic markers (77.4%). Among these polymorphic PKAMs, 1043 were polymorphic exclusively in wild species. Data obtained from 24 genotypes were used to calculate the PIC value of each PKAM marker, and PIC values ranged from 0.04 to 0.38 with an average of 0.09 (Supplementary Table S2). In terms of the parental polymorphism of different mapping populations, polymorphism rates varied considerably, depending on the crossing parental lines under comparison, from a low of 14 polymorphic PKAMs (ICP 8863 × ICPL 20097) to a high of 1094 polymorphic PKAMs (ICP 28 × ICPW 94) (Supplementary Table S1).
3.4. SNP-based genetic map
With a goal of developing an SNP-based genetic map in pigeonpea, genotyping data were obtained on 167 progenies for all the 1094 polymorphic PKAMs. However, high-quality data obtained for 1008 PKAMs were considered for further analysis. Genotyping data obtained for all 1008 PKAMs were tested for the Mendelian/non-Mendelian segregation pattern. As a result, the 33 PKAMs showing non-Mendelian inheritance were removed from further analysis. Subsequently, genotyping data for 975 PKAMs (470 PKAMs based on GAIIx-SNPs and 505 PKAMs from TOG-SNPs) were used for linkage analysis, with JoinMap4.0 used to construct the genetic linkage map.32
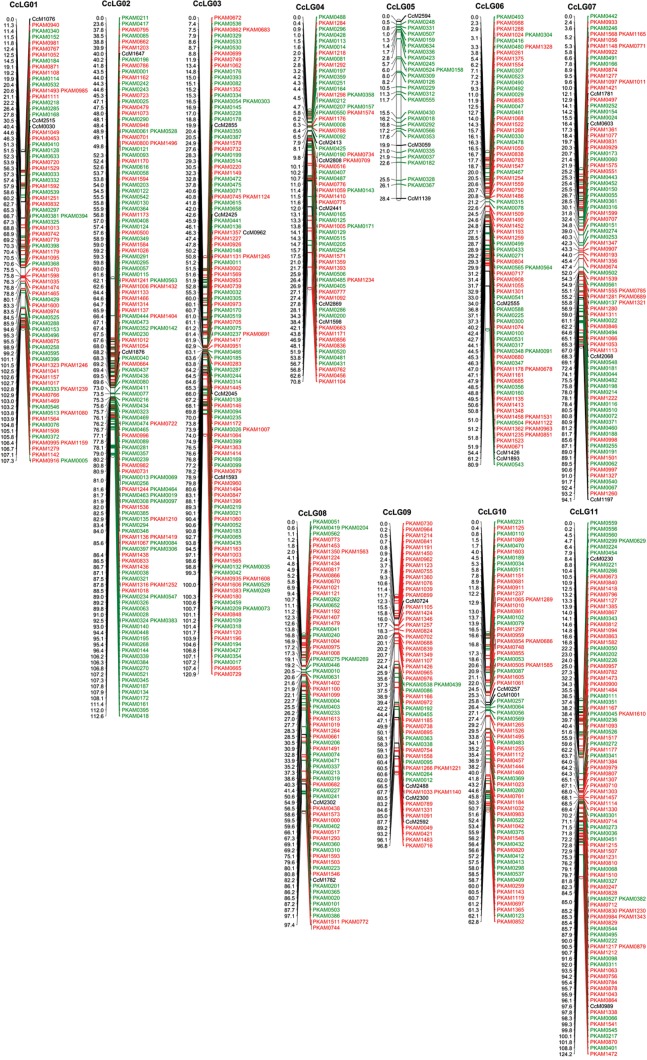
In summary, 11 LGs were generated using an LOD threshold value of 5.0, which is in agreement with the haploid chromosome number (11) in pigeonpea. A total of 55 PKAMs that failed to be assigned to these 11 LGs were not incorporated in further analyses. While assigning the order of PKAMs in different individual LGs, map positions could not be assigned for 45 PKAMs. As a result, the developed genetic map contains 875 (444 GAIIx-SNP and 431 TOG-SNP) loci on 11 LGs (Supplementary Fig. S1), ranging from 25.7 cM (CcLG05) to 124.25 cM (CcLG11) in length, and a total map length of 967.03 cM (Supplementary Table S3). The number of markers per LG ranges from 25 (CcLG05) to 134 (CcLG02), with an average of 79.54 (Table 2 and Supplementary Fig. S1). The highest marker density with an inter-marker distance of 0.84 cM was observed on CcLG02, while CcLG09 had the lowest marker density with average marker spacing 1.79 cM. Most of the spaces between two markers were smaller than 20 cM on the genetic linkage map. However, there were only two spaces where the distances between markers were large, i.e. 23.56 cM between PKAM0211 and PKAM0417 on CcLG02 and 28.85 cM between PKAM0671 and PKAM0543 on CcLG06.
Table 2.
Features of SNP-based and integrated pigeonpea genetic maps
| Pigeonpea LG | SNP-based map |
Integrated map (SNPs + SSRs) |
||||
|---|---|---|---|---|---|---|
| Size (cM) | Number of loci | Average inter- loci distance | Size (cM) | Number of loci | Average inter-loci distance | |
| CcLG01 | 95.83 | 79 | 1.21 | 107.29 | 82 | 1.31 |
| CcLG02 | 112.55 | 134 | 0.84 | 112.55 | 136 | 0.83 |
| CcLG03 | 107.85 | 118 | 0.91 | 120.88 | 123 | 0.98 |
| CcLG04 | 70.29 | 65 | 1.08 | 70.77 | 70 | 1.01 |
| CcLG05 | 25.7 | 25 | 1.03 | 28.41 | 28 | 1.01 |
| CcLG06 | 81.28 | 82 | 0.99 | 80.93 | 85 | 0.95 |
| CcLG07 | 93.22 | 84 | 1.11 | 94.14 | 88 | 1.07 |
| CcLG08 | 96.48 | 71 | 1.36 | 97.43 | 73 | 1.33 |
| CcLG09 | 96.75 | 54 | 1.79 | 96.81 | 58 | 1.67 |
| CcLG10 | 62.83 | 69 | 0.91 | 62.83 | 71 | 0.88 |
| CcLG11 | 124.25 | 94 | 1.32 | 124.18 | 96 | 1.29 |
| Total | 967.03 | 875 | 1.11 | 996.21 | 910 | 1.09 |
3.5. Integrated genetic map
With an objective to provide anchor points to integrate an SSR-based genetic linkage map with the SNP-based genetic linkage map, the newly developed mapping population (ICP 28 × ICPW 94) was also genotyped with targeted SSRs (2–5) from each LG of the SSR-based genetic linkage map previously developed by Bohra et al.9 In this context, genotyping data generated for a total of 35 SSRs. All the 35 SSRs were mapped onto 11 LGs (CcLG01–CcLG11). The number of SSR markers per LG varied from two (CcLG02, CcLG08, CcLG10 and CcLG11) to five (CcLG03 and CcLG04) (Table 2). After integration of the SSR markers, the total distance of the integrated genetic linkage map increased by 29.18 cM (Fig. 1). Subsequently, the integrated genetic map was compared with the SSR-based genetic linkage map and the marker order was consistent between similar LGs. However, in the case of CcLG03, out of five common markers, two markers, namely CcM1593 and CcM2045, had different positions. All pairwise comparisons between the integrated LGs and SSR-based LGs have been shown in Supplementary Fig. S2. As expected, the marker order and distances were well conserved between the integrated genetic map and the SNP-based genetic map.
Figure 1.
An integrated genetic map of pigeonpea. This genetic map is developed on the F2 mapping population derived from ICP 28 × ICPW 94. The map comprises 910 loci (875 PKAMs and 35 SSRs) in which 444 PKAMs were developed based on GAIIx-SNPs, shown in red; 431 PKAMs were developed based on TOGs-SNPs, shown in green, and 35 SSRs previously mapped by Bohra et al.9 shown in black.
3.6. Genome relationships of pigeonpea with closely related legume species
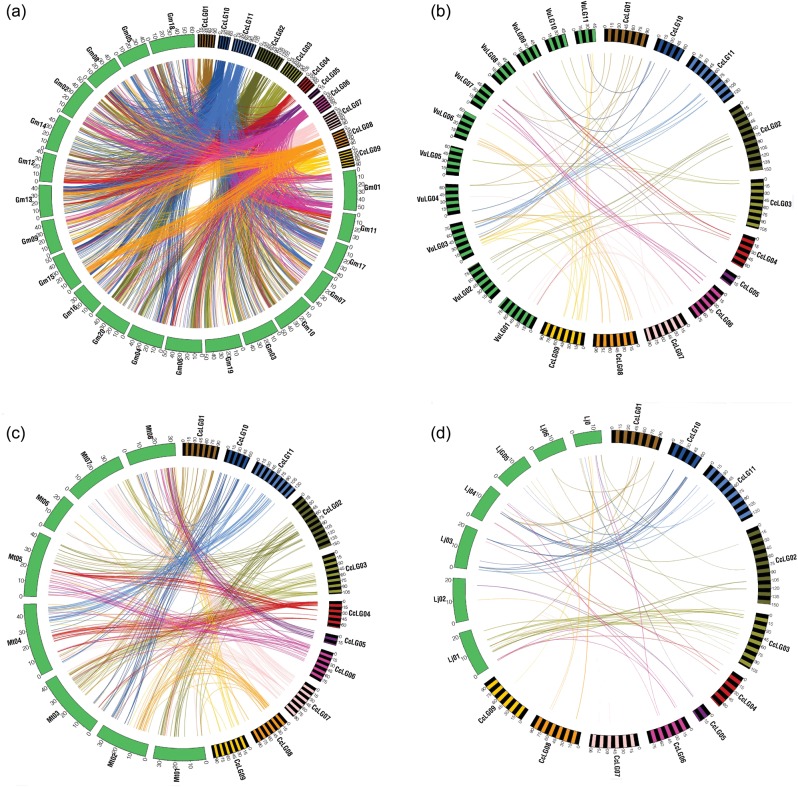
For comparative genome analysis and to identify conserved synteny between genomes of pigeonpea and other related legume species, we combined both the genetic map position information for pigeonpea loci and genome sequence information of closely related species of different clades. A set of 875 mapped loci for which both genetic map positions and sequence information were available (Supplementary Table S3) was compared with the genome assemblies of soybean (Glyma1), Medicago (Mt 3.5) and Lotus (Lj 2.5 pseudomolecules) and with the transcript-specific genetic map of cowpea.23 In the comparison of pigeonpea with soybean, the highest percentage of the sequence similarity was identified. As expected, each pigeonpea LG shows extensive synteny with two or more chromosomes in soybean (Fig. 2a), probably due to the independent duplication event in the soybean genome.39 A total of 687 pigeonpea unique loci matched with 2480 soybean sequence stretches distributed on different chromosomes of soybean (Glyma1 assembly; Table 3). Maximum similarity was identified between CcLG04 with Gm13 followed by CcLG02 with Gm10, CcLG03 with Gm06 and Gm04, CcLG03 with Gm06, CcLG06 with Gm01, CcLG10 with Gm15, CcLG07 with Gm18, CcLG08 with Gm15, CcLG11 with Gm17, CcLG01 with Gm18, CcLG09 with Gm08 and CcLG05 with Gm08.
Figure 2.
Syntenic relationships of individual LGs of pigeonpea with other legumes. Each line radiating from a pigeonpea LG represents a similarity match found in a block between pigeonpea and other legumes. (a) Pigeonpea LGs showing synteny with the genome assembly of soybean, (b) pigeonpea LGs showing synteny with the cowpea transcript map, (c) pigeonpea LGs showing synteny with the genome assembly of Medicago and (d) pigeonpea LGs showing synteny with the genome assembly of Lotus.
Table 3.
Detailed results on the comparison of mapped marker loci of pigeonpea with the soybean (G. max) genome
| LGs | Pigeonpea unique loci (no.) |
Glycine max chromosomes/Scaffolds |
|||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gm01 | Gm02 | Gm03 | Gm04 | Gm05 | Gm06 | Gm07 | Gm08 | Gm09 | Gm10 | Gm11 | Gm12 | Gm13 | Gm14 | Gm15 | Gm16 | Gm17 | Gm18 | Gm19 | Gm20 | Scaffold_1129 | Scaffold_41 | Scaffold_42 | Scaffold_317 | Scaffold_96 | Scaffold_23 | Scaffold_1337 | Scaffold_1655 | Scaffold_90 | Total | ||
| CcLG01 | 52 | 7 | 10 | 15 | 3 | 2 | 1 | 16 | 18 | 2 | 8 | 32 | 2 | 2 | 4 | 2 | 7 | 2 | 45 | — | 5 | — | — | — | — | — | — | — | — | — | 183 |
| CcLG02 | 101 | 4 | 42 | 6 | 3 | — | 10 | 3 | 3 | 4 | 83 | 1 | 6 | 14 | 41 | 5 | 2 | 11 | 2 | 7 | 59 | 2 | 2 | — | — | — | — | — | — | — | 310 |
| CcLG03 | 99 | 5 | 6 | 48 | 78 | 6 | 78 | 7 | 5 | 2 | 7 | 3 | 3 | 7 | 7 | 4 | 6 | 3 | 2 | 44 | 6 | — | 1 | — | — | — | — | — | — | — | 328 |
| CcLG04 | 52 | 2 | 2 | — | 2 | 2 | 12 | — | 3 | 1 | — | 53 | 54 | 106 | — | 5 | 1 | — | 5 | 0 | 1 | — | — | 1 | — | — | — | — | — | — | 250 |
| CcLG05 | 23 | 2 | — | — | 7 | 27 | 4 | 10 | 27 | 2 | 1 | — | — | — | 1 | 3 | — | — | 1 | 2 | — | — | — | — | — | — | — | — | — | — | 87 |
| CcLG06 | 66 | 69 | 34 | 3 | 5 | 2 | 5 | 8 | 4 | 20 | 6 | 35 | 3 | 3 | 4 | 3 | 6 | 8 | 4 | 6 | 5 | — | — | — | 2 | 2 | — | — | — | — | 237 |
| CcLG07 | 66 | 5 | 8 | 6 | 19 | 4 | 12 | 14 | 16 | 23 | 8 | 4 | 2 | 3 | — | 1 | 14 | 2 | 57 | 20 | 4 | — | — | — | — | — | 2 | — | — | — | 224 |
| CcLG08 | 54 | 5 | 5 | 2 | 1 | 4 | 1 | 12 | 10 | 49 | 6 | 3 | 5 | 18 | 3 | 51 | 16 | 5 | 1 | 2 | 5 | — | — | — | — | — | — | 1 | 1 | 206 | |
| CcLG09 | 41 | — | — | 2 | 1 | 26 | — | 5 | 31 | 10 | — | 2 | 2 | 11 | — | 2 | 4 | 1 | 3 | 12 | 2 | — | — | — | — | — | — | — | — | — | 114 |
| CcLG10 | 56 | 9 | 7 | 8 | 24 | 7 | 18 | 8 | 33 | 5 | 3 | 11 | 16 | 40 | 6 | 64 | 3 | 5 | 17 | 4 | 7 | — | — | — | — | — | — | — | — | 1 | 296 |
| CcLG11 | 77 | 6 | 5 | 8 | 5 | 30 | 1 | 26 | 21 | 10 | 2 | 6 | 1 | 20 | 8 | 6 | 21 | 47 | 13 | 5 | 4 | — | — | — | — | — | — | — | — | — | 245 |
| Total | 687 | 114 | 119 | 98 | 148 | 110 | 142 | 109 | 171 | 128 | 124 | 150 | 94 | 224 | 74 | 146 | 80 | 84 | 150 | 102 | 98 | 2 | 3 | 1 | 2 | 2 | 2 | 1 | 1 | 1 | 2480 |
The numbers shown in bold represent the highest matches between pigeonpea and soybean.
In the case of cowpea in which the genetic map was used for the comparison, least matches were observed between pigeonpea and cowpea genomes. Only 57 unique pigeonpea loci showed synteny with 62 loci on the cowpea map (Fig. 2b, Supplementary Table S4). In the case of pigeonpea and Medicago, 228 unique pigeonpea loci showed significant matches with 349 genomic regions on the Medicago chromosome (Fig. 2c, Supplementary Table S5). A total of 20 pigeonpea loci from CaLG02 showed similarity to MtChr01 genomic regions. A similar number of loci from CcLG03 showed similarity to MtChr03 genomic regions. CcLG04 showed almost equal similarity to MtChr04 (21) and MtChr05 (18). Similarly, loci from CcLG07 showed maximum matches to MtChr07 followed by CcLG08 with MtChr02, CcLG05 with MtChr08, CcLG06 with MtChr05 and CcLG11 with MtChr04. In the comparison of pigeonpea with Lotus, 216 pigeonpea unique loci matched with 303 different genomic regions on the Lotus chromosomes (Fig. 2d, Supplementary Table S6). In brief, each LG of pigeonpea showed considerable synteny with one or more chromosomes of Medicago and Lotus. The distribution of similarity hits across eight pigeonpea LGs was varied from 2 (CcLG05) to 9 (CcLG09) while comparing with cowpea, from 14 (CcLG09) to 55 (CcLG04) with Medicago and from 18 (CcLG01) to 74 (CcL03) with Lotus.
4. Discussion
The current availability of more than 3000 PCR-based markers in pigeonpea8,9 could not provide high or significant marker density in any of the populations to be adequate to allow a thorough scan of the genome for QTL discovery, association analysis, map-based cloning and anchoring of the genome sequence with the genetic map. This can be attributed to the low level of polymorphism in Cajanus spp. as well as the small number of lines in the mapping populations used for developing the genetic maps. To overcome the above-mentioned problems to some extent, a new mapping population with 167 F2 lines, compared with 72 lines used in the map of Bohra et al.9 , was developed and used for developing the genetic map. Furthermore, SNP markers were targeted for developing the cost-effective genotyping platform and developing the genetic map. As the SNP markers were derived from genes, the comparison of the SNP-based genetic map of pigeonpea with the genome sequence assemblies of soybean, Medicago and Lotus and the transcript map of cowpea provided the genome relationship of pigeonpea with the genomes of these legumes.
When compared with the other marker systems, SNP markers are unique with regard to their amenability to high-throughput and low-cost (per data point) genotyping platforms.17 In the case of pigeonpea, a total of 17 113 SNPs were discovered after comparing the transcript sequence reads from 12 parental lines of six different mapping populations with the transcriptome and/or genome sequence of pigeonpea.6,13 To prove the usefulness of these predicted polymorphisms for practical plant breeding applications, validation of these SNPs is required. For this purpose, a number of SNP genotyping platforms such as GoldenGate, Infinium and KASPar assays are available. However, in this study, due to its cost- effective and flexible nature, the KASPar assay was developed for 1616 SNPs. KASPar assays can be flexibly used to validate any number of SNP markers on a desired range of accessions, unlike many other SNP genotyping platforms such as GoldenGate or Infinium assay, to produce a sufficient number of polymorphic markers in a given population to obtain a better coverage. In the published literature, three reports are available on the development of KASPar assays in crop plants. For instance, in the case of wheat, KASPar assays were developed for 1114 SNPs and validated on 23 wheat varieties and also used for integrating SNP markers into the genetic map of wheat.26 In the case of common bean, KASpar assays have been developed for 94 SNPs and used for analysing genetic diversity in 70 accessions.27 Very recently, Hiremath et al.28 developed KASpar assays for 2005 SNPs in chickpea and used these for genetic diversity analysis and genetic mapping in chickpea and comparative mapping in legumes. A comparison of genotyping ∼100 chickpea lines with ∼500 SNPs on GoldenGate and KASPar assays shows the superiority of KASPar assays over GoldenGate assays in terms of cost as well as time used. In summary, all these three studies underline the importance of KASPar assays for SNP genotyping on a large scale for genetics and breeding applications. In the present study, though 1827 SNPs were attempted for conversion into KASPar assays, only 1616 (88.4%) markers could be successfully converted. The failure of the remaining SNP markers (11.6%) to be validated is likely due to the presence of paralogous sequences, incorrect primer designing and/or the need to optimize PCR conditions. This conversion rate is higher than that of the other KASPar studies on wheat (67%)26 and chickpea (80.6%).28 This rate of conversion from selected SNPs to functional KASPar assays could probably be increased with optimization of primer designing and amplification conditions. However, we have made no attempt to optimize the failed assays. For ease of use, developed KASPar assay-based markers were designated as PKAMs.
Screening of a new set of SNP markers (KASPar assays) on a set of 24 diverse pigeonpea genotypes representing the parents of 14 mapping populations segregating for various economically important traits provides readily available polymorphic markers for developing genetic maps and undertaking trait mapping in the respective mapping populations. In fact, these crossing combinations were selected based on diversity revealed through trait phenotyping and SSR profiling data.40 Although KASPar assays were developed for SNPs identified to be polymorphic between ICP 28 and ICPW 94, only 71.4% (1154) markers showed polymorphism in the tested genotypes. The remaining 28.6% (462) markers did not show polymorphism at all, including between the ICP 28 and ICPW 94 genotypes, indicating incorrect prediction. Out of 462 monomorphic markers, 379 (82.1%) markers were identified based on the Illumina GA IIx transcript sequence data, and the remaining 83 (17.9%) SNPs were identified based on allele re-sequencing of TOGs. This emphasizes the need of stringent selection criteria and the validation of in silico identified SNPs via allele re-sequencing.28 In brief, this study provides a list of polymorphic markers for different mapping populations that segregate for a number of important traits like Fusarium wilt, sterility mosaic disease and fertility restoration etc. that are important for pigeonpea improvement. The number of polymorphic markers identified in this study in intra-specific mapping populations was less (up to 55 markers in a given cross; Supplementary Table S1); however, these polymorphic markers would be helpful in enriching the recently developed SSR-based genetic linkage maps of intra-specific mapping populations.14
The present study reports a comprehensive genetic map comprising 875 loci by using 167 F2 individuals derived from ICP 28 × ICPW 94. Initial construction of a skeletal map with un-skewed markers and followed by the integration of distorted markers helped in minimizing the possibility for spurious assignments of markers.9 Eight hundred and seventy-five marker loci were mapped on 11 LGs corresponding to the 11 chromosome pairs of the pigeonpea genome. The total length of the map was 967.03 cM and the average marker spacing was 1.11 cM. The current pigeonpea linkage map is a considerable improvement over the previous pigeonpea genetic linkage maps using SSR and DArT markers.9,12 The marker density in the current map is almost three times higher than that in the previous maps. This higher marker density would be useful in determining double recombinants affecting a single marker. SNP genotyping using KASPar assays resulted in a much lower genotyping error rate than that obtained with markers like SSRs. In addition, SNP markers showing null alleles or an excess number of double recombinants were removed from the analyses. Owing to this careful error checking, the current map shows an increase in the total marker density compared with the previous maps developed by using SSR and DArT markers.9,12 However, we have noticed that in two LGs (i.e. CcLG02 and CcLG06), a few marker spaces were larger than 20 cM. Therefore, it is required to develop more markers and fill the gaps in the large marker space to increase the density of the linkage map. Earlier to this map, Bohra et al.9 developed an SSR marker-based map comprising 239 loci by using 72 F2 lines derived from ICP 28 × ICPW 94. To develop a consensus map, the newly developed mapping population (ICP 28 × ICPW 94) was also genotyped with targeted SSRs and an integrated genetic linkage map covering 996.21 cM was constructed. With the help of a recently available pigeonpea draft genome sequence,13 efforts are underway to develop a large number of SSR and SNP markers. Therefore, this map should serve as a ‘reference map’ for other future genetic maps of pigeonpea. Moreover, as the SNP markers are derived from the transcriptome sequences, these markers and the map would be very useful for linking the future genetic maps and the genome sequence of pigeonpea. In addition to the polymorphic markers in parental combinations of intra-specific mapping populations, most of the mapped PKAM markers in the inter-specific mapping population were monomorphic in cultivated parental lines. However, these mapped loci have provided a resource that can be used for conducting association analysis and linkage disequilibrium estimation in pigeonpea germplasm.
The recent development of a large data set of crop genomic sequences has aided in global gene predictions as well as in the identification of sequences important in gene regulation. In addition, comparative sequence analysis of crops is poised to contribute to the exploration of the genetic bases for differences and similarities among species. We are likely at last to understand the genetic explanation of how species have adapted to perform their shared or unique biological functions. Analysis of the sequences containing mapped SNPs onto pigeonpea genetic maps against two model legume species (Lotus and Medicago) and two crop legume species (soybean and cowpea) showed maximum similarity to soybean sequences (2480 soybean sequences). In general, markers mapped on one LG hit sequences from several chromosomes of soybean, suggesting the occurrence of chromosomal rearrangement in both the genomes. As soybean has under gone recent complete genome duplication,39 almost all the pigeonpea sequences have shown two to three different hits with soybean genome sequences. It also proves the taxonomical classification of pigeonpea and soybean, which lie in close proximity under subfamily Papilionoideae.41 With the model legume species 39.8% (Medicago) and 34.6% (Lotus) sequence similarities were identified whereas in the case of cowpea, 7.1% pigeonpea sequences could provide similarity results. This may be attributed to the fact that for sequence similarity analysis with pigeonpea sequences, genome assemblies of soybean, Medicago and Lotus were used, and during the time of analysis, genome sequence information was not available for cowpea; hence, the analysis was done by comparing with the high-density linkage map developed by Muchero et al.23 We discovered that synteny blocks in each of the 11 pigeonpea LGs were syntenic to their counterparts of the chromosomes of four legumes, implying certain colinearity for the syntenic chromosome/linkage pairs. The conserved sequences identified among five legumes (pigeonpea, soybean, cowpea, Medicago and Lotus) and the data from the comparative genome analysis should facilitate studies on genome evolution and analysis of the structural genome, but more importantly should facilitate the functional inference of genes in pigeonpea. The determination of gene functions is difficult in non-model species including pigeonpea; thus, functional genome analysis will have to rely heavily on the establishment of orthologies from model species.
In summary, this study provides an extensive resource of SNPs, their conversion in cost-effective KASPar assays and their application in constructing a dense genetic map in pigeonpea and in similarity analysis across five legumes. The developed genetic map is the most comprehensive genetic map for pigeonpea based on a single mapping population. Through mapped SNPs, we have identified complex syntenic relationships between soybean and pigeonpea by comparative genomics analysis. We consider that it will be possible for pigeonpea breeders to attain one of their most important goals, to rapidly and economically genotype thousands of accessions with a large and flexible number of markers. Although this is the first reported sizeable scale SNP mapping effort in pigeonpea, a larger number of informative SNPs mapped at minimum intervals will be necessary for broader applications. The extensive genomics resources developed by the whole-genome sequencing of pigeonpea, coupled to future re-sequencing multiple breeding lines, promise to considerably increase the number of informative SNPs, permitting exceptional levels of precision genetic analysis in pigeonpea breeding.
Supplementary data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Funding
The authors are thankful to CGIAR Generation Challenge Programme for SP2 commissioned project (G4008.47) and Theme Leader Discretionary Funds for support in carrying out this study.
Supplementary Material
Acknowledgements
Thanks are also due to Anuja Dubey, Pavana Hiremath, Sarwar Azam, A Bhanuprakash, Trushar Shah and Poornima Reddy for their help in some analysis and useful discussions while preparing the manuscript.
Footnotes
Edited by Satoshi Tabata
References
- 1.Varshney R.K., Graner A., Sorrells M.E. Genomics-assisted breeding for crop improvement. Trends Plant Sci. 2005;10:621–30. doi: 10.1016/j.tplants.2005.10.004. doi:10.1016/j.tplants.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 2.Varshney R.K., Hoisington D.A., Tyagi A.K. Advances in cereal genomics and applications in crop breeding. Trends Biotech. 2006;24:490–9. doi: 10.1016/j.tibtech.2006.08.006. doi:10.1016/j.tibtech.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 3.Varshney R.K., Thudi M., May G.D., Jackson S.A. Legume genomics and breeding. Plant Breed. Rev. 2010;33:257–304. doi:10.1002/9780470535486.ch6. [Google Scholar]
- 4.Raju N.L., Gnanesh B.N., Lekha P.T., et al. The first set of EST resource for gene discovery and marker development in pigeonpea (Cajanus cajan L.) BMC Plant Biol. 2010;10:45. doi: 10.1186/1471-2229-10-45. doi:10.1186/1471-2229-10-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dutta S., Kumawat G., Singh B.P., et al. Development of genic-SSR markers by deep transcriptome sequencing in pigeonpea [Cajanus cajan (L.) Millspaugh] BMC Plant Biol. 2011;11:17. doi: 10.1186/1471-2229-11-17. doi:10.1186/1471-2229-11-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dubey A., Farmer A., Schlueter J., et al. Defining the transcriptome assembly and its use for genome dynamics and transcriptome profiling studies in pigeonpea (Cajanus cajan L.) DNA Res. 2011;18:153–64. doi: 10.1093/dnares/dsr007. doi:10.1093/dnares/dsr007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kudapa H., Bharti A.K., Cannon S.B., et al. A comprehensive transcriptome assembly of pigeonpea (Cajanus cajan L.) using Sanger and second-generation sequencing platforms. Mol. Plant. 2012;5:1020–1028. doi: 10.1093/mp/ssr111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saxena R.K., Prathima C., Saxena K.B., Hoisington D.A., Singh N.K., Varshney R.K. Novel SSR markers for polymorphism detection in pigeonpea (Cajanus spp.) Plant Breed. 2010;129:142–8. doi:10.1111/j.1439-0523.2009.01680.x. [Google Scholar]
- 9.Bohra A., Dubey A., Saxena R.K., et al. Analysis of BAC-end sequences (BESs) and development of BES-SSR markers for genetic mapping and hybrid purity assessment in pigeonpea (Cajanus spp.) BMC Plant Biol. 2011;11:56. doi: 10.1186/1471-2229-11-56. doi:10.1186/1471-2229-11-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saxena R.K., Cui X., Thakur V., Walter B., Close T.J., Varshney R.K. Single feature polymorphisms (SFPs) for drought tolerance in pigeonpea [Cajanus cajan (L.) Millspaugh] Funct. Integr. Genomics. 2011;11:651–7. doi: 10.1007/s10142-011-0227-2. doi:10.1007/s10142-011-0227-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang S., Pang W., Ash G., et al. Low level of genetic diversity in cultivated pigeonpea compared to its wild relatives is revealed by diversity arrays technology. Theor. Appl. Genet. 2006;113:585–95. doi: 10.1007/s00122-006-0317-z. doi:10.1007/s00122-006-0317-z. [DOI] [PubMed] [Google Scholar]
- 12.Yang S., Saxena R.K., Kulwal P.L., et al. First genetic map of pigeonpea based on Diversity Array Technology (DArT) markers. J. Genet. 2011;90:103–9. doi: 10.1007/s12041-011-0050-5. doi:10.1007/s12041-011-0050-5. [DOI] [PubMed] [Google Scholar]
- 13.Varshney R.K., Chen W., Li Y., et al. Draft genome sequence of pigeonpea (Cajanus cajan), an orphan legume crop of resource-poor farmers. Nat. Biotechnol. 2012;30:83–9. doi: 10.1038/nbt.2022. doi:10.1038/nbt.2022. [DOI] [PubMed] [Google Scholar]
- 14.Bohra A., Saxena R.K., Gnanesh B.N., et al. An intra-specific consensus genetic map of pigeonpea [Cajanus cajan (L.) Millspaugh] derived from six mapping populations. Theor. Appl. Genet. 2012;125:1325–1338. doi: 10.1007/s00122-012-1916-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gnanesh B.N., Bohra A., Sharma M., et al. Genetic mapping and quantitative trait locus analysis of resistance to sterility mosaic disease in pigeonpea [Cajanus cajan (L.) Millsp.] Field Crops Res. 2011;123:53–61. doi:10.1016/j.fcr.2011.04.011. [Google Scholar]
- 16.Rafalski A. Applications of single nucleotide polymorphisms in crop genetics. Curr. Opin. Plant Biol. 2002;5:94–100. doi: 10.1016/s1369-5266(02)00240-6. doi:10.1016/S1369-5266(02)00240-6. [DOI] [PubMed] [Google Scholar]
- 17.Mammadov J., Chen W., Mingus J., Thompson S., Kumpatla S. Development of versatile gene-based SNP assays in maize (Zea mays L.) Mol. Breed. 2011;3:779–790. [Google Scholar]
- 18.Close T.J., Bhat P.R., Lonardi S., et al. Development and implementation of high-throughput SNP genotyping in barley. BMC Genomics. 2009;10:582. doi: 10.1186/1471-2164-10-582. doi:10.1186/1471-2164-10-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akhunov E., Nicolet C., Dvorak J. Single nucleotide polymorphism genotyping in polyploidy wheat with the Illumina GoldenGate assay. Theor. App. Genet. 2009;119:507–17. doi: 10.1007/s00122-009-1059-5. doi:10.1007/s00122-009-1059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan J., Yang X., Shah T., et al. High-throughput SNP genotyping with the GoldenGate assay in maize. Mol. Breed. 2009;25:441–51. doi:10.1007/s11032-009-9343-2. [Google Scholar]
- 21.Durstewitz G., Polley A., Plieske J., Luerssen H., Graner E.M., Wieseke R., Ganal M.W. SNP discovery by amplicon sequencing and multiplex SNP genotyping in the allopolyploid species Brassica napus. Genome. 2010;53:948–56. doi: 10.1139/G10-079. doi:10.1139/G10-079. [DOI] [PubMed] [Google Scholar]
- 22.Hyten D.L., Song Q., Choi I.Y., et al. High-throughput genotyping with the GoldenGate assay in the complex genome of soybean. Theor. Appl. Genet. 2008;116:945–52. doi: 10.1007/s00122-008-0726-2. doi:10.1007/s00122-008-0726-2. [DOI] [PubMed] [Google Scholar]
- 23.Muchero W., Diop N., Bhat P., et al. A consensus genetic map of cowpea [Vigna unguiculata (L) Walp.] and synteny based on EST-derived SNPs. Proc. Natl Acad. Sci. USA. 2009;106:18159–64. doi: 10.1073/pnas.0905886106. doi:10.1073/pnas.0905886106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deulvot C., Charrel H., Marty A., et al. Highly-multiplexed SNP genotyping for genetic mapping and germplasm diversity studies in pea. BMC Genomics. 2010;11:468. doi: 10.1186/1471-2164-11-468. doi:10.1186/1471-2164-11-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ganal M.W., Durstewitz G., Polley A., et al. A large maize (Zea mays L.) SNP genotyping array: development and germplasm genotyping, and genetic mapping to compare with the B73 reference genome. PLoS One. 2011;6:e28334. doi: 10.1371/journal.pone.0028334. doi:10.1371/journal.pone.0028334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Allen A.M., Barker G.L.A., Berry S.T., et al. Transcript-specific, single-nucleotide polymorphism discovery and linkage analysis in hexaploid bread wheat (Triticum aestivum L.) Plant Biotechnol. J. 2011;9:1086–99. doi: 10.1111/j.1467-7652.2011.00628.x. doi:10.1111/j.1467-7652.2011.00628.x. [DOI] [PubMed] [Google Scholar]
- 27.Cortés A.J., Chavarro M.C., Blair M.W. SNP marker diversity in common bean (Phaseolus vulgaris L.) Theor. Appl. Genet. 2011;123:827–45. doi: 10.1007/s00122-011-1630-8. doi:10.1007/s00122-011-1630-8. [DOI] [PubMed] [Google Scholar]
- 28.Hiremath P.J., Kumar A., Penmetsa R.V., et al. Large-scale development of cost-effective SNP marker assays for diversity assessment and genetic mapping in chickpea and comparative mapping in legumes. Plant Biotechnol. J. 2012;10:716–732. doi: 10.1111/j.1467-7652.2012.00710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cuc L.M., Mace E.S., Crouch J.H., Quang V.D., Long T.D., Varshney R.K. Isolation and characterization of novel microsatellite markers and their application for diversity assessment in cultivated groundnut (Arachis hypogaea) BMC Plant Biol. 2008;8:55. doi: 10.1186/1471-2229-8-55. doi:10.1186/1471-2229-8-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller N.A., Kingsmore S.F., Farmer A., et al. Management of high-throughput DNA sequencing projects: Alpheus. J. Comput. Sci. Syst. Biol. 2008;1:132–48. doi: 10.4172/jcsb.1000013. doi:10.4172/jcsb.1000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orru L., Catillo G., Napolitano F., De Matteis G., Scata M.C., Signorelli F., Moioli B. Characterization of a SNPs panel for meat traceability in six cattle breeds. Food Control. 2009;20:856–60. doi:10.1016/j.foodcont.2008.10.015. [Google Scholar]
- 32.Van Ooijen J.W. JoinMap4. Software for the calculation of genetic linkage maps in experimental populations. Plant Res. Int., Wageningen: The Netherlands. 2006 [Google Scholar]
- 33.Kosambi D.D. The estimation of map distances from recombination values. Ann. Eugen. 1943;12:172–5. [Google Scholar]
- 34.Voorrips R.E. MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002;93:77–8. doi: 10.1093/jhered/93.1.77. doi:10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]
- 35.Beckstette M., Homann R., Giegerich R., Kurtz S. Fast index based algorithms and software for matching position specific scoring matrices. BMC Bioinformatics. 2006;7:389. doi: 10.1186/1471-2105-7-389. doi:10.1186/1471-2105-7-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vandepoele K., Saeys Y., Simillion C., Raes J., Van de Peer Y. The automatic detection of homologous regions (ADHoRe) and its application to microlinearity between Arabidopsis and rice. Genome Res. 2002;12:1792–801. doi: 10.1101/gr.400202. doi:10.1101/gr.400202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krzywinski M., Schein J., Birol I., et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–45. doi: 10.1101/gr.092759.109. doi:10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kassa M.T., Penmetsa R.V., Carrasquilla-Garcia N., et al. Genetic patterns of domestication in pigeonpea (Cajanus cajan (L.) Millsp.) and wild Cajanus relatives. PLoS One. 2012;7:e39563. doi: 10.1371/journal.pone.0039563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmutz J., Cannon S.B., Schlueter J., et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463:178–83. doi: 10.1038/nature08670. doi:10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- 40.Saxena R.K., Saxena K.B., Kumar R.V., Hoisington D.A., Varshney R.K. SSR-based diversity in elite pigeonpea genotypes for developing mapping populations to map resistance to Fusarium wilt and sterility mosaic disease. Plant Breed. 2010;129:135–41. doi:10.1111/j.1439-0523.2009.01698.x. [Google Scholar]
- 41.Cannon S.B., May G.D., Jackson S.A. Three sequenced legume genomes and many crop species: Rich opportunities for translational genomics. Plant Physiol. 2009;151:970–7. doi: 10.1104/pp.109.144659. doi:10.1104/pp.109.144659. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.