Abstract
When, ∼20 years ago, investigators first determined that components of the genome considered nonfunctional had, in fact, gene regulatory capacity, they probably had no idea of their potential in controlling cell fate and were forced to revise and somehow reorganize their view of the molecular biology.
Indeed, it is currently well documented how a class of small non-coding RNAs, microRNAs, are conserved among the species, expressed in different tissues and cell types and involved in almost every biological process, including cell cycle, growth, apoptosis, differentiation and stress response, exerting a finely tuned regulation of gene expression by targeting multiple molecules.
As a consequence of the widespread range of processes they are able to influence, it is not surprising that miRNA deregulation is a hallmark of several pathological conditions, including cancer. Indeed, the aberrant expression of these tiny molecules in human tumors is not just a casual association, but they can exert a causal role, as oncogenes or tumor suppressors, in different steps of the tumorigenic process, from initiation and development to progression toward the acquisition of a metastatic phenotype.
An increasing body of evidence has indeed proved the importance of miRNAs in cancer, suggesting their possible use as diagnostic, prognostic and predictive biomarkers and leading to exploit miRNA-based anticancer therapies, either alone or in combination with current targeted therapies, with the goal to improve disease response and increase cure rates. Here, we review our current knowledge about miRNA involvement in cancer.
Introduction
microRNAs (miRNAs or miRs) are endogenous, small non-coding single-stranded RNAs of ∼22 nucleotides in length, found in both plants and animals, which function at posttranscriptional level as negative regulators of gene expression.
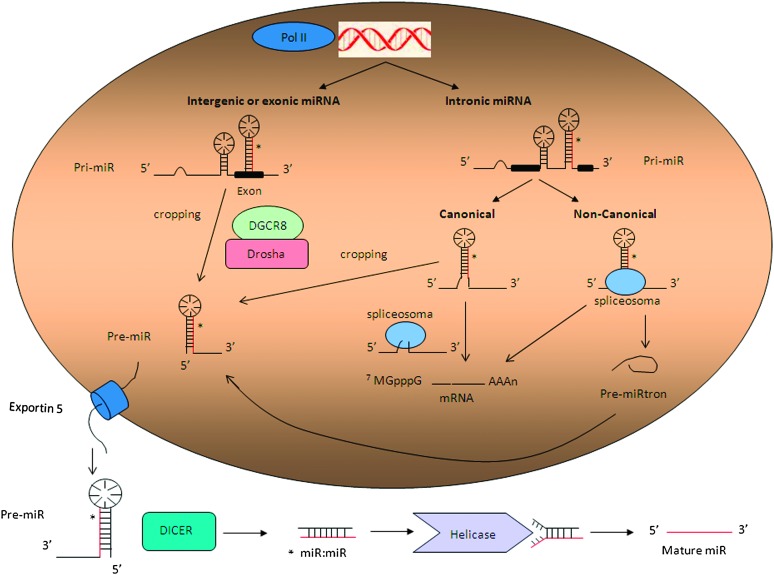
Annotation of genomic positions of miRNAs indicates that most miRNAs genes are located in intergenic regions, but they are also found within exonic or intronic regions in either sense or antisense orientation. The miRNAs localized within introns of protein-encoding or -non-encoding genes have been denominated ‘mirtrons’ (1). miRNAs can be organized as individual genes or localized as clusters representing miRNAs families, which are commonly related in sequence and function. miRNAs are mainly transcribed by RNA polymerase II (RNA pol II) from their own promoter or from promoter of the host gene in which they reside. RNA pol II synthesized large miRNA precursors called primary-miRNAs (pri-miRNAs) (2), which contain both 5′-cap structure (7MGpppG) as well as 3′-end poly(A) tail (3). Clustered miRNAs might be transcribed from a single transcription unit as polycistronic primary-miRNA.
microRNA biogenesis can be then summarized in two main processing steps, taking place, respectively, in the nucleus and in the cytoplasm: pri-microRNAs are first processed into the nucleus by RNAse III Drosha, associated to a double stranded RNA-binding protein DGCR8 (DiGeorge syndrome critical region gene 8; Pasha in flies) known as the microprocessor complex, that generates ∼70 nucleotides precursor miRNA products, which locally fold into stable secondary stem-loop structures (4).
The short stem plus a ∼2 nt 3′ overhang of the originated precursor molecules are recognized by the Ran-GTP-dependent transporter Exportin 5, which mediates the translocation to the cytoplasm (5). Here the second cropping process (dicing) takes place, performed by the RNAse III enzyme Dicer (Dicer 1 in flies) associated to TRBP (TAR RNA-binding protein) or protein activator of the interferon induced protein kinase (also known as PRKRA), and Argonaute (AGO1-4), which cleave the miRNA precursor hairpin generating a transitory miRNA/miRNA* duplex (also named miR-3p/miR-5p), which includes the mature miRNA guide, generally selected according to thermodynamic properties, and the complementary passenger strand, usually subjected to degradation. However, whereas the so called miRNA* was initially thought to be the strand subjected to degradation, instead more recent evidence suggests that it does not simply represent a non-functional bioproduct of miRNA biogenesis, but it can be selected as a functional strand and play significant biological roles (6).
This duplex is then loaded into the miRNA-associated RNA-induced silencing complex, including the mature single-stranded miRNA molecule and AGO proteins, where the mature miRNA is able to regulate gene expression at posttranscriptional level, binding for the most part through partial complementarity to target messenger RNAs (mRNAs) [generally the 3′ untranslated regions (UTR)], and mainly leading to mRNA degradation or translation inhibition, depending on the sequence complementarity between the small RNA and the target mRNA (7).
Additional findings suggested, however, that both miRNA biogenesis and function are more complex than previously expected. Canonical and non-canonical intronic miRNAs (mirtrons) seem to follow alternative biogenesis pathways (8). Canonical mirtrons are processed co-transcriptionally before splicing, and the splicing commitment complex is thought to tether the introns while Drosha cleaves the miRNA hairpin. At this point, the precursor miRNA enters the classical miRNA pathway, whereas the rest of the transcript undergoes precursor mRNA splicing and produces mature protein-coding mRNA. In non-canonical pathway, mirtrons are produced from spliced introns as debranched introns that mimic the structural features of precursor miRNAs and enter to miRNA-processing pathway without Drosha-mediated cleavage (1,9,10). (Figure 1).
Fig. 1.
microRNA biogenesis.
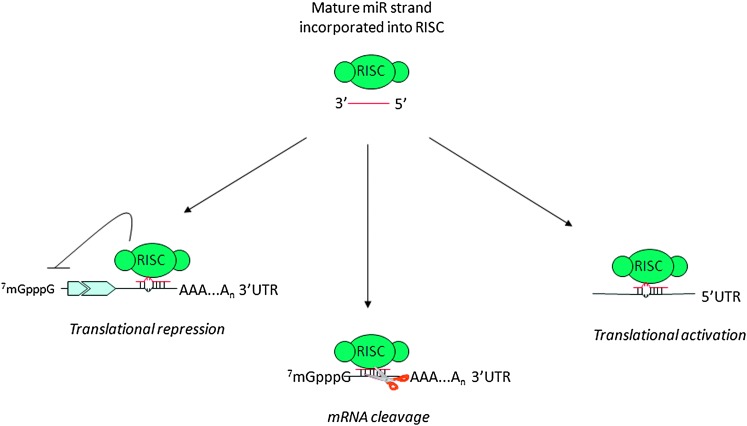
Beside biogenesis, recent reports have shed more light into the complex mechanisms regulating microRNA function on target mRNAs. Indeed, microRNAs mainly recognize complementary sequences in the 3′ UTRs of their target mRNAs; however, more recent studies have reported that they can also bind to the 5′ UTR or the open reading frame (11–14) and, even more surprisingly, they can upregulate translation upon growth arrest conditions (7) (Figure 2).
Fig. 2.
microRNA function on target molecules.
In addition, it has been evidenced that mature miRNAs may also be localized in nucleus, through a specific hexanucleotide (AGUGUU) sequence which acts as a transferable nuclear localization element (15). Moreover, it has been shown that vesicles of endocytic origin, known as exosomes, may contain both mRNA and microRNAs, which can be delivered to another cell and be functional in this new location. These RNA molecules were denominated exosomal shuttle RNAs as they mediate exchange of miRNAs with other cells, which represents a novel mechanism of genetic exchange (16).
Overall, these data show the complexity and widespread regulation exerted by miRNAs and motivate the searches for their currently still unidentified functions.
microRNA dysregulation in cancer: how the story began
The first evidence of the involvement of microRNAs in human cancer derived from studies on chronic lymphocitic leukemia (CLL): Dr Croce’s group found that critical region at chromosome 13q14, frequently deleted in CLL, does not contain a protein-coding tumor suppressor gene, but two microRNA genes, miR-15a and miR-16-1, expressed in the same polycistronic RNA. A couple of years later, they mapped all the known microRNA genes and found that many of them are located in regions of the genome involved in chromosomal alterations, such as deletion or amplification, in many different human tumors (17). After these early studies indicating the role of microRNA genes in the pathogenesis of human cancer, platforms to assess the global expression of microRNA genes in normal and diseased tissues have been developed, as an effort to establish whether microRNA profiling could be used for tumor classification, diagnosis and prognosis (18): after an extensive use of custom-made (19) and then commercial miRNA microarrays and bead-based flow cytometric miRNA analysis methods (20), the last generation of large-scale profiling method is represented by the high-throughput deep sequencing (21,22).
miRNA profiles can distinguish not only between normal and cancerous tissue and identify tissues of origin but they can also discriminate different subtypes of a particular cancer or even specific oncogenic abnormalities: miRNAs, for example, are differentially expressed between basal and luminal breast cancer subtypes (23,24) and can specifically classify estrogen receptor, progesterone receptor and HER2/neu receptor status (25–27).
Even more importantly, several groups in recent years have reported how microRNA profiling can predict disease outcome or response to therapy. After the first evidence in CLL, where a unique microRNA signature was associated with prognostic factors and disease progression in CLL (28), and lung cancer, where miR-155 overexpression and let-7a downregulation were able to predict poor disease outcome (29), several other reports have supported the significance of microRNAs as prognostic biomarkers (30,31).
Extremely important is also the possibility to evaluate miRNA expression to predict the response to specific drugs since it might be useful for a more accurate selection of patients potentially responsive to a specific therapy. miR-21, for example, is sufficient to predict poor response to adjuvant chemotherapy in adenocarcinomas (32) and in pancreatic cancer patients treated with gemcitabine (33).
In summary, the potential of miRNA signatures to distinguish between tumor and normal tissue, to discriminate between different subgroups of tumors and to predict outcome or response to therapy have focused scientist attention on these small molecules as potential clinical biomarkers, either diagnostic, predictive or prognostic.
Multiple layers of microRNA expression regulation
Several mechanisms can control microRNA expression and result to be altered in human diseases, including cancer: chromosomal abnormalities, as first suggested by the pioneer study on miR-15a and 16-1 (34), supported by the evidence that microRNAs are frequently located in regions of the genome involved in alterations in cancer (17) and then confirmed by several studies (35,36); mutations, as the inherited mutations in the primary transcripts of miR-15a and miR-16-1 responsible for reduced expression of the two microRNAs in vitro and in vivo in CLL (37); single nucleotide polymorphisms, as described in lung cancer (38).
In addition to structural genetic alterations, the deregulated microRNA expression in cancer can also be due to epigenetic changes, as altered DNA methylation, as suggested by the evidence that half of the genomic sequences of miRNA genes are associated with CpG islands (39) and then proved by several experimental reports. Most part of the currently published studies have used the approach to unmask epigenetically silenced oncosuppressor microRNAs inducing chromatin-remodeling by drug (as the DNA methyltransferase inhibitor 5-aza-2′-deoxycytidine) treatment, as observed for miR-127 (40), miR-9-1 (41) as well as the clustered miR-34b and miR-34c (42); alternatively, the evaluation of miRNA profiling analysis in DNMT1- and DNMT3b-deficient colorectal cancer cells allowed the identification of another hypermethylated oncosuppressor microRNA, miR-124a (43).
Conversely, the upmodulation of putative oncogenic microRNAs in cancer can be due to DNA hypomethylation (44,45).
Beside DNA methylation, another epigenetic mechanism that can affect microRNAs expression is represented by histone acetylation: histone deacetylase inhibition is indeed followed by the extensive and rapid alteration of microRNA levels (46–50).
However, the scenario connecting microRNAs and epigenetics is even more complicated, being microRNAs themselves able to regulate the expression of components of the epigenetic machinery, both DNMT and histone deacetylase enzymes, creating a highly controlled feedback mechanism (51–56).
This evidence raises the intriguing idea to apply as possible therapeutic approach the modulation of microRNA expression by targeting components of these regulatory networks.
microRNA expression can be also modulated as a consequence of defects in the microRNA biogenesis machinery: deregulation of different cofactors, for example, can affect miRNA expression with important biological implications (57,58).
In particular, it seems that Dicer or Drosha silencing promotes cellular transformation and tumorigenesis in vivo: conditional loss of Dicer1 in mice lung tissues enhances the development of lung tumors in a K-ras mouse model (59). Finally, loss of Dicer and/or Drosha has also been inversely correlated with outcome in lung cancer (60), cancers of the ovarian epithelium (61) and more recently in other tumor types as nasopharyngeal carcinoma (62), neuroblastoma (63) and breast (64).
It may sound surprising that reprogramming of the whole ‘microRNome’, including both oncomiRNAs and tumor suppressor miRNAs, can lead to a specific effect: how is the equilibrium shifted in favor of an antitumoral effect? This might be related to the possibility that most microRNAs seem to exert a role as oncosuppressors and consequently are mostly downregulated in human neoplasia (20).
A few reports, however, describe a positive correlation between Dicer expression and poor outcome in colorectal cancer (65) and in prostate cancer (66) or the overexpression of Drosha in cervical cancer (67), thus raising the important issue to validate this still debated question and verify whether the effect of targeting the microRNA machinery might be tissue-related.
microRNA processing can be also affected by other microRNAs, directly or indirectly, thus creating a complex level of reciprocal interaction and regulation. Piccolo’s group (68) have described a microRNA family, miR-103-107, able to empower the metastatic potential targeting Dicer thus attenuating the global microRNA biosynthesis, with a particular effect mediated by the downregulation of miR-200 family, and the consequent switch to a more mesenchymal and aggressive phenotype. A more recent report by Tang et al. (69) demonstrates that microRNAs can have nuclear functions, directly regulating other microRNA processing: mouse miR-709 is indeed predominantly located in the nucleus, where it directly binds to a recognition element on pri-miR-15a/16-1 preventing its processing.
Finally, a deregulation of miRNA expression can be a result of increased or decreased transcription due to an altered transcription factor activity: microRNA can indeed be either positively or negatively regulated by transcription factor with oncosuppressive, as p53 activating miR-34a (70,71) or miR-205 (72), or oncogenic functions, as MYC, activating miR-17-92 cluster and repressing let-7 (73) and miR-29 family members (74), or ZEB1, directly repressing the transcription of members of the miR-200 family (75), which are in turn able to directly target ZEB1 and ZEB2 (76).
microRNA function: a wide network of molecular interactions
It is currently well documented how cancers develop sophisticated networks of biological activities that contribute to their ability to progress and, in some cases, evade treatment.
Gain-of-function and loss-of-function experiments, in combination with target prediction analyses, have demonstrated that microRNAs can affect different steps of the tumorigenic process.
ApoptomiRs and microRNAs regulating proliferation
Among the pioneer studies demonstrating that microRNAs can impair cell proliferation or induce apoptosis through oncogene targeting, it is still worth citing the first pioneer studies: miR-15a-16-1 targeting BCL-2 (77) or let-7 targeting RAS (78) and MYC (79). Vice versa, miRNAs with oncogenic properties can negatively regulate tumor suppressor proteins as the well-described miR-21: the expression of this miRNA has been reported at high levels in breast (27), glioblastomas (80) and pancreas (81) among others. Chan et al. blocked miR-21 expression in glioblastoma cell lines and reported an increased activation of caspases and of apoptosis (82). Additional studies showed that miR-21 exerts its anti-apoptotic effects by targeting the tumor suppressors phosphatase and tensin homolog and programmed cell death 4 (83,84). More recently, Slack’s group (85) has shown that mice conditionally expressing miR-21 develop a pre-B malignant lymphoid-like phenotype, thus demonstrating that miR-21 is a genuine oncogene.
Unfortunately, this is one of the few microRNA engineered animal models developed up to date, models that can provide the genetic demonstration of the causative involvement of a specific microRNA in a biological phenomenon through knock out or transgene introduction. miR-17-92 cluster and miR-155, both overexpressed in lymphoproliferative disorders, including lymphomas and leukemia (86,87), were the first examples of miRNAs with oncogenic activity validated in engineered animal models (87–90). Notably, overexpression of miR-155 alone in the lymphoid compartment was sufficient to cause cancer and did not require any other cooperative mutation or oncogene expression (89).
miRs as a tool to tackle drug resistance
Being able to affect practically all biological processes, including proliferation and apoptosis, it is not surprising that microRNAs can impact response to specific drugs, including chemotherapy.
This hypothesis mostly derives from in vitro studies of gain or loss of function, where candidate miRNAs are initially identified in tumor cell lines with different degrees of resistance to specific therapeutic drugs and then targeted in order to overcome drug resistance. One of the first reports describing the involvement of microRNAs in chemoresistance was performed in cholangiocarcinoma cell lines (90), where inhibition of miR-21 and miR-200b increased sensitivity to gemcitabine. This first evidence was followed by many other studies (91–93).
However, beside chemotherapy, microRNAs can also improve the responsiveness to targeted therapies: overexpression of miR-221 and miR-222 is responsible for resistance to anti-estrogenic therapies, as Tamoxifen (94,95) and Fulvestran (96), whereas ectopic expression of oncosuppressor miR-205 is able to improve the responsiveness to Tyrosin Kinase Inhibitors through direct targeting of HER3 (97).
These associations highlight not only the importance of microRNAs as predictive biomarkers but also the possibility to use them as an alternative approach for tackling drug resistance.
MetastamiRs
To successfully metastasize, a tumor cell must complete a complex set of processes, including invasion, survival and arrest in the circulatory system and colonization of foreign organs. Despite great advancements in the knowledge of metastasis biology, the molecular mechanisms are still not completely understood. Remarkably, a number of miRNAs have shown a regulatory role in the metastatic program, thus giving raise to the term metastamiRs: this expression was indeed recently introduced by Welch et al. to refer to those regulatory miRNAs which promote or suppress various steps in migration and metastasis of cancer cells (98), affecting key steps as epithelial-mesenchymal transition (EMT), migration and angiogenesis.
Among the metastasis promoters, we can cite:
miR-10b. Originally reported as downregulated in most breast cancers in comparison with normal mammary tissues (27), surprisingly miR-10b resulted also highly expressed in ∼50% of metastatic tumors. In 2007, Ma et al. (99) from Robert Weinberg’s group evidenced that upregulation of miR-10b suppressed homeobox D10 (HOXD10) expression, thus allowing the activation of pro-metastatic gene RHOC and initiation of breast cancer invasion and metastasis. A few years later, the same group has exploited a possible therapeutic application, reporting that systemic treatment of tumor-bearing mice with miR-10b antagomirs suppresses breast cancer metastasis (100).
miR-21 stimulates invasation, extravasation and metastasis in different tumor types, included colorectal cancer (101) and breast cancer (102).
miR-373 and miR-520c. In 2006, through a genetic screen Agami’s laboratory identified miR-373 as a potential oncogene in testicular germ cell tumors, where it suppressed p53-induced pathway, thus cooperating with oncogenic RAS to promote cellular transformation (103). A few years later, the same group (104) used a similar approach on the non-metastatic MCF7 cell line and found that miR-373 and miR-520c stimulated cell migration and invasion in vitro and in vivo regulating the cell surface glycoprotein CD44 (cell surface receptor for hyaluronan). It is important to underline that, in breast cancer cells, the metastatic potential is indeed strongly correlated to EMT and the CD44+/CD24− stem cell phenotype.
A few relevant examples of metastasis inhibitors:
miR-34a, lost in several tumor types and involved into the network mediated by the well-known ‘genome guardian’ p53 (70), inhibits migration and invasion by downregulation of MET expression in human hepatocellular carcinoma cells (105).
miR-200 family members and miR-205 have been shown to reduce cell migration and invasiveness targeting ZEB transcription factors, known inducers of EMT (76). Moreover, miR-205 inhibits in vitro migration and experimental lung metastasis in MDA-MB-231 cells targeting VEGFA (106).
miR-126 and miR-335 act as negative regulators of tumor invasion and metastasis in human breast and lung cancer (107). Notably, the same group has more recently revealed that endogenous miR-126 non-cell autonomously regulates endothelial cell recruitment to metastatic breast cancer cells, in vitro and in vivo. It suppresses metastatic endothelial recruitment, metastatic angiogenesis and metastatic colonization through coordinate targeting of IGFBP2, PITPNC1 and MERTK, pro-angiogenic genes and biomarkers of human metastasis (108). Thus, miRNAs can exert their function influencing interaction between different cell types. Another example is represented by the acquirement of a metastatic phenotype following miR-320 loss in cancer-associated fibroblasts: miR-320 is indeed a crucial component of a phosphatase and tensin homolog-controlled tumor–suppressive axis in stromal fibroblasts and loss of phosphatase and tensin homolog and miR-320 induces an oncogenic secretome that reprogrammes the tumor microenvironment to promote invasion and angiogenesis (109).
Indeed, one of the crucial steps of the metastatic process is represented by neo-angiogenesis, which allows cells to reach and disseminate through the systemic circulation. microRNAs can control tumor progression also at this level either promoting, as miR-210 (110) and miR-17-92 (111), or inhibiting, as miR-221 and miR-222 or miR-16, miR-15b, miR-20a and miR-20b (112), proliferation of endothelial cells.
Always focusing on metastasis-inducers microRNAs, Ma et al. (113) described how miR-9 increases the metastatic potential by targeting E-cadherin and thus activating the beta-catenin signaling, which contributes to upregulate the expression of vascular endothelial growth factor.
Interestingly, supporting the correlation between microRNAs and metastases, it has been reported that primary tumors and metastasis from the same tissue show a similar pattern of microRNAs expression (114). Being a more accurate classifier than mRNA expression studies, miRNA profiling has thus revealed the potential to solve one of the most demanding issues in cancer diagnostic: the origin of metastasis of unknown primary tumors.
microRNAs enter the stem cell world
miRNAs have emerged as important regulators of embryonic development (115) and stem cell functions (116) in mammals.
Interestingly, miRNA expression patterns in embryonic stem cells often bear resemblance to those observed tumor cells, especially the most undifferentiated and aggressive subtypes, and this evidence is not surprising if we consider that miRNAs can control EMT process, differentiation and pluripotency. Moreover, it has been reported that microRNAs can affect CSCs (cancer stem cells) biology: let-7, for example, regulates stem cell properties as self-renewal and differentiation. Indeed, lentiviral-mediated overexpression of let-7a inhibited cell proliferation, mammosphere formation, tumor formation and metastasis in NOD-SCID mice and reduced the proportion of undifferentiated cells in vitro (117).
In keeping with the described previously oncosuppressive role of miR-200, it has been reported that this family of microRNAs are significantly downregulated in both breast CSCs and normal mammary stem and/or progenitor cells (118). Functional studies showed that overexpression of miR-200c reduced the clonogenic and tumor-initiation activities in BCSCs and suppressed formation of mammary ducts by normal mammary stem cells. The stem cell factor BMI-1 was directly modulated by miR-200c (119).
A never-ending puzzle: factors influencing microRNA function
Considering the different rules regulating miRNA/target interaction, and the evidence that microRNAs can target multiple molecules, it is unlikely that miRNAs will be responsible for a specific phenotype by aiming at a single target. Instead, it is thought that miRNAs engage in complex interactions with the machinery that controls the transcriptome and concurrently target multiple mRNAs. This is probably the most intriguing rational supporting the idea of using microRNAs as anticancer drugs.
Computational algorithms for target identification, mainly based on the free binding energy between a miRNA and a putative target mRNA sequence are by definition prediction tools, which need an experimental validation. The most commonly used validation method is represented by a reporter assay: the co-transfection of the miRNA of interest and the 3′UTR of the target mRNA cloned downstream the luciferase gene leads to reduction of the reporter activity due to the binding of the miRNA to the recognition site on the target sequence. This inhibitory effect is impaired by mutating the miRNA binding sequence of the target 3′UTR. However, even though this method suggests a physical and functional interaction between a miRNA and its target, it does not prove it directly. To this aim, more rigorous pull-down assays have been designed, as immunoprecipitation of labeled miRNA/mRNA complexes and consequent target identification by reverse transcription–PCR and sequencing (120) or immunoprecipitation with Ago2 antibody, thus isolating the ternary, presumably functional, miRNA/mRNA/Ago2 complex (121).
However, the rules of miRNA/target mRNA regulation are even more complicated than previously thought. As example, the discovery of other functional non-coding RNAs (ncRNAs), interconnected with each other, has revealed a network of regulatory molecules definitely more intricate than expected.
One of the first studies reporting the existence of other ncRNAs involved in tumorigenesis and connected to microRNAs was reported by our group (122): more in detail, Calin et al. observed that a large fraction of genomic ultraconserved regions encode a particular set of ncRNAs whose expression is altered in human cancers, and which can be regulated by microRNAs.
A more recent report by Pandolfi’s group (123) has introduced the revolutionary concept that miRNA effect on mRNA containing common miRNA recognition elements can be affected by competing endogenous RNAs: RNA transcripts, both protein coding and non-coding, can compete for miRNA binding, thus co-regulating each other.
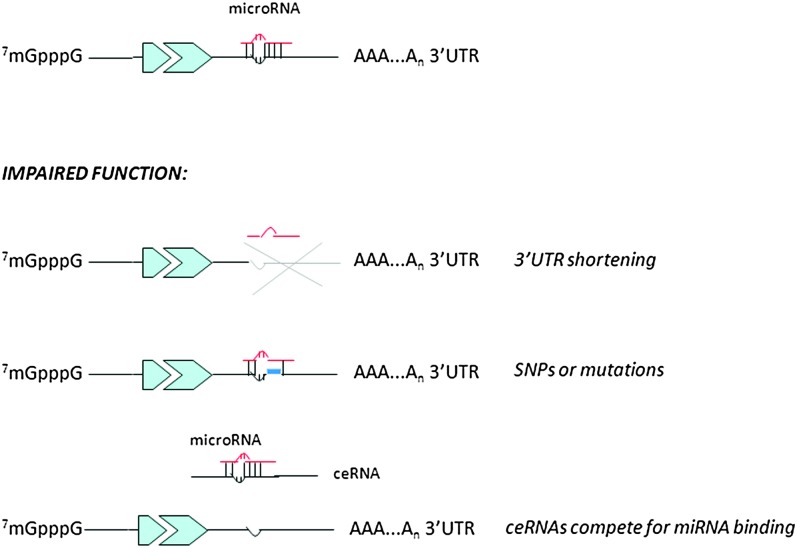
Beside the existence of other RNAs able to interfere with miRNA function, other mechanisms can affect their regulatory action on target molecules: one example is represented by the evidence that mRNAs can present or develop specific alterations to escape miRNA control.
Different studies have indeed reported the existence of oncogenic mRNAs carrying mutations or single nucleotide polymorphisms in their 3′ UTR allowing them to avoid miRNA binding and consequent negative control, as demonstrated, for example for let-7 and RAS interaction in lung cancer, where an single nucleotide polymorphism in the let-7 binding site on RAS 3′ UTR alters RAS expression and is associated with higher occurrence risk (124). Another very interesting report is the study published by Sandberg et al. (125), who discovered how proliferating cells express mRNAs with shortened 3′ UTRs and fewer microRNA target sites. It would be of extreme interest to evaluate the selection for oncogenes with shortened 3′ UTRs in different tumor types (Figure 3).
Fig. 3.
Mechanisms controlling microRNA function: how cells can escape miRNA-mediated control
Future perspectives
The past decade has witnessed an explosion of research focused on small ncRNAs: conserved among the species and involved in every biological process examined, these tiny RNA molecules have been demonstrated to be crucial regulators of gene expression.
Cancer is defined by abnormal and uncontrolled cell division, a phenotype that arises from the alteration of different mechanisms, leading not only to misregulation of several protein coding genes but also to a global change in miRNA profile. Being microRNAs major regulators of gene expression, with roles in nearly every area of cell behavior, development and survival and able to regulate multiple targets acting as oncogenes or tumor suppressor genes, it is not surprising that their altered expression contributes to a substantial cell re-organization and is causally involved in so many different human tumors.
However, although significant advances have been made for the future role of miRNAs in diagnostics, there have been so far fewer reported successes in the development of miRNAs for use in therapy. Indeed, even though a number of reports have described the possibility to reintroduce or inhibit microRNAs (reviewed in ref. 126), there are still many issues that need to be addressed for an effective translation in clinics, as the development of efficient methods of a specific drug delivery and the accurate prevision of putative unwanted off target effects.
Nevertheless, the results obtained up to date seem quite promising and encouraging, and even though we still have to improve the knowledge in microRNA field to even think of future therapeutic applications, we might be not so far from there.
Funding
National Cancer Institute to C.M.C.; Start Up AIRC Grant to M.V.I.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- CLL
chronic lymphocytic leukemia
- EMT
epithelial–mesenchymal transition
- mRNA
messenger RNA
- miRNA or miR
microRNA
- ncRNA
non-coding RNA
References
- 1.Ruby JG, et al. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee Y, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cai X, et al. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee Y, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 5.Yi R, et al. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhayani MK, et al. Functional relevance of miRNA* sequences in human disease. Mutat. Res. 2012;731:14–19. doi: 10.1016/j.mrfmmm.2011.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vasudevan S, et al. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 8.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 9.Berezikov E, et al. Mammalian mirtron genes. Mol. Cell. 2007;28:328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okamura K, et al. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila . Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ørom UA, et al. MicroRNA-10a binds the 5'UTR of ribosomal protein mRNAs and enhances their translation. Mol. Cell. 2008;30:460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 12.Lytle JR, et al. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5' UTR as in the 3' UTR. Proc. Natl Acad. Sci. USA. 2007;104:9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moretti F, et al. Mechanism of translational regulation by miR-2 from sites in the 5' untranslated region or the open reading frame. RNA. 2010;16:2493–2502. doi: 10.1261/rna.2384610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qin W, et al. miR-24 regulates apoptosis by targeting the open reading frame (ORF) region of FAF1 in cancer cells. PLoS One. 2010;5:e9429. doi: 10.1371/journal.pone.0009429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hwang HW, et al. A hexanucleotide element directs microRNA nuclear import. Science. 2007;315:97–100. doi: 10.1126/science.1136235. [DOI] [PubMed] [Google Scholar]
- 16.Valadi H, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 17.Calin GA, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl Acad. Sci. USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Calin GA, et al. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 19.Liu CG, et al. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc. Natl Acad. Sci. USA. 2004;101:9740–9744. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 21.Creighton CJ, et al. Expression profiling of microRNAs by deep sequencing. Brief. Bioinform. 2009;10:490–497. doi: 10.1093/bib/bbp019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farazi TA, et al. MicroRNA sequence and expression analysis in breast tumors by deep sequencing. Cancer Res. 2011;71:4443–4453. doi: 10.1158/0008-5472.CAN-11-0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blenkiron C, et al. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 2007;8:R21. doi: 10.1186/gb-2007-8-10-r214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sempere LF, et al. Altered MicroRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res. 2007;67:11612–11620. doi: 10.1158/0008-5472.CAN-07-5019. [DOI] [PubMed] [Google Scholar]
- 25.Mattie MD, et al. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol. Cancer. 2006;5:24. doi: 10.1186/1476-4598-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lowery AJ, et al. MicroRNA signatures predict oestrogen receptor, progesterone receptor and HER2/neu receptor status in breast cancer. Breast Cancer Res. 2009;11:R27. doi: 10.1186/bcr2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iorio MV, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 28.Calin GA, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N. Engl. J. Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 29.Yanaihara N, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 30.Li X, et al. Survival prediction of gastric cancer by a seven-microRNA signature. Gut. 2010;59:579–585. doi: 10.1136/gut.2008.175497. [DOI] [PubMed] [Google Scholar]
- 31.Caramuta S, et al. MicroRNA expression profiles associated with mutational status and survival in malignant melanoma. J. Invest. Dermatol. 2010;130:2062–2070. doi: 10.1038/jid.2010.63. [DOI] [PubMed] [Google Scholar]
- 32.Schetter AJ, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giovannetti E, et al. MicroRNA-21 in pancreatic cancer: correlation with clinical outcome and pharmacologic aspects underlying its role in the modulation of gemcitabine activity. Cancer Res. 2010;70:4528–4538. doi: 10.1158/0008-5472.CAN-09-4467. [DOI] [PubMed] [Google Scholar]
- 34.Calin GA, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl Acad. Sci. USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang L, et al. microRNAs exhibit high frequency genomic alterations in human cancer. Proc. Natl Acad. Sci. USA. 2006;103:9136–9141. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tagawa H, et al. A microRNA cluster as a target of genomic amplification in malignant lymphoma. Leukemia. 2005;19:2013–2016. doi: 10.1038/sj.leu.2403942. [DOI] [PubMed] [Google Scholar]
- 37.Raveche ES, et al. Abnormal microRNA-16 locus with synteny to human 13q14 linked to CLL in NZB mice. Blood. 2007;109:5079–5086. doi: 10.1182/blood-2007-02-071225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu Z, et al. Genetic variants of miRNA sequences and non-small cell lung cancer survival. J. Clin. Invest. 2008;118:2600–2608. doi: 10.1172/JCI34934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weber B, et al. Methylation of human microRNA genes in normal and neoplastic cells. Cell Cycle. 2007;6:1001–1005. doi: 10.4161/cc.6.9.4209. [DOI] [PubMed] [Google Scholar]
- 40.Saito Y, et al. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 41.Lehmann U, et al. Epigenetic inactivation of microRNA gene hsa-mir-9-1 in human breast cancer. J. Pathol. 2008;214:17–24. doi: 10.1002/path.2251. [DOI] [PubMed] [Google Scholar]
- 42.Toyota M, et al. Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer Res. 2008;68:4123–4132. doi: 10.1158/0008-5472.CAN-08-0325. [DOI] [PubMed] [Google Scholar]
- 43.Lujambio A, et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 2007;67:1424–1429. doi: 10.1158/0008-5472.CAN-06-4218. [DOI] [PubMed] [Google Scholar]
- 44.Brueckner B, et al. The human let-7a-3 locus contains an epigenetically regulated microRNA gene with oncogenic function. Cancer Res. 2007;67:1419–1423. doi: 10.1158/0008-5472.CAN-06-4074. [DOI] [PubMed] [Google Scholar]
- 45.Iorio MV, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 46.Scott GK, et al. Rapid alteration of microRNA levels by histone deacetylase inhibition. Cancer Res. 2006;66:1277–1281. doi: 10.1158/0008-5472.CAN-05-3632. [DOI] [PubMed] [Google Scholar]
- 47.Shin S, et al. MicroRNAs that respond to histone deacetylase inhibitor SAHA and p.53 in HCT116 human colon carcinoma cells. Int. J. Oncol. 2009;35:1343–1352. doi: 10.3892/ijo_00000452. [DOI] [PubMed] [Google Scholar]
- 48.Bandres E, et al. Epigenetic regulation of microRNA expression in colorectal cancer. Int. J. Cancer. 2009;125:2737–2743. doi: 10.1002/ijc.24638. [DOI] [PubMed] [Google Scholar]
- 49.Saito Y, et al. Chromatin remodeling at Alu repeats by epigenetic treatment activates silenced microRNA-512-5p with downregulation of Mcl-1 in human gastric cancer cells. Oncogene. 2009;28:2738–2744. doi: 10.1038/onc.2009.140. [DOI] [PubMed] [Google Scholar]
- 50.Rhodes LV, et al. The histone deacetylase inhibitor trichostatin A alters microRNA expression profiles in apoptosis-resistant breast cancer cells. Oncol. Rep. 2012;27:10–16. doi: 10.3892/or.2011.1488. doi: 10.3892/or.2011.1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fabbri M, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc. Natl Acad. Sci. USA. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Garzon R, et al. MicroRNA 29b functions in acute myeloid leukemia. Blood. 2009;114:5331–5341. doi: 10.1182/blood-2009-03-211938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Benetti R, et al. A mammalian microRNA cluster controls DNA methylation and telomere recombination via Rbl2-dependent regulation of DNA methyltransferases. Nat. Struct. Mol. Biol. 2008;15:268–279. doi: 10.1038/nsmb.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sinkkonen L, et al. MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 2008;15:259–267. doi: 10.1038/nsmb.1391. [DOI] [PubMed] [Google Scholar]
- 55.Chen JF, et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roccaro AM, et al. microRNA-dependent modulation of histone acetylation in Waldenstrom macroglobulinemia. Blood. 2010;116:1506–1514. doi: 10.1182/blood-2010-01-265686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Y, et al. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat. Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Viswanathan SR, et al. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kumar MS, et al. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev. 2009;23:2700–2704. doi: 10.1101/gad.1848209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Karube Y, et al. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci. 2005;96:111–115. doi: 10.1111/j.1349-7006.2005.00015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Merritt WM, et al. Dicer, Drosha, and outcomes in patients with ovarian cancer. N. Engl. J. Med. 2008;359:2641–2650. doi: 10.1056/NEJMoa0803785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guo X, et al. The microRNA-processing enzymes: Drosha and Dicer can predict prognosis of nasopharyngeal carcinoma. J. Cancer Res. Clin. Oncol. 2012;138:49–56. doi: 10.1007/s00432-011-1058-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lin RJ, et al. microRNA signature and expression of Dicer and Drosha can predict prognosis and delineate risk groups in neuroblastoma. Cancer Res. 2010;70:7841–7850. doi: 10.1158/0008-5472.CAN-10-0970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Grelier G, et al. Prognostic value of Dicer expression in human breast cancers and association with the mesenchymal phenotype. Br. J. Cancer. 2009;101:673–683. doi: 10.1038/sj.bjc.6605193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Faber C, et al. Overexpression of Dicer predicts poor survival in colorectal cancer. Eur. J. Cancer. 2011;47:1414–1419. doi: 10.1016/j.ejca.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 66.Chiosea S, et al. Up-regulation of dicer, a component of the MicroRNA machinery, in prostate adenocarcinoma. Am. J. Pathol. 2006;169:1812–1820. doi: 10.2353/ajpath.2006.060480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Muralidhar B, et al. Functional evidence that Drosha overexpression in cervical squamous cell carcinoma affects cell phenotype and microRNA profiles. J. Pathol. 2011;224:496–507. doi: 10.1002/path.2898. doi: 10.1002/path. 2898. [DOI] [PubMed] [Google Scholar]
- 68.Martello G, et al. A MicroRNA targeting dicer for metastasis control. Cell. 2010;141:1195–1207. doi: 10.1016/j.cell.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 69.Tang R, et al. Mouse miRNA-709 directly regulates miRNA-15a/16-1 biogenesis at the posttranscriptional level in the nucleus: evidence for a microRNA hierarchy system. Cell Res. 2012;22:504–515. doi: 10.1038/cr.2011.137. doi: 10.1038/cr.2011.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.He L, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chang TC, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol. Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Piovan C, et al. Oncosuppressive role of p53-induced miR-205 in triple negative breast cancer. Mol. Oncol. doi: 10.1016/j.molonc.2012.03.003. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chang TC, et al. Lin-28B transactivation is necessary for Myc-mediated let-7 repression and proliferation. Proc. Natl Acad. Sci. USA. 2009;106:3384–3389. doi: 10.1073/pnas.0808300106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mott JL, et al. Transcriptional suppression of mir-29b-1/mir-29a promoter by c-Myc, hedgehog, and NF-kappaB. J. Cell. Biochem. 2010;110:1155–1164. doi: 10.1002/jcb.22630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Burk U, et al. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO. 2008;9:582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gregory PA, et al. MicroRNAs as regulators of epithelial-mesenchymal transition. Cell Cycle. 2008;7:3112–3118. doi: 10.4161/cc.7.20.6851. [DOI] [PubMed] [Google Scholar]
- 77.Cimmino A, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl Acad. Sci. USA. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Johnson SM, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 79.Sampson VB, et al. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res. 2007;67:9762–9770. doi: 10.1158/0008-5472.CAN-07-2462. [DOI] [PubMed] [Google Scholar]
- 80.Ciafre SA, et al. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem. Biophys. Res. Commun. 2005;334:1351–1358. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 81.Bloomston M, et al. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297:1901–1908. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- 82.Chan JA, et al. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 83.Frankel LB, et al. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J. Biol. Chem. 2008;283:1026–1033. doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]
- 84.Medina PP, et al. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467:86–90. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- 85.Garzon R, et al. MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood. 2008;111:3183–3189. doi: 10.1182/blood-2007-07-098749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.He L, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xiao C, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat. Immunol. 2008;9:405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mu P, et al. Genetic dissection of the miR-17∼92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 2009;23:2806–2811. doi: 10.1101/gad.1872909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Costinean S, et al. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc. Natl Acad. Sci. USA. 2006;103:7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Meng F, et al. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology. 2006;130:2113–2129. doi: 10.1053/j.gastro.2006.02.057. [DOI] [PubMed] [Google Scholar]
- 91.Kovalchuk O, et al. Involvement of microRNA-451 in resistance of the MCF-7 breast cancer cells to chemotherapeutic drug doxorubicin. Mol. Cancer Ther. 2008;7:2152–2159. doi: 10.1158/1535-7163.MCT-08-0021. [DOI] [PubMed] [Google Scholar]
- 92.Xia L, et al. miR-15b and miR-16 modulate multidrug resistance by targeting BCL2 in human gastric cancer cells. Int. J. Cancer. 2008;123:372–379. doi: 10.1002/ijc.23501. [DOI] [PubMed] [Google Scholar]
- 93.Chen F, et al. Inhibition of c-FLIP expression by miR-512-3p contributes to taxol-induced apoptosis in hepatocellular carcinoma cells. Oncol. Rep. 2010;23:1457–1462. doi: 10.3892/or_00000784. [DOI] [PubMed] [Google Scholar]
- 94.Zhao JJ, et al. MicroRNA-221/222 negatively regulates estrogen receptor alpha and is associated with tamoxifen resistance in breast cancer. J. Biol. Chem. 2008;283:31079–31086. doi: 10.1074/jbc.M806041200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 95.Miller TE, et al. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J. Biol. Chem. 2008;283:29897–29903. doi: 10.1074/jbc.M804612200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rao X, et al. MicroRNA-221/222 confers breast cancer fulvestrant resistance by regulating multiple signaling pathways. Oncogene. 2011;30:1082–1097. doi: 10.1038/onc.2010.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Iorio MV, et al. microRNA-205 regulates HER3 in human breast cancer. Cancer Res. 2009;69:2195–2200. doi: 10.1158/0008-5472.CAN-08-2920. [DOI] [PubMed] [Google Scholar]
- 98.Hurst DR, et al. Metastamir: the field of metastasis-regulatory microRNA is spreading. Cancer Res. 2009;69:7495–7749. doi: 10.1158/0008-5472.CAN-09-2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ma L, et al. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 100.Ma L, et al. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat. Biotechnol. 2010;28:341–347. doi: 10.1038/nbt.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Asangani IA, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 102.Zhu S, et al. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008;18:350–359. doi: 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]
- 103.Voorhoeve PM, et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 2006;124:1169–1181. doi: 10.1016/j.cell.2006.02.037. [DOI] [PubMed] [Google Scholar]
- 104.Huang Q, et al. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat. Cell Biol. 2008;10:202–210. doi: 10.1038/ncb1681. [DOI] [PubMed] [Google Scholar]
- 105.Li N, et al. miR-34a inhibits migration and invasion by down-regulation of c-Met expression in human hepatocellular carcinoma cells. Cancer Lett. 2009;275:44–53. doi: 10.1016/j.canlet.2008.09.035. [DOI] [PubMed] [Google Scholar]
- 106.Wu H, et al. Suppression of cell growth and invasion by miR-205 in breast cancer. Cell Res. 2009;19:439–448. doi: 10.1038/cr.2009.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tavazoie SF, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Png KJ, et al. A microRNA regulon that mediates endothelial recruitment and metastasis by cancer cells. Nature. 2011;481:190–194. doi: 10.1038/nature10661. . doi: 10.1038/nature10661. [DOI] [PubMed] [Google Scholar]
- 109.Khew-Goodall Y, et al. Stromal miR-320 keeps an oncogenic secretome in check. Nat. Cell Biol. 2012;14:124–125. doi: 10.1038/ncb2431. . doi: 10.1038/ncb2431. [DOI] [PubMed] [Google Scholar]
- 110.Pulkkinen K, et al. Hypoxia induces microRNA miR-210 in vitro and in vivo ephrin-A3 and neuronal pentraxin 1 are potentially regulated by miR-210. FEBS Lett. 2008;582:2397–2401. doi: 10.1016/j.febslet.2008.05.048. [DOI] [PubMed] [Google Scholar]
- 111.Dews M, et al. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat. Genet. 2006;38:1060–1065. doi: 10.1038/ng1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Poliseno L, et al. MicroRNAs modulate the angiogenic properties of HUVECs. Blood. 2006;108:3068–3071. doi: 10.1182/blood-2006-01-012369. [DOI] [PubMed] [Google Scholar]
- 113.Ma L, et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat. Cell Biol. 2010;12:247–256. doi: 10.1038/ncb2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rosenfeld N, et al. MicroRNAs accurately identify cancer tissue origin. Nat. Biotechnol. 2008;26:462–469. doi: 10.1038/nbt1392. [DOI] [PubMed] [Google Scholar]
- 115.Bernstein E, et al. Dicer is essential for mouse development. Nat. Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 116.Calabrese JM, et al. RNA sequence analysis defines Dicer's role in mouse embryonic stem cells. Proc. Natl Acad. Sci. USA. 2007;104:18097–18102. doi: 10.1073/pnas.0709193104. Erratum in: Proc. Natl Acad. Sci. USA. 2007, 104, 21021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yu F, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 118.Shimono Y, et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138:592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wellner U, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat. Cell Biol. 2009;11:1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 120.Hsu PW, et al. miRNAMap: genomic maps of microRNA genes and their target genes in mammalian genomes. Nucleic Acids Res. 2006;34:D135–D139. doi: 10.1093/nar/gkj135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chi SW, et al. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Calin GA, et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007;12:215–229. doi: 10.1016/j.ccr.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 123.Salmena L, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chin LJ, et al. A SNP in a let-7 microRNA complementary site in the KRAS 3' untranslated region increases non-small cell lung cancer risk. Cancer Res. 2008;68:8535–8540. doi: 10.1158/0008-5472.CAN-08-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sandberg R, et al. Proliferating cells express mRNAs with shortened 3' untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Iorio MV, et al. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol. Med. 2012;4:143–159. doi: 10.1002/emmm.201100209. . doi: 10.1002/emmm.201100209. [DOI] [PMC free article] [PubMed] [Google Scholar]