Summary
Regulation of secondary metabolite (SM) gene clusters in Aspergillus nidulans has been shown to occur through cluster specific transcription factors or through global regulators of chromatin structure such as histone methyltransferases, histone deacetylases, or the putative methyltransferase LaeA. A multi-copy suppressor screen for genes capable of returning SM production to the SM deficient ΔlaeA mutant resulted in identification of the essential histone acetyltransferase EsaA, able to complement an esa1 deletion in Saccharomyces cereviseae. Here we report that EsaA plays a novel role in SM cluster activation through histone 4 lysine 12 (H4K12) acetylation in four examined SM gene clusters (sterigmatocystin, penicillin, terrequinone, and orsellinic acid), in contrast to no increase in H4K12 acetylation of the housekeeping tubA promoter. This augmented SM cluster acetylation requires LaeA for full effect and correlates with both increased transcript levels and metabolite production relative to wild type. H4K12 levels may thus represent a unique indicator of relative production potential, notably of SMs.
Introduction
Filamentous fungi are known for producing many bioactive compounds known as secondary metabolites (SMs). Many of these compounds have potential as pharmaceutical or agriculture agents, but are normally hypothesized to provide a selective advantage for the fungus in its natural environment. In agreement with this, many compounds are produced specifically in response to external cues, or at different stages of development (reviewed in Bennett, 1987; Hoffmeister and Keller, 2007).
Activation of production of SMs is regulated by a number of environmental stimuli, including carbon source, nitrogen source, temperature, and pH (reviewed in Calvo et al., 2002). Detection of these stimuli may be shunted through regulatory cascades resulting in the transcriptional activation of genes required for SM biosynthesis (reviewed in Yin and Keller, 2011). Towards this end, a critical and global activator of secondary metabolism was identified as the Velvet complex, a heterotrimeric complex consisting of the proteins Velvet (VeA), Velvet-like B (VelB), and Loss of aflR Expression A (LaeA; Bayram et al., 2008; Sarikaya Bayram et al., 2010). Formation of this complex occurs in the nucleus under conditions promoting sexual development and SM production, including growth in the dark. LaeA was isolated independently in a screen aimed at identifying genes required for sterigmatocystin (ST) biosynthesis (Butchko et al., 1999), and has since been shown to be required for biosynthesis of multiple compounds in the Aspergilli and other fungal genera, including ST, penicillin, terrequinone, lovastatin, gliotoxin, aflatoxin, kojic acid, and many others (Bok et al., 2004; Kale et al., 2008; Oda et al., 2011). Less is known about the requirement of veA for SM production, but it too has been shown to be essential for sterigmatocystin and penicillin biosynthesis (Kato et al., 2003). Historically researchers have used a strain of A. nidulans containing a mutated allele of veA known as veA1. This allele was favored for use at one time as veA1 strains are able to produce asexual spores in growth chambers without the need for lights. It now known that VeA is a light regulated protein that enters the nucleus in dark conditions and that the veA1 mutation truncates the nuclear localization signal and prevents active import of VeA into the nucleus, leading to decreased sexual development and SM production, mimicking growth under light conditions (Stinnett et al., 2007; Bayram et al., 2008). A comparison of veA and veA1 strains has proven useful in elucidating pathways important for both sporulation and SM (Shabaan et al. 2010). Although the exact mechanism of Velvet complex metabolite regulation is currently unknown, mutations in complex members and the resulting metabolism defects have been partially remediated through several pathways, including modulation of chromatin modifications, discussed in further detail below.
Genes required for biosynthesis of SMs are located adjacent to one another within clusters in the genome, leading to the hypothesis that co-regulation of these gene clusters may be achieved through alterations in chromatin structure of the entire cluster (reviewed in Keller and Hohn, 1997, Hoffmeister and Keller, 2007). Initial evidence supporting this idea came from studies of the aflatoxin cluster in Aspergillus parasiticus, where translocation of two different biosynthetic genes outside of the cluster resulted in dramatically reduced gene expression (Liang et al., 1997; Chiou et al., 2002). In A. nidulans, removal of a component of the COMPASS complex component cclA, responsible for histone 3 lysine 4 (H3K4) trimethylation, resulted in induction of the monodictyphenone cluster, producing emodin and several derivatives which were not previously believed to be produced by this species (Bok et al., 2009). The characterization of an antimicrobial activity in the ΔcclA strain, attributed to the emodin derivative 2-hydroxyemodin (Giles et al., 2011), illustrates the newly valued property of drug discovery via manipulation of the histone code of fungal SM clusters (reviewed in Strauss and Reyes-Dominguez, 2011; Cichewicz, 2010).
Interest in modifying histones to increase SM production - or even to awaken silent SM clusters - arose from an initial study where specific chromatin modifiers were found to partially restore the SM defect in strains lacking the aforementioned global regulator LaeA. A. nidulans LaeA deletion strains produce little or no detectable sterigmatocystin (ST), however, deletions of the histone deacetylase (HDAC) hdaA or the heterochromatin protein 1 homolog hepA were shown to individually increase ST as well as restore ST production to the laeA mutant (Shwab et al., 2007; Reyes-Dominguez et al., 2010). Furthermore, this same study showed that treatment with HDAC inhibitors increased production of several unknown SMs in Alternaria alternata and Penicillium expansum (Shwab et al., 2007), leading to series of additional studies employing this same tactic to up regulate SM production in A. niger (Fisch et al., 2009) and A. fumigatus (Nishida et al., 2010; reviewed in Cichewicz, 2010).
As in other organisms, modulation of histone acetylation affects gene expression in the Aspergilli. An initial study examining the aflatoxin gene cluster in A. parasiticus found a temporal link between H4 acetylation and transcription of genes in the cluster. Here, the order in which genes were transcribed reflected the order in which H4 acetylation was detected (Roze et al., 2007). Further examination of this system showed a decrease in transcript of the MYST family acetyltransferase MYST3 under conditions where aflatoxin production was repressed by treatment with volatiles from willow bark, leading the authors to hypothesize that the decrease in MYST3 was implicated in the defect of aflatoxin production (Roze et al., 2011).
Further evidence supporting the role of acetyltransferases in regulation of SM clusters was found for the orsellinic acid gene cluster in A. nidulans. The production of orsellinic acid and derivatives has previously been shown under multiple manipulations (Bok et al., 2009; Sanchez et al., 2009), including co-culture with the bacterium Streptomyces rapamycinicus (Schroeckh, et al., 2009). In order to investigate requirements for induction of this cluster under conditions of co-culture, researchers examined orsellinic acid production under these conditions by strains containing individual deletions of 36 of the 40 acetyltransferases (Nuetzmann et al., 2011). This screen uncovered a requirement for the SAGA complex histone acetyltransferase (HAT) GcnE (Reyes-Dominguez et al., 2008) for induction of this cluster under the conditions tested. Significantly, an increase in both H3K9 and H3K14 acetylation within the cluster, mediated by GcnE, was seen under conditions of co-culture, and thus induction of orsellinic acid. An early increase in acetylation and transcript levels has also been seen for several SM genes involved in fumonisin biosynthesis in Fusarium verticilloides after treatment with the HDAC inhibitor TSA (Visentin et al., 2011).
In light of the evidence supporting a role of histone acetylation in regulation of SM gene clusters, it was compelling for our lab to find a homolog of the Saccharomyces cerevisiae MYST family acetyltransferase Esa1 in a multi-copy suppressor screen of inducers of secondary metabolism. Esa1 and its homologs have been shown to be essential in all eukaryotes examined, including S. cerevisiae (Smith et al., 1998; Clarke et al., 1999), Schizosaccharomyces pombe (Gomez et al., 2008), Drosophila melanogaster (Zhu et al., 2007), and Mus musculus (Hu et al., 2009). Esa1 in yeast has been shown to localize to promoters of active protein encoding genes (Reid et al., 2000; Robert et al., 2004), where it is capable of acetylating multiple sites, most notably histone 4 (H4) lysines 5, 8, and 12 (K5, K8, and K12), and H2B K5 (Smith et al., 1998; Clarke et al., 1999; Suka et al., 2001). As might be expected of an essential protein, this enzyme has been implicated in critical cellular processes, including DNA double strand break repair (Bird et al., 2002), and chromosomal segregation (Clarke et al., 1999; Gomez et al,. 2008). Esa1 is also involved in transcriptional induction (Reid et al., 2000; Nourani et al., 2004) and elongation (Ginsburg et al., 2009), and plays an important role in regulation of subtelomeric genes (Clarke et al., 2006; Zhou et al., 2011).
Since many of the Aspergilli SM gene clusters are positioned in subtelomeric regions, implicated in virulence in the pathogens A. flavus and A. fumigatus and regulated by LaeA (Perrin et al., 2007; Bok et al., 2006; Georgianna et al., 2010), we were particularly interested to see whether alterations in esaA levels would selectively effect expression of SM clusters at different locations across the genome. Here we find a strong correlation of H4K12 levels with SM cluster activation of normally expressed metabolites.
Results
A ΔlaeA suppressor contains a homolog of yeast acetyltransferase ESA1
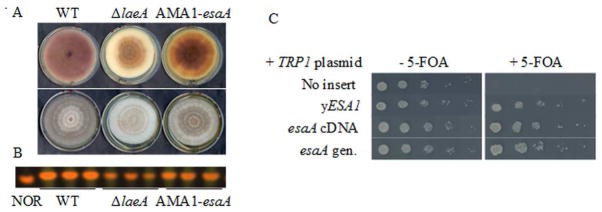
Fourteen unique inserts were previously identified in a multicopy suppressor screen aimed at identifying genes capable of restoring the ST precursor norsolorinic acid (NOR) production to a strain lacking the global SM regulator laeA in a sensitized background (ΔstcE::argB; veA1) (Shaaban et al., 2010). Deletion of stcE not only blocks ST production at a step where the easily visualized precursor NOR accumulates, but also results in a slight leakiness of NOR production in a laeA deletion background, likely due to the inclusion of the primary metabolism gene argB used for stcE disruption (Bok et al., 2009; Shaaban et al., 2010; Reyes-Dominguez et al., 2010). This background therefore allowed us to identify not only genes capable of suppressing the laeA deletion, but also those which increased NOR production independently from LaeA. Genes recovered in this screen covered a number of functional categories, including but not limited to: transcription factors (e.g. RsmA, Shaaban et al., 2010), RNA splicing factors, and chromatin modifiers. The latter group was of particular interest, as reverse genetics had previously identified several chromatin modifiers capable of increasing SM, some of them partially restoring aspects of the ΔlaeA SM defect. These included deletions of homologs of the H3K9 methyltransferase ClrD, the protein binding to H3K9me3 and thereby promoting formation of heterochromatin, HepA heterochromatin protein-1 (Reyes-Dominguez et al., 2010), and the histone deacetylase (HDAC) HdaA (Shwab et al., 2007). One of the inserts identified in the multicopy suppressor screen contained a single gene, AN10956.4, homologous to the Saccharomyces cerevisiae histone acetyltransferase Esa1 (Fig. 1A and 1B, Smith et al., 1998; Clarke et al., 1999).
Figure 1.
A. A multicopy suppressor screen searching for restoration of metabolite production to a ΔlaeA strain uncovered a plasmid containing a homolog of yeast ESA1. Strains containing this plasmid have increased pigmentation relative to the ΔlaeA parent. Strains are: WT - TJMP32.1, ΔlaeA - RJHO26, AMA1 - Suppressor 8. B. Thin layer chromatography of culture extracts confirms increased NOR production in ΔlaeA strains with AMA1-esaA. C. A plasmid shuffle assay confirms the ability of A. nidulans esaA to complement the lethal phenotype of a yeast ESA1 deletion strain. Strains lacking a functional copy of ESA1 or equivalent are unable to grow on media with 5-FOA.
AN10956.4, hereafter referred to as esaA, has 65% identity at the protein level to S. cerevisiae Esa1, and 53% identity to Schizosaccharomyces pombe Mst1, two previously characterized fungal homologs (Fig. S1, Smith et al., 1998; Clarke et al., 1999, Gomez et al., 2008). To determine whether A. nidulans esaA could functionally compensate for S.c.ESA1, a plasmid shuffle assay testing complementation was performed. As deletions of ESA1 homologs are lethal in all model organisms tested, including budding yeast (Smith et al., 1998; Clarke et al., 1999), fission yeast (Gomez et al., 2008), flies (Zhu et al., 2007), and mice (Hu et al., 2009), some form of this enzyme must be present in the cell to allow for survival.
To address this issue, a S. cerevisiae strain lacking genomic ESA1 and harboring a CEN-URA3-S.c.ESA1 plasmid was transformed with plasmids containing a TRP3 auxotrophic marker and either esaA cDNA or genomic DNA. After overnight growth, serial dilutions were transferred to media with or without 5-FOA to select against the CEN-URA3-S.c.ESA1 plasmid. Strains lacking an equivalent copy of S.c.ESA1 failed to growth on media containing 5-FOA, while strains containing S.c.ESA1 or a functional equivalent were able to grow on both media (Fig. 1C). As expected, both forms of esaA vectors were able to rescue the lethality of the yeast deletion, proving not only is esaA able to substitute for the native Esa1 function in this strain, but suggesting the genomic esaA transcript is processed sufficiently subsequent to transcription.
EsaA mutant effects in A. nidulans
In order to confirm the essential nature of EsaA in A. nidulans, a segregation assay was performed. Initially, a strain overexpressing esaA was constructed using pJW53 to target a fusion of esaA under the control of the highly active gpdA promoter to the pyroA locus of the recipient strain RJMP1.59 (Table 1). The resulting strain TAAS16.13 (gpdA(p)::esaA::pyroA4::pyroA; pyrG89) was confirmed by Southern and northern blotting (Figure S2). TAAS16.13 was then mated with DVARI (pabaA1; trpC801; ΔargB::trpC; yA2; ΔveA::argB) to isolate progeny with an additional auxotrophic marker (TAAS44.33: pyroA4::pyroA::gpdA(p)::esaA; ΔargB::trpC; pyrG89). Subsequent deletion of the native esaA copy using A. parasiticus pyrG was performed, yielding strain TAAS48.7 (pyroA4::pyroA::gpdA(p)::esaA; ΔesaA::pyrG; ΔargB::trpC; pyrG89) and mutants were confirmed by PCR and Southern blot (Figure S3). Mating of TAAS48.7 with RJMP101.5 (pyrG89; wA3) resulted in all prototrophic progeny containing a deletion of the native copy of esaA. Further analysis by PCR screening showed that all progeny containing the ΔesaA allele also contained the overexpression allele at the pyroA locus (100/100). In contrast, 48/100 progeny contained the wA3 allele, which is defective in production of conidial pigment, leading to white spores (Mayorga and Timberlake, 1990), confirming recombination between the parental strains. As this confirms the requirement for EsaA for normal growth of A. nidulans, strains overexpressing esaA were selected for further analysis. Desired prototrophs were obtained by PCR screening recombinants between TAAS16.13 and RJW34.1.
Table 1.
Aspergillus nidulans strains used in this study
| Strain name | Genotype | Source |
|---|---|---|
| Suppressor 8 | pyrG89;AMA1::pyr-4::AN10956.4; wA3; argB2; methG1; ΔstcE::argB; trpC801; Δ laeA::methG veA1 | This study |
| RJHO126 | ΔstcE::argB; trpC801; biA1; wA3; veA1 | J. Hicks and N.P. Keller |
| TJMP32.1 | pyrG89; AMA1:pyr-4; wA3; argB2; methG1; ΔstcE::argB; trpC801; ΔlaeA::methG; veA1 | J. Palmer and N.P. Keller |
| RJMP1.59 | pyrG89; pyroA4 | Shaaban et al., 2010 |
| TAAS16.13* | gpdA(p)::esaA::pyroA4::pyroA; pyrG89 | This study |
| DVARI* | pabaA1; trpC801; ΔargB::trpC; yA2; ΔveA::argB | Kim et al., 2002 |
| RAAS44.33 | pyrG89; ΔargB::trpC; pyroA4::pyroA::gpdA(p)::esaA | This study |
| TAAS48.7* | pyrG89; ΔesaA::pyrG; ΔargB::trpC; pyroA4::pyroA::gpdA(p)::esaA | This study |
| RJMP101.5* | pyrG89; wA3 | J. Palmer and N.P. Keller |
| RJW65 | ΔstcE::argB; veA1 | Bok et al., 2009 |
| RAAS22.170 | gpdA(p)::esaA::pyroA4::pyroA; ΔstcE::argB; veA1 | This study |
| RJW81 | ΔstcE::argB; ΔlaeA::methG; veA1 | Bok et al., 2009 |
| RAAS22.174 | gpdA(p)::esaA::pyroA4::pyroA; ΔstcE::argB; ΔlaeA::methG; veA1 | This study |
| RJMP103.5 | J. Palmer and N.P. Keller | |
| RAAS22.98 | gpdA(p)::esaA::pyroA4::pyroA | This study |
| RAAS22.5 | gpdA(p)::esaA::pyroA4::pyroA; ΔlaeA::metG | This study |
| RAAS22.135 | gpdA(p)::esaA::pyroA4::pyroA; veA1 | This study |
| RAAS22.100 | gpdA(p)::esaA::pyroA4::pyroA; ΔlaeA::metG; veA1 | This study |
| RJW34.1* | pyrG1; wA3; argB2; metG1; ΔstcE::argB; trpC801; ΔlaeA::metG; veA1 | Shaaban et al., 2010 |
| RJW41.A | ΔlaeA::metG; metG1 | Bayram et al., 2008 |
| RJW46.4 | ΔlaeA::metG; metG1; veA1 | Bok and Keller, 2004 |
| RDIT2.3 | veA1 | Shaaban et al., 2010 |
| RJMP116.3 | pyrG89; ΔtlhA::A fumigatus pyrG | Palmer et al., 2010 |
| TDP1-1* | pyrG89; riboB2; pyroA4 ΔtlhA::A fumigatus pyrG; veA1 | Palmer et al., 2010 |
| RAAS141.4 | pyrG89; gpdA(p)::esaA::pyroA4::pyroA ΔtlhA::A fumigatus pyrG | This study |
Strains denoted with * were used for sexual crosses to generate additional strains.
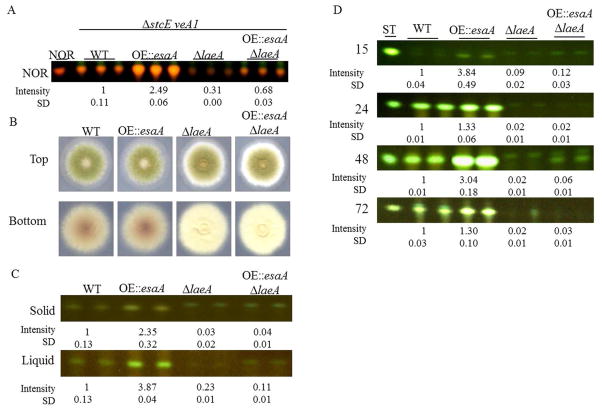
As the gpdA(p)::esaA (OE::esaA) construct used for our overexpression strain may not express esaA as highly as the multicopy AMA1 plasmid identified in the original suppressor screen, we first confirmed the ability of the OE::esaA allele to moderate the ΔlaeA SM defect in the sensitized ΔstcE::argB; veA1 background as used for the original suppressor. TLC analysis confirmed the ability of OE::esaA to not only increase NOR production to WT levels in a ΔlaeA background, but to also significantly increase NOR production in strains with WT laeA (Fig. 2A). As noted earlier, this increase may be due to effects occurring either dependently or independently from LaeA. We were therefore interested in determining the effects of OE::esaA in a wild type background without any mutations which may influence development and SM (such as ΔstcE::argB and veA1). Thus all additional studies utilize the wild type stcE and veA backgrounds unless otherwise noted.
Figure 2.
A. Overexpression of esaA suppresses ΔlaeA NOR production defects in a ΔstcE veA1 background, mimicking the effects of Suppressor 8. Strains are WT – RJW81; OE::esaA – RAAS22.170; ΔlaeA – RJW65; OE::esaA ΔlaeA – RAAS22.174. B. In a veA background, overexpression of esaA has only mild phenotypic effects when incubated in constant light. C. TLC of extracts from cultures from solid GMM after 3 days incubation and liquid GMM after 48 hours under constant light. Shown is the band corresponding to sterigmatocystin (ST). OE::esaA results in increased ST when incubated under light conditions, particularly in liquid shaking culture. Under the conditions tested, OE::esaA is unable to rescue ST production in the ΔlaeA strains. D. Production of sterigmatocystin from liquid shake GMM cultures at 15, 24, 48, and 72 hours. ST is increased at early timepoints under light incubation in liquid shake, and levels remain elevated through longer incubations. Strains used are WT – RJMP103.5; OE::esaA – RAAS22.98; ΔlaeA – RJW41.A; OE::esaA ΔlaeA – RAAS22.5.
Phenotypic analysis of these OE::esaA strains showed little deviation from wild type (Figure 2B). To assess the effects of overexpressing esaA on SM production, TLC analysis was performed under a number of conditions, including solid and liquid minimal media under both constant light and constant dark (Fig. 2C and S4). Under constant light, the OE::esaA strain increased production of ST by 2–4 fold. Under constant dark, this increase was nearly abolished. We had noted similar phenotypes in other SM inducers, e.g. higher induction in light (Shabaan et al. 2010), and found this particular phenomenon was veA dependent. Therefore, for this parameter, we also assessed esaA overexpression in both light and dark in the veA1 background. Interestingly, this increase in ST production occurs regardless of illumination in a veA1 mutant background (Fig. S4), suggesting that efficient increases in ST production by OE::esaA only occur under conditions where VeA is not effectively imported into the nucleus. In contrast to the suppression seen in the leaky ΔstcE background, ST production is not increased in the double mutant relative to ΔlaeA in strains lacking this mutation, independent of veA allele, suggesting that the mechanism of SM production increase is operating separately from LaeA.
Because loss of hdaA had resulted in early ST production (Schwab et al. 2007), we examined the production of this metabolite over several time points in the OE::esaA strains. As shown in Figure 2D for liquid shake growth, ST synthesis was strongly upregulated at all timepoints in the first two days of fungal growth, although the degree of induction varied among timepoints. Increased ST production was correlated with a slight premature induction of sexual development, which was no longer significant at later timepoints (Fig. S4C).
Increasing H4 acetylation leads to increased activation of SM gene clusters that is not location specific
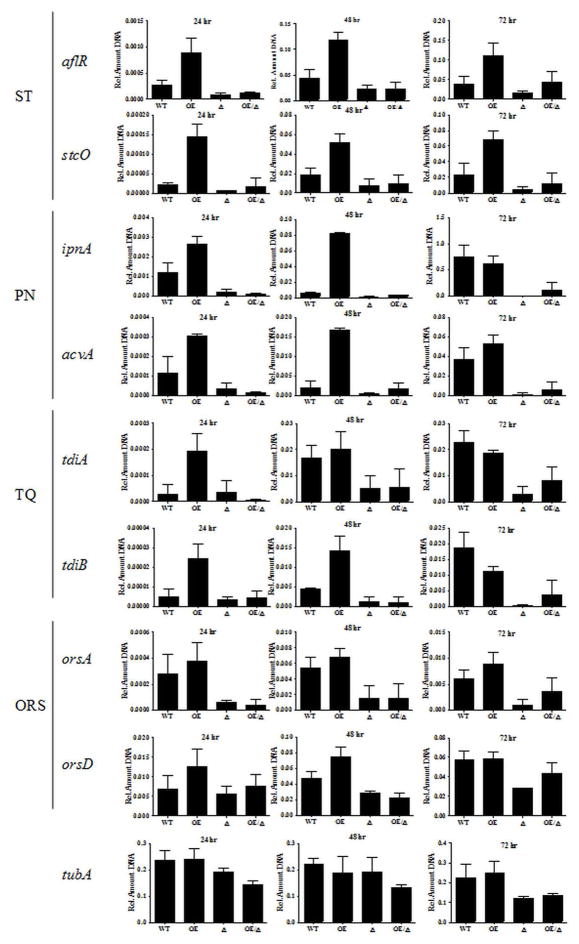
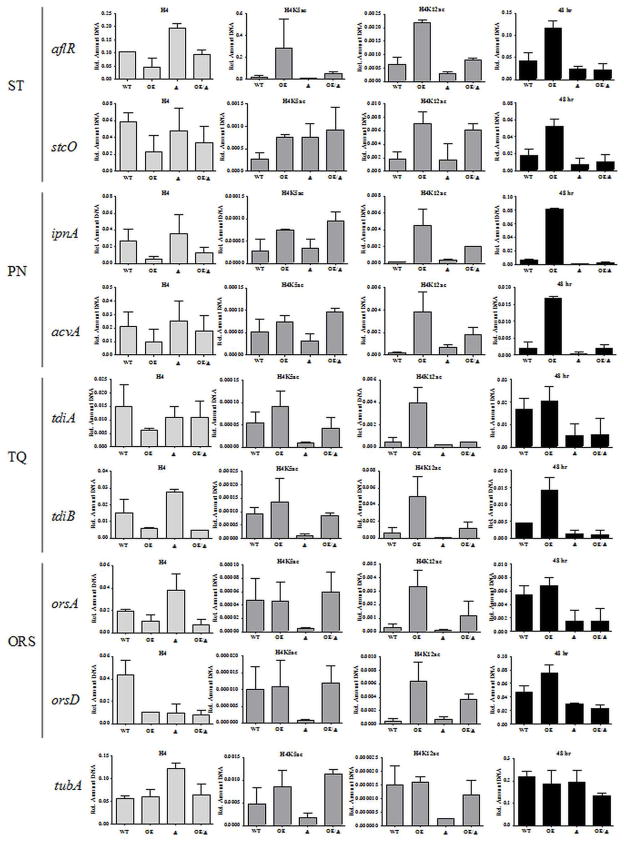
In order to investigate the early increase of ST production seen in OE::esaA, and to further investigate any remediation in the LaeA-null background, qRT-PCR was used to quantify amount of transcript produced at 24, 48, and 72 hour liquid shake (Figure 3). We examined 8 different genes from 4 different SM gene clusters, responsible for producing the compounds sterigmatocystin (ST; aflR and stcO; Keller and Adams, 1995), penicillin (PN; ipnA and acvA; MacCabe et al., 1990), terrequinone (TQ; tdiA and tdiB; Bouhired et al., 2007), and orsellinic acid (ORS; orsA and orsD; Sanchez et al., 2009). These genes were chosen as they had been characterized and assessed in other studies (Fig. S5; Nützmann et al., 2011).
Figure 3.
OE::esaA single mutant strains show increased transcript levels of genes required for secondary metabolite biosynthesis. WT: WT; OE:OE::esaA; Δ:ΔlaeA; OE/Δ:OE::esaA ΔlaeA. Transcript levels of aflR, stcO, ipnA, acvA, tdiA, and tdiB are all increased at 24 hours relative to wild type. At 48 hours this increased has abated for tdiA. At 72 hours, increased transcripts are only seen for aflR and stcO. All genes but those required for orsellinic acid biosynthesis (orsA and orsD) are normally expressed in WT under these conditions. OE::esaA increases transcript levels of these genes at early timepoints, but is unable to stimulate transcription in ΔlaeA mutants. In contrast, OE::esaA single mutant strains show no change in transcript levels of the alpha-tubulin gene tubA. Transcript levels are not significantly different among strains at either 24 or 48 hours. At 72 hours, slightly decreased transcript levels are seen in the ΔlaeA strains. Transcript levels displayed are relative to actin (actA). Error bars represent standard deviation of biological duplicates.
The former 3 clusters are all expressed in wild type under the conditions tested, and have previously been shown to require laeA for induction (Bok et al., 2004). In contrast, orsellinic acid is not produced under standard laboratory conditions and is only activated upon exposure to altered media conditions, chromatin modifiers, or competing organisms (Sanchez et al., 2009; Bok et al., 2009; Schroeckh et al., 2009). These gene clusters range from 3 to 25 genes in size, and are located on 4 of the 8 chromosomes present in A. nidulans. Additionally, 3 of these clusters (ST, PEN, and ORS) are located within 150 kB from the end of the sequence assembly, suggesting that they may be subject to subtelomeric regulation.
Interestingly, increased transcript levels were seen at 6 of the 8 SM genes examined (all except orsA and orsD) at the 24 hour timepoint in the OE::esaA strain. At 48 hours, increased transcript levels in this strain were not apparent for tdiA, but tdiB, the PN genes, and ST genes were still upregulated relative to wild type. For PN and TQ genes, no significant difference was seen at 72 hours, although both aflR and stcO are still overrepresented, suggesting that overexpression of esaA results in further activation of the SM gene clusters that are normally expressed. In agreement with this, no remediation of transcript levels was seen in the double mutant. As a control for normally expressed genes outside of SM clusters, the alpha-tubulin gene tubA was used (Doshi et al., 1991). Expression of tubA was constant among strains at all timepoints examined, although there was a slight decrease in tubA transcript levels in the laeA strains at the 72 hour timepoint.
These results did not support a role for EsaA in transcriptional activation dependent on cluster location, unlike in yeast where ESA1 has been implicated in modulating regulation of certain subtelomeric genes (Clarke et al., 2006; Zhou et al., 2011). To garner further insight into any relationship of EsaA activity and telomere gene regulation, we next assessed any role for EsaA expression in telomere position effect (TPE). A. nidulans possesses a putative TPE system involved in gene repression in areas adjacent to telomere caps that can be remediated by loss of several heterochromatin-associated proteins, particularly HdaA (Palmer et al., 2010). In contrast, introduction of the OE::esaA allele was unable to suppress the growth and sexual defects of a telomere position effect mutant (Fig. S6). This result is consistent with the ability of OE::esaA but not ΔhdaA to upregulate the telomere distal terrequinone gene cluster.
OE::esaA has increased H4 acetylation
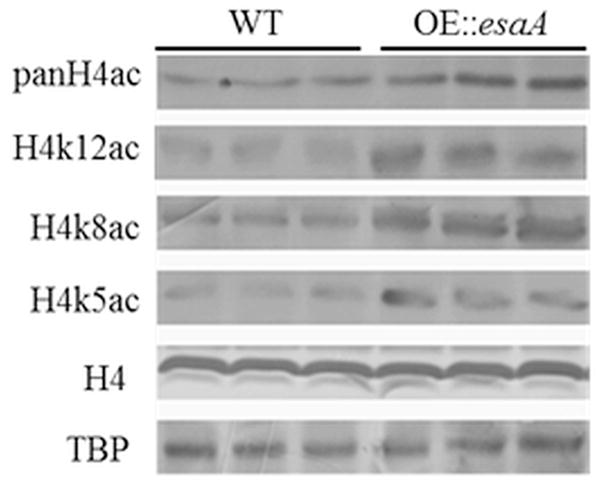
In order to investigate whether the aforementioned increase in transcription was the result of an increase in acetylation, western blotting was performed to determine acetylation levels across the genome. S. cerevisiae Esa1 has previously been described to acetylate histone 4 on residues K5, K8, K12, and K16 in vivo (Smith et al. 1998; Clarke et al., 1999). To address whether the OE::esaA strains showed increased acetylation at these loci, western blotting of total nuclear extracts was performed, examining putative H4 targets of EsaA (Fig. 4). Under constant light and liquid shake conditions, where the OE::esaA showed the largest increase in ST production relative to WT, total acetylation is increased as measured with a pan-acetyl H4 anitbody. H4K5, H4K8, and H4K12 also showed increases, confirming EsaA target specificity. As this method cannot differentiate between large increases of acetylation at specific loci and subtle increases across the whole genome, chromatin immunoprecipitation (ChIP) was used to examine acetylation levels at specific loci.
Figure 4.
Western blotting shows an increase in acetylation of at least 3 predicted targets of EsaA. OE::esaA shows an increased level of total H4 acetylation as measured by anti-pan acetyl H4. This is confirmed by increased in 3 predicted targets of EsaA H4 K5, K8, and K12. H4 and TATA-binding protein (TBP) are used as loading controls.
Overexpression of esaA increases acetylation of SM gene promoters
To further determine if acetylation is specific to distinct regions, ChIP technology was employed to examine the acetylation levels of two different H4 residues at the promoter regions of the 8 previously examined genes (Fig. 5). Promoter regions were examined due to the recruitment of yeast Esa1 to the promoters of protein coding genes and its subsequent acetylation (Robert et al., 2004).
Figure 5.
OE::esaA strains have decreased H4 occupancy and a relative increase in H4K12 acetylation at all SM loci tested. H4K5 acetylation is only increased at the stcO promoter. Acetylation data has been normalized to total H4 in all cases. Transcript levels at the same timepoint are shown for reference. In contrast, tubA acetylation levels are unaffected in the OE::esaA single mutant strain. ΔlaeA single mutant strains have increased levels of H4 at this promoter, and a relative decrease in acetylation. These effects are remediated in the double OE::esaA ΔlaeA strain. Note than in ΔlaeA strains, an increase in acetylation is unable to restore transcript levels. Error bars represent standard deviation of biological duplicates.
Although yeast acetylation of H4 lysine residues K5, K8, and K12 is not differentially targeted by Esa1 (Arnold et al., 2011), our data suggests acetylation levels of H4K5 and H4K12 do not always correlate at the loci examined, with only levels of K12 acetylation consistently being increased relative to wild type. The larger standard deviations associated with H4K5 acetylation levels made these results more difficult to interpret. Most strikingly, OE::esaA strains showed decreased levels of total H4 at all SM gene promoters tested.
Although acetylation levels were increased in the double OE::esaA ΔlaeA strain at most loci examined, there was at best a mild remediation of transcript levels (Fig. 3), supportive of the poor remediation of product formation (Fig. 2 and 6). This suggests that LaeA is acting downstream of or in a different pathway than acetylation during activation of these clusters, and is required for efficient activation of transcription.
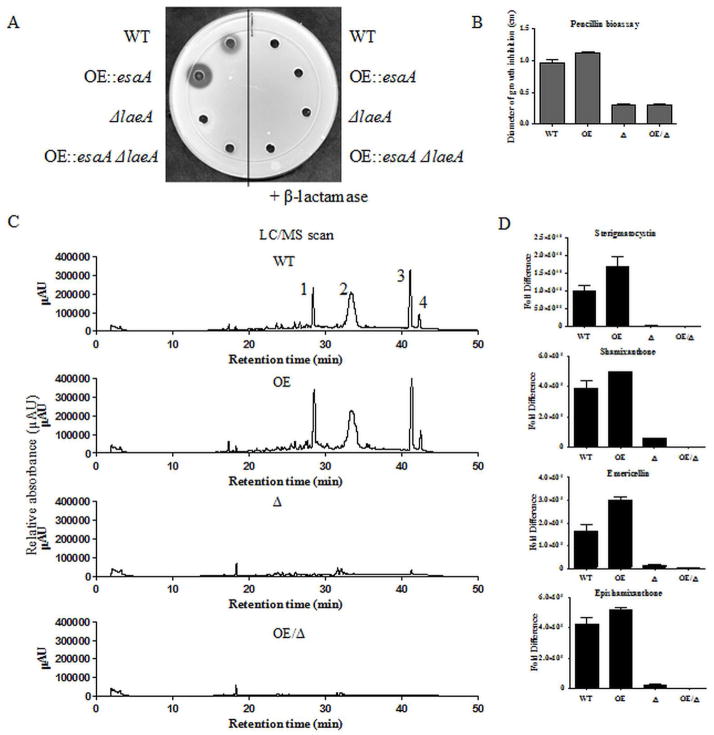
Figure 6.
A. OE::esaA strains show an increased penicillin titer as determined by growth inhibition of the indicator organism Micrococcus luteus. Extracts from 72 hour liquid shake culture were used. No remediation of penicillin production is seen in the double mutant, as was suggested from transcript data. B. Quantification of the zones of inhibition from the penicillin bioassay confirms these results. C. LC data from chloroform extracts of 72 hour liquid shake cultures. OE::esaA shows an increased production of compounds that are normally expressed in the wild type under these conditions. 1. Sterigmatocystin 2. Unknown 3. Shamixanthone and Emericellin 4. Epishamixanthone. D. Quantification of the metabolites seen in C.
As might be expected from a gene encoding a critical structural protein, neither promoter occupancy nor acetylation levels differ drastically at the tubA locus among all 4 strains examined (Fig. 5). A mild increase in H4 levels, and resulting decrease in acetylation, was seen in the ΔlaeA strain, although transcript levels do not differ between this and any other strain.
Increased activation in the OE::esaA strains is correlated with increased metabolite production
As the clusters analyzed range in size from 3 to 25 genes, and only two genes from each cluster were examined, we were interested in determining whether the increases in acetylation and transcript level were reflected by increased metabolite production. Previous TLC analysis supports this case for ST (See Fig. 2), but levels of other metabolites were unknown. In order to determine relative penicillin levels, a penicillin bioassay was performed against the indicator organism Micrococcus luteus. As penicillin is normally detectable from wild type extracts after 72 hours in liquid shaking conditions (Bok and Keller, 2004), all 4 strains were analyzed under this condition. Aqueous extracts from the wild type strain showed a moderate inhibition of M. luteus growth, exhibited by a clear zone of inhibition around the extract containing well (Figure 6A, B). As predicted from our transcript data, the OE::esaA strain produced a larger zone of inhibition, indicating a greater amount of penicillin produced. Both the ΔlaeA and double mutant OE::esaA ΔlaeA strains yielded a barely noticeable ring of inhibition, indicating a low level of penicillin production. For both ST and penicillin, transcript levels at the genes examined were indicative of metabolite production.
To examine whether these early increases in transcript levels resulted in accumulation of other SMs, LC-MS was used to quantify these metabolites. Seventy-two hour cultures were used to allow for accumulation of metabolite from WT cultures. At this timepoint, the previously noted increases in ST levels were quantified, confirming our TLC results (Fig. 6C, D). In addition, the xanthones emericellin, shamixanthone, and epishamixanthone (Sanchez et al., 2011), as well as an uncharacterized metabolite, were also increased in the OE::esaA strain. As predicted, SM levels were not increased in the double mutant relative to the single ΔlaeA strain. Although we were curious if the levels of terrequinone and orsellinic acid also reflected our previous data, these metabolites were below the level of detection for all strains tested under the conditions used, similar to those used for both qRT-PCR and ChIP analysis.
Discussion
Although the field of regulation of SMs in fungal species is developing at a rapid rate, much is still unknown about the mechanisms governing these processes. Perhaps the best characterized class of activators are the SM cluster specific transcription factors. Overexpression of cluster specific transcription factors is often able to activate cluster expression independent of other conditions, as shown for the ST cluster TF aflR, the aspyridone cluster TF apdR, or the asperfuranone TF afoA (Yu et al., 1996; Bergmann et al., 2007; Chiang et al., 2009). Less certain is the mechanism of action of global regulators, such as the Velvet complex, containing VeA, VelB, and LaeA (Bayram et al., 2008; Sarikaya Bayram et al., 2010). For historical and practical reasons (e.g. increased asexual sporulation in dark conditions), many researchers work with strains of A. nidulans containing a mutated veA allele called veA1. This allele has been shown to alter secondary metabolite production, likely due to misplaced cellular placement of the VeA1 protein (Stinnett et al. ,2007, Bayram et al., 2008). Fortuitously, use of this allele improves screening ability in mutagenesis schemes to identify secondary metabolite regulators (Shabaan et al., 2010). Therefore, to gain further insight into possible pathways acting downstream or in addition to the Velvet complex, a multicopy suppressor screen was used to identify genes capable of increasing metabolite production in a laeA deletant created in a highly SM sensitized background (ΔstcE::argB, veA1) where subtle changes in the SM norsolorinic acid could be visualized by eye.
Visual screening of over 30,000 mutants ultimately uncovered numerous classes of genes capable of partially restoring SM production to the laeA mutant under the conditions tested, including the previously published bZIP transcription factor RsmA (Shaaban et al., 2010). Another of these genes was the putative acetyltransferase EsaA, of particular interest because of the link of other chromatin modifiers to ΔlaeA remediation (Shwab et al., 2007; Reyes-Dominguez et al., 2010). As EsaA is essential in all organisms examined, we then chose to focus on a strain with increased expression of this gene. Overexpressing esaA resulted in an approximately 2 fold increase in NOR production in the sensitized ΔstcE::argB; veA1 background. The OE::esaA allele also increased ST production by this factor relative to wild type, although it was unable to restore significant metabolite production to the ΔlaeA mutant in a background lacking the stcE deletion. This is likely due to the leaky SM phenotype of the ΔstcE::argB allele where insertion of the primary metabolite gene argB in the ST cluster modestly increases activation of the cluster region (Bok et al. 2006). This suggests that EsaA is able to modulate the degree of activation of NOR production, although LaeA is likely required for full effect.
The impact of esaA overexpression on ST was similar to those of overexpression of rsmA, where remediation of the ΔlaeA phenotype did not occur under all conditions or timepoints tested, and was particularly sensitive to veA allele (e.g. veA versus veA1) or light vs dark incubation (Shaaban et al., 2010). These results were also reminiscent to those of ClrD where loss of this protein rescued ST production in ΔlaeA background dependent on culture conditions (Reyes-Dominguez et al., 2010). Differences in genetic background, type of overexpression (multi-copy vs. gpdA promoter fusion), and different environmental conditions all may contribute to the ability of esaA and other SM regulators to suppress the laeA SM defect.
Further analysis of our mutant series showed an early increase in ST production (interestingly associated with precocious sexual production, Fig. S4C, also regulated by the Velvet complex), leading us to investigate expression of multiple SM gene clusters at different timepoints, selected based on both location and normal expression. Our data show the OE::esaA strain exhibiting increased transcript levels of normally transcribed SM genes than wild type at early timepoints, independent of genome location. Sterigmatocystin, penicillin, and terrequinone all followed this pattern, with slight differences regarding the time at which this increase was no longer significant. These compounds are all normally produced under the culture conditions used in our experiments, with temporal differences in accumulation which could reflect the times at which the OE::esaA no longer stimulated increased transcript levels. In contrast, orsellinic acid, which is not normally expressed under our culture conditions, showed no change in transcript levels relative to wild type.
While S. cereviseae Esa1 plays an important role in regulation of subtelomeric genes (Clarke et al., 2006; Zhou et al., 2011), our data suggests A. nidulans EsaA displays no genome location specificity, at least in regards to SM clusters, contrasting with data from loss of HdaA, where HdaA was important in regulation of the two subtelomeric clusters, ST and penicillin, but not the telomere distal terrequinone cluster. Consistent with these secondary metabolism effects, ΔhdaA, but not OE::esaA, mediates TPE activity in A. nidulans. These findings continue to support differences in chromatin regulation of gene activity in budding yeast versus filamentous fungi.
As EsaA is a predicted histone 4 acetyltransferase, we then investigated whether we could detect increased acetylation at the expected target residues (Smith et al., 1998, Clarke et al., 1999). Global acetylation analysis by western blotting showed increased acetylation of lysine residues on histone 4, confirming this hypothesis. We then chose to examine the acetylation levels at promoters of the previously studied genes. Interestingly, at all SM loci examined, including the poorly expressed orsellinic acid cluster, histone 4 occupancy was decreased in the OE::esaA strain. However, this was not true of the housekeeping tubA gene. In many other characterized systems, high levels of transcription have previously been associated with nucleosome depletion at promoter regions (reviewed in Bai and Morozov, 2010), suggesting that this may be a contributing factor to the changes seen in transcript levels of SM genes. These differences are no longer significant at the highly transcribed gene tubA, suggesting that transcription may already be optimized. At the SM loci examined, a large relative increase in H4K12 acetylation in the OE::esaA strains was also seen. H4K5 acetylation levels were more varied, but appeared to follow similar acetylation patterns as H4K12. Because of these differences, we propose H4K12 levels may represent a unique indicator of relative induction potential in fungi, particularly of SMs. It is interesting that this same mark, H4K12ac, contributes to telomere plasticity in budding yeast (Zhou et al., 2011). Further studies may uncover additional genes also subject to stimulation of transcription by increased H4 acetylation.
In yeast, Esa1 dependent histone acetylation has been shown to promote binding of the RSC and SWI/SNF chromatin remodeling complexes, as well as facilitate recruitment of TFIID (Hassan et al., 2001; Hassan et al., 2002; Carey et al., 2006; Uprety et al., 2011). In a similar fashion, A. nidulans EsaA may be depositing acetylation marks to loosen chromatin structure and aid in the recruitment of effector complexes required for transcription. When these effector complexes, which may include chromatin remodeling complexes, the Velvet complex, or cluster specific transcription factors, are present, increased acetylation aids in their recruitment and facilitates increased transcription. In contrast, for genes not normally transcribed, due to inappropriate culture conditions or lack of necessary effector components, deposition of acetyl groups still occurs, but transcriptional activation is blocked at a downstream step (Fig. 7). This is supported by the fact that overexpression of esaA is unable to rescue the SM defects of either ΔveA or ΔaflR strains (data not shown), and the poor restoration of ΔlaeA SM defects (Figure 2), confirming the action of these regulators either downstream of acetylation, or within an additional pathway essential for SM activation.
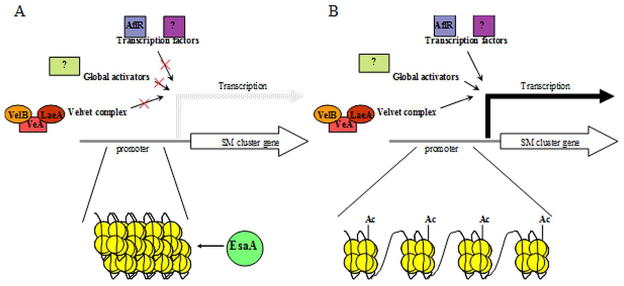
Figure 7.
Model for action of EsaA acetylation. A. Prior to induction of gene clusters, condensed chromatin structure similar to heterochromatin is present, preventing access of effectors facilitating downstream activation of SM gene clusters, including the Velvet complex and other global effectors and transcription factors. B. After acetylation by EsaA, chromatin structure relaxes, allowing the induction of clusters by other effectors.
Evidence from our experiments supports this model, with the normally expressed sterigmatocystin, penicillin, and terrequinone clusters exhibiting increased histone acetylation and increased transcript levels relative to wild type, particularly at earlier timepoints. In ΔlaeA strains, this increase in acetylation is insufficient to restore transcripts to wild type levels. As seen from analysis of SMs by both a penicillin bioassay and LC-MS, normally produced metabolites are present at markedly higher levels in the OE::esaA strain, suggesting increased expression and action of the respective gene clusters and enzymes.
The orsellinic acid cluster, in contrast, is not normally activated under standard laboratory conditions. When a wild type strain was subjected to 20 culture conditions, only incubation in stationary liquid Czapek media resulted in detectable production of orsellinic acid and its derivatives (Sanchez et al., 2009). Removal of an enzyme encoding a component of the H3K4 methyltransferase complex, cclA, resulted in similar production of orsellinic acid. Activation of this cluster could also be achieved by co-culture with the bacterium Streptomyces rapamycinicus, and was shown to be dependent on the H3 acetyltransferase Gcn5 (Schroeckh et al., 2009; Nützmann et al., 2011), suggesting that this cluster may be turned on in response to external environmental stimuli in a mechanism mediated by different chromatin modifiers. As the exact mechanism of ORS cluster regulation remains unknown, it is difficult to postulate which factors may be required for induction of orsellinic acid production under standard culture conditions. We can predict if these factors were present in sufficient levels to allow orsellinic acid production, activation would also be facilitated by the esaA overexpession, as seen for the sterigmatocystin, penicillin, and terrequinone clusters.
Similarly, OE::esaA was able to suppress the ΔlaeA metabolism defect in a ΔstcE mutant background (where the primary metabolism gene argB has replaced the stcE gene, Bok et al. 2006), which shows low levels of NOR accumulation, but not a WT background, which does not show accumulation of ST. Taken together, these results suggest that increasing H4 acetylation is capable of enhancing inherent activation of genes. This increase in acetylation is not sufficient to activate silent genes, but facilitates activation in the face of proper stimuli. Supporting this, OE::esaA increases in ST production are not significant under dark conditions, where the Velvet complex is efficiently imported into the nucleus to stimulate SM production.
To summarize, we have uncovered a pathway involving the histone 4 acetyltransferase EsaA in increased activation of SM gene clusters. Increased acetylation as deposited by this enzyme results in increased transcript levels at expressed loci, relative to wild type. Increased acetylation may also facilitate activation of additional genes relative to wild type, increasing products which may contribute to fungal fitness. Although the exact effectors recognizing the alterations in acetylation and acting downstream are still currently unknown, evidence from yeast suggests the recruitment of additional chromatin modifying complexes or other effectors to be necessary (Hassan et al., 2001B). In agreement with this, an intact Velvet complex, including LaeA, most efficiently translates the acetylation signal into the noted increased transcript levels. Further experiments will shed light onto additional mechanisms governing the noted changes.
Experimental Procedures
esaA sequence analysis
Genomic DNA sequence and translation of AN10956.4 (esaA) gene was obtained from the Aspergillus Comparative Sequencing Project, Broad Institute of Harvard and MIT (http://www.broadinstitute.org/). Protein sequence alignment of EsaA to Saccharomyces cerevisiae and Schizosaccharomyces pombe Mst1 was done using the Clustal W method (Thompson et al., 1994) in the MegAlign module (v. 8.0.2) of Lasergene sequence analysis package (DNASTAR INC, Madison, WI).
Culture conditions, Southern, and northern analysis
All strains (Table 1) were propagated at 37 °C on glucose minimum medium (GMM) with appropriate supplements. Fungal DNA was isolated as previously described (Shimizu and Keller 2001). DNA manipulations, Southern, and northern analysis were conducted according to standard procedures (Sambrook and Russell 2001).
Complementation of yESA1 esaA cDNA was amplified from an A. nidulans vegetative cDNA library (Cho et al., 2003) using primers esaA cDNA 5′ EcoRI and esaA 3′ EcoRI. Genomic esaA was amplified from genomic DNA using primers esaA w esaA UTR 5′ and esaA 3′ EcoRI. Double joint PCR (Yu et al., 2004) was used to fuse each fragment to the yeast ESA1 promoter sequence previously amplified from S. cerevisiae genomic DNA using either ESA1(p) EcoRI and ESA1(p) 3′ esaA (esaA cDNA) or ESA1(p) EcoRI and ESA1(p) w esaA UTR 3′ (genomic esaA). Resulting double joint fragments were cloned into pLP61 (Clarke et al., 1999) using EcoRI. The resulting plasmids pAAS23 and pAAS29 were confirmed by restriction enzyme digestions and sequencing.
pLP61, pLP798, pAAS23, and pAAS29 were transformed into LPY2641 (Clarke et al., 1999). Transformants were grown overnight at 30°C in liquid shaking culture in SD medium (Sherman, 2002). Tenfold dilutions were plated on both solid SD and SD + 5-FOA (0.1%) + uracil (20 mg/L) and grown for 3 days at 30°C.
Construction of mutant esaA strains
esaA coding sequence and approximately 500 bp downstream was amplified from genomic DNA using primers esaA 5′ for and esaA 3′ flank rev NotI. Double joint PCR (Yu et al., 2004) was used to fuse the resulting fragment to a 1.5 kb fragment of the gpdA promoter, previously amplified using gpdF and gpdA(p) 3′ esaA. The resulting fragment was cloned using NotI into pJW53 (Tsitsigiannis et al., 2004), which contains ¾ of the A. nidulans pyroA gene. Plasmids were confirmed by restriction enzyme digestion and sequencing before transformation of RJMP1.59 as previously described (Miller et al., 1985), with the minor modification of protoplasts being plated in 0.75% molten top agar. Transformants were examined for targeted integration at the pyroA locus using PCR and Southern blotting (Fig. S2A). Overexpression of esaA was confirmed by northern blot (Fig. S2B). Prototrophic overexpression strains were obtained by crossing the transformant TAAS16.13 with RJW34.1. Desired recombinants were confirmed by PCR screening.
1 kb of esaA flanking regions were amplified and fused to A. parasiticus pyrG from pJW24 (Calvo et al., 2004) using double joint PCR (Yu et al., 2004). The resulting knockout construct was transformed into RAAS44.33 (pyrG89; ΔargB::trpC; pyroA4::pyroA::gpdA(p)::esaA obtained from a mating of TAAS16.13 and DVAR1 (Kim et al., 2004)). Transformants were examined for targeted replacement of the native esaA locus by PCR and Southern blotting (Fig. S3). Prototrophic esaA deletion strains were obtained by crossing the transformant TAAS48.7 with RJMP101.5. The genotypes of 100 recombinant progeny were examined by PCR, confirming 100% co-segregation of the ectopic overexpression allele with the native esaA knockout. Segregation of the wA3 allele was tested phenotypically as a control for successful recombination.
Phenotypic characterization and SM analysis on solid media
SM production was assessed by thin-layer chromatography (TLC). For TLC, 10 μl of 1 × 103 spore/μl was point-inoculated on the center of glucose minimal medium (GMM) and cultured for 72 hr at 37°C. An agar plug of the center of colonies was removed and SMs extracted with ethyl acetate according to the Smedsgaard’s method (Smedsgaard, 1997). Extracts (10 μl/sample) were loaded onto silica TLC plates (Whatman, PE SIL, Maidstone, Kent, England) and metabolites were separated in the developing solvent toluene:ethyl acetate:glacial acetic acid (TEA, 8:1:1). Images were taken following exposure to UV radiation at 366 nm.
Analysis of sexual spore production
Prototrophic strains in a wild type veA genetic background were used for quantification of ascospores. Quantification was performed on overlay inoculated cultures set up by pipetting 1 × 106 conidia into CHAMPS medium with 0.75% molten agar that was subsequently poured over 1.5% solid agar petri dishes. Cultures were incubated at 37°C in the dark for 4 days and agar cores were taken from the plates with a 1 cm cork borer. After homogenization, ascospores were quantified using a hemacytometer and represented as ascospores per square millimeter. 4 replicates were performed for each strain and condition.
Northern and qRT-PCR analysis
Fifty milliliter cultures of liquid GMM were inoculated with 1 × 106 spores per ml and incubated at 250 rpm and 37°C for 24, 48, and 72 hours under light. Mycelia were harvested, lyophilized overnight, and total RNA was extracted using Isol-RNA Lysis Reagent (5 Prime) according to manufacturer’s recommendations. Subsequent northern analysis was done using radiolabeled probes for the corresponding transcript (primers are listed in Table S1).
For qRT-PCR analysis, 10 Pg RNA was digested with DNAse I (NEB Cat# M0303L) to remove any contaminating genomic DNA. cDNA synthesis reactions were performed using the Bio-Rad iScript™ cDNA Synthesis Kit (Cat# 170-8891) according to manufacturer’s protocols. Amplification and detection of cDNA in real-time qPCR was performed with iQ™ SYBR® Green Supermix #170-8880 (Bio-Rad) following the manufacturer’s instructions, using 50ng cDNA per reaction. To verify that the qPCR products were of the correct size, 10 μl of each reaction was run on a 2% agarose gel stained with ethidium bromide and visualized under UV light. All transcripts were normalized relative to actin (actA; Upadhyay et al., 2008) transcript quantities. qPCR primers are listed in Table S2.
Western analysis of histone modifications
250 milliliter cultures of liquid GMM were inoculated with 1 × 106 spores per ml and incubated at 250 rpm and 37°C for 48 hours, after which nuclear extracts were isolated as previously described (Palmer, 2008). Approximately 50 μg of nuclear protein extract was electrophoresed on a 10% Tricine-SDS-PAGE gel (Shaegger, 2006) and subsequently electroblotted to nitrocellulose membranes. Detection of histone modifications was conducted using the following primary antibodies from Abcam, Cambridge, UK: rabbit polyclonal to histone H4 acetyl K5 (ab51997, 1:2,500), rabbit polyclonal to histone H4 acetyl K8 (ab15823, 1:2,500), rabbit polyclonal to histone H4 acetyl K12 (ab1761, 1:2,500), rabbit polyclonal to human C-terminus histone H4 antibody (ab10158, 1:2,500), and mouse monoclonal to TBP (ab61411, 1:1000), and rabbit polyclonal to anti pan acetyl H4 (Active Motif, #39925, 1:1000), Colorimetric detection was employed using 1-Step NBT/BCIP (Thermo Scientific, #34042) and a secondary goat anti-rabbit alkaline phosphatase conjugated antibody (Pierce, #31342) diluted 1:5,000 or secondary goat anti-mouse alkaline phosphatase conjugated antibody (Gibco, #13864-012) diluted 1:5,000. Triplicate biological cultures were performed.
Chromatin immunoprecipitation and real time qPCR analysis
250 milliliter cultures of liquid GMM were inoculated with 1 × 106 spores per ml and incubated at 250 rpm and 37°C for 48 hours under light. Duplicate cultures were performed for each strain. Chromatin immunoprecipitation was carried out as described previously (Bernreiter et al., 2007). Antibodies used for ChIP were purchased from Abcam, Cambridge, UK (rabbit polyclonal to histone H4 acetyl K5, ab51997, rabbit polyclonal to histone H4 acetyl K12, ab1761, and rabbit polyclonal to human C-terminus histone H4 antibody, ab10158). Two micrograms of antibody was used per reaction of 200 mg total protein. Amplification and detection of precipitated DNA in real-time qPCR was performed with iQ™ SYBR® Green Supermix (Bio-Rad, Cat#170-8880) following the manufacturer’s instructions. Relative amounts of DNA were calculated by dividing the immunoprecipitated DNA by the input DNA. Each PCR reaction was replicated. To normalize the amount of DNA precipitated with histone H4-acetyl K5 and H4-acetyl K12, the quantities from precipitation with these antibodies was divided by the previously calculated ratio of the anti-C-terminus histone H4 precipitation to input DNA. Two biological repeats were performed for each strain and standard deviation was calculated from these. qPCR primers are listed in Table S2.
Liquid culture SM and quantification of metabolites
Fifty milliliter cultures of liquid GMM were inoculated with 1 × 106 spores per ml and incubated at 250 rpm and 37°C for 15, 24, 48, and 72 hours. For small scale SM extraction, 30 ml ethyl acetate was added to 50 ml whole culture and the mixture was agitated for 30 min at room temperature. The ethyl acetate extraction was then dried completely at room temperature and resuspended in 0.1 ml ethyl acetate. TLCs were performed as described above for solid culture. ST intensity was quantified using ImageJ software.
For the penicillin bioassay, 50 milliliter cultures of liquid GMM were inoculated with 1 × 106 spores per ml and incubated at 250 rpm and 37°C for 72 hours under light. 10 ml culture was used to perform the bioassay against Micrococcus luteus as described in Bok and Keller, 2004. For large scale extraction for quantification, 250 milliliter cultures of liquid GMM were inoculated with 1 × 106 spores per ml and incubated at 250 rpm and 37°C for 72 hours under light. For SM extraction, 250 ml chloroform was added to 250 ml whole culture and the mixture was agitated for 30 min at room temperature. The chloroform layer was removed and concentrated using a Buchi Rotavapor R-210. The dried residue was then re-dissolved in 0.2 ml of DMSO:MeOH (1:4). After filtration, 10 μl of DMSO/MeOH extract was injected for high performance liquid chromatography-photodiode array detection- mass spectrometry (HPLC-DAD-MS) analysis as described previously (Bok et al., 2009). For quantitation fold differences of metabolites, electrospray ionization (ESI) in positive mode was used for the detection of sterigmatocystin, emericellin, shamixanthone, and epishamixanthone by using extracted ion chromatogram at m/z 325, 409, 389, and 389, respectively. The fold differences were calculated according to the following formula:
Supplementary Material
Table 2.
Yeast strains and plasmids used in this study
| LPY2641 | MATa his3Δ200 leu2-3,112 trp1Δ1 ura3-52 esa1Δ::HIS3 bearing an ESA1-URA3-CEN plasmid | Clarke et al., 1999 |
| pLP61 | TRP1-2μm | Clarke et al., 1999 |
| pLP798 | TRP1-2μm-ESA1 | Clarke et al., 1999 |
| pAAS29 | TRP1-2 μm-esaA (genomic) | This study |
| pAAS30 | TRP1-2 μm-esaA (cDNA) | This study |
| pJW24 | A. parasiticus pyrG in pBSSK- | Calvo et al., 2004 |
| pJW53 | 3/4 pyroA in pBSSK- | Tsitsigiannis et al., 2004 |
S. cerevisiae strains and additional plasmids used for these studies.
Acknowledgments
This research was funded by NIH PO1 GM084077 to B.R.O., C.C.C.W., and N.P.K., NIH T32 GM07133 to A.A.S., and by NIH NRSA AI55397 to A.A.S.. Work in Vienna was funded by grant LS09-042 of the Vienna Science and Technology Fund (WWTF) and grant S-10003-B17 of the Austrian Science Fund to J.S.. A.A.S. would like to thank Lorraine Pillus for graciously providing the yeast vectors and strain used.
References
- Allard S, Utley RT, Savard J, Clarke A, Grant P, Brandl CJ, Pillus L, Workman JL, Côté J. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J. 1999;18:5108–5119. doi: 10.1093/emboj/18.18.5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold KM, Lee S, Denu JM. Processing mechanism and substrate selectivity of the core NuA4 histone acetyltransferase complex. Biochemistry. 2011;50:727–737. doi: 10.1021/bi101355a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L, Morozov AV. Gene regulation by nucleosome positioning. Trends Genet. 2010;26:476–483. doi: 10.1016/j.tig.2010.08.003. [DOI] [PubMed] [Google Scholar]
- Bayram O, Krappmann S, Ni M, Bok JW, Helmstaedt K, Valerius O, Braus-Stromeyer S, Kwon NJ, Keller NP, Yu JH, Braus GH. VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science. 2008;320:1504–1506. doi: 10.1126/science.1155888. [DOI] [PubMed] [Google Scholar]
- Bennett JW. Mycotoxins, mycotoxicoses, mycotoxicology and Mycopathologia. Mycopathologia. 1987;100:3–5. doi: 10.1007/BF00769561. [DOI] [PubMed] [Google Scholar]
- Bergmann S, Schümann J, Scherlach K, Lange C, Brakhage AA, Hertweck C. Genomics-driven discovery of PKS-NRPS hybrid metabolites from Aspergillus nidulans. Nat Chem Biol. 2007;3:213–217. doi: 10.1038/nchembio869. [DOI] [PubMed] [Google Scholar]
- Bird AW, Yu DY, Pray-Grant MG, Qiu Q, Harmon KE, Megee PC, Grant PA, Smith MM, Christman MF. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature. 2002;419:411–415. doi: 10.1038/nature01035. [DOI] [PubMed] [Google Scholar]
- Bok JW, Chiang YM, Szewczyk E, Reyes-Dominguez Y, Davidson AD, Sanchez JF, Lo HC, Watanabe K, Strauss J, Oakley BR, Wang CC, Keller NP. Chromatin-level regulation of biosynthetic gene clusters. Nat Chem Biol. 2009;5:462–464. doi: 10.1038/nchembio.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bok JW, Keller NP. LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot Cell. 2004;3:527–535. doi: 10.1128/EC.3.2.527-535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bok JW, Noordermeer D, Kale SP, Keller NP. Secondary metabolic gene cluster silencing in Aspergillus nidulans. Mol Microbiol. 2006;61:1636–1645. doi: 10.1111/j.1365-2958.2006.05330.x. [DOI] [PubMed] [Google Scholar]
- Butchko RA, Adams TH, Keller NP. Aspergillus nidulans mutants defective in stc gene cluster regulation. Genetics. 1999;153:715–720. doi: 10.1093/genetics/153.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo AM, Bok J, Brooks W, Keller NP. veA is required for toxin and sclerotial production in Aspergillus parasiticus. Appl Environ Microbiol. 2004;70:4733–4739. doi: 10.1128/AEM.70.8.4733-4739.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo AM, Wilson RA, Bok JW, Keller NP. Relationship between secondary metabolism and fungal development. Microbiol Mol Biol Rev. 2002;66:447–459. doi: 10.1128/MMBR.66.3.447-459.2002. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey M, Li B, Workman JL. RSC exploits histone acetylation to abrogate the nucleosomal block to RNA polymerase II elongation. Mol Cell. 2006;24:481–487. doi: 10.1016/j.molcel.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang YM, Szewczyk E, Davidson AD, Keller N, Oakley BR, Wang CC. A gene cluster containing two fungal polyketide synthases encodes the biosynthetic pathway for a polyketide, asperfuranone, in Aspergillus nidulans. J Am Chem Soc. 2009;131:2965–2970. doi: 10.1021/ja8088185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou CH, Miller M, Wilson DL, Trail F, Linz JE. Chromosomal location plays a role in regulation of aflatoxin gene expression in Aspergillus parasiticus. Appl Environ Microbiol. 2002;68:306–315. doi: 10.1128/AEM.68.1.306-315.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho JH, Yun SS, Jang YK, Cha MJ, Kwon NJ, Chae SK. Identification and Cloning of jipA Encoding a Polypeptide That Interacts with a Homolog of Yeast Rad6, UVSJ in Aspergillus nidulans. The Journal of Microbiology. 2003;41:46–51. [Google Scholar]
- Cichewicz RH. Epigenome manipulation as a pathway to new natural product scaffolds and their congeners. Nat Prod Rep. 2010;27:11–22. doi: 10.1039/b920860g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke AS, Samal E, Pillus L. Distinct roles for the essential MYST family HAT Esa1p in transcriptional silencing. Mol Biol Cell. 2006;17:1744–1757. doi: 10.1091/mbc.E05-07-0613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doshi P, Bossie CA, Doonan JH, May GS, Morris NR. Two alpha-tubulin genes of Aspergillus nidulans encode divergent proteins. Mol Gen Genet. 1991;225:129–141. doi: 10.1007/BF00282651. [DOI] [PubMed] [Google Scholar]
- Fisch KM, Gillaspy AF, Gipson M, Henrikson JC, Hoover AR, Jackson L, Najar FZ, Wägele H, Cichewicz RH. Chemical induction of silent biosynthetic pathway transcription in Aspergillus niger. J Ind Microbiol Biotechnol. 2009;36:1199–1213. doi: 10.1007/s10295-009-0601-4. [DOI] [PubMed] [Google Scholar]
- Georgianna DR, Fedorova ND, Burroughs JL, Dolezal AL, Bok JW, Horowitz-Brown S, Woloshuk CP, Yu J, Keller NP, Payne GA. Beyond aflatoxin: four distinct expression patterns and functional roles associated with Aspergillus flavus secondary metabolism gene clusters. Mol Plant Pathol. 2010;11:213–226. doi: 10.1111/j.1364-3703.2009.00594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles SS, Soukup AA, Lauer C, Shaaban M, Lin A, Oakley BR, Wang CC, Keller NP. Cryptic Aspergillus nidulans antimicrobials. Appl Environ Microbiol. 2011;77:3669–3675. doi: 10.1128/AEM.02000-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsburg DS, Govind CK, Hinnebusch AG. NuA4 lysine acetyltransferase Esa1 is targeted to coding regions and stimulates transcription elongation with Gcn5. Mol Cell Biol. 2009;29:6473–6487. doi: 10.1128/MCB.01033-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene AV, Keller N, Haas H, Bell-Pedersen D. A circadian oscillator in Aspergillus spp. regulates daily development and gene expression. Eukaryot Cell. 2003;2:231–237. doi: 10.1128/EC.2.2.231-237.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez EB, Nugent RL, Laria S, Forsburg SL. Schizosaccharomyces pombe histone acetyltransferase Mst1 (KAT5) is an essential protein required for damage response and chromosome segregation. Genetics. 2008;179:757–771. doi: 10.1534/genetics.107.085779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock LC, Behta RP, Lopes JM. Genomic analysis of the Opi- phenotype. Genetics. 2006;173:621–634. doi: 10.1534/genetics.106.057489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan AH, Neely KE, Workman JL. Histone acetyltransferase complexes stabilize swi/snf binding to promoter nucleosomes. Cell. 2001;104:817–827. doi: 10.1016/s0092-8674(01)00279-3. [DOI] [PubMed] [Google Scholar]
- Hassan AH, Prochasson P, Neely KE, Galasinski SC, Chandy M, Carrozza MJ, Workman JL. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell. 2002;111:369–379. doi: 10.1016/s0092-8674(02)01005-x. [DOI] [PubMed] [Google Scholar]
- Hu Y, Fisher JB, Koprowski S, McAllister D, Kim MS, Lough J. Homozygous disruption of the Tip60 gene causes early embryonic lethality. Dev Dyn. 2009;238:2912–2921. doi: 10.1002/dvdy.22110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kale SP, Milde L, Trapp MK, Frisvad JC, Keller NP, Bok JW. Requirement of LaeA for secondary metabolism and sclerotial production in Aspergillus flavus. Fungal Genet Biol. 2008;45:1422–1429. doi: 10.1016/j.fgb.2008.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato N, Brooks W, Calvo AM. The expression of sterigmatocystin and penicillin genes in Aspergillus nidulans is controlled by veA, a gene required for sexual development. Eukaryot Cell. 2003;2:1178–1186. doi: 10.1128/EC.2.6.1178-1186.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller NP, Adams TH. Analysis of a mycotoxin gene cluster in Aspergillus nidulans. SAAS Bull Biochem Biotechnol. 1995;8:14–21. [PubMed] [Google Scholar]
- Keller NP, Hohn TM. Metabolic Pathway Gene Clusters in Filamentous Fungi. Fungal Genet Biol. 1997;21:17–29. [PubMed] [Google Scholar]
- Kim H, Han K, Kim K, Han D, Jahng K, Chae K. The veA gene activates sexual development in Aspergillus nidulans. Fungal Genet Biol. 2002;37:72–80. doi: 10.1016/s1087-1845(02)00029-4. [DOI] [PubMed] [Google Scholar]
- Lafon A, Chang CS, Scott EM, Jacobson SJ, Pillus L. MYST opportunities for growth control: yeast genes illuminate human cancer gene functions. Oncogene. 2007;26:5373–5384. doi: 10.1038/sj.onc.1210606. [DOI] [PubMed] [Google Scholar]
- Liang SH, Wu TS, Lee R, Chu FS, Linz JE. Analysis of mechanisms regulating expression of the ver-1 gene, involved in aflatoxin biosynthesis. Appl Environ Microbiol. 1997;63:1058–1065. doi: 10.1128/aem.63.3.1058-1065.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayorga ME, Timberlake WE. Isolation and molecular characterization of the Aspergillus nidulans wA gene. Genetics. 1990;126:73–79. doi: 10.1093/genetics/126.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCabe AP, Riach MB, Unkles SE, Kinghorn JR. The Aspergillus nidulans npeA locus consists of three contiguous genes required for penicillin biosynthesis. EMBO J. 1990;9:279–287. doi: 10.1002/j.1460-2075.1990.tb08106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida H, Motoyama T, Yamamoto S, Aburatani H, Osada H. Genome-wide maps of mono- and di-nucleosomes of Aspergillus fumigatus. Bioinformatics. 2009;25:2295–2297. doi: 10.1093/bioinformatics/btp413. [DOI] [PubMed] [Google Scholar]
- Nourani A, Utley RT, Allard S, Côté J. Recruitment of the NuA4 complex poises the PHO5 promoter for chromatin remodeling and activation. EMBO J. 2004;23:2597–2607. doi: 10.1038/sj.emboj.7600230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nützmann HW, Reyes-Dominguez Y, Scherlach K, Schroeckh V, Horn F, Gacek A, Schümann J, Hertweck C, Strauss J, Brakhage AA. Bacteria-induced natural product formation in the fungus Aspergillus nidulans requires Saga/Ada-mediated histone acetylation. Proc Natl Acad Sci U S A. 2011;108:14282–14287. doi: 10.1073/pnas.1103523108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda K, Kobayashi A, Ohashi S, Sano M. Aspergillus oryzae laeA regulates kojic acid synthesis genes. Biosci Biotechnol Biochem. 2011;75:1832–1834. doi: 10.1271/bbb.110235. [DOI] [PubMed] [Google Scholar]
- Palmer JM, Mallaredy S, Perry DW, Sanchez JF, Theisen JM, Szewczyk E, Oakley BR, Wang CC, Keller NP, Mirabito PM. Telomere position effect is regulated by heterochromatin-associated proteins and NkuA in Aspergillus nidulans. Microbiology. 2010;156:3522–3531. doi: 10.1099/mic.0.039255-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer JM, Perrin RM, Dagenais TR, Keller NP. H3K9 methylation regulates growth and development in Aspergillus fumigatus. Eukaryot Cell. 2008;7:2052–2060. doi: 10.1128/EC.00224-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin RM, Fedorova ND, Bok JW, Cramer RA, Wortman JR, Kim HS, Nierman WC, Keller NP. Transcriptional regulation of chemical diversity in Aspergillus fumigatus by LaeA. PLoS Pathog. 2007;3:e50. doi: 10.1371/journal.ppat.0030050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Dominguez Y, Bok JW, Berger H, Shwab EK, Basheer A, Gallmetzer A, Scazzocchio C, Keller N, Strauss J. Heterochromatic marks are associated with the repression of secondary metabolism clusters in Aspergillus nidulans. Mol Microbiol. 2010;76:1376–1386. doi: 10.1111/j.1365-2958.2010.07051.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Dominguez Y, Narendja F, Berger H, Gallmetzer A, Fernandez-Martin R, Garcia I, Scazzocchio C, Strauss J. Nucleosome positioning and histone H3 acetylation are independent processes in the Aspergillus nidulans prnD-prnB bidirectional promoter. Eukaryot Cell. 2008;7:656–663. doi: 10.1128/EC.00184-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert F, Pokholok DK, Hannett NM, Rinaldi NJ, Chandy M, Rolfe A, Workman JL, Gifford DK, Young RA. Global position and recruitment of HATs and HDACs in the yeast genome. Mol Cell. 2004;16:199–209. doi: 10.1016/j.molcel.2004.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roze LV, Arthur AE, Hong SY, Chanda A, Linz JE. The initiation and pattern of spread of histone H4 acetylation parallel the order of transcriptional activation of genes in the aflatoxin cluster. Mol Microbiol. 2007;66:713–726. doi: 10.1111/j.1365-2958.2007.05952.x. [DOI] [PubMed] [Google Scholar]
- Roze LV, Koptina AV, Laivenieks M, Beaudry RM, Jones DA, Kanarsky AV, Linz JE. Willow volatiles influence growth, development, and secondary metabolism in Aspergillus parasiticus. Appl Microbiol Biotechnol. 2011;92:359–370. doi: 10.1007/s00253-011-3339-7. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning : a laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y: 2001. [Google Scholar]
- Sanchez JF, Chiang YM, Szewczyk E, Davidson AD, Ahuja M, Elizabeth Oakley C, Woo Bok J, Keller N, Oakley BR, Wang CC. Molecular genetic analysis of the orsellinic acid/F9775 gene cluster of Aspergillus nidulans. Mol Biosyst. 2010;6:587–593. doi: 10.1039/b904541d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez JF, Entwistle R, Hung JH, Yaegashi J, Jain S, Chiang YM, Wang CC, Oakley BR. Genome-based deletion analysis reveals the prenyl xanthone biosynthesis pathway in Aspergillus nidulans. J Am Chem Soc. 2011;133:4010–4017. doi: 10.1021/ja1096682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarikaya Bayram O, Bayram O, Valerius O, Park HS, Irniger S, Gerke J, Ni M, Han KH, Yu JH, Braus GH. LaeA control of velvet family regulatory proteins for light-dependent development and fungal cell-type specificity. PLoS Genet. 2010;6:e1001226. doi: 10.1371/journal.pgen.1001226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeckh V, Scherlach K, Nützmann HW, Shelest E, Schmidt-Heck W, Schuemann J, Martin K, Hertweck C, Brakhage AA. Intimate bacterial-fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans. Proc Natl Acad Sci U S A. 2009;106:14558–14563. doi: 10.1073/pnas.0901870106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H. Tricine-SDS-PAGE. Nat Protoc. 2006;1:16–22. doi: 10.1038/nprot.2006.4. [DOI] [PubMed] [Google Scholar]
- Shaaban M, Palmer JM, El-Naggar WA, El-Sokkary MA, Habib e-S, Keller NP. Involvement of transposon-like elements in penicillin gene cluster regulation. Fungal Genet Biol. 2010a;47:423–432. doi: 10.1016/j.fgb.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaaban MI, Bok JW, Lauer C, Keller NP. Suppressor mutagenesis identifies a velvet complex remediator of Aspergillus nidulans secondary metabolism. Eukaryot Cell. 2010b;9:1816–1824. doi: 10.1128/EC.00189-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 2002;350:3–41. doi: 10.1016/s0076-6879(02)50954-x. [DOI] [PubMed] [Google Scholar]
- Shimizu K, Keller NP. Genetic involvement of a cAMP-dependent protein kinase in a G protein signaling pathway regulating morphological and chemical transitions in Aspergillus nidulans. Genetics. 2001;157:591–600. doi: 10.1093/genetics/157.2.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shwab EK, Bok JW, Tribus M, Galehr J, Graessle S, Keller NP. Histone deacetylase activity regulates chemical diversity in Aspergillus. Eukaryot Cell. 2007;6:1656–1664. doi: 10.1128/EC.00186-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smedsgaard J. Micro-scale extraction procedure for standardized screening of fungal metabolite production in cultures. J Chromatogr A. 1997;760:264–270. doi: 10.1016/s0021-9673(96)00803-5. [DOI] [PubMed] [Google Scholar]
- Smith ER, Eisen A, Gu W, Sattah M, Pannuti A, Zhou J, Cook RG, Lucchesi JC, Allis CD. ESA1 is a histone acetyltransferase that is essential for growth in yeast. Proc Natl Acad Sci U S A. 1998;95:3561–3565. doi: 10.1073/pnas.95.7.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinnett SM, Espeso EA, Cobeño L, Araújo-Bazán L, Calvo AM. Aspergillus nidulans VeA subcellular localization is dependent on the importin alpha carrier and on light. Mol Microbiol. 2007;63:242–55. doi: 10.1111/j.1365-2958.2006.05506.x. [DOI] [PubMed] [Google Scholar]
- Strauss J, Reyes-Dominguez Y. Regulation of secondary metabolism by chromatin structure and epigenetic codes. Fungal Genet Biol. 2011;48:62–69. doi: 10.1016/j.fgb.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suka N, Suka Y, Carmen AA, Wu J, Grunstein M. Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol Cell. 2001;8:473–479. doi: 10.1016/s1097-2765(01)00301-x. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsitsigiannis DI, Zarnowski R, Keller NP. The lipid body protein, PpoA, coordinates sexual and asexual sporulation in Aspergillus nidulans. J Biol Chem. 2004;279:11344–11353. doi: 10.1074/jbc.M310840200. [DOI] [PubMed] [Google Scholar]
- Upadhyay S, Shaw BD. The role of actin, fimbrin and endocytosis in growth of hyphae in Aspergillus nidulans. Mol Microbiol. 2008;68:690–705. doi: 10.1111/j.1365-2958.2008.06178.x. [DOI] [PubMed] [Google Scholar]
- Uprety B, Lahudkar S, Malik S, Bhaumik SR. The 19S proteasome subcomplex promotes the targeting of NuA4 HAT to the promoters of ribosomal protein genes to facilitate the recruitment of TFIID for transcriptional initiation in vivo. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visentin I, Montis V, Döll K, Alabouvette C, Tamietti G, Karlovsky P, Cardinale F. The transcription of genes in the biosynthetic pathway for fumonisin mycotoxins is epigenetically and differentially regulated in the fungal maize pathogen Fusarium verticillioides. Eukaryot Cell. 2011 doi: 10.1128/EC.05159-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W, Keller NP. Transcriptional regulatory elements in fungal secondary metabolism. J Microbiol. 2011;49:329–339. doi: 10.1007/s12275-011-1009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JH, Butchko RA, Fernandes M, Keller NP, Leonard TJ, Adams TH. Conservation of structure and function of the aflatoxin regulatory gene aflR from Aspergillus nidulans and A. flavus. Curr Genet. 1996;29:549–555. doi: 10.1007/BF02426959. [DOI] [PubMed] [Google Scholar]
- Yu JH, Hamari Z, Han KH, Seo JA, Reyes-Domínguez Y, Scazzocchio C. Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet Biol. 2004;41:973–981. doi: 10.1016/j.fgb.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Zhou BO, Wang SS, Zhang Y, Fu XH, Dang W, Lenzmeier BA, Zhou JQ. Histone H4 lysine 12 acetylation regulates telomeric heterochromatin plasticity in Saccharomyces cerevisiae. PLoS Genet. 2011;7:e1001272. doi: 10.1371/journal.pgen.1001272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Singh N, Donnelly C, Boimel P, Elefant F. The cloning and characterization of the histone acetyltransferase human homolog Dmel\TIP60 in Drosophila melanogaster: Dmel\TIP60 is essential for multicellular development. Genetics. 2007;175:1229–1240. doi: 10.1534/genetics.106.063685. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.