Abstract
We demonstrate that a common laboratory filter paper uniformly adsorbed with biofunctionalized plasmonic nanostructures can serve as a highly sensitive transduction platform for rapid detection of trace bioanalytes in physiological fluids. In particular, we demonstrate that bioplasmonic paper enables rapid urinalysis for the detection of kidney cancer biomarkers in artificial urine down to a concentration of 10 ng/ml. Compared to conventional rigid substrates, bioplasmonic paper offers numerous advantages such as high specific surface area (resulting in large dynamic range), excellent wicking properties (naturally microfluidic), mechanical flexibility, compatibility with conventional printing approaches (enabling multiplexed detection and multi-marker biochips), and significant cost reduction.
Introduction
Renal cancer is generally silent, frequently fatal and accounts for 3% of adult malignancies. It is estimated that in 2012 alone 64,770 new cases will be diagnosed with and 13,570 deaths will occur of cancer of the kidney and renal pelvis. 1 Altogether, this disease represents the sixth leading cause of death due to cancer. In a recent study, it has been demonstrated that the proteins aquaporin-1 (AQP1) and adipophilin (ADFP) in urine form excellent candidates for the non-invasive and early detection of renal cancer carcinoma (RCC).2 While this study clearly demonstrated the possibility of using these proteins as potential biomarkers for early and non-invasive detection of RCC, an inexpensive and high throughput biodetection platform is required to translate these biomarkers to clinical setting and incorporation into routine health physical. Conventional labeled assays such as enzyme-linked immunosorbent assay (ELISA) are time consuming and expensive, and require tedious labeling procedures making them unsuitable for rapid and routine urinalysis. The above considerations clearly suggest the need for a label-free approach, for rapid and quantitative detection of the proteins in urine at physiologically relevant concentrations (ng/ml).
Localized surface plasmon resonance (LSPR) of metal nanostructures, which involves collective coherent oscillation of dielectrically confined conduction electrons, is sensitive to numerous factors such as composition, size, shape, surrounding dielectric medium, and proximity to other nanostructures.3,4,5,6 The sensitivity of LSPR to local changes in dielectric environment renders it an attractive transduction platform for chemical and biological sensing. 7, 8, 9, 10, 11, 12 LSPR of metal nanostructures has been shown to be sensitive enough to differentiate inert gases with refractive index contrast (δn) on the order of 3×10−4, probe the conformational changes of biomacromolecules, detect single biomolecule binding events, monitor the kinetics of catalytic activity of single nanoparticles, and even optically detect single electron.13,14,15,16,17 Detection of various biomolecules such as proteins18,19,20,21,22 and DNA, 23, 24, 25 have been demonstrated in the past few years making the transduction platform promising for the development of simple, highly sensitive, label-free, and cost-effective diagnostics.
Most of the LSPR sensors demonstrated so far rely on rigid planar substrates (e.g., glass, silicon) owing to their compatibility with various well-established lithographic approaches (e.g., photolithography, e-beam lithography and nanosphere lithography), which are routinely employed for either fabrication or assembly of plasmonic nanotransducers.26,27,28,29 Compared to these conventional substrates, paper has numerous advantages such as high specific surface area, excellent wicking properties, compatibility with conventional printing approaches, significant cost reduction and easy disposability. For all these reasons, paper is gaining increased attention as a potential substrate for various applications including biodiagnostics, food quality testing as well as environmental monitoring. 30,31,32, 33,34,35,36 Paper substrates functionalized with inorganic nanoparticles such as gold, silver, and titania have been investigated for a wide range of applications including self-cleaning,37 disinfection,38 colorimetric sensing,39 surface enhanced Raman scattering (SERS) based trace analyte detection,40,41,42 and antimicrobial activity.43
Here, we report for the first time a bioplasmonic paper that comprises of a common laboratory filter paper uniformly adsorbed with biofunctionalized plasmonic nanostructures for the detection of target bioanalytes from model physiological fluids. The sensitivity and detection limit of the paper based LSPR substrate is on par or better compared to the conventional rigid substrates using similar plasmonic nanostructures. Furthermore, we demonstrate the deployment of such bioplasmonic paper as a flexible swab to collect and detect trace quantities (few µg spread over cm2 area) of model bioanalytes on real-world surfaces.
Experimental Section
Synthesis of gold nanorods (AuNRs)
Gold nanorods were synthesized using a seed-mediated approach.46,47 Seed solution was prepared by adding 0.6 mL of an ice-cold sodium borohydride solution (10 mM) into 10 mL of 0.1 M cetyltrimethylammonium bromide (CTAB) and 2.5×10−4 M chloroauric acid (HAuCl4) solution under magnetic stirring at room temperature. The color of the seed solution changed from yellow to brown. Growth solution was prepared by mixing 95 mL of CTAB (0.1 M), 0.5 mL of silver nitrate (10 mM), 5 mL of HAuCl4 (10 mM), and 0.55 mL of ascorbic acid (0.1 M) in the same order. The solution was homogenized by gentle stirring. To the resulting colorless solution, 0.12 mL of freshly prepared seed solution was added and set aside in dark for 14 h. Prior to use, the AuNR solution was centrifuged at 10,000 rpm for 10 min to remove excess CTAB and redispersed in nanopure water (18.2 MΩ-cm). The centrifugation procedure was repeated twice.
AuNR-IgG conjugates preparation
To a solution of heterobifunctional polyethylene glycol (SH-PEG-COOH) in water (37.5 µl, 20 µM, Mw=5000 g/mol, Jenkem Technology), 1-Ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC, Thermo Scientific) and N-hydroxy succinimide (NHS, Thermo Scientific) with the same molar ratio as SH-PEG-COOH were added followed by shaking for 1 h. The pH of the above reaction mixture was adjusted to 7.4 by adding 10× concentrated phosphate buffered saline (PBS), followed by the addition of Rabbit Immunoglobulin G (IgG) (10 µl, 75 µM, Mw=150 kDa, Thermo Scientific). The reaction mixture was incubated for 2 h, and then filtered to remove any byproduct during the reaction using a 50 kDa filter. The final SH-PEG-IgG conjugates solution (0.75 µM) was obtained after washing with PBS buffer (pH 7.4) twice. AuNR-IgG conjugates solution was prepared by adding 50 µl SH-PEG-IgG conjugates solution to 1 ml twice centrifuged AuNR solution with incubation for 1 h. The amount of SH-PEG-IgG was optimized to obtain maximum coverage of IgG on AuNR surface (Fig. S4).
AuNR and AuNR-IgG conjugates on rigid planar substrates
Glass substrates were cut into approximately 1×2 cm rectangular slides and cleaned in piranha solution (3:1 (v/v) mixture of H2SO4 and 30% H2O2) followed by extensive rinsing with nanopure water. AuNR or AuNR-IgG conjugates were adsorbed onto glass substrates following the modification of the surface with (3-mercaptopropyl)-trimethoxy-silane (MPTMS) by exposing the glass surface to 5% MPTMS in ethanol for 15 min followed by ultrasonication in ethanol for 30 min and rinsing with water. Subsequently, the glass surface was exposed to AuNR or AuNR-IgG conjugates solution for 1 h, followed by rinsing with water to remove the loosely bound nanorods. Similar approach was employed for biofunctionalization of AuNR with anti-AQP1 (Sigma), which were employed for the detection of AQP1 (Origene) from artificial urine (Surine, Cerilliant).
Plasmonic paper substrates preparation
Adsorption of AuNR-IgG conjugates on paper substrate was achieved by immersing a 1×1 cm2 paper in AuNR-IgG conjugates solution (2 ml) for 12 h. After removing from the solution, the paper was thoroughly rinsed with water, and then exposed to SH-PEG solution to block non-specific binding. LSPR measurements were performed by exposing the plasmonic paper to various concentrations of anti-Rabbit IgG (Thermo Scientific) in PBS for 1 h, followed by thorough rinsing with PBS and water and drying with a stream of nitrogen. Six UV-Vis extinction spectra were collected for each substrate before and after anti-IgG exposure. Each spectrum represented a different spot within the same substrate.
Characterization techniques
Transmission electron microscopy (TEM) micrographs were recorded on a JEM-2100F (JEOL) field emission instrument. Samples were prepared by drying a drop of the solution on a carbon-coated grid, which had been previously made hydrophilic by glow discharge. Scanning electron microscope (SEM) images were obtained using a FEI Nova 2300 Field Emission SEM at an accelerating voltage of 10 kV. Atomic force microscopy (AFM) images were obtained using Dimension 3000 (Digital instruments) AFM in light tapping mode.44 DLS measurements were performed using Malvern Zetasizer (Nano ZS).
Extinction spectra and Raman spectra measurements
Extinction spectra from paper substrates were collected using a CRAIC microspectrophotometer (QDI 302) coupled to a Leica optical microscope (DM 4000M) with 100× objective in the range of 450–800 nm with 10 accumulations and 0.8 sec exposure time in reflection mode. The spectral resolution of the spectrophotometer is 0.2 nm. Shimadzu UV-1800 spectrophotometer was employed for collecting UV-vis extinction spectra from solution and glass samples. Raman spectra were obtained using a Renishaw inVia confocal Raman spectrometer mounted on a Leica microscope with 20× objective (NA=0.4) and 785nm wavelength diode laser (0.5 mW). The spectra were obtained in the range of 400–1800 cm−1 with one accumulation and 10 s exposure time.
Results and Discussion
Laboratory filter paper (Whatman # 1) employed as a plasmonic substrate in this study is characterized by cellulose fibers with a diameter of ~10µm (Supporting information, Fig. S1). AFM images revealed the hierarchical fibrous morphology of the filter paper with cellulose nanofibers braided into microfibers (average diameter of ~ 400 nm) (Fig. S1). The root mean square (RMS) surface roughness of the paper was found to be 72 nm over 5 × 5 µm2 area, which indicates the large surface area of the paper substrate available for adsorption of biofunctionalized plasmonic nanostructures.
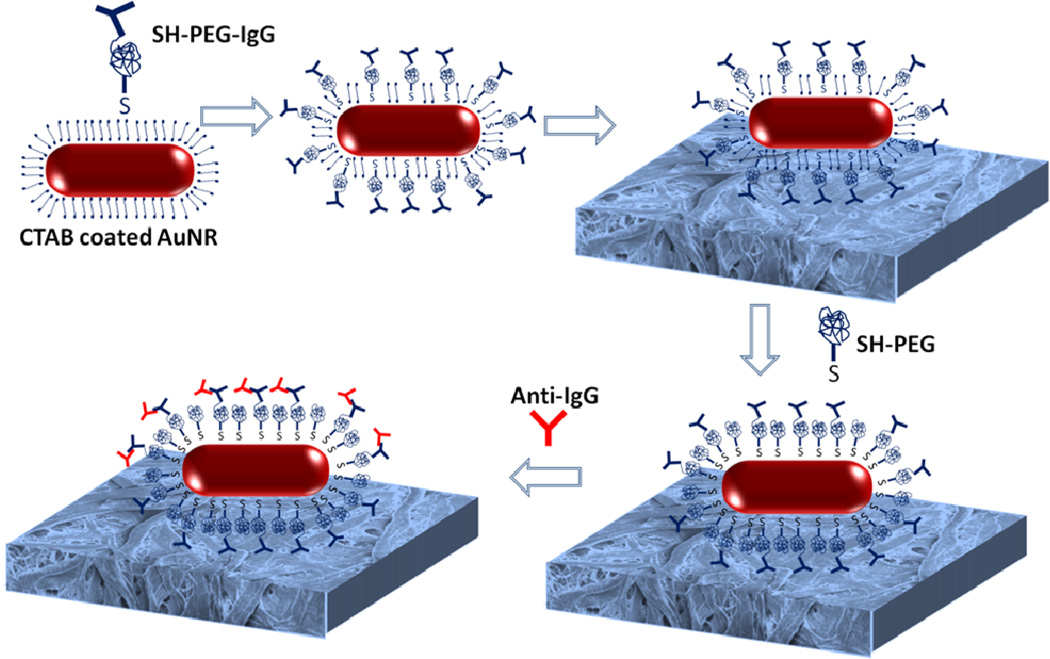
We employed Goat Rabbit IgG (IgG henceforth) as model capture biomolecule and Goat anti-Rabbit IgG (anti-IgG) as model target bioanalyte demonstrate the LSPR biosensor. Bioplasmonic paper, which serves as a LSPR biosensor, is comprised of biofunctionalized gold nanorods (AuNRs) adsorbed on the filter paper. Figure 1 illustrates the steps involved in the fabrication and deployment of bioplasmonic paper as a LSPR sensor for the detection of target bioanalytes. Thiol-terminated polyethylene glycol (SH-PEG), a hydrophilic polymer, serves as a flexible linker to increase the accessibility of IgG to target biomolecules and forms a stable protective layer around AuNRs to reduce non-specific binding. AuNR-IgG conjugates were prepared by functionalizing AuNRs with SH-PEG-IgG molecules (see experimental section for details). Subsequently, AuNR-IgG conjugates were adsorbed onto paper substrate by exposing the paper substrates to AuNR-IgG conjugate solution followed by thorough rinsing with water to remove the weakly adsorbed nanorods. Following the adsorption of the AuNR-IgG conjugates, the paper substrates were exposed to SH-PEG solution to block non-specific binding. Exposure to anti-IgG solution resulted in its specific binding to IgG, which can be detected as a spectral shift of the LSPR wavelength of AuNRs.
Figure 1.
Schematic illustration representing the fabrication steps involved in the fabrication of bioplasmonic paper for anti-IgG detection.
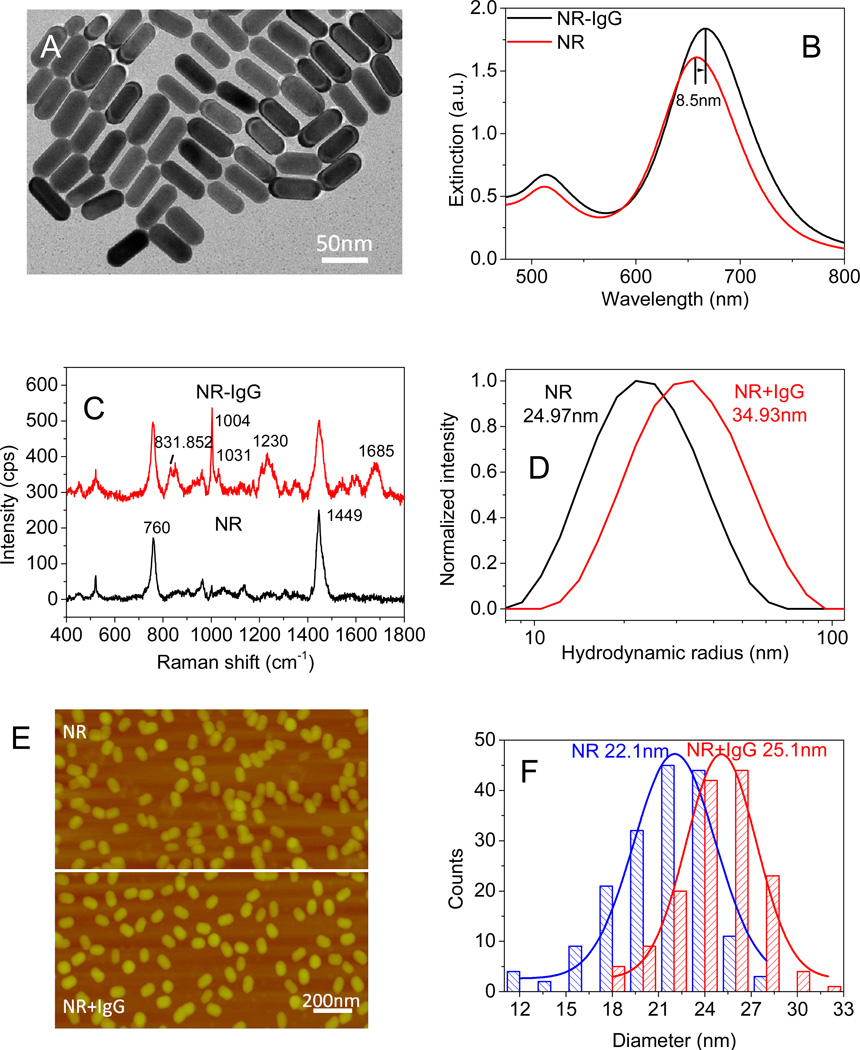
AuNRs are particularly attractive as plasmonic nanotransducers considering the large refractive index sensitivity of longitudinal LSPR, facile and large tunability of the LSPR wavelength of nanorods with aspect ratio and the electromagnetic (EM) hot-spots at the edges.45 AuNRs, synthesized using a seed-mediated approach,46,47 are positively charged with a length of 55.8±3.6 nm and a diameter of 22.0±2.1 nm (TEM image in Fig. 2A). UV-vis extinction spectra of the AuNRs are characterized by two distinct bands corresponding to the transverse (lower wavelength) and longitudinal (higher wavelength) oscillation of electrons with incident EM field. The longitudinal plasmon band of AuNRs, which is known to be more sensitive to the refractive index change of the surrounding medium compared to the transverse band, exhibited a red shift of ~8.5 nm and an increase in extinction intensity upon SH-PEG-IgG appendage (Fig. 2B). The red shift and increase in intensity correspond to the increase in the refractive index around the AuNRs with SH-PEG-IgG binding. SERS spectra collected from AuNRs and AuNR-IgG conjugates also confirm the biofunctionalization of AuNRs (Fig. 2C). The successful bioconjugation of IgG is evidenced by the Raman bands at 831 cm−1 and 852 cm−1, 1004 cm−1 and 1031 cm−1, 1230 cm−1, and 1685 cm−1 corresponding to tyrosine, phenylalanine, amide III, and amide I, respectively.
Figure 2.
(A) TEM image of AuNRs. (B) UV-vis extinction spectra confirming AuNR and SH-PEG-IgG conjugation. The UV-vis extinction spectrum of 56×22 nm AuNRs shows a longitudinal LSPR wavelength of 658 nm (AuNRs, red). After incubation with SH-PEG-IgG, the λmax red shifts 8.5 nm (AuNR-IgG, black). (C) Hydrodynamic diameters obtained from DLS show that the average hydrodynamic diameter increased by 10nm following IgG conjugation. (D) Surface enhanced Raman spectra before and after the conjugation of antibody on gold nanorods reveal the Raman bands corresponding to IgG. (E-F) AFM images of AuNRs and AuNR+IgG conjugates, revealing the increase in the average diameter of the nanorods following the bioconjugation.
The average hydrodynamic diameter of AuNR and AuNR-IgG measured using dynamic light scattering (DLS) was found to be ~25.0 and ~35.0 nm, respectively (Fig. 2D). The increase in hydrodynamic diameter of AuNR can be primarily attributed to the successful appendage of IgG (Mw=150 KDa) to AuNR surface. Only a small increase of ~0.2 nm was observed for the conjugation of SH-PEG (Mw=5 KDa) (see Fig. S2). Figure 2E shows the AFM images of the AuNR and AuNR-IgG adsorbed on silicon surfaces. Apart from indicating the stability of AuNR-IgG in solution (absence of any large scale aggregates), a small increase in the average diameter (~ 3nm) of nanorods was observed following SH-PEG-IgG functionalization, which closely corresponds to the size of IgG (Fig. 2F).48,49 The discrepancy between the DLS and AFM measurements can be attributed to two factors: (i) AFM imaging was performed in dry state as opposed to DLS measurements, which reveals the hydrodynamic radius of AuNR (ii) possible deformation (compression under AFM tip) of the biomolecules and polymer during AFM imaging although the imaging was performed in light tapping mode.
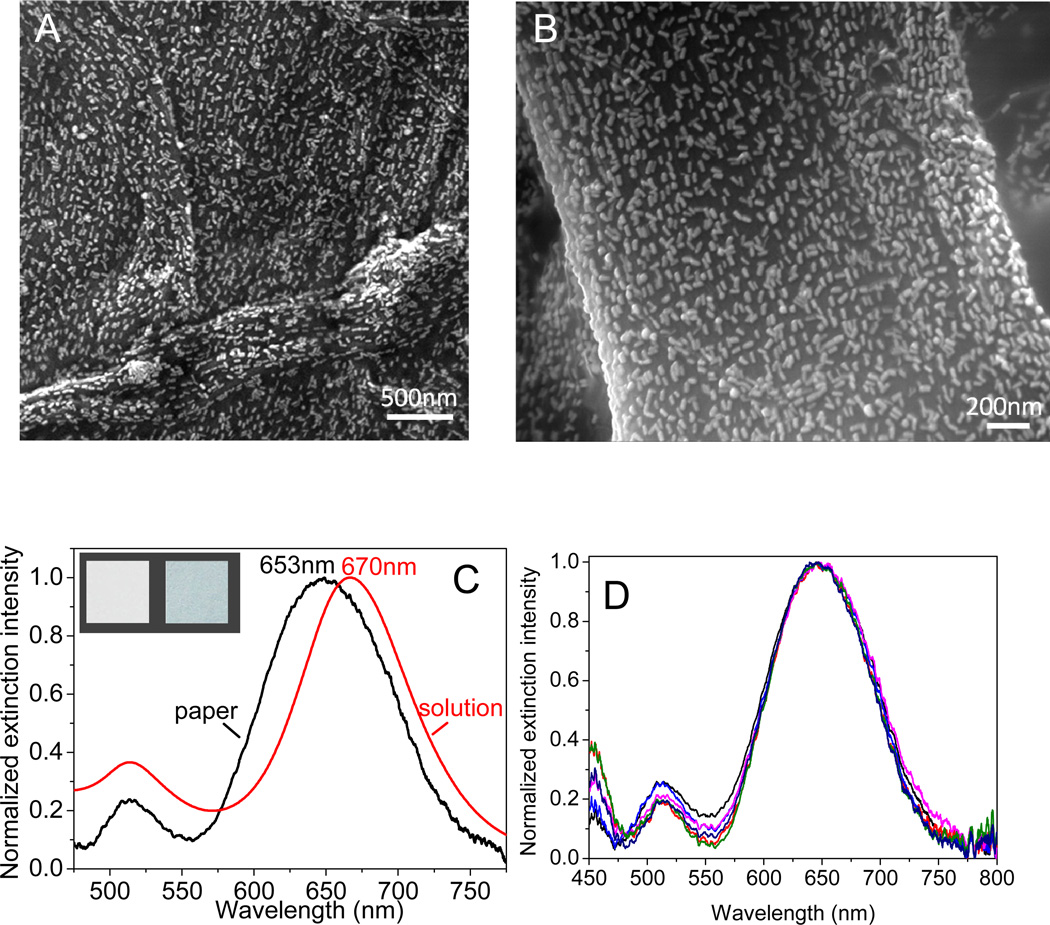
An important consideration in the design of bioplasmonic paper substrates is the optical homogeneity of the substrate. The homogeneity determines the noise floor of the spectral shift, which in turn determines the resolution of the bioplasmonic paper. SEM images revealed highly uniform distribution of AuNR-IgG conjugates on paper surface with no signs of aggregation or patchiness on the substrate (Fig. 3A). Higher magnification image reveals the preferential alignment of AuNR-IgG conjugates along cellulose fibers. The surface density of the AuNR-IgG conjugates adsorbed on the paper substrate was found to be 351±11/µm2, which can be controlled by incubation time (Fig. 3B). SERS spectra were collected from AuNRs and AuNR-IgG conjugates paper to confirm the preservation of IgG conjugation during incubation process (Fig. S5).
Figure 3.
(A-B) SEM images of paper adsorbed with AuNR-IgG conjugates. (C) Extinction spectra of AuNR-IgG conjugates in solution and on paper (inset: shows photographs of the bare filter paper (left) and filter paper after adsorption of AuNR+IgG conjugates (right)). (D) Six extinction spectra collected from different spots on a plasmonic paper of 0.5×0.5 cm2 area showing the remarkable spectral homogeneity of bioplasmonic paper.
Longitudinal LSPR wavelength of AuNR-IgG conjugates adsorbed on paper substrate exhibited a significant blue shift (~17nm) compared to that in solution due to the change in the refractive index of the surrounding medium (aqueous to air+substrate) (Fig. 3C).50 The extinction spectrum from the paper substrate was collected from a 2×2 µm2 area using a microspectrometer mounted on an optical microscope, which corresponds to ~1400 nanorods. The narrow and symmetric extinction band (FWHM of 115 nm) corresponding to the longitudinal LSPR of AuNR indicates the absence of AuNR aggregates on paper surface, which is in agreement with the SEM images discussed above. Extinction spectra collected across 0.5×0.5 cm2 area revealed excellent optical uniformity of the bioplasmonic paper substrate. LSPR wavelength measured from six different spots over such large area exhibited a small standard deviation of ~1 nm (Fig. 3D). The excellent spectral homogeneity is due to the highly uniform adsorption of AuNR-IgG conjugates as evidenced by the SEM images. The spectral homogeneity observed here is quite remarkable considering the simplicity of the fabrication process and the inherent heterogeneity of the paper substrates (large surface roughness and hierarchical nature of the fibrous mat).
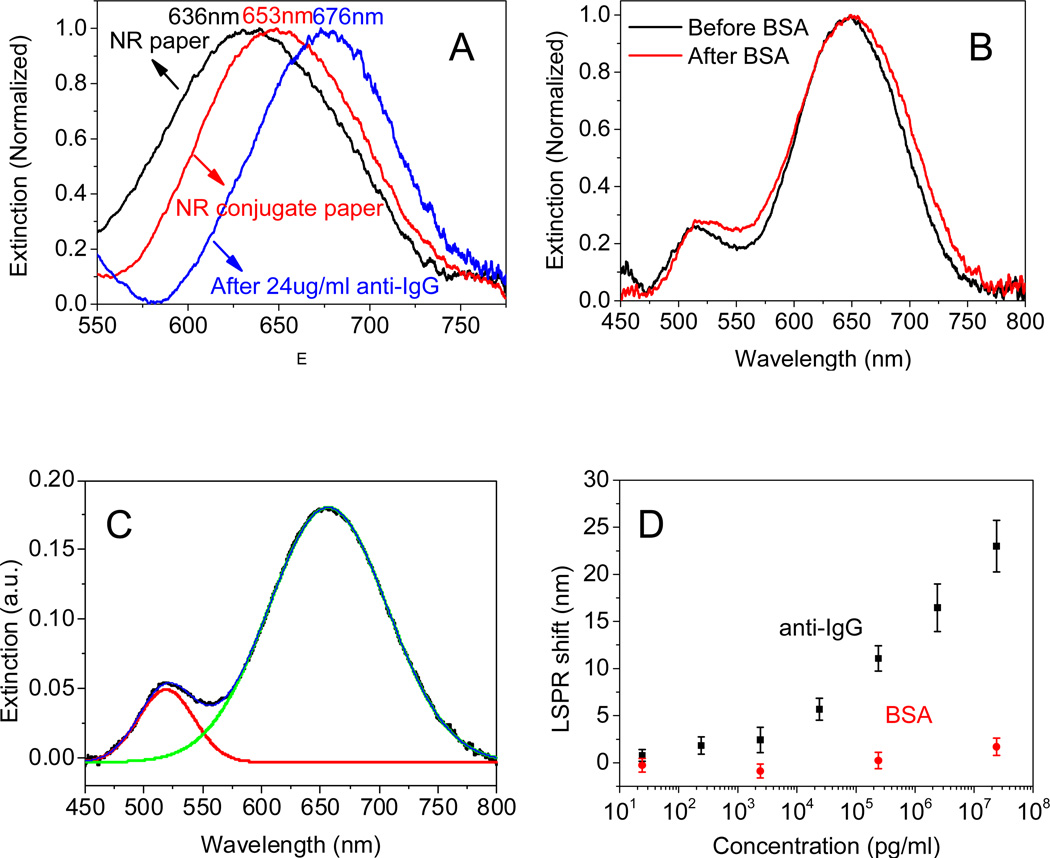
The biosensing capability of the paper substrates is demonstrated by using anti-IgG as a model bioanalyte, which is known to exhibit strong and specific binding the heavy chains of IgG.51 Extinction spectra were obtained from paper substrates adsorbed with bare AuNRs, AuNR-IgG conjugates with SH-PEG (for blocking non-specific binding) and subsequently exposed to 24 µg/ml of anti-IgG (Fig. 4A). LSPR wavelength exhibited a ~17 nm red shift with the partial replacement of CTAB layer with AuNR-IgG conjugates and SH-PEG molecules, and a further red shift of ~23 nm upon specific binding of anti-IgG (24 µg/ml) to IgG. On the other hand, a small red shift of ~2 nm was observed upon exposure to bovine serum albumin (BSA) (24 µg/ml), which indicates small non-specific binding of the biofunctionalized nanorods (Fig. 4B).
Figure 4.
(A) Extinction spectra of AuNR paper substrate (black), AuNR-IgG conjugates on the paper substrate before (red) and after binding of anti-IgG (blue). (B) Control experiment showing the small non-specific binding of BSA on bioplasmonic paper substrate. (C) LSPR spectra deconvoluted using two Gaussian peaks. (D) Plot showing the LSPR peak shift of bioplasmonic paper for various concentrations of anti-IgG and BSA.
The extinction spectrum was deconvoluted by fitting the extinction spectrum with two Gaussian peaks, from which the longitudinal LSPR wavelength of AuNRs was employed to monitor the biomolecule binding (Fig. 4C). A semi-log plot of the longitudinal LSPR wavelength shift for different concentrations of anti-IgG revealed that LSPR shift monotonically increases with increase in the concentration of anti-IgG. The plot also reveals extremely small non-specific binding of BSA for various concentrations (Fig. 4D). Conventional glass substrates exhibited smaller shift (20 nm) compared to the bioplasmonic paper (24 nm) under similar conditions (Fig. S3). The larger shift in the case of paper substrate compared to glass substrate is possibly due to the 3D porous structure of paper substrate, which enables better uptake and transport of the bioanalytes to the plasmonic nanostructures. A detection limit of 24 pg/ml (~0.16 pM) was noted in the case of bioplasmonic paper, which is on par with that observed in the case of other rigid substrates.52
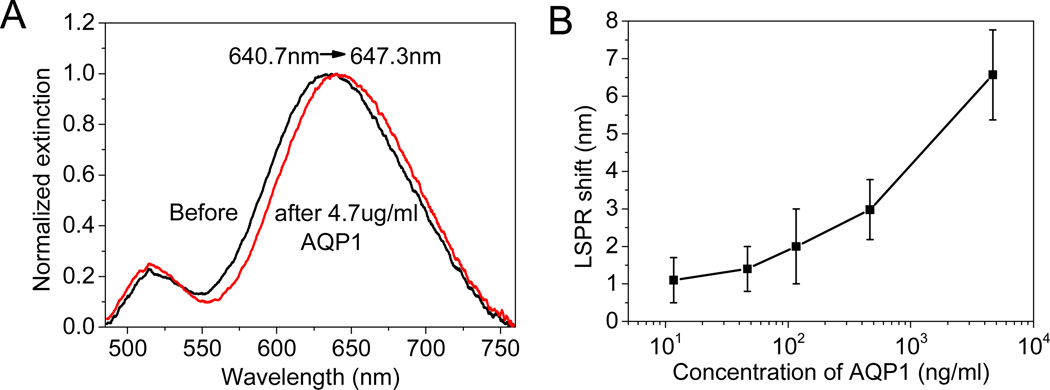
Similar approach was employed to detect AQP1, a biomarker for RCC in urine. We employed AuNR appended with anti-AQP1 as biofunctionalized transducers for the detection of AQP1 in artificial urine. Figure 5A shows the shift in the longitudinal LSPR of AuNR upon binding of AQP1 (4.67 µg/ml) from artificial urine sample (Fig. 5A). A monotonic increase in the LSPR shift is observed with increasing the concentration of AQP1 in artificial urine (Fig. 5B). The measured LSPR shift for 10 ng/ml (0.35 nM) of AQP1 is ~1.1 nm. This shift is significantly higher than the noise floor of the bioplasmonic paper (~0.6 nm). The detection limit matches with the lower limit of the range of AQP1 in patients with RCC.53 The detection limit of AQP1 is higher than anti-IgG, which is possibly due to: (i) the molecular weight of AQP1 is 28.3 kDa, much smaller than model bioanalyte anti-IgG 150 kDa, causing a smaller change in refractive index; and (ii) limited binding sites accessible to AQP1 compared to that available for anti-IgG, which binds to the heavy chain of IgG. The detection limit can be lowered by optimizing the ratio of SH-IgG and AuNRs, and selecting nanostructures or their organized assemblies with higher LSPR sensitivity.
Figure 5.
(A) Extinction spectra of AuNR appended with anti-AQP1 adsorbed on a paper substrate before (black) and after exposing to 4.7µ/ml AQP1 in artificial urine (red) (B) Plot showing the LSPR peak shift of bioplasmonic paper for various concentrations of AQP1.
One of the distinct advantages of the bioplasmonic paper is the ability to collect trace amount of bioanalytes from real-world surfaces by simply swabbing across the surface. To demonstrate such unique ability, 12 µg of anti-IgG was deposited on a tomato as purchased from a grocery store. A slightly wet (in PBS buffer) bioplasmonic paper substrate was swabbed on the surface of the tomato to collect the bioanalytes (Fig. S6A). The longitudinal LSPR wavelength exhibited a red shift (~21 nm) upon specific binding of anti-IgG, which was collected from six different spots in the center of the plasmonic paper (Fig. S6B). The inset photographs of Figure 6B shows that the color of the bioplasmonic paper turned from blue to grey upon binding of the target biomolecules to the biofunctionalized nanorods. A control experiment was performed by swabbing a slightly wetted plasmonic paper on the surface of as purchased tomato (i.e., no prior cleaning). Longitudinal LSPR wavelength exhibited a ~3 nm red shift, which can be attributed to the surface contaminants non-specifically adsorbing to AuNR on the unwashed tomato (Fig. S6C). Considering that the swabbing of the surface results in only a fraction of the biomolecules being absorbed onto the paper, the much higher LSPR shift upon specific binding of anti-IgG on the surface clearly indicates the plasmonic paper substrate to be excellent candidate for collecting trace amount of bioanalytes from real-world surfaces as LSPR biosensors.
Conclusions
In conclusion, we have demonstrated a bioplasmonic paper for the rapid and label-free detection of AQP1, a biomarker for early detection of RCC. Bioplasmonic paper exhibited excellent spectral homogeneity, sensitivity, dynamic range and selectivity, comparable to or better than the conventional rigid LSPR substrates. Furthermore, owing to its unique flexible and conformal nature, bioplasmonic paper enables simultaneous collection and detection of trace amount of bioanalytes from real-world surfaces. Paper based LSPR substrates offer numerous advantages such as low cost, easy storage, transport and disposability, which lead to simple, inexpensive plasmonic biochips for point-of-care diagnostics. Besides being highly cost-effective and environmental-friendly, the use of paper-based LSPR substrates when transformed into printable microfluidic devices will enable the detection of multiple bioanalytes in complex physiological fluids. The synergism of paper-based microfluidics and LSPR based biosensing is expected to be truly transformative by opening up novel avenues in multi-analyte biological detection.
Supplementary Material
Acknowledgements
This work was supported by Office of Congressionally Directed Medical Research Programs under contract No. W81XWH-11-1-0439 to SS, and National Institutes of Health under grant No. R01CA14152 and the Barnes-Jewish Hospital Foundation to JJM.
Footnotes
Supporting Information Available
This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Howlader N, Noone AM, Krapcho M, Neyman N, Aminou R, Altekruse SF, Kosary CL, Ruhl J, Tatalovich Z, Cho H, Mariotto A, Eisner MP, Lewis DR, Chen HS, Feuer EJ, Cronin KA, editors. SEER Cancer Statistics Review, 1975–2009 (Vintage 2009 Populations) Bethesda, MD: National Cancer Institute; [Google Scholar]
- 2.Morrissey JJ, London AN, Luo J, Kharasch ED. Mayo Clin Proc. 2010;85:413–421. doi: 10.4065/mcp.2009.0709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosi NL, Mirkin CA. Chem. Rev. 2005;105:1547–1562. doi: 10.1021/cr030067f. [DOI] [PubMed] [Google Scholar]
- 4.Maier SA, Atwater HA. App. Phys. Rev. 2005;98:011101. [Google Scholar]
- 5.Anker JN, Hall WP, Lyandres O, Shah NC, Zhao J, Van Duyne RP. Nat. Mater. 2008;7:442–453. doi: 10.1038/nmat2162. [DOI] [PubMed] [Google Scholar]
- 6.Mayer KM, Hafner JH. Chem. Rev. 2011;111:3828–3857. doi: 10.1021/cr100313v. [DOI] [PubMed] [Google Scholar]
- 7.Sepulveda B, Angelome PC, Lechuga LM, Liz-Marzán LM. Nano Today. 2009;4:244–251. [Google Scholar]
- 8.Haes AJ, Van Duyne RP. J. Am.Chem. Soc. 2002;124:10596–10604. doi: 10.1021/ja020393x. [DOI] [PubMed] [Google Scholar]
- 9.Riboh JC, Haes AJ, McFarland AD, Yonzon CR, Van Duyne RP. J. Phys. Chem B. 2003;107:1772–1780. [Google Scholar]
- 10.Yonzon CR, Jeoung E, Zou S, Schatz GC, Mrksich M, Van Duyne RP. J. Am. Chem. Soc. 2004;126:12669–12676. doi: 10.1021/ja047118q. [DOI] [PubMed] [Google Scholar]
- 11.Haes AJ, Chang L, Klein WL, Van Duyne RP. J. Am. Chem. Soc. 2005;127:2264–2271. doi: 10.1021/ja044087q. [DOI] [PubMed] [Google Scholar]
- 12.Svedendahl M, Chen S, Dmitriev A, Kall M. Nano Lett. 2009;9:4428–4433. doi: 10.1021/nl902721z. [DOI] [PubMed] [Google Scholar]
- 13.Bingham JM, Anker JN, Kreno LE, Van Duyne RP. J. Am.Chem. Soc. 2010;132:17358–17359. doi: 10.1021/ja1074272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall WP, Modica J, Anker J, Lin Y, Mrksich M, Van Duyne RP. Nano Lett. 2011;11:1098–1105. doi: 10.1021/nl103994w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mayer KM, Hao F, Lee S, Nordlander P, Hafner JH. Nanotechnology. 2010;21:255503. doi: 10.1088/0957-4484/21/25/255503. [DOI] [PubMed] [Google Scholar]
- 16.Novo C, Funston AM, Gooding AK, Mulvaney P. J. Am. Chem. Soc. 2009;131:14664–14666. doi: 10.1021/ja905216h. [DOI] [PubMed] [Google Scholar]
- 17.Novo C, Funston AM, Mulvaney P. Nat. Nanotechnol. 2008;3:598. doi: 10.1038/nnano.2008.246. [DOI] [PubMed] [Google Scholar]
- 18.Anker JN, Hall WP, Lyandres O, Shah NC, Zhao J, Van Duyne RP. Nat. Mater. 2008;7:442–453. doi: 10.1038/nmat2162. [DOI] [PubMed] [Google Scholar]
- 19.Olkhov RV, Fowke JD, Shaw AM. Anal. Biochem. 2009;385:234–241. doi: 10.1016/j.ab.2008.10.042. [DOI] [PubMed] [Google Scholar]
- 20.Nusz GJ, Marinakos SM, Curry AC, Dahlin A, Hook F, Wax A, Chilkoti A. Anal. Chem. 2008;80:984–989. doi: 10.1021/ac7017348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marinakos SM, Chen S, Chilkoti A. Anal. Chem. 2007;79:5278–5283. doi: 10.1021/ac0706527. [DOI] [PubMed] [Google Scholar]
- 22.Mayer KM, Lee S, Liao H, Rostro BC, Fuentes A, Scully PT, Nehl CL, Hafner JH. ACS Nano. 2008;2:687–692. doi: 10.1021/nn7003734. [DOI] [PubMed] [Google Scholar]
- 23.Sönnichsen C, Reinhard BM, Liphardt J, Alivisatos AP. Nat. Biotechnol. 2005;23:741–745. doi: 10.1038/nbt1100. [DOI] [PubMed] [Google Scholar]
- 24.Schultz S, Smith DR, Mock JJ, Schultz DA. Proc. Natl. Acad. Sci. U.S.A. 2000;97:996–1001. doi: 10.1073/pnas.97.3.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sannomiya T, Hafner C, Voros J. Nano Lett. 2008;8:3450–3455. doi: 10.1021/nl802317d. [DOI] [PubMed] [Google Scholar]
- 26.Mayer KM, Hafner JH. Chem. Rev. 2011;111:3828–3857. doi: 10.1021/cr100313v. [DOI] [PubMed] [Google Scholar]
- 27.Haes AJ, Zou S, Schatz GC, Van Duyne RP. J. Phys. Chem. B. 2004;108:109–11. doi: 10.1021/jp051178g. [DOI] [PubMed] [Google Scholar]
- 28.Cinel NA, Butun S, Ozbay E. Opt Express. 2012;20:2587–2597. doi: 10.1364/OE.20.002587. [DOI] [PubMed] [Google Scholar]
- 29.Kim S, Jung J, Choi D, Jung H, Yang S. Langmuir. 2006;22:7109–7112. doi: 10.1021/la0605844. [DOI] [PubMed] [Google Scholar]
- 30.Parolo C, Merkoçi A. Chem. Soc. Rev. 2013 doi: 10.1039/c2cs35255a. [DOI] [PubMed] [Google Scholar]
- 31.Li X, Ballerini DR, Shen W. Biomicrofluidics. 2012;6:011301. doi: 10.1063/1.3687398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X, Tian JF, Shen W. Cellulose. 2010;17:649–659. [Google Scholar]
- 33.Martinez AW, Phillips ST, Butte MJ, Whitesides GM. Angew. Chem. Int. Ed. 2007;46:1318–1320. doi: 10.1002/anie.200603817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao W, van der Berg A. Lab Chip. 2008;8:1988–1991. doi: 10.1039/b814043j. [DOI] [PubMed] [Google Scholar]
- 35.Cheng C-M, Martinez AW, Gong J, Mace CR, Phillips ST, Carrilho E, Mirica KA, Whitesides GM. Angew. Chem. Int. Ed. 2010;49:4771–4774. doi: 10.1002/anie.201001005. [DOI] [PubMed] [Google Scholar]
- 36.Martinez AW, Phillips ST, Whitesides GM. Anal. Chem. 2010;82:3–10. doi: 10.1021/ac9013989. [DOI] [PubMed] [Google Scholar]
- 37.Uddin MJ, Cesano F, Scarano D, Bonino F, Agostini G, Spoto G, Bordiga S, Zecchina A. J. Photochem. Photobiol. A. 2008;199:64–72. [Google Scholar]
- 38.Geng X, Filipe C, Pelton R. Appita J. 2008;61:456–460. [Google Scholar]
- 39.Zhao W, Ali MM, Aguirre SD, Brook MA, Li Y. Anal. Chem. 2008;80:8431–8437. doi: 10.1021/ac801008q. [DOI] [PubMed] [Google Scholar]
- 40.Lee CH, Tian L, Singamaneni S. ACS Appl. Mater. Interfaces. 2010;2:3429–3435. doi: 10.1021/am1009875. [DOI] [PubMed] [Google Scholar]
- 41.Lee CH, Hankus ME, Tian L, Pellegrino PM, Singamaneni S. Anal. Chem. 2011;83:8953–8958. doi: 10.1021/ac2016882. [DOI] [PubMed] [Google Scholar]
- 42.Yu WW, White M. Anal. Chem. 2010;82:9626–9630. doi: 10.1021/ac102475k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tankhiwale R, Bajpai SK. Colloids Surf. B. 2009;69:164–168. doi: 10.1016/j.colsurfb.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 44.Tsukruk VV, Singamaneni S. Scanning Probe Microscopy of Soft Matter: Fundamentals and Practices. Wiley-VCH; 2012. [Google Scholar]
- 45.Nusz GJ, Curry AC, Marinakos SM, Wax A, Chilkoti A. ACS Nano. 2009;3:795–806. doi: 10.1021/nn8006465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee KS, EI-Sayed MA. J. Phys. Chem. B. 2005;109:20331. doi: 10.1021/jp054385p. [DOI] [PubMed] [Google Scholar]
- 47.Gole A, Murphy CJ. Langmuir. 2008;24:266. doi: 10.1021/la7026303. [DOI] [PubMed] [Google Scholar]
- 48.Jones VW, Kenseth JR, Porter MD, Mosher CL, Henderson E. Anal. Chem. 1998;70:1233–1241. doi: 10.1021/ac971125y. [DOI] [PubMed] [Google Scholar]
- 49.Ouerghi O, Touhami A, Othmane A, Ben Ouada H, Martelet C, Fretigny C, Jaffrezic-Renault N. Sensors and Actuators B. 2002;84:167–175. [Google Scholar]
- 50.Chen C-D, Cheng S-F, Chau L-K, Chris Wang CR. Biosens. Bioelec. 2007;22:926–932. doi: 10.1016/j.bios.2006.03.021. [DOI] [PubMed] [Google Scholar]
- 51.Product data sheet of ImmunoPure Goat Anti-Ribbit IgG. Thermo Scientific [Google Scholar]
- 52.Mayer KM, Hafner JH. Chem. Rev. 2011;111:3828–3857. doi: 10.1021/cr100313v. [DOI] [PubMed] [Google Scholar]
- 53.Morrissey JJ, Kharasch ED. Unpublished. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.