Abstract
The origin and evolution of “ORFans” (suspected genes without known relatives) remain unclear. Here, we take advantage of a unique opportunity to examine the population diversity of thousands of ORFans, based on a collection of 35 complete genomes of isolates of Escherichia coli and Shigella (which is included phylogenetically within E. coli). As expected from previous studies, ORFans are shorter and AT-richer in sequence than non-ORFans. We find that ORFans often are very narrowly distributed: the most common pattern is for an ORFan to be found in only one genome. We compared within-species population diversity of ORFan genes with those of two control groups of non-ORFan genes. Patterns of population variation suggest that most ORFans are not artifacts, but encode real genes whose protein-coding capacity is conserved, reflecting selection against nonsynonymous mutations. Nevertheless, nonsynonymous nucleotide diversity is higher than for non-ORFans, whereas synonymous diversity is roughly the same. In particular, there is a several-fold excess of ORFans in the highest decile of diversity relative to controls, which might be due to weaker purifying selection, positive selection, or a subclass of ORFans that are decaying.
Keywords: ORFan, lineage-specific genes, evolution, population genetics, positive selection, negative selection
Introduction
Two decades ago, protein biochemists supposed that the universe of proteins would be limited to a modest number of distinctive types, for example, a thousand different folds (Chothia 1992). However, new kinds of proteins continued to appear, often in the form of “orphans” or “singletons” that could not be matched to any known proteins. In the past decade, sequencing of bacterial genomes led to the discovery of huge numbers of so-called ORFans or ORFan genes, open reading frames (ORFs) encoding orphan proteins, that is, proteins with no known relatives (Wilson et al. 2005). The high frequency of ORFans in newly determined bacterial genomes originally was attributed to limited sampling of genomes. Indeed, as more genomes are sequenced, the fraction of genes in a given genome that are designated as ORFans can never increase, but only decrease when a relative is discovered. However, a comparative analysis of 122 bacterial genomes indicated that the total number of different kinds of ORFans was increasing linearly (Wilson et al. 2005).
Some descriptive generalities about these ORFans emerge from previous studies. ORFans are found in almost all the bacterial genomes, except some small genomes of intracellular parasites or endosymbionts (Wilson et al. 2005). ORFans tend to be more AT-rich than sequences with a broader phylogenetic distribution (Charlebois et al. 2003; Daubin and Ochman 2004). On average, ORFans tend to be shorter than genes with a broader phylogenetic distribution (Daubin and Ochman 2004).
Numerous studies have addressed the possible origins of ORFans (Ohno 1984; Logsdon and Doolittle 1997; Ochman et al. 2000; Long 2001; Long et al. 2003; Daubin and Ochman 2004; Wang et al. 2004; Hahn and Lee 2005; Yin and Fischer 2006, 2008; Rancurel et al. 2009; Toll-Riera et al. 2009). One type of explanation is that ORFans are not real genes, but artifacts of annotation, resulting from the algorithms used to detect coding sequences in genomes (Daubin and Ochman 2004). Another possibility is that ORFan genes, although real, are not true orphans, but have evolved so rapidly that their family affinity cannot be discerned. Although rapid evolution by amino acid replacements alone seems unlikely to explain the status of ORFans, they may have undergone more radical evolutionary changes, such as frameshifts or rearrangements (Hahn and Lee 2005; Cai and Petrov 2010). Finally, it may be that ORFans represent newly acquired genes, having arisen either de novo from noncoding sequences or having been acquired by lateral gene transfer from an uncharacterized source, for example, unknown prokaryotes or bacteriophages (Ochman et al. 2000; Yin and Fischer 2006; Knowles and McLysaght 2009; Toll-Riera et al. 2009).
Recent studies suggest that a small proportion of human-specific genes or primate-specific genes originated from noncoding sequences (Knowles and McLysaght 2009; Toll-Riera et al. 2009). Recent studies in archaeal, bacterial, and viral genomes suggest that a proportion of ORFans likely originated via horizontal gene transfer from distant cellular sources (Yin and Fischer 2008; Cortez et al. 2009). It has been argued that the AT-richness of ORFans reflects lateral transfer from AT-rich bacteriophage genomes (Daubin et al. 2003), or that it reflects misannotation of intergenic regions, on the grounds that such regions often are AT-rich (Charlebois et al. 2003). The shortness of ORFans also suggests the possibility of misannotation (Ochman 2002).
The emerging consensus seems to be that ORFans largely represent an influx of relatively impermanent sequences from bacteriophages and other mobile elements. Whether this consensus will prove correct, it leaves open the question of how ORFans are evolving within a species. On the one hand, they might be undergoing slow regression if their similarity to real genes is merely accidental (if they are annotation artifacts) or merely historical (e.g., if they are nonfunctional remnants of past bacteriophage infections). On the other hand, they might be rapidly evolving genes responsible for lineage-specific adaptation; or they might be conserved genes responsible for lineage-specific features of morphology or physiology (Khalturin et al. 2008).
In this study, we begin to address the evolution of ORFans, as distinct from the origins of ORFans, by taking advantage of the fact that there are thousands of ORFans available in several dozens of complete Escherichia coli genomes. Although conventional taxonomy associates a number of these genomes with the genus Shigella, known Shigella genomes cluster phylogenetically within a much larger set of known set of E. coli genomes, with other Escherichia species as outgroups (Touchon et al. 2009). For this reason, when we refer to E. coli later, we mean to include isolates designated conventionally as E. coli or Shigella while excluding other Escherichia species (e.g., E. fergusoni).
Here, we examine the population diversity of ORFans in E. coli (i.e., E. coli plus Shigella) as compared with that of two sets of progressively older and more widely distributed non-ORFan genes. This approach allows us to control for the idiosyncratic ages and distributions of ORFans by comparing them only with clusters pruned to have a matching set of strains. As expected, we find that ORFans are shorter and more AT-rich than non-ORFans. ORFans tend to have a very narrow distribution among E. coli genomes, with most of them being found in only a single genome. The majority of ORFan genes appear to be conserved, at least in the short term, as judged by the overall tendency toward lower nonsynonymous diversity than synonymous diversity among those ORFans present in more than one genome. However, nonsynonymous diversity is higher than for non-ORFans, and in particular shows an excess of high-diversity clusters of ORFan genes.
Materials and Methods
Sequences, Clusters, Alignments, and Function Annotations
Clusters of homologous sequences used in this study ultimately derive from the Protein Clusters database (Klimke et al. 2009), a collection of automatically clustered Reference Sequence proteins from complete genomes (of prokaryotes, plasmids, viruses, organelles, and complete and incomplete genomes of protozoa and plants). Some clusters include curated information on protein function (Klimke et al. 2009); those without known function are annotated as “hypothetical protein.”
Data on protein clusters in prokaryotes obtained from NCBI represent the January 2010 version of the clusters database. Specifically, the PRK_summary, PRK_AllProteins.bcp, Clusters.bcp, and NonCuratedClusters.bcp files were downloaded from ftp://ftp.ncbi.nih.gov/genomes/CLUSTERS/Jan_2010/PRK. These clusters include proteins from 35 E. coli genomes and 88 E. coli plasmid genomes (including Shigella and Shigella plasmid genomes). The E. coli genomes used in the study are shown in table 1. The plasmid genome ids used in the study are listed in the supplementary data, Supplementary Material online.
Table 1.
Size and Other Compositional Features of Escherichia coli Genomes Used in This Study
| E. coli Strain Name | Genome Size (Mb) | Protein-Coding Genes | ORFans | Genome-Specific ORFans | ORFans (%) | Genome-Specific ORFans (%) | Functionally Annotated ORFans (%) | Functionally Annotated Non-ORFans (%) |
|---|---|---|---|---|---|---|---|---|
| Shigella dysenteriae Sd197 | 4.56 | 4,086 | 1,511 | 416 | 37.0 | 10.2 | 2.1 | 50.7 |
| S. flexneri 5 str. 8401 | 4.57 | 3,938 | 879 | 177 | 22.3 | 4.5 | 3.2 | 56.4 |
| E. coli BW2952 | 4.58 | 4,067 | 415 | 17 | 10.2 | 0.4 | 12.5 | 62.0 |
| E. coli BL21 (DE3) | 4.60 | 4,198 | 547 | 37 | 13.0 | 0.9 | 9.1 | 58.7 |
| S. flexneri 2a str. 2457T | 4.60 | 3,971 | 858 | 90 | 21.6 | 2.3 | 4.3 | 56.1 |
| E. coli B str. REL606 | 4.63 | 4,180 | 523 | 25 | 12.5 | 0.6 | 9.6 | 58.5 |
| S. boydii Sb227 | 4.63 | 4,057 | 1,040 | 225 | 25.6 | 5.5 | 3.4 | 54.6 |
| E. coli str. K-12 substr. MG1655 | 4.64 | 4,127 | 415 | 22 | 10.1 | 0.5 | 12.3 | 62.0 |
| E. coli HS | 4.64 | 4,275 | 687 | 103 | 16.1 | 2.4 | 7.1 | 56.5 |
| E. coli IAI1 | 4.70 | 4,295 | 608 | 58 | 14.2 | 1.4 | 8.1 | 57.3 |
| E. coli str. K-12 substr. DH10B | 4.70 | 4,095 | 452 | 31 | 11.0 | 0.8 | 11.7 | 61.4 |
| E. coli ATCC 8739 | 4.75 | 4,174 | 446 | 26 | 10.7 | 0.6 | 10.1 | 59.7 |
| S. flexneri 2a str. 301 | 4.82 | 4,288 | 1,112 | 152 | 25.9 | 3.5 | 3.7 | 52.2 |
| S. boydii CDC 3083–94 | 4.86 | 4,228 | 1,257 | 329 | 29.7 | 7.8 | 2.4 | 50.0 |
| E. coli 536 | 4.94 | 4,414 | 824 | 206 | 18.7 | 4.7 | 4.2 | 54.0 |
| S. sonnei Ss046 | 5.05 | 4,315 | 975 | 156 | 22.6 | 3.6 | 3.4 | 54.0 |
| E. coli O127:H6 str. E2348/69 | 5.07 | 4,518 | 687 | 135 | 15.2 | 3.0 | 6.0 | 53.8 |
| E. coli IAI39 | 5.13 | 4,626 | 778 | 106 | 16.8 | 2.3 | 5.9 | 52.5 |
| E. coli 55989 | 5.15 | 4,736 | 853 | 130 | 18.0 | 2.7 | 6.1 | 52.8 |
| E. coli S88 | 5.16 | 4,777 | 726 | 55 | 15.2 | 1.2 | 5.1 | 51.5 |
| E. coli SE11 | 5.17 | 4,805 | 937 | 197 | 19.5 | 4.1 | 5.3 | 52.5 |
| E. coli UTI89 | 5.18 | 5,072 | 996 | 94 | 19.6 | 1.9 | 4.0 | 48.7 |
| E. coli ED1a | 5.20 | 4,842 | 1,008 | 211 | 20.8 | 4.4 | 3.6 | 49.6 |
| E. coli SMS-3–5 | 5.21 | 4,750 | 846 | 163 | 17.8 | 3.4 | 4.5 | 53.0 |
| E. coli CFT073 | 5.23 | 5,014 | 1,297 | 325 | 25.9 | 6.5 | 2.7 | 47.4 |
| E. coli E24377A | 5.25 | 4,743 | 1,027 | 248 | 21.7 | 5.2 | 4.5 | 52.3 |
| E. coli UMN026 | 5.35 | 4,903 | 823 | 114 | 16.8 | 2.3 | 5.5 | 51.5 |
| E. coli O103:H2 str. 12009 | 5.48 | 5,052 | 980 | 69 | 19.4 | 1.4 | 5.1 | 49.2 |
| E. coli APEC O1 | 5.51 | 4,742 | 737 | 109 | 15.5 | 2.3 | 4.1 | 49.2 |
| E. coli O157:H7 str. Sakai | 5.60 | 5,201 | 1,204 | 117 | 23.1 | 2.2 | 3.7 | 47.2 |
| E. coli O157:H7 EDL933 | 5.62 | 5,246 | 1,283 | 165 | 24.5 | 3.1 | 3.6 | 47.2 |
| E. coli O157:H7 str. TW14359 | 5.62 | 5,315 | 1,243 | 57 | 23.4 | 1.1 | 3.5 | 46.2 |
| E. coli O157:H7 str. EC4115 | 5.70 | 5,308 | 1,390 | 169 | 26.2 | 3.2 | 2.8 | 45.7 |
| E. coli O111:H str. 11128 | 5.79 | 5,257 | 1,045 | 150 | 19.9 | 2.9 | 4.9 | 47.5 |
| E. coli O26:H11 str. 11368 | 5.86 | 5,438 | 1,187 | 78 | 21.8 | 1.4 | 4.2 | 46.9 |
A local database was created using mySQL (http://dev.mysql.com/ [cited 2011 January]) and populated with data from the earlier mentioned files, so as to contain data on cluster ID, protein GIs, taxon ID, scientific name of each taxon, and genome ID. Three groups of E. coli clusters (t1, t2, and ORFan) were designated, as described in the next section. The DNA sequences were downloaded from NCBI according to their protein GI number, and added to the mySQL database. The DNA sequence FASTA file for each protein cluster was prepared from the mySQL database and aligned by MUSCLE (Edgar 2004).
Some of the clusters originally from the NCBI clusters database have paralogous subfamilies (880 ORFan clusters, 148 t1 clusters, and 32 t2 clusters). To remove paralogy, we redefined these as the largest subcluster within each cluster that includes only one gene copy from each E. coli accession. The clustering method is the unweighted pair group method with arithmetic mean, performed in MUSCLE (Edgar 2004)
Definition of ORFan and Non-ORFan Groups of Gene Clusters
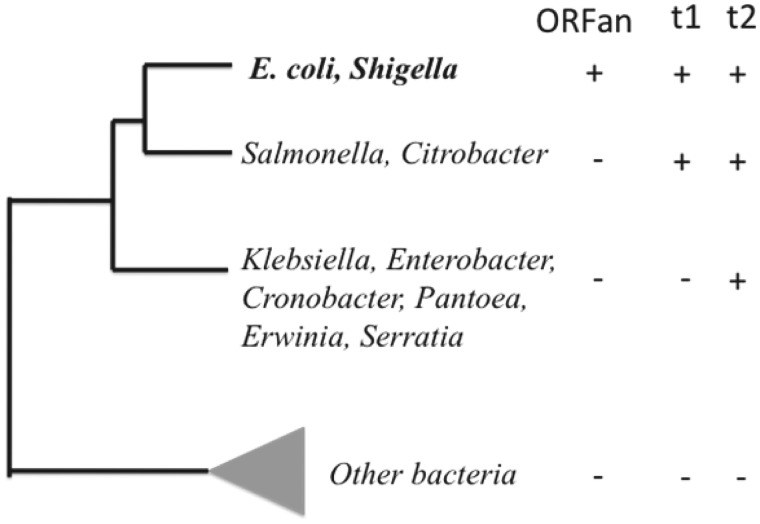
Non-ORFan control groups representing different phylogenetic depths were identified as shown in figure 1. If a protein cluster has members distributed beyond E. coli, but within the t1 group of species, it falls in the t1 group. More widely distributed members fall into the t2 group. These two groups represent robust clades in a more detailed phylogenetic tree computed from the data of Wu et al. (2009). Because the original tree of Wu et al. did not include bootstrap support values, a new tree was computed from a pruned alignment using the RAXML (Stamatakis 2006) and consense (Felsenstein 1995) software available via the CIPRES server (http://www.phylo.org/ [cited 2011 January]). The consense program was used to compute a consensus tree from 1000 RAXML bootstrap replicates performed using the WAG model. The phylogeny is provided as a PDF image (Fig S1) and as a NEXUS file in the supplementary data, Supplementary Material online. From the resulting tree, we identified two monophyletic clades with strong support (>96% bootstrap support), corresponding to the t1 (younger) and t2 (older) control groups.
Fig. 1.—
Clades used to define comparison groups with different phylogenetic depths. The t1 and t2 clades were chosen due to high bootstrap support (>96%) in a phylogeny of species computed as described (Materials and Methods) and available as supplementary figure S1, Supplementary Material online.
Population Statistics and Between-Group Comparisons
Patterns of nucleotide variation in each gene cluster were analyzed using conventional approaches of molecular population genetics (Nei and Gojobori 1986; Nei 1987; Hey and Wakeley 1997). The dN (dS) value is the average of all pairwise nonsynonymous (synonymous) differences among sequences, calculated according to Nei and Gojobori (1986). Given that some clusters have a dS value of zero, the difference between dN and dS was calculated, instead of the conventional dN/dS ratio.
Recognizing that ORFans are systematically more narrowly distributed within E. coli than non-ORFans (see Results), we created customized control clusters according to the schema in figure 2. As a result, every ORFan gene cluster has its own matching sets of t1 and t2 clusters pruned to have precisely the same strain composition, for example, if a particular ORFan cluster has the four sequences from genomes A, B, C, and D, then all of its non-ORFan control clusters are pruned to contain only the four sequences from A, B, C, and D. If a control cluster could not be pruned due to the absence of a matching strain, it was discarded.
Fig. 2.—
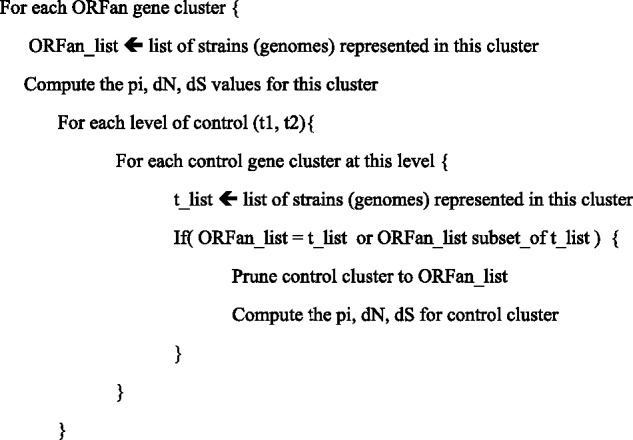
Scheme for creating matching control clusters for each ORFan cluster. The pseudocode shown here describes the method used to generate customized control clusters. If ORFan_list is not equal to the intersection of t_list and ORFan_list, then the putative control cluster cannot be used because it does not have the right set of strains.
We do not attempt to identify individual clusters with significant dN − dS values, because these values have enormous uncertainty (due to the small size and high similarity of the sequences). Instead, the distribution of values for ORFans was compared with those of non-ORFans in two ways. One mode of comparison uses Student’s t-test or the Wilcoxon test (Moore and McCabe 2002), with a significance level of 5%. Each ORFan has a (typically different) number of control clusters, and thus the number of compared values is unequal, with ORFans being compared with a larger number of non-ORFan controls that are hierarchically structured (one set of t1 or t2 control clusters for each ORFan cluster). Thus, when comparing ORFan clusters to non-ORFan controls, we randomly pick a single control cluster for each ORFan to compute a mean and variance, so that the t-test or Wilcoxon test is carried out on a nonstructured sample. Then, this calculation is repeated 1,000 times. If the majority of replicates indicate a significant difference, then the expected value of the test statistic is significant, and thus we treat the difference as significant.
The second mode of comparison is by ranking the statistics of each ORFan gene in its customized non-ORFan control group. The number of control clusters is not large enough in many cases to create smooth distributions for purposes of percentile ranking; therefore, we divide the control distribution into 10 bins, that is, deciles rather than percentiles. In order not to sacrifice statistical power, decile-based ranking is performed only for ORFans that have a customized non-ORFan control group with more than 99 clusters. Note that the first decile bin is the top 10% of the non-ORFan control distribution. An ORFan value ranked in the first decile bin of the t1 or t2 reference distribution is in the highest 10% of values, whereas an ORFan value ranked in the 10th decile bin is in the lowest 10% of values.
Results
ORFans as a Feature of Genome Composition
A summary of the composition of 35 genomes is given in table 1. On average, 19.5% of genes in a genome are species-specific ORFans, found only in strains of the species E. coli. This frequency ranges from 10.1% to 37%. On average, 3% of genes in a genome (range, 0.4% to 10.2%) are found only in that genome. The K-12 strain MG1655 has the lowest proportion of ORFans, whereas Shigella dysenteriae Sd197 has the highest proportion. As shown in table 2, in total, there are 5,101 genome-specific ORFans. These ORFan clusters cannot be used for the analysis of population diversity, because each has only one member. The remaining ORFans fall into 3,773 clusters, each with two or more sequences. The t1 group includes 610 clusters with two or more sequences, and 225 clusters with only one sequence. There are 373 clusters in t2 group, all of which include two or more sequences in each cluster.
Table 2.
Comparison Groups Used to Evaluate Diversity of ORFans in Escherichia coli
| Comparison Group | Definition | Sizea |
|---|---|---|
| ORFan | Clusters of E. coli (Shigella) sequences that are putative genes with representatives found only in E. coli (Shigella), not in any other genome. | 3,773 (5,101) |
| t1 | Clusters of E. coli (Shigella) sequences found in some of Salmonella, Citrobacter, but not in any other genomes. | 610 (225) |
| t2 | Clusters of E. coli (Shigella) sequences found in some of Salmonella, Citrobacter, and some of Klebsiella, Enterobacter, Cronobacter, Pantoea, Erwinia, Serratia but not found in other genomes. | 373 (0) |
aThe number of clusters with more than one representative among E. coli genomes; the number in parenthesis is the number of clusters with only a single E. coli sequence, thus not useful for population analysis.
Little is known about the functions of these ORFans. On average, only 5.5% of ORFans in a genome (range, 2.1% to 12.3%) have protein functions assigned by the annotation approach used in the Protein Clusters database (which may implicate a very specific class of enzyme-catalyzed reaction, or only a particular family of functions). By contrast, among all protein-coding genes in a genome, 53% (on average) have information on function.
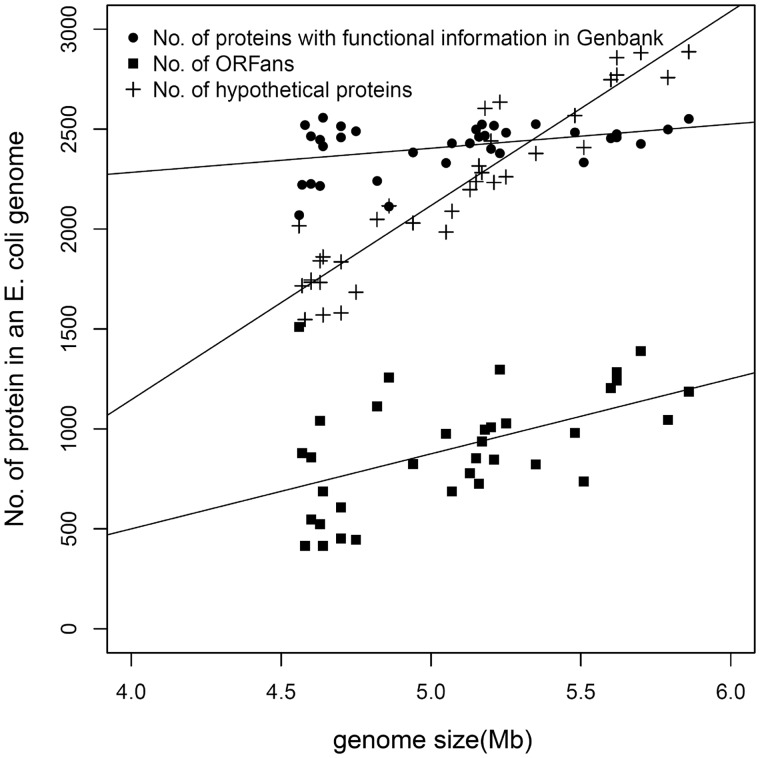
The frequencies of these different categories of proteins, as a function of genome size, are shown in figure 3. This reveals a clear relationship. The number of proteins for which functions have been inferred [in the original Protein Clusters database (Klimke et al. 2009)] remains relatively constant despite a 40% increase in size from the smallest to the largest genomes. Instead, the increase in genome size is attributable to genes (or putative genes) that lack functional annotations. Thus, differences in the complement of genes carried by an E. coli strain are not due primarily to duplication of long-established genes that are widely distributed and well annotated, but are overwhelmingly due to gain of genes that are poorly known. A minority of these genes are ORFans, found only in E. coli (Shigella).
Fig. 3.—
ORFan composition as a function of genome size in 35 Escherichia coli strains. For each genome, counts are shown for three categories of putative protein-coding genes, along with regression lines. Two of the categories are mutually exclusive: each gene in a genome is either from a cluster (in the NCBI Protein Clusters database) that has a curated functional annotation (solid circle), or it is from a cluster annotated as “hypothetical protein” (plus symbols). The solid squares show the counts of ORFans, the vast majority of which are noncurated (see text). As genome size increases, the number of proteins with assigned functions remains nearly constant. The increase in genome size is not mainly attributable to ORFans, but is attributable to other genes for which functions are unknown.
Distinctive Features of ORFan Genes
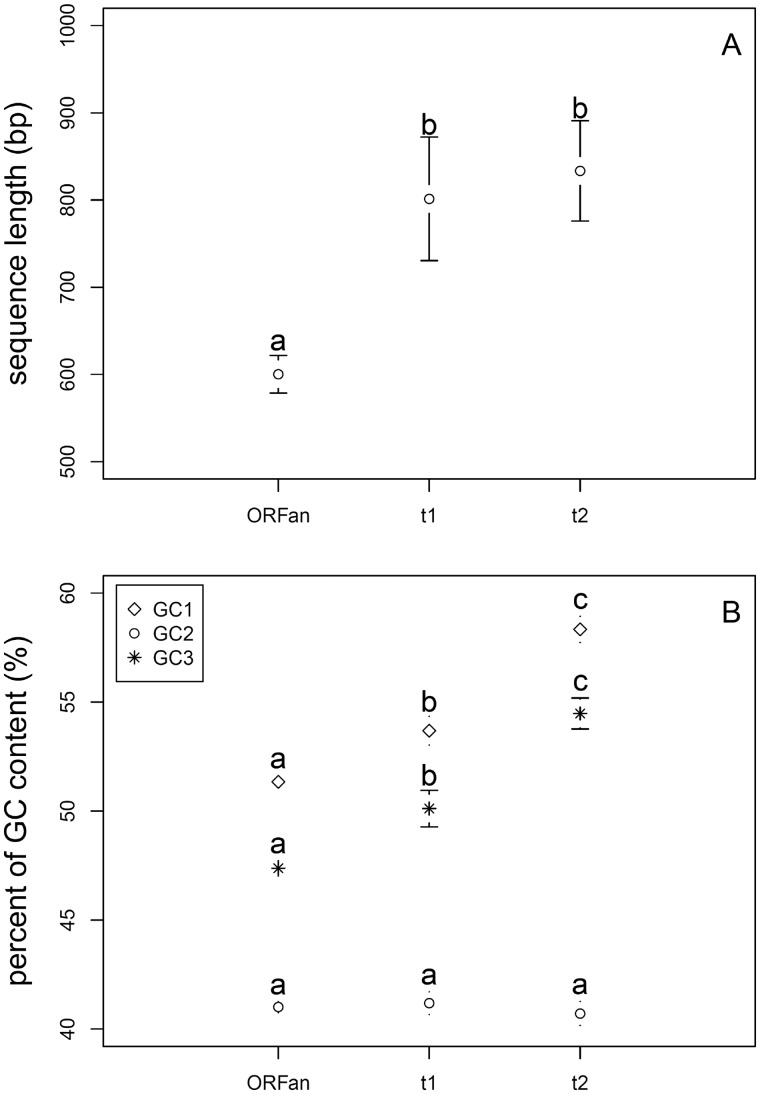
ORFans show some unique features when compared with non-ORFans (fig. 4). The average sequence length of ORFans (∼600 nucleotides or 200 codons) is significantly shorter than the t1 and t2 controls according to both Student’s t-test and Wilcoxon test (P < 0.01) (Moore and McCabe 2002). ORFan genes have the lowest average GC content at the first and third position of codons, followed by the t1 group, then the t2 group. GC content at the second position of codons is not significantly different among these three groups. The most obvious interpretation of this result is that, when new genes are acquired, they are initially more AT-rich, and gradually change toward the composition of long-established genes, as suggested by Lawrence and Ochman (1997).
Fig. 4.—
Genic features of ORFans compared with non-ORFans. (A), average size (in base pairs). ORFans are shorter than non-ORFans. (B), average percent of GC at first (GC1), second (GC2), and third (GC3) position of codons. Except for GC2 in (B), the three classes of clusters ORFans differ significantly in genic features. ORFans have lower GC content at first and third positions of codons. Bars represent 95% confidence intervals. The letters denote significantly different results by the Wilcoxon test (results from Student’s t-test are the same).
Population Distribution of ORFans
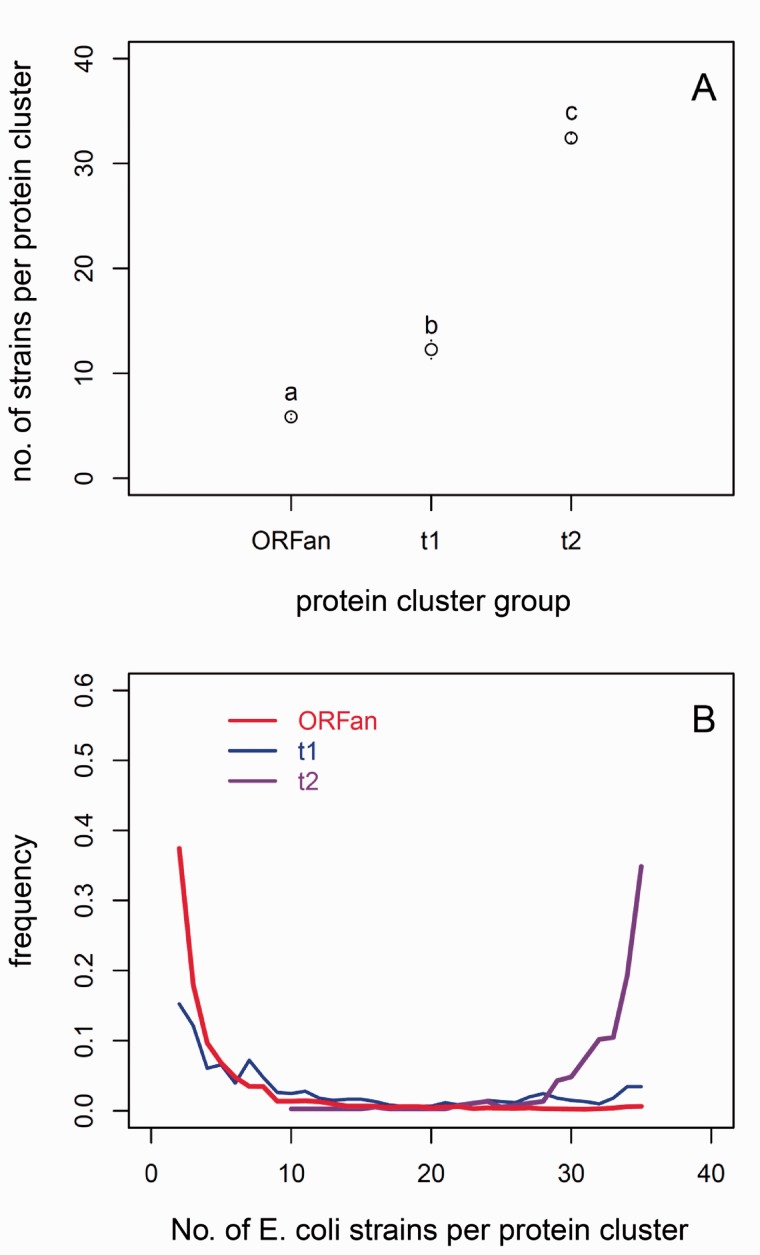
The typical ORFan gene has a restricted distribution among E. coli strains, being present in 6.2 of the 35 genomes, on average. By comparison, t1 and t2 genes are represented in 13.2 and 32.4 (respectively) strains, on average (fig. 5A). The difference among these three groups is significant according to both Student’s t-test and Wilcoxon test (P < 0.001). The frequency distributions for all three groups are shown in figure 5B. For ORFans, genes represented in only a single genome represent the largest frequency class (57%). The 1-gene clusters of ORFans and t1 genes are excluded from figure 5B, which shows only those clusters used in population analysis, that is, clusters with at least two members.
Fig. 5.—
Distribution of ORFan and non-ORFan genes among genomes of Escherichia coli strains. (A) The average number of E. coli stains per protein cluster in the ORFan and non-ORFan cluster groups; (B) the frequency distribution of number of E. coli stains per cluster used in the ORFan and non-ORFan comparison groups (this excludes ORFan clusters with only one member, which is the most common size of a cluster). ORFans typically have narrow distributions, while non-ORFans in the t2 comparison group are present in most genomes. Non-ORFans in the t1 group have an intermediate distribution. The letters in (A) denote significantly different results by the Wilcoxon test (results from Student’s t-test are the same).
Sequence Diversity of ORFans
The sequence diversity of ORFan clusters is very low, due to their small size and recent origins, and this affects the kinds of analysis that are possible. Specifically, the distribution of numbers of nonsynonymous polymorphisms in ORFan clusters is as follows: 0 (polymorphisms): 792 (clusters); 1: 264; 2: 187; 3: 128; 4: 86; 5: 83; 6: 68; 7: 59; 8: 44; 9: 37; and >9: 472. The counts for synonymous polymorphisms are 0: 938; 1: 271; 2: 133; 3: 76; 4: 76; 5: 51; 6: 52; 7: 33; 8: 27; 9: 31; >9: 532.
Given that some clusters have a dS value of zero, the difference between dN and dS was calculated, instead of the more conventional dN/dS ratio. ORFans that represent real protein-coding genes typically would be expected to show the pattern typical of real protein-coding genes, which is that dS exceeds dN; for ORFans that represent pseudogenes or misannotated regions, the expected values of dN and dS are the same; ORFans undergoing adaptive evolution are expected to have higher dN value than dS (Hurst 2002).
As shown in table 3, most ORFan, t1 and t2 clusters have dN less than dS. However, compared with t1 and t2, ORFan clusters are less likely to have a negative dN – dS, and more likely to have a positive value (χ2 = 66.06, 292.94; df = 2; P < 0.001). It is interesting to note that, relative to t2, the t1 group also shows a significantly higher proportion of genes with dN − dS greater than 0 (χ2 = 119.35, df = 2, P < 0.001).
Table 3.
Proportions of Clusters in Each Comparison Group that Fall into Different Categories with Respect to the Relationship of dN to dS
| Comparison Group | dN – dS > 0 | dN – dS < 0 | dN = dS = 0 | Number |
|---|---|---|---|---|
| ORFan | 944 (25) | 1,953 (52) | 876 (23) | 3,773 |
| t1 | 104 (17) | 423 (69) | 83 (14) | 610 |
| t2 | 8 (2) | 365 (98) | 0 (0) | 373 |
Note.—The numbers are number of genes in the genes in the category. The number in the parenthesis is the percentage of genes in the category.
However, the differences between the categories show a large effect of cluster size (fig. 5). This is shown by the fact that approximately 23% of ORFan clusters and 17% of t1 clusters have equal dN and dS. These clusters are nonpolymorphic in E. coli and have dN = dS = 0, presumably because the clusters are very small. Most ORFan clusters have fewer than five sequences, while clusters of such small size are vanishingly rare in the t2 group of genes that have been present for millions of years in the lineage leading to E. coli.
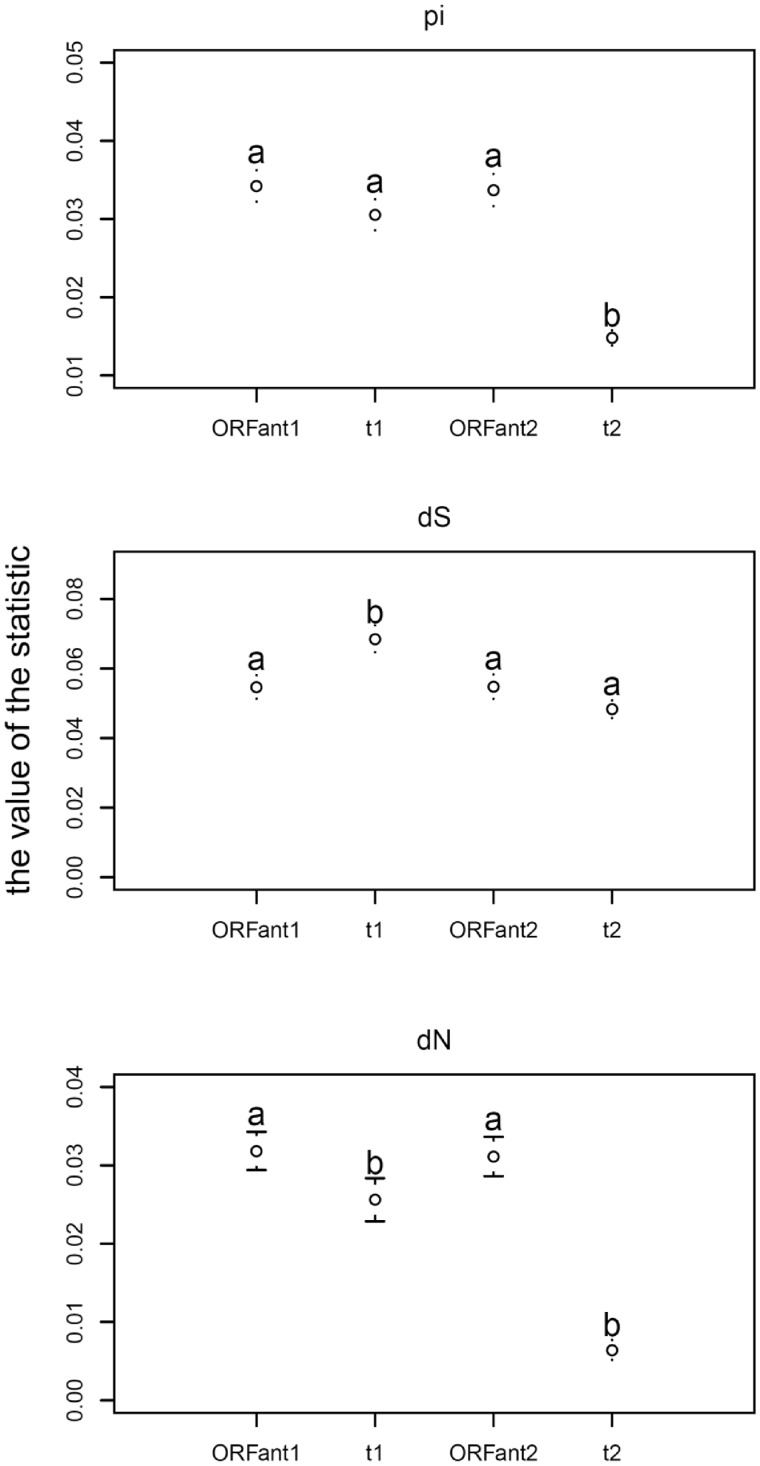
To correct for these differences in cluster size, we created matched comparison clusters as described (earlier, and fig. 2). The results of the cluster-size-corrected comparisons of diversity statistics are shown in figure 6, and with more detail in figure 7. ORFans show a higher mean value of Pi than genes in the t1 and t2 groups, though only the difference with t2 is significant. Both Student’s t-test and the Wilcoxon test indicate that the average dS value of ORFan gene group is significantly lower than t1 group and not significantly different from t2 group. Average dN value of ORFan gene group is higher than t1 and t2 group. According to the Wilcoxon test, the difference between ORFans and t1 group, ORFans and t2 group is significant. According to Student’s t-test, the difference between ORFans and t1 group is not significant; the difference between ORFans and t2 group is significant. The result suggests that ORFan gene group show similar or lower dS value, but higher dN value than t1 or t2 gene group.
Fig. 6.—
Mean population statistics compared between ORFans and non-ORFans. This figure shows a comparison of means for pi (upper panel), dS (middle panel), and dN (lower panel), whereas the complete distributions are compared, via deciles, in figure 7. There are two different sets of mean values for ORFan clusters, because the comparisons with t1 and t2 use overlapping but nonidentical sets of ORFan clusters, due to the need to create matching controls with the same strain composition (see Materials and Methods). Although synonymous diversity is not much different, nonsynonymous diversity, as well as total diversity (Pi), is significantly different between ORFans and non-ORFans in the t2 control group. The letters denote significantly different results by the Wilcoxon test; Student’s t-test gives a slightly different result for dN, not shown, with no significant distinction between t1 and the ORFan clusters (i.e., the pattern is a–a–a–b).
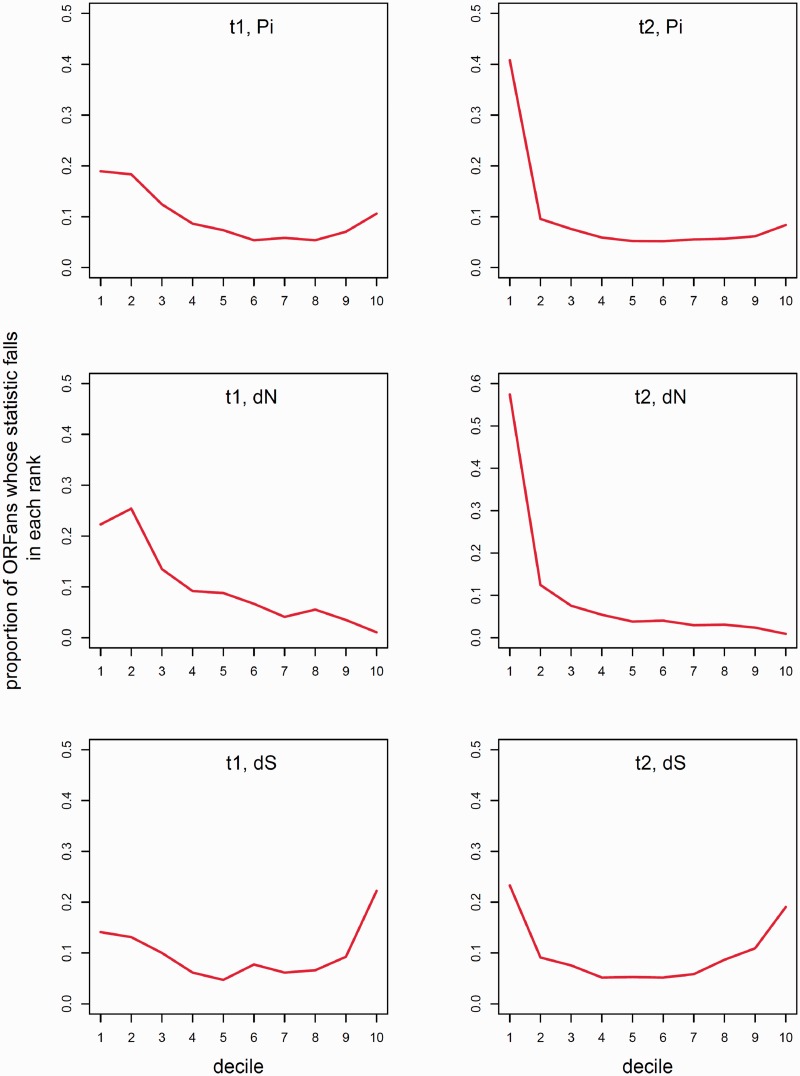
Fig. 7.—
Distribution of diversity statistics compared between ORFans and non-ORFan controls. The three rows show the distributions for pi (upper), dS (middle) and dN (lower), for both t1 (left column) and t2 (right column) control sets. To understand the shape of the distribution of population statistics for ORFans, values for ORFans were gathered into decile bins defined by the non-ORFan control clusters, that is, each bin comprises 10% of the distribution of non-ORFan values. The value on the Y axis for the first decile bin in (A), for instance, represents the frequency with which the Pi value for an ORFan ranks in the top 10% of the values in its customized t1 control group. (C, E) The same comparison for dN and dS; (B, D, and F) the distribution of Pi, dN, and dS (respectively) relative to the t2 comparison group. The null expectation is a straight line at a value of 10%, with a slight anomaly at the low (right) end of the distribution due to zero values (in cases where zero values exceed 10% of the control distribution, zero values in the ORFan distribution will be placed in whichever bin is counted first, which in this case tends to leave a shortage in the last bin). The symmetric but slightly U-shaped distribution of dS values indicates that ORFans exhibit greater variance, but otherwise have the same distribution of synonymous differences as non-ORFans. However, the deviation from the distribution of dN (and Pi) values is asymmetric, with a 2-fold or more excess of ORFan clusters with diversity in the top 10% or 20% of the distribution relative to non-ORFan controls.
To dissect these differences in more detail, we ranked ORFan values relative to their matched controls, using deciles (rather than percentiles, due to the limited number of control clusters for each ORFan). This allows the data from all ORFans to be combined via rankings, even though each ORFan has a different control distribution. The results are shown in figure 7. The symmetric but slightly U-shaped distributions in figure 7E and 7F suggest that synonymous variation in ORFs is distributed very similarly to that in non-ORFans, and differs only in having somewhat greater stochastic variance, as expected given that ORFans are approximately three-fourth the size of non-ORFans (fig. 4A). The difference between ORFans and non-ORFans for dN (and Pi) is much more marked and asymmetric, being skewed to the left, toward higher values. The ORFan distribution has a substantial excess of values in the top one or two deciles, relative to the distribution expected from examining non-ORFans. For the comparison of dN values with the t2 comparison group, there is approximately a 5-fold excess of ORFans in the first decile. That is, the most unusual feature of ORFans, considered as a class of probable genes, is the presence of a large subclass of ORFans with high nonsynonymous diversity.
Discussion
The results given above on the genic features of ORFans, and the composition of genomes with respect to ORFans, are largely confirmatory. Our results show that approximately 19% of genes per genome are E. coli lineage-specific genes. As expected from previous studies (Charlebois et al. 2003; Daubin and Ochman 2004), ORFans, as a class, are slightly shorter and more AT-rich than older and more widely distributed genes.
What is more distinctive about the present study is the use of a population sample to study within-species diversity in ORFans. The set of available genomes, of course, is not in any sense a random sample of the E. coli population, but instead is heavily biased toward strains of relevance to technology (i.e., laboratory strains) and medicine (i.e., pathogenic strains). Nevertheless, the set of available genomes is a subset of the population that includes considerable diversity, and is worthy of study. Our analysis has some bearing on two issues, the origins of ORFans, and their subsequent evolution in E. coli.
Origins of ORFans
A few tentative conclusions are possible regarding the possible origins of ORFans, which might emerge as annotation artifacts, horizontally acquired genes, de novo genes, or highly diverged homologs of much older genes. The distribution of ORFans among genomes (fig. 3) argues against the idea that ORFans may represent members of well known protein families whose affinities remain undetected due to rapid divergence. Specifically, the number of curated proteins remains roughly constant as the number of ORFans increases, whereas if ORFans frequently represent highly diverged genes, we would expect this number to decrease (if old genes diverge in situ into unrecognizable ORFans) or to increase (if old genes frequently spawn duplicates that eventually diverge unrecognizably into ORFans, we would expect to see a contingent of rapidly diverging duplicates that are not yet unrecognizably diverged).
The distribution of sequence diversity statistics holds other clues that we would interpret as follows. The t2 group consists (presumably) entirely of real genes, and nearly all clusters have dN < dS. Most ORFan clusters also have dN < dS. Specifically, approximately two-thirds of the time, the dN – dS value is negative (table 3). This might suggest that at least two-third of the ORFans are legitimate genes. However, no such simple conclusion is possible given the potential for statistical noise in dN – dS. For the ORFan group, most clusters have fewer than six sequences (fig. 5), and the resulting small chance of finding polymorphisms results in many cases of dN = dS = 0, whereas for the t2 group, most clusters have over two dozen sequences, and there are no cases of dN = dS = 0. If there is some fraction of ORFans that are pseudogenes for which the expected values of dN and dS are equal, then due to the low frequency of polymorphisms, there will be some cases of dN = dS = 0, along with cases in which the dN – dS value deviates stochastically in the positive or negative direction. For instance, if we look at the ORFan and t2 rows in table 3, one interpretation is that one-fourth of the ORFans are real genes (like t2 genes) with negative dN – dS, while the other three-fourths are misannotated genes with dN – dS values distributed equally among positive, negative, and zero values.
This interpretation is difficult to sustain if one considers the t1 group. The t1 group includes genes that, albeit young compared with t2 genes, have been around for tens of millions of years, and thus the t1 group presumably includes no artifacts. Yet, t1 clusters do not have the same size distribution as t2 clusters, but instead are intermediate, with most t1 clusters being in only a minority of the E. coli genomes (fig. 5). This suggests that genes in the t1 group are more frequently lost than those in the t2 group. That the t1 clusters are different in character from the t2 clusters is clear in figure 6. Figures 6 and 7 show that, when we correct for the sizes of clusters, ORFan clusters are very much like t1 clusters, and are rather unlike t2 clusters, in their distribution of dN and Pi.
Whatever the reason for the narrower distributions of t1 clusters, the dN – dS value is negative only four-fifths of the time, whereas for t2 genes it is nearly always negative. If ORFans are composed of a fraction f of artifacts for which the distribution of positive and negative dN – dS values is 1:1 (i.e., assuming for the sake of approximation that random deviations from 0 are symmetric), and some fraction 1 − f of genes with a 1:4 ratio like t1 genes, f could not be greater than one-half, otherwise the ratio of positive to negative values would be more than the observed 1:2 ratio (i.e., it would be more like the 1:1 ratio expected for pseudogenes).
Thus, although the interpretation is not straightforward, the results presented here suggest that most ORFans are functional genes rather than artifacts of misannotation, and that they do not arise by radical divergence from well-known sequence families. This suggestion that most ORFans encode functional proteins, if correct, is corroborated by evidence from some other organisms (Benson et al. 2001; Awano et al. 2006; Cai et al. 2008; Chen et al. 2010). For instance, gene BSC4 in Saccharomyces cerevisiae, which is found de novo created from noncoding sequence, is found to be involved in the DNA repair pathway during the stationary phase of S. cerevisiae and contribute to the robustness of S. cerevisiae, when shifted to a nutrient-poor environment.
Diversification of ORFans in E. coli
Both in bacteria and eukaryotes, lineage-specific genes are seen to evolve at a faster rate than broadly distributed genes (Domazet-Loso and Tautz 2003; Daubin and Ochman 2004; Wang et al. 2005; Kuo et al. 2008). For example, Daubin and Ochman (2004) found that genes that are restricted to E. coli MG1655 and Salmonella enterica evolve faster than more widely distributed genes. Similar results were found in Drosophila, parasitic protozoa, and rodents (Domazet-Loso and Tautz 2003; Wang et al. 2005; Kuo et al. 2008). This pattern of the relationship between evolutionary rate and apparent age (phylogenetic breadth of a gene) could be due to the variation in strength of purifying selection or due to the variation in the rate of adaptive evolution. Weaker purifying selection in young genes would imply that these genes have less effect on fitness and therefore are less selectively constrained. The alternative explanation, not mutually exclusive, is that lineage-specific genes participate more in lineage-specific adaptation, and therefore evolve faster (Cai and Petrov 2010).
Our results show that ORFans have a distribution of synonymous diversity similar to non-ORFans. The main difference between ORFans and non-ORFans is an excess of clusters with high dN values. As shown in figure 7D, about one-half of the ORFan dN values fall into the top 10% of the t1 control distribution, and the remainder are distributed with slightly decreasing frequency throughout the remainder of the distribution. It might be argued that the more recent the evolutionary acquisition of a gene sampled from within a population, the greater the extent to which observed differences will reflect short-term population dynamics, dominated by mutation, rather than the longer time-scale over which the effects of selective filtering accumulate. By this argument, following Kryazhimskiy and Plotkin (2008), one expects the ratio dN/dS to converge on 1 for the youngest genes. However, if we may assume vertical inheritance, this argument does not apply to the interpretation of our results, because every comparison between an ORFan cluster and a non-ORFan cluster is based on a matched set of strains.
Several other factors might account for the higher distribution of dN values in ORFans. Although we argued above that most ORFans are not artifacts, nevertheless, this conclusion still allows for a substantial minority of ORFans to be artifacts, for example, misannotated noncoding regions, or former genes that no longer contribute to fitness (e.g., acquired bacteriophage genes). This fraction of ORFans would be diversifying at the rate of maximal neutral change for a noncoding DNA sequence in E. coli.
Another possibility is that ORFans, as a class of functional protein-coding genes, have a higher rate of change due to stochastic acceptance of mutations, that is, reduced selective “constraint.” As noted earlier, t1 genes of (apparent) intermediate age also have a higher rate of nonsynonymous change than the older t2 genes; ORFan genes are much like t1 genes in their diversity statistics. A higher rate of evolution might be argued on various empirical or logical grounds. ORFans are shorter, thus the encoded proteins have a higher surface-to-volume ratio, which entails a higher rate of evolution (given the higher rate of evolution of surface sites in proteins). ORFan genes may be expressed at low levels (or infrequently), which may make them more tolerant of changes relative to genes expressed at high (and consistent) levels. ORFans are not acquired gradualistically, one codon at a time, but arrive in large pieces, and—even without presuming anything about how these new pieces fit into the economy of the cell—it stands to reason that they do not fit as precisely as pieces that have been around for many millions of years, which again suggests that ORFans will be more tolerant of arbitrary changes. A previous study in humans showed that older genes carry fewer and less frequent nonsynonymous single-nucleotide polymorphisms than younger primate-specific genes (Cai and Petrov 2010).
A third possibility would be that relative to non-ORFans, there is an excess of ORFans experiencing rapid evolution via beneficial mutations. A previous study showed an excess of primate-specific genes with a signal of positive selection, relative to genes with a broader distribution (Cai and Petrov 2010). Currently, these possibilities cannot be distinguished without a deeper examination of ORFan diversity.
Further Questions
In this study, we have applied relatively conventional methods of analysis to a difficult set of data, to obtain preliminary interpretations. Much more could be done to evaluate the evolution of ORFans in E. coli with additional data or specialized methods. For instance, a more integrated analysis that includes expression data (e.g., RNASeq data) and proteomics data might clarify the picture of ORFan genomics. Unfortunately such data are not available systematically for the whole set of E. coli strains. A further type of analysis that is possible and that represents an obvious next step would be to analyze in more detail the likelihood that the map locations of ORFans will reveal clues about their origin. Specifically, a variety of studies cited earlier have suggested that ORFan genes are frequently acquired via bacteriophage infections, which implies that the ORFans should arrive in clusters (representing inserted prophages) and will tend to be found at prophage insertion sites in the genome.
If most ORFans are showing a conservative pattern of evolution (i.e., dN < dS, as argued earlier), the results presented here have an interesting implication with respect to the above view that ORFans are mainly acquired from phage genomes. Specifically, the conservative pattern of divergence would indicate that the ORFans are not merely silent prophage genes. To the extent that horizontal propagation of phage kills the host cell, but ORFans are propagated vertically (after acquisition) and show a conservative pattern of divergence, this conservation cannot reflect selection for phage propagation, but only for host (lysogen) propagation, indicating a role for these genes in host propagation. Thus, the biological significance of these results ultimately will depend on distinguishing the contributions of 1) vertical inheritance of an ORFan acquired in an ancestral strain, 2) within-population events of chromosome recombination that may transfer genes between strains, and 3) repeated introgression of the ORFan from sources outside the E. coli population. This would seem to be difficult without having some model of the evolution of ORFan sources outside of E. coli, currently a mystery.
Supplementary Material
A list of plasmid identifiers, as well as the phylogeny used to define control groups (in PDF and NEXUS formats), are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank John Moult for discussions throughout the project, and Howard Ochman and Zvi Kelman for comments. This work was supported by a grant from the National Institutes of Health grant (GM081511) to John Moult and A.S. The identification of any specific commercial software products is for the purpose of specifying a protocol, and does not imply a recommendation or endorsement by the National Institute of Standards and Technology.
Literature Cited
- Awano T, et al. A frame shift mutation in canine TPP1 (the ortholog of human CLN2) in a juvenile dachshund with neuronal ceroid lipofuscinosis. Mol Genet Metabol. 2006;89:254–260. doi: 10.1016/j.ymgme.2006.02.016. [DOI] [PubMed] [Google Scholar]
- Benson MD, et al. A new human hereditary amyloidosis: the result of a stop-codon mutation in the apolipoprotein AII gene. Genomics. 2001;72:272–277. doi: 10.1006/geno.2000.6499. [DOI] [PubMed] [Google Scholar]
- Cai JJ, Petrov DA. Relaxed purifying selection and possibly high rate of adaptation in primate lineage-specific genes. Genome Biol Evol. 2010;2:393–409. doi: 10.1093/gbe/evq019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Zhao RP, Jiang HF, Wang W. De novo origination of a new protein-coding gene in Saccharomyces cerevisiae. Genetics. 2008;179:487–496. doi: 10.1534/genetics.107.084491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlebois RL, Clarke GD, Beiko RG, Jean A. Characterization of species-specific genes using a flexible, Web-based querying system. FEMS Microbiol Lett. 2003;225:213–220. doi: 10.1016/S0378-1097(03)00512-3. [DOI] [PubMed] [Google Scholar]
- Chen SD, Zhang YE, Long MY. New genes in Drosophila quickly become essential. Science. 2010;330:1682–1685. doi: 10.1126/science.1196380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chothia C. Proteins. One thousand families for the molecular biologist. Nature. 1992;357:543–544. doi: 10.1038/357543a0. [DOI] [PubMed] [Google Scholar]
- Cortez D, Forterre P, Gribaldo S. A hidden reservoir of integrative elements is the major source of recently acquired foreign genes and ORFans in archaeal and bacterial genomes. Genome Biol. 2009;10:R65. doi: 10.1186/gb-2009-10-6-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubin V, Lerat E, Perriere G. The source of laterally transferred genes in bacterial genomes. Genome Biol. 2003;4: R57. doi: 10.1186/gb-2003-4-9-r57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubin V, Ochman H. Bacterial genomes as new gene homes: the genealogy of ORFans in E coli. Genome Res. 2004;14:1036–1042. doi: 10.1101/gr.2231904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domazet-Loso T, Tautz D. An evolutionary analysis of orphan genes in Drosophila. Genome Res. 2003;13:2213–2219. doi: 10.1101/gr.1311003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP (Phylogenetic inference package) Seattle: Department of Genetics, University of Washington; 1995. [Google Scholar]
- Hahn Y, Lee B. Identification of nine human-specific frameshift mutations by comparative analysis of the human and the chimpanzee genome sequences. Bioinformatics. 2005;21:I186–I194. doi: 10.1093/bioinformatics/bti1000. [DOI] [PubMed] [Google Scholar]
- Hey J, Wakeley J. A coalescent estimator of the population recombination rate. Genetics. 1997;145:833–846. doi: 10.1093/genetics/145.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst L. The Ka/Ks ratio: diagnosing the form of sequence evolution. Trends Genet. 2002;18:486–489. doi: 10.1016/s0168-9525(02)02722-1. [DOI] [PubMed] [Google Scholar]
- Khalturin K, et al. A novel gene family controls species-specific morphological traits in hydra. PLoS Biol. 2008;6:2436–2449. doi: 10.1371/journal.pbio.0060278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimke W, et al. The national center for biotechnology information's protein clusters database. Nucleic Acids Res. 2009;37:D216–D223. doi: 10.1093/nar/gkn734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles DG, McLysaght A. Recent de novo origin of human protein-coding genes. Genome Res. 2009;19:1752–1759. doi: 10.1101/gr.095026.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryazhimskiy S, Plotkin JB. The population genetics of dN/dS. PLoS Genet. 2008;4:e1000304. doi: 10.1371/journal.pgen.1000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo MC, Chou LF, Chang HY. Evolution of exceptionally large genes in prokaryotes. J Mol Evol. 2008;66:333–349. doi: 10.1007/s00239-008-9081-9. [DOI] [PubMed] [Google Scholar]
- Lawrence JG, Ochman H. Amelioration of bacterial genomes: rates of change and exchange. J Mol Evol. 1997;44:383–397. doi: 10.1007/pl00006158. [DOI] [PubMed] [Google Scholar]
- Logsdon JM, Jr, Doolittle WF. Origin of antifreeze protein genes: a cool tale in molecular evolution. Proc Natl Acad Sci U S A. 1997;94:3485–3487. doi: 10.1073/pnas.94.8.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M. Evolution of novel genes. Curr Opin Genet Dev. 2001;11:673–680. doi: 10.1016/s0959-437x(00)00252-5. [DOI] [PubMed] [Google Scholar]
- Long M, et al. Origin of new genes: evidence from experimental and computational analyses. Genetica. 2003;118:171–182. [PubMed] [Google Scholar]
- Moore DS, McCabe GP. New York: W.H. Freeman; 2002. Introduction to the practice of statistics. [Google Scholar]
- Nei M. New York: Columbia University Press; 1987. Molecular evolutionary genetics. [Google Scholar]
- Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Ochman H. Distinguishing the ORFs from the ELFs: short bacterial genes and the annotation of genomes. Trends Genet. 2002;18:335–337. doi: 10.1016/s0168-9525(02)02668-9. [DOI] [PubMed] [Google Scholar]
- Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- Ohno S. Birth of a unique enzyme from an alternative reading frame of the preexisted, internally repetitious coding sequence. Proc Natl Acad Sci U S A. 1984;81:2421–2425. doi: 10.1073/pnas.81.8.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rancurel C, Khosravi M, Dunker AK, Romero PR, Karlin D. Overlapping genes produce proteins with unusual sequence properties and offer insight into de novo protein creation. J Virol. 2009;83:10719–10736. doi: 10.1128/JVI.00595-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Toll-Riera M, et al. Origin of primate orphan genes: a comparative genomics approach. Mol Biol Evol. 2009;26:603–612. doi: 10.1093/molbev/msn281. [DOI] [PubMed] [Google Scholar]
- Touchon M, et al. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 2009;5:e1000344. doi: 10.1371/journal.pgen.1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Yu H, Long M. Duplication-degeneration as a mechanism of gene fission and the origin of new genes in Drosophila species. Nat Genet. 2004;36:523–527. doi: 10.1038/ng1338. [DOI] [PubMed] [Google Scholar]
- Wang W, et al. Origin and evolution of new exons in rodents. Genome Res. 2005;15:1258–1264. doi: 10.1101/gr.3929705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson GA, Bertrand N, Patel Y, Hughes JB, Feil EJ, Field D. Orphans as taxonomically restricted and ecologically important genes. Microbiology. 2005;151:2499–2501. doi: 10.1099/mic.0.28146-0. [DOI] [PubMed] [Google Scholar]
- Wu D, et al. A phylogeny-driven genomic encyclopaedia of bacteria and archaea. Nature. 2009;462:1056–1060. doi: 10.1038/nature08656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Fischer D. On the origin of microbial ORFans: quantifying the strength of the evidence for viral lateral transfer. BMC Evol Biol. 2006;6:63. doi: 10.1186/1471-2148-6-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Fischer D. Identification and investigation of ORFans in the viral world. BMC Genom. 2008;9:24. doi: 10.1186/1471-2164-9-24. [DOI] [PMC free article] [PubMed] [Google Scholar]