Abstract
Males and females experience differences in gene dose for loci in the nonrecombining region of heteromorphic sex chromosomes. If not compensated, this leads to expression imbalances, with the homogametic sex on average exhibiting greater expression due to the doubled gene dose. Many organisms with heteromorphic sex chromosomes display global dosage compensation mechanisms, which equalize gene expression levels between the sexes. However, birds and Schistosoma have been previously shown to lack chromosome-wide dosage compensation mechanisms, and the status in other female heterogametic taxa including Lepidoptera remains unresolved. To further our understanding of dosage compensation in female heterogametic taxa and to resolve its status in the lepidopterans, we assessed the Indian meal moth, Plodia interpunctella. As P. interpunctella lacks a complete reference genome, we conducted de novo transcriptome assembly combined with orthologous genomic location prediction from the related silkworm genome, Bombyx mori, to compare Z-linked and autosomal gene expression levels for each sex. We demonstrate that P. interpunctella lacks complete Z chromosome dosage compensation, female Z-linked genes having just over half the expression level of males and autosomal genes. This finding suggests that the Lepidoptera and possibly all female heterogametic taxa lack global dosage compensation, although more species will need to be sampled to confirm this assertion.
Keywords: dosage compensation, Lepidoptera, sex chromosomes, de novo transcriptome assembly, orthology
Introduction
There is an inherent imbalance in gene dosage for sex-linked genes between the sexes (Bachtrog et al. 2011). In female heterogametic taxa, such as birds, snakes, and lepidopterans, ZW females have one fewer copy of all Z-linked genes compared with ZZ males. A converse situation exists in male heterogametic taxa, such as mammals and Drosophila, where XY males have half the gene dose for X-linked genes compared with XX females. Differences in gene dose result in differences in expression for many genes (Schlattl et al. 2011; Zhou et al. 2011), and this could produce differences in expression for many hundreds of genes in systems with highly differentiated sex chromosomes.
Although the relationship between dose and expression is complex (Papp et al. 2003; Birchler et al. 2005, 2007), it is clear that some genes are dosage sensitive (Redon et al. 2006; Estivill and Armengol 2007; Makino and McLysaght 2010), and for these genes, changes in gene dose can result in severe phenotypes. Ohno (1967) suggested that this imbalance in gene dosage would result in selection for a dosage compensation mechanism to restore expression of the single X or Z chromosome in the heterogametic sex to the diploid level that existed before degradation of the Y or W gene content. Originally envisaged to apply to the entirety of the sex chromosome, it is clear that animals actually exhibit a range of sex chromosome dosage compensation (Mank 2009; Mank et al. 2011). At one end of the spectrum are organisms with nearly complete chromosome-wide compensating mechanisms, such as Drosophila and Caenorhabditis elegans (Lucchesi 1973; McDonel et al. 2006; Ercan et al. 2007), where the average expression of the X chromosome between the sexes, and between the single X and the diploid autosomes in the heterogametic sex, is equal. At the other end of the spectrum are species with only partial compensation, where the majority of genes on the X or Z are uncompensated in the heterogametic sex. In these cases, only a subset of sex-linked genes shows balanced expression between the sexes, and the average expression of the single X or Z in the heterogametic sex is less than the autosomal average and also less than the X or Z chromosome average in the homogametic sex. Examples of incomplete dosage compensation have been documented in birds (Itoh et al. 2007; Naurin et al. 2011; Wolf and Bryk 2011), platypus (Deakin et al. 2008), and Schistosoma (Vicoso and Bachtog 2011). There are also species in the middle of the spectrum, such as Tribolium (Prince et al. 2010), and potentially the eutherian mammals (Julien et al. 2012, Pessia et al. 2012), though there is still an unresolved debate regarding the status of dosage compensation in the placental mammals, with some studies indicating complete dosage compensation and others showing only partial compensation (Xiong et al. 2010; Deng et al. 2011; Kharchenko et al. 2011; Lin et al. 2012).
This variation in sex chromosome dosage compensation begs interesting questions about why some clades evolve complex mechanisms of dosage compensation for whole chromosomes and others simply compensate dosage sensitive genes (Mank 2009; Mank and Ellegren 2009; Wright and Mank 2012). To address these questions, we require information about the status of dosage compensation in a range of animals, both male and female heterogametic. The Lepidoptera, comprising butterflies and moths, share an orthologous Z chromosome, although the W is not conserved (Sahara et al. 2012). The status of dosage compensation of the Lepidopteran Z chromosome is currently unclear. Initial evidence from the silkworm, Bombyx mori, indicates incomplete dosage compensation, similar to that found in birds (Arunkumar et al. 2009; Zha et al. 2009). However, a subsequent reanalysis of the same data set, which corrected for a number of microarray biases, called this result into question, suggesting that Z:A ratios are less than 1 in both females and males (Walters and Hardcastle 2011). Unfortunately, the data set was ultimately deemed too noisy to be conclusive.
As a large clade with ancient and highly distinct Z and W sex chromosomes, the Lepidoptera represent a valuable group to investigate the status of sex chromosome dosage compensation. To resolve whether chromosome-wide Z-linked gene dosage compensation occurs in Lepidoptera, we built a de novo transcriptome assembly from Illumina RNA-Seq data from the Indian meal moth, Plodia interpunctella, which lacks a complete draft reference genome. The three fully sequenced Lepidopteran genomes, Bombyx (Mita et al. 2004), Monarch (Zhan et al. 2011), and Heliconius (The Heliconius Genome Consortium 2012), show a remarkable conservation of Z chromosome gene content across both butterflies and moths. We were therefore able to use orthology from the fully sequenced B. mori genome to assign putative genomic location for assembled transcripts as either Z-linked or autosomal, and we then estimated male and female expression levels for all putative 1:1 orthologs. The results show that not only is this strategy effective for assessing genomic location and expression level in the absence of full genome sequences, but more importantly that Plodia lacks complete sex chromosome dosage compensation.
Materials and Methods
Sample Preparation and Sequencing
We collected whole-body samples from 20 male and 20 virgin female Indian meal moths (P. interpunctella) at 1 day of age. Larvae were reared at a standard density (10 larvae in 150 ml of medium) using standard protocols on a diet of bran midlings, yeast, honey, and glycerol and were maintained at 28°C with a 16:8 h light–dark cycle (Ingleby et al. 2010). These samples were divided into four nonoverlapping sex-specific pools, each consisting of 10 same-sex individuals, thus creating two replicate pools for each sex. Pooling is useful in that it reduces replicate variance, and therefore increases statistical power, although at the cost of the ability to track individuals. RNA was extracted using an RNeasy Qiagen kit and prepared for sequencing by the University of Exeter Biosciences Sequencing Service using standard Illumina protocols.
Whole transcriptome shotgun sequencing (RNA-Seq) was performed on an Illumina Genome Analyzer II, with a separate lane for each of the four pools. The resulting 76-bp paired-end reads were assessed for quality using FastQC (http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc, last accessed November 2, 2011) and filtered with FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/index.html, last accessed August 5, 2011) to exclude reads where less than 90% of positions base calls had quality scores of Q20 or above. Additionally, any reads identified as containing residual adaptor sequence were removed before transcriptome assembly. Postfiltering, there were on average more than 24 million reads per pool.
De Novo Transcriptome Assembly
We built a de novo transcriptome assembly for P. interpunctella with the filtered RNA-Seq reads using Trinity (Grabherr et al. 2011). Trinity does not rely on orthology but rather builds the transcriptome de novo based on read information alone. Trinity clusters the sequence data into a series of de Brujin graphs, used to process similar reads that correspond to each gene, locus, or set of analogous genes within the transcriptome. These graphs are processed to determine full-length transcripts, identifying alternatively spliced isoforms and paralogous genes. We combined the four pools of filtered paired-end reads in a single de novo transcriptome assembly to enable orthology determination and comparisons of the generated contigs across the four samples. We used RSEM (Li and Dewey 2011) to map the filtered read sets back to the resulting transcriptome assembly to obtain read count estimates for each of the 58,979 contigs for each of the four pools.
Previous microarray studies typically filtered out all genes with expression below the background hybridization threshold, effectively removing loci with low or no expression. The digital nature of RNA-Seq data raises the potential for inappropriate filtering thresholds. In particular to this study, because gene silencing rates have been shown to be different between sex chromosomes and autosomes, and because this can affect estimates of dosage compensation (Kharchenko et al. 2011), we discarded all contigs that did not have at least four reads per million mappable reads in at least two samples. This avoids any potential problems with different proportions of contigs on the Z and autosomes without significant expression and also reduces the number of partial and erroneous contigs that typically have low read coverage. This resulted in 17,246 contigs. The absence of filtering in a previous study of sex chromosome dosage compensation (Xiong et al. 2010) was heavily criticized, and this called into question their conclusions based on their low-stringency filters (Castagné et al. 2011; Deng et al. 2011; Kharchenko et al. 2011; Lin et al. 2011). However, although filtering to remove genes with minimal expression is still debated (He et al. 2011), filtering to remove nonexpressed and inactivated genes is an important step when comparing sex chromosomes with autosomes. This controversy highlights the significance of the type of filtering that is performed and the effect this can have on the conclusions drawn from this type of expression study.
Orthology and Chromosome Location
We compared Plodia contigs with the genes from the annotated genome of the silkworm B. mori (Mita et al. 2004) obtained from KAIKObase version 3.2.1 (Shimomura et al. 2009). Putative orthology was determined using a reciprocal best-hit approach with a Basic Local Alignment Search Tool (BLAST) e-value cutoff of 1 × 10−10 (Altschul et al. 1990). The 1:1 orthology prediction between Bombyx and Plodia allowed us to use the chromosome positions of Bombyx genes as a putative chromosome location for each of the Plodia contigs. Plodia contigs that reciprocally matched Bombyx Z genes were classed as Z-borne contigs and contigs that matched Bombyx autosomal genes were termed autosomal in Plodia. All subsequent analyses were based solely on inferred 1:1 orthologs, with the remaining contigs discarded. This allowed us to identify 350 Plodia contigs with putative Z-linkage and 7,997 putative autosomal contigs. Although contigs do not necessarily correspond directly to single genes, the methods of the Trinity assembly (Grabherr et al. 2011), the fact that we removed all contigs <200 bp and the exclusive use of 1:1 orthologs means that most Plodia contigs for the expression study represent single genes.
Expression Comparisons of Z versus Autosomes in Both Sexes
We calculated fragments per kilobase per million mappable reads (FPKM), a factor that corrects for contig length and variations in read depth across samples (Mortazavi et al. 2008) for 1:1 Plodia orthologs for each pool individually. Because the correlation between same-sex replicates was very high (male r2 = 0.830, female r2 = 0.869), we combined replicates and calculated mean male and female FPKM for each ortholog by averaging across same-sex replicate pools. Repeatability of FPKM estimates for same-sex replicate pools was also estimated with Spearman’s ρ, an estimate of rank order correlation, and replicates showed high similarity for both male (Spearman’s ρ = 0.906; P < 2.2 × 10−16) and female (Spearman’s ρ = 0.928; P < 2.2 × 10−16) pools.
The FPKM-corrected expression levels of Z-linked and autosomal contigs were compared within each sex. Mann–Whitney U (MWU) was employed to test for significant differences between average Z and autosome expression levels. A resampling test was also employed to determine the probability that a subset of the autosomal contigs could yield the significant differences observed between Z and autosome contigs in females. Random samples of 350 female autosomal contigs (corresponding to the number of Z-linked orthologs in our data set) were selected as pseudo-Z linked sets, to compare to the remaining autosomal contigs. Average expression for the two sets was calculated and MWU employed to test for significance between the groups. This additional evidence supports the predictions of genomic location and orthology, was replicated 1,000 times, and the number of significant results was recorded with exact permutation P values (Phipson and Smyth 2010).
In addition to Z to autosome ratios within each sex, we plotted the differences in expression profile between male and female Z-linked versus autosomal expression, first log2 transforming the data to aid visualization, then fitting linear regression lines. Z-linked contig expression was further analyzed by separating Z-linked contigs into four quartiles based on male expression levels. Male-to-female Z-linked expression was compared, and MWU was employed to test for significant differences between the sexes Z-linked expression within each quartile.
Gene Ontology
We tested whether there was any over- or underabundance of functional Gene Ontology (GO) terms (Ashburner et al. 2000) for the Z chromosome compared with the autosomes and for contigs exhibiting male- or female-limited expression using the Drosophila melanogaster GO. We used BLAST to compare each Plodia contig to the annotated D. melanogaster genome annotation, taking the highest ranked BLAST hit [release 5.44 (Adams et al. 2000)] for all BLASTs where the highest hit passed a threshold of P < 1 × 10−5. Z-chromosome, male-limited, and female-limited target lists were separately compared with all expressed contigs using GOrilla to identify enriched GO terms with a threshold of P < 1 × 10−5 with multiple hypothesis correction (Eden et al. 2007, 2009).
Results
Using RNA-Seq data from four nonoverlapping sex-specific pools, each containing 10 moth individuals, we constructed a de novo transcriptome assembly for the non-model Lepidopteran, the Indian meal moth, P. interpunctella, using Trinity (Grabherr et al. 2011). Synteny was inferred from the silk moth, B. mori, annotated genome sequence (Mita et al. 2004; Shimomura et al. 2009) for 1:1 orthologs, yielding 350 putative Z-linked contigs and 7,997 putative autosomal contigs. Correlations between FPKM-corrected expression levels in same-sex replicate pools were highly significant for both the male (r2 = 0.830; Spearman’s ρ = 0.906; P < 2.2 × 10−16) and female (r2 = 0.869; Spearman’s ρ = 0.928; P < 2.2 × 10−16) pools.
Replicate pools were combined to provide average autosomal and Z-linked expression (table 1 and fig. 1). Dosage compensation is a mechanism by which the heterogametic sex, in the case of P. interpunctella, the female, would hypertranscribe Z-linked loci to balance expression of the single Z chromosome with the diploid autosomes (Ohno 1967; Mank 2009; Mank et al. 2011). It is therefore most appropriate to test for dosage compensation by comparing average Z expression with average autosomal expression (Z:A) for males and females separately. Male average Z:A of 0.954 was not significantly different from one (MWU; P = ns), in stark contrast to female Z:A of 0.534 (MWU; P = 2.2 × 10−16; table 1), as would be expected in the absence of dosage compensation mechanisms.
Table 1.
Average FPKM Levels for Male and Female Plodia Z-linked and Autosomal Contigs
| Male | Female | |
|---|---|---|
| Z | 56.107 | 32.540 |
| A | 58.786 | 60.983 |
| Z:A | 0.954 | 0.534 |
| MWU | 8.96 × 10−1 | 2.2 × 10−16 |
Note.—Z:A expression ratios for each sex and the P value for a Mann–Whitney U (MWU) test of significant difference between average Z and autosomal expression are listed.
Fig. 1.—
FPKM levels of autosomal (green) and Z-linked (orange) contigs in sex-averaged pools. Genomic location is based on inferred synteny with Bombyx mori. Plot boxes represent the median and interquartile range of FPKM expression level. Box notches are a visual aid that are approximate to the 95% confidence interval of the median and signify strong evidence that the medians differ for sample pairs if the notches do not overlap (Chambers et al. 1983). The plot tails extend to the most extreme data point that is no more than 1.5× the interquartile range.
To support the orthology predictions of genomic location in Plodia, we used a resampling approach to determine the probability that a random subsample of 350 female autosomal contigs would yield at least as much expression difference as we observed between female Z-linked and autosomal contigs. We compared average expression between autosomal and 1,000 resampled pseudo-Z contig sets, and in these comparisons, none of the resamples showed as much expression difference as our inferred Z and autosomal data set (P = 1 × 10−4, 1,000 replicates). Comparing the expression of the pseudo-Z sets with the observed expression difference alone does not provide sufficient statistical rigor, as even if the resampling results are not as extreme as the real data, the differences between the pseudo-Z contig sets and the remaining female autosomal contigs could still be significant. Therefore, we also assessed statistical difference between our autosomal and resampled pseudo-Z replicates with MWU tests using a cutoff of P < 5 × 10−2. Few of the resampled pseudo-Z sets showed any significant difference to the autosomal data (P = 4.4 × 10−2, 1,000 replicates), again indicating that Z contigs on average had been inferred correctly and that our data were sufficient to assess dosage compensation differences between the Z chromosome and autosomes. The high level of synteny between two other Lepidopteran genomes, the Monarch (Zhan et al. 2011) and Heliconius (The Heliconius Genome Consortium 2012) butterflies with Bombyx, adds further support to our methodology of using orthology to predict Z-linkage. These combined lines of evidence add support for our genomic location predictions.
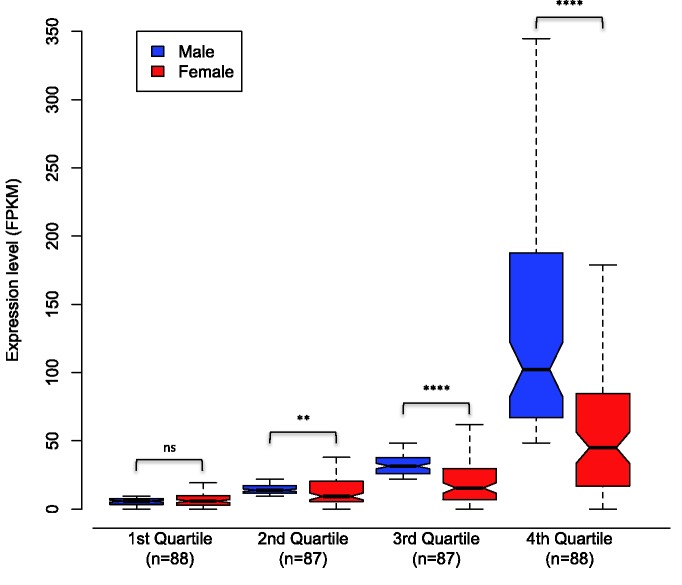
Although it is most appropriate to compare Z:A ratios within each sex separately, comparing male and female expression is useful in estimating the degree of dosage effect. Figure 2 illustrates the dosage effects of the Z chromosome, with female expression of Z-linked contigs averaging 0.58× that of males due to their halved gene dose. Figure 2 also suggests that the degree of dosage effect is different for lowly versus highly expressed contigs. We investigated this further by comparing male and female FPKM for Z-linked contigs based on average expression in males (fig. 3). This analysis, which separated Z-linked contigs into quartiles of increasing male expression, demonstrates that dosage effects are least manifest for lowly expressed contigs (MWU; P = ns) and increase for each quartile, with significant differences between males and females for the second (MWU; P = 4.41 × 10−3), third (MWU; P = 4.22 × 10−9), and fourth (MWU; P = 6.77 × 10−11) quartiles. No functional difference could be detected using GO analyses between Z-linked and autosomal contigs or between sex-limited contigs and autosomes (supplementary material S1, Supplementary Material online). Interestingly, sex-limited autosomal contigs showed high expression levels, with male-limited autosomal contigs averaging 110.72 FPKM and female-limited autosomal contigs averaging 375.78 FPKM, compared with the autosomal average (58.79 FPKM in males and 60.98 FPKM in females).
Fig. 2.—
Sex-average log2 FPKM values for autosomal (green) and Z-linked (orange) contigs based on inferred synteny with Bombyx. In both classes, a linear regression line has been fitted.
Fig. 3.—
FPKM-normalized average expression levels from male and female Plodia Z-linked contigs, identified by inferred synteny with Bombyx mori. Contigs were divided into four quartiles based on male expression level. Mann–Whitney U tests of significance of average Z-linked expression identified no significant difference between sexes in quartile 1 (P = 2.78 × 10−1) but significant differences in quartiles 2 (P = 4.41 × 10−3), 3 (P = 4.22 × 10−9), and 4 (P = 6.77 × 10−11). Plot boxes represent the median and interquartile range of FPKM expression level. Box notches are a visual aid that are approximate to the 95% confidence interval of the median and signify strong evidence that the medians differ for sample pairs if the notches do not overlap (Chambers et al. 1983). The plot tails extend to the most extreme data point that is no more than the 1.5× interquartile range.
Discussion
Our results indicate that Plodia lacks complete Z chromosome dosage compensation, with average female expression of Z-linked contigs slightly more than half that of male Z chromosome expression and slightly more than half of the average autosomal expression levels. This suggests that incomplete dosage compensation may be the norm in Lepidoptera, although data on additional species, and a resolution of the debate surrounding the Bombyx data (Zha et al. 2009; Walters and Hardcastle 2011), will be needed to confirm the generality of this pattern throughout the clade.
Plodia exhibits a similar pattern of partial, gene-by-gene compensation observed in both birds (Arnold et al. 2008) and Schistosoma (Vicoso and Bachtrog 2011), the female heterogametic animal taxa that have been previously assessed. This indicates that Z chromosomes may not generally be associated with global chromosome-wide mechanisms of dosage compensation that exist in some male heterogametic systems. The effects of Z chromosome dose in Plodia, the imbalance in Z to autosome expression levels, are somewhat greater (0.53) than the dosage effects observed in birds, which range from 0.6 to 0.75 (Mank and Ellegren 2009; Wolf and Bryk 2011). Our estimate of dosage effect in Plodia is also lower than the estimate from Bombyx, which ranges from 0.67 to 0.83 depending on the tissue sampled (Zha et al. 2009). However, this estimate may be confounded by data filtering and noise or difference in sampling strategy (Walters and Hardcastle 2011). It is worth noting that the majority of lepidopterans lack recombination in females (Marec and Traut 1993; Yoshido et al. 2005; Wang 2011), just as recombination is absent in heterogametic XY males in Drosophila (Morgan 1914).
Methodological Remarks
To better understand the evolution of sex chromosomes and dosage compensation mechanisms, we require a broader understanding of the status of dosage compensation in a diverse range of male and female heterogametic taxa. However, the desired diversity in organisms does not always match the availability of complete reference genomes with which to assess dosage compensation. Genome sequencing has focused on particular groups, and although more and more genomes are becoming available, there are notable gaps. Additionally, a large number of genome projects have either only sequenced the homogametic sex or have not sequenced in sufficient depth to accurately reconstruct the sex chromosomes in enough detail to enable mapping and quantification of RNA-Seq data. Our study has overcome these barriers by conducting de novo transcriptomic sequence assembly and inferring genomic location based on synteny with the related silk moth, B. mori. Using this method, we have been able to assess dosage compensation in an organism without having to first construct a complete reference genome, saving considerable time and resources. This study presents an effective strategy for significantly increasing the number of species for which transcriptome analysis in general, and dosage compensation in particular, can be determined. However, this may not be a satisfactory solution in all taxa if a representative genome has not yet been completed or synteny is low. Although this approach is subject to biases of orthology-based studies, it allows data points from multiple species to be gathered from homologous chromosomes to strengthen our understanding of dosage compensation in each clade and will help to resolve the status of dosage compensation in contested taxa. Future studies could also use RNA-seq barcoding information to track male and female individuals (Craig et al. 2008; Cronn et al. 2008) and then verify Z-linkage by comparing male and female heterozygosity levels.
Gene Expression and Dosage Effects
In addition to an absence of global dosage compensation in Plodia, the actual dosage difference between the sexes is not uniform across all types of contigs. Our comparison of male and female expression levels for Z-linked contigs showed that dosage differences between the sexes are more pronounced for highly expressed contigs. Detailed quartile-based analysis of expression level showed that dosage effects for contigs with the lowest expression levels were not significantly different between the sexes and that there was a trend for greater dosage effect between the sexes as expression level increased. This demonstrates the multiplying effect of Z copy number in highly expressed contigs when a global dosage compensation mechanism is absent. The largest dosage differences are observed in highly expressed contigs, which is intriguing, particularly considering that it is believed that highly expressed genes are generally more functionally important (having larger fitness effects), as shown by their elevated levels of purifying selection, conservation, and slower evolution (Pál et al. 2001; Drummond et al. 2005; Liao and Zhang 2006; Resch et al. 2007). The large dosage differences in highly expressed contigs may also be indicative of the importance of sex-specific fitness effects, discussed further later.
Unlike Plodia, birds (Arnold et al. 2008) and Schistosoma (Vicoso and Bachtrog 2011), some male heterogametic systems possess global compensation mechanisms. For example, Drosophila males hypertranscribe the X chromosome to equalize expression levels, achieving parity between the sexes, as well as between the single male X and the diploid autosomes (Bachtrog et al. 2010). This means that male-biased X-linked genes in Drosophila must be expressed over and above the already hypertranscribed levels associated with dosage compensation. At high expression levels, the ability to hypertranscribe from the single X chromosome may become saturated, explaining the deficit of male-biased highly expressed genes on the Drosophila X chromosome (Vicoso and Charlesworth 2009). Applied to our data in Plodia, the higher dosage difference for highly expressed contigs suggests that the ability of regulatory networks to buffer dosage effects of Z-linked genes decreases with expression level, possibly because the transcriptional process becomes similarly saturated.
Sexual Conflict over Dosage Compensation
The lack of complete dosage compensation in Plodia raises questions about sex chromosome evolution and the role of genes on the sex chromosomes. The single copy of the Z chromosome possessed by females equates to on average slightly more than half the expression level of males, as observed for the majority of female Z-linked contigs in Plodia. The differing expression levels presumably translate into differences in protein levels, and because Z-linked loci interact with the remainder of the genome through genetic networks, dosage differences between males and females have the potential to affect a large proportion of autosomal loci. This difference in expression of the Z chromosome in female heterogametic species and the X chromosome in male heterogametic systems creates conflict between the sexes over optimal transcription level (Mank et al. 2011). Selection to hypertranscribe X- or Z-linked genes in the heterogametic sex can lead to overexpression in the homogametic sex, which for dosage-sensitive genes could be as harmful as underexpression. Dosage compensation effectively decouples the correlation between male and female transcription rates, thereby resolving sexual conflict related to Z chromosome dosage differences.
Sexual conflict over transcription rates is believed to have brought about X chromosome inactivation in therian mammals. Therian X inactivation, previously thought to be a mechanism of dosage compensation, is in fact the result of conflict over optimal transcription of X-linked dosage-sensitive genes (Pessia et al. 2012). In this clade, although a number of dosage-sensitive genes have been compensated to the levels of autosomal genes in both sexes, the majority of X-linked genes remain expressed at lower levels than the autosomal genes in both sexes. However, the status of dosage compensation in the eutherians remains contentious, with several recent conflicting studies (Xiong et al. 2010; Deng et al. 2011; Kharchenko et al. 2011; Julien et al. 2012; Lin et al. 2012).
In addition to Plodia, there are a number of species, including birds (Itoh et al. 2007; Mank 2009), Schistosoma (Vicoso and Bachtrog 2011), and platypus (Deakin et al. 2008) that only compensate a minority of X or Z chromosome genes. Birds and Schistosoma are female heterogametic, whereas platypus males have five XY sex chromosome pairs. The fitness effects of incomplete dosage compensation in these organisms are unclear, as sexual conflict over optimal transcription rates of Z- or X-linked genes is unresolved. It may be that the majority of uncompensated genes are insensitive to dosage effects, although the correlation between dosage effects and expression level that we observed would suggest otherwise. Alternatively, there may yet be unknown mechanisms of minimizing dosage effects in these systems.
Concluding Remarks
Here, we employed de novo transcriptome assembly methods, in conjunction with genomic positional location inference from the related B. mori reference genome, to demonstrate that the Indian meal moth, P. interpunctella, lacks complete Z chromosome dosage compensation. This approach expands the number of data points that can be obtained for each clade, increasing the scope for studies of sex chromosome dosage compensation and other transcriptomic analyses.
Our analysis shows that P. interpunctella lacks complete Z chromosome dosage compensation, showing approximately half the expression in females compared with males for Z-linked contigs, the largest dose effect related to sex chromosomes yet observed. This work suggests the entire Lepidopteran clade may lack complete Z chromosome dosage compensation, with previous studies on B. mori proving inconclusive (Zha et al. 2009; Walters and Hardcastle 2011), though studies on a range of species will be required to confirm this assertion. This finding adds further weight to the observation that complete sex chromosome dosage compensation may be less common in female- compared with male-heterogametic species (Mank 2009; Naurin et al. 2011), although this is still based on relatively few data points. It is hoped that the status of dosage compensation in an expanded range of species with old and highly diverged sex chromosomes will soon be determined, allowing for the synthesis and understanding of the evolution of complete versus incomplete sex chromosome dosage compensation and the sexual conflict over expression that underlies it.
Supplementary Material
Supplementary material S1 is available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank JC Perry, MA Pointer, AE Wright, F Zimmer, and S Montgomery for helpful comments on this manuscript. They acknowledge the use of the UCL Unity SMP Facility, and associated support services, in the completion of this work. This work was supported by a Royal Society Wolfson Award to N.W. and the European Research Council under the Framework 7 Agreement to J.E.M. [grant agreement 260233].
Literature Cited
- Adams MD, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EF, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Arnold AP, Itoh Y, Melamed E. A bird's-eye view of sex chromosome dosage compensation. Annu Rev Genom Hum Genet. 2008;9:109–127. doi: 10.1146/annurev.genom.9.081307.164220. [DOI] [PubMed] [Google Scholar]
- Arunkumar KP, Mita K, Nagaraju J. The silkworm Z chromosome is enriched in testis-specific genes. Genetics. 2009;182:493–501. doi: 10.1534/genetics.108.099994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, et al. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D, Toda NRT, Lockton S. Dosage compensation and demasculinization of X chromosomes in Drosophila. Curr Biol. 2010;20:1476–1481. doi: 10.1016/j.cub.2010.06.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D, et al. Are all sex chromosomes created equal? Trends Genet. 2011;27:350–357. doi: 10.1016/j.tig.2011.05.005. [DOI] [PubMed] [Google Scholar]
- Birchler JA, Riddle NC, Auger DL, Veitia RA. Dosage balance in gene regulation: biological implications. Trends Genet. 2005;21:219–226. doi: 10.1016/j.tig.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Birchler JA, Yao H, Chudalayandi S. Biological consequences of dosage dependent gene regulatory systems. Biochim Biophys Acta. 2007;1769:422–428. doi: 10.1016/j.bbaexp.2006.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castagné R, et al. The choice of the filtering method in microarrays affects the inference regarding dosage compensation of the active X-chromosome. PLoS One. 2011;6:e23956. doi: 10.1371/journal.pone.0023956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers JM, Cleveland WS, Kleiner B, Tukey PA. Graphical methods for data analysis. Belmont (CA): Wadsworth & Brooks/Cole; 1983. [Google Scholar]
- Craig DW, et al. Identification of genetic variants using bar-coded multiplexed sequencing. Nat Methods. 2008;5:887–893. doi: 10.1038/nmeth.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronn R, et al. Multiplex sequencing of plant chloroplast genomes using Solexa sequencing-by-synthesis technology. Nucleic Acids Res. 2008;36:e122. doi: 10.1093/nar/gkn502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deakin JE, Hore TA, Koina E, Graves JAM. The status of dosage compensation in the multiple X chromosomes of platypus. PLoS Genet. 2008;4:e1000140. doi: 10.1371/journal.pgen.1000140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng XX, et al. Evidence for compensatory upregulation of expressed X-linked genes in mammals, Caenorhabditis elegans and Drosophila melanogaster. Nat Genet. 2011;43:1179–1185. doi: 10.1038/ng.948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DA, et al. Why highly expressed proteins evolve slowly. Proc Natl Acad Sci U S A. 2005;102:14338–14343. doi: 10.1073/pnas.0504070102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E, et al. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E, Lipson D, Yogev S, Yakhini Z. Discovering motifs in ranked lists of DNA sequences. PLoS Comput Biol. 2007;3:e39. doi: 10.1371/journal.pcbi.0030039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ercan S, et al. X chromosome repression by localization of the C. elegans dosage compensation machinery to sites of transcription initiation. Nat Genet. 2007;39:403–408. doi: 10.1038/ng1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estivill X, Armengol L. Copy number variants and common disorders: filling the gaps and exploring complexity in genome-wide association studies. PLoS Genet. 2007;3:1787–1799. doi: 10.1371/journal.pgen.0030190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, et al. He et al. reply. Nat Genet. 2011;43:1171–1172. [Google Scholar]
- Heliconius Genome Consortium. Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature. 2012;487:94–98. doi: 10.1038/nature11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingleby FC, Lewis Z, Wedell N. Level of sperm competition promotes evolution of male ejaculate allocation patterns in a moth. Anim Behav. 2010;80:37–43. [Google Scholar]
- Itoh Y, et al. Dosage compensation is less effective in birds than in mammals. J Biol. 2007;6:2. doi: 10.1186/jbiol53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien P, et al. Mechanisms and evolutionary patterns of mammalian and avian dosage compensation. PLoS Biol. 2012;10:e1001328. doi: 10.1371/journal.pbio.1001328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Xi RB, Park PJ. Evidence for dosage compensation between the X chromosome and autosomes in mammals. Nat Genet. 2011;43:1167–1169. doi: 10.1038/ng.991. [DOI] [PubMed] [Google Scholar]
- Li B, Dewey C. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao B-Y, Zhang J. Low rates of expression profile divergence in highly expressed genes and tissue-specific genes during mammalian evolution. Mol Biol Evol. 2006;23:1119–1128. doi: 10.1093/molbev/msj119. [DOI] [PubMed] [Google Scholar]
- Lin F, Xing K, Zhang J, He X. Expression reduction in mammalian X chromosome evolution refutes Ohno’s hypothesis of dosage compensation. Proc Natl Acad Sci U S A. 2012;109:11752–11757. doi: 10.1073/pnas.1201816109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, et al. Relative overexpression of X-linked genes in mouse embryonic stem cells is consistent with Ohno's hypothesis. Nat Genet. 2011;43:1169–1170. doi: 10.1038/ng.992. [DOI] [PubMed] [Google Scholar]
- Lucchesi JC. Dosage compensation in Drosophila. Annu Rev Genet. 1973;7:225–237. doi: 10.1146/annurev.ge.07.120173.001301. [DOI] [PubMed] [Google Scholar]
- Makino T, McLysaght A. Ohnologs in the human genome are dosage balanced and frequently associated with disease. Proc Natl Acad Sci U S A. 2010;107:9270–9274. doi: 10.1073/pnas.0914697107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank JE. The W, X, Y, and Z of sex chromosome dosage compensation. Trends Genet. 2009;25:226–233. doi: 10.1016/j.tig.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank JE, Ellegren H. All dosage compensation is local: gene by gene regulation of sex-biased expression on the chicken Z chromosome. Heredity. 2009;102:312–320. doi: 10.1038/hdy.2008.116. [DOI] [PubMed] [Google Scholar]
- Mank JE, Hosken DJ, Wedell N. Some inconvenient truths about sex chromosome dosage compensation and the potential role of sexual conflict. Evolution. 2011;65:2133–2144. doi: 10.1111/j.1558-5646.2011.01316.x. [DOI] [PubMed] [Google Scholar]
- Marec F, Traut W. Synaptonemal complexes in female and male meiotic prophase of Ephestia kuehniella (Lepidoptera) Heredity. 1993;71:394–404. [Google Scholar]
- McDonel P, Jans J, Peterson BK, Meyer BJ. Clustered DNA motifs mark X chromosomes for repression by a dosage compensation complex. Nature. 2006;444:614–618. doi: 10.1038/nature05338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita K, et al. The genome sequence of silkworm, Bombyx mori. DNA Res. 2004;11:27–35. doi: 10.1093/dnares/11.1.27. [DOI] [PubMed] [Google Scholar]
- Morgan TH. No crossing over the male Drosophila of genes in the second and third pair of chromosomes. Biol Bull. 1914;26:195–204. [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Naurin S, Hansson B, Hasselquist D, Kim YH, Bensch S. The sex-biased brain: sexual dimorphism in gene expression in two species of songbirds. BMC Genomics. 2011;12:37. doi: 10.1186/1471-2164-12-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. Sex chromosomes and sex linked genes. Berlin (Germany): Springer-Verlag; 1967. [Google Scholar]
- Pál C, Papp B, Hurst LD. Highly expressed genes in yeast evolve slowly. Genetics. 2001;158:927–931. doi: 10.1093/genetics/158.2.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp B, Pal C, Hurst LD. Dosage sensitivity and the evolution of gene families in yeast. Nature. 2003;424:194–197. doi: 10.1038/nature01771. [DOI] [PubMed] [Google Scholar]
- Pessia E, et al. Mammalian X chromosome inactivation evolved as a dosage-compensation mechanism for dosage-sensitive genes on the X chromosome. Proc Natl Acad Sci U S A. 2012;109:5346–5351. doi: 10.1073/pnas.1116763109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phipson B, Smyth GK. Permutation P-values should never be zero. Calculating exact P-values when permutations are randomly drawn. Stat Appl Genet Mol Biol. 2010;9 doi: 10.2202/1544-6115.1585. Article39. [DOI] [PubMed] [Google Scholar]
- Prince EG, Kirkland D, Demuth JP. Hyperexpression of the X chromosome in both sexes results in extensive female bias on X-linked genes in the flour beetle. Genome Biol Evol. 2010;2:336–346. doi: 10.1093/gbe/evq024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon R, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resch AM, et al. Widespread positive selection in synonymous sites of mammalian genes. Mol Biol Evol. 2007;24:1821–1831. doi: 10.1093/molbev/msm100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahara k, Yoshido A, Traut W. Sex chromosome evolution in moths and butterflies. Chromosome Res. 2012;20:83–94. doi: 10.1007/s10577-011-9262-z. [DOI] [PubMed] [Google Scholar]
- Schlattl A, Anders S, Waszak SM, Huber W, Korbel JO. Relating CNVs to transcriptome data at fine resolution: assessment of the effect of variant size, type, and overlap with functional regions. Genome Res. 2011;21:2004–2013. doi: 10.1101/gr.122614.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura M, et al. KAIKObase: an integrated silkworm genome database and data mining tool. BMC Genomics. 2009;10:486. doi: 10.1186/1471-2164-10-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B, Bachtrog D. Lack of global dosage compensation in Schistosoma mansoni, a female-heterogametic parasite. Genome Biol Evol. 2011;3:230–235. doi: 10.1093/gbe/evr010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B, Charlesworth B. The deficit of male-biased genes on the D. melanogaster X chromosome is expression-dependent: a consequence of dosage compensation? J Mol Evol. 2009;68:576–583. doi: 10.1007/s00239-009-9235-4. [DOI] [PubMed] [Google Scholar]
- Walters JR, Hardcastle TJ. Getting a full dose? Reconsidering sex chromosome dosage compensation in the silkworm, Bombyx mori. Genome Biol Evol. 2011;3:491–504. doi: 10.1093/gbe/evr036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B. Achiasmy or heterochiasmy: does meiotic recombination occur in female Lepidoptera? J Res Lepidoptera. 2011;44:43–45. [Google Scholar]
- Wolf JBW, Bryk J. General lack of global dosage compensation in ZZ/ZW systems? Broadening the perspective with RNA-Seq. BMC Genomics. 2011;12:91. doi: 10.1186/1471-2164-12-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AE, Mank JE. Battle of the sexes: conflict over dosage-sensitive genes and the origin of X chromosome inactivation. Proc Natl Acad Sci U S A. 2012;109:5144–5145. doi: 10.1073/pnas.1202905109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong YY, et al. RNA sequencing shows no dosage compensation of the active X-chromosome. Nat Genet. 2010;42:1043–1047. doi: 10.1038/ng.711. [DOI] [PubMed] [Google Scholar]
- Yoshido A, Bando H, Yasukochi H, Sahara K. The Bombyx mori karyotype and the assignment of linkage groups. Genetics. 2005;170:675–685. doi: 10.1534/genetics.104.040352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zha X, et al. Dosage analysis of Z chromosome genes using microarray in silkworm, Bombyx mori. Insect Biochem Mol. 2009;35:315–321. doi: 10.1016/j.ibmb.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Zhan S, Merlin C, Boore JL, Reppert SM. The monarch butterfly genome yields insights into long-distance migration. Cell. 2011;147:1171–1185. doi: 10.1016/j.cell.2011.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Lemos B, Dopman EB, Hartl DL. Copy-number variation: the balance between gene dosage and expression in Drosophila melanogaster. Genome Biol Evol. 2011;3:1014–1024. doi: 10.1093/gbe/evr023. [DOI] [PMC free article] [PubMed] [Google Scholar]