Abstract
The presence of spliceosomal introns in eukaryotic genes poses a major puzzle for the study of genome evolution. Intron densities vary enormously among distant lineages. However, the mechanisms driving intron gains are poorly understood and very few intron gains and losses have been documented over short evolutionary time spans. Fungi emerged recently as excellent models to study intron evolution and “reverse splicing” was found to be a major driver of recent intron gains in a clade of ascomycete fungi. We screened a total of 38 genomes from two fungal clades important in medicine and agriculture to identify intron gains and losses both within and between species. We detected 86 and 198 variable intron positions in the Cryptococcus and Fusarium clades, respectively. Some genes underwent extensive changes in their exon–intron structure, with up to six variable intron positions per gene. We identified a very recently gained intron in a group of tomato-infecting strains belonging to the F. oxysporum species complex. In the human pathogen C. gattii, we found recent intron losses in subtypes of the species. The two studied fungal clades provided evidence for extensive changes in their exon–intron structure within and among closely related species. We show that both intronization of previously coding DNA and insertion of exogenous DNA are the major drivers of intron gains.
Keywords: spliceosomal introns, intron gains, Fusarium, Cryptococcus, population genomics
Introduction
The presence of spliceosomal introns in genes is a major puzzle of eukaryotic genome evolution (Lynch and Richardson 2002). Many eukaryotic genes contain at least one intron that must be removed by the spliceosomal machinery from RNA transcripts. No universal advantage of introns has been shown to date. Introns of some genes may contain highly conserved regulatory sequences (Duret and Bucher 1997; Sorek and Ast 2003). In others, alternative splicing may generate adaptive protein diversity (Dibb 1993). However, none of the proposed benefits appear general enough to explain the ubiquity of introns. Conversely, the presence of introns in genes presents several burdens to the organism. The splicing sites required for the removal of intron sequence from RNA transcripts must be conserved, otherwise mutations would lead to the transcription of nonsense alleles. In particularly intron-dense genomes such as in mammals, there must be a significant metabolic cost to the replication of noncoding DNA and the transcription of intron sequences into RNA.
The emergence of introns likely coincided with the first eukaryotic cells, as all sequenced eukaryotic genomes contain at least some introns and a presumably fully functional spliceosomal machinery (Collins and Penny 2005; Roy 2006; Roy and Gilbert 2006; Koonin 2009). Whether the emergence of introns predates the evolution of eukaryotes was a matter of considerable debate, however the currently favored view is that prokaryotic genes were always devoid of introns. Recent studies suggest that the genome of the last common ancestor of eukaryotes was rapidly invaded by a very large number of introns (reviewed in Koonin 2009). In extant lineages of eukaryotes, intron densities vary extensively, from a few introns per genome in the yeast Saccharomyces cerevisiae and some protozoans to eight introns per gene in several mammalian genomes (Jeffares et al. 2006). Across the eukaryotic tree, highly variable intron densities are found within many major lineages, whereas plants and animals were found to have generally high intron densities. The extreme differences in intron densities must, therefore, have been caused by large rate variations in intron gains and losses among particular phylogenetic clades. Lynch (2007) proposed that genomic intron densities are largely governed by the effective population size of the corresponding organisms. Intron insertions are assumed to be on average slightly deleterious and driven to fixation due to genetic drift (Lynch 2002). Empirical evidence for a role of population structure was found for newly arisen introns in Daphnia (Li et al. 2009). As many multicellular eukaryotes such as plants and animals tend to have small effective population sizes, the strength of selection against deleterious intron insertions is expected to be relatively weak and introns may proliferate (Lynch and Richardson 2002; Lynch 2006, 2007). However, empirical support for the direct effect of genetic drift is still weak.
Fungi present a unique diversity in intron densities, varying from 0.05 introns per gene in S. cerevisiae to 5.5 introns per gene in Cryptococcus neoformans (Jeffares et al. 2006). Intron gains and losses were shown to be in an approximate balance over long evolutionary timeframes in Euascomycota (Dibb 1993; Nielsen et al. 2004). However, in other fungal lineages losses predominate (Stajich et al. 2007). Most fungi likely have large effective population sizes and fungal introns tend to be very short compared with introns found in animal genomes, as expected by the stronger purifying selection operating in large fungal populations (Lynch 2007). However, purifying selection does not seem to uniformly depress intron densities in fungi.
Despite the enormous differences in intron densities among genomes, only few mechanisms of intron gains or losses have been convincingly established and almost no data exist on the relative importance of these mechanisms across eukaryotes. Proposed mechanisms for intron gains are highly diverse and range from genomic duplication or transposable element insertion (reviewed in Koonin 2009; Roy and Irimia 2009), to mutations leading to the creation of functional splicing sites and hence new introns or insertions occurring as a result of double-strand breaks in exons (Lonberg and Gilbert 1985; Jeffares et al. 2006; Koren et al. 2007; Irimia et al. 2008; Roy 2009). The “reverse splicing” mechanism (Cavalier-Smith 1985) for the creation of introns requires the reversibility of the splicing reaction (the re-insertion of an intron sequence into a different RNA transcript), subsequent retrotranscription into cDNA and homologous recombination with the genomic gene copy. The key step, the reversibility of the splicing reaction, was recently demonstrated in yeast (Tseng and Cheng 2008). The major mechanism thought to be responsible for intron loss is the recombination of a genomic gene copy with a homologous reverse transcribed RNA transcript that was at least partially spliced (reviewed in Roy and Irimia 2009).
To gain a broad understanding of the relative importance of mechanisms of intron gain and loss that contribute to the enormous variation in genomic intron densities, a large number of transient stages of intron evolution need to be examined. The comparison of sets of closely related genomes is especially promising, as a large number of orthologous intron positions can be reliably screened. Furthermore, closely related genomes may retain the signatures predicted by the different mechanisms of intron gain and loss. Intraspecific comparisons of the planktonic crustacean Daphnia revealed 24 transitory stages of intron gains and losses. Short repeat sequences found close to the splicing sites of new introns suggested that double-strand breaks were associated with intron gains (Li et al. 2009). In the ascomycete fungus Mycosphaerella, 52 transitory intron positions were found to be segregating within populations of the species (Torriani et al. 2011). Two related species also harbored a substantial number of transitory intron positions. The major mechanism driving intron gains was proposed to be intron transposition mediated by “reverse splicing,” as numerous intron sequences shared high sequence similarities despite being located in unrelated genes.
Fungi emerged as especially suitable model organisms to study intron evolution because large population sizes likely retain different stages in the gain or loss of an intron within species as shown in M. graminicola (Torriani et al. 2011). Rates of intron gain and loss in fungi were shown to be relatively high (Nielsen et al. 2004). However, up until now comparisons of genomes within and among closely related species were restricted to different serotypes of Cryptococcus neoformans and the Mycosphaerella clade. In Fusarium and Cryptococcus, two fungal clades comprising major plant and human pathogens, major advances in the availability of genome sequences were made recently. The newly available data provides a unique opportunity to significantly expand our knowledge of the evolution of intron positions.
The ascomycete Fusarium clade comprises highly diverse pathogenic fungi found in a vast range of hosts and environments causing significant economic damage to crops (Agrios 2005). Fusarium belongs to the order of Hypocreales that emerged 150–200 Ma (Sung et al. 2008); however, members of the Fusarium clade are likely to be substantially younger. F. oxysporum is a species complex of asexual fungi causing wilt and root rot diseases on over 120 plant species (Michielse and Rep 2009). The genome of the tomato wilt strain FOL 4287 was used to describe the extensive lineage-specific genomic regions conferring virulence on the host (Ma et al. 2010). An additional 10 genomes of the F. oxysporum species complex (FOSC) were made available by the Broad Institute comprising two further strains infecting tomato and strains infecting cabbage, Arabidopsis, banana, melon, and cotton. One strain was isolated from human blood. F. oxysporum infections can be life threatening in susceptible neutropenic individuals (O'Donnell et al. 2004). Genomes of two related species in the Fusarium clade infecting maize (F. verticillioides) and wheat and barley (F. graminearum) were also fully sequenced (Ma et al. 2010).
The basidiomycete C. neoformans species complex is common human pathogens causing significant numbers of deaths, especially in immunodeficient patients (Park et al. 2009). The C. neoformans species are divided into three varieties and four serotypes. The major serotypes diverged 37–80 Ma depending on the estimates (Xu et al. 2000; Sharpton et al. 2008). C. neoformans var. grubii (serotype A) is found to cause infection in a large number of AIDS patients. C. neoformans var. neoformans (serotype D) is rarely pathogenic. C. gattii (serotype B) causes cryptococcosis even in otherwise healthy patients and has a high death rate despite antifungal therapy (Park et al. 2009). An outbreak of cryptococcosis that occurred in British Columbia, Canada, and subsequently spread to neighboring regions of the Pacific Northwest was caused mostly by a molecular subtype of C. gattii termed VGII (Bartlett et al. 2008). The three other known subtypes (VGI, VGIII, and VGIV) were found mostly in environmental samples with VGI being the most frequently sampled strain globally. The molecular characterization of the different VG types was recently expanded through genome resequencing of 20 VGI, VGII, and VGIII strains in addition to the two reference genomes available for C. gattii WM276 and R265 (Gillece et al. 2011). There is evidence for a significant number of intron losses among the major serotypes of Cryptococcus (Stajich and Dietrich 2006; Sharpton et al. 2008).
Fungi have emerged as promising models to study mechanisms underlying intron gain and loss in sets of closely related organisms. A large number of closely related C. gattii and Fusarium genomes were made available very recently and provided a unique opportunity to identify recent examples of intron gains and losses among closely genomes. Through comparative genomic analyses, we aim to identify common themes of recent intron gains to provide a comprehensive view on intron evolution across a broad range of fungi.
Materials and Methods
Fusarium Genome Sequences
Genome assemblies from 11 F. oxysporum, one F. verticillioides and one F. graminearum strains were obtained from the Broad Institute of Harvard and MIT in January 2012 (http://www.broadinstitute.org, last accessed February 25, 2012). The strains of the FOSC cover a wide range of environments and hosts (table 1) and infect a wide variety of crops, fruits, and vegetables. The FOSC lineage 3-a was obtained from human blood and is known to cause localized necrotic diseases in immunocompetent individuals (O'Donnell et al. 2004) and was shown to infect contact lens users (Chang et al. 2006). The strain Fo47 colonizes plant roots and shows biological control properties by suppressing wilt diseases (Fravel et al. 2003). The sequenced F. verticillioides strain 7600 is widely used in molecular and pathological studies and is known to cause kernel and ear rot on maize. F. graminearum causes head blight (scab) on wheat and barley and the genome of strain PH-1 was included in our analysis.
Table 1.
Fusarium Strains Included in the Comparative Genomics Study
| Species | NRRLa | Strain | Forma Specialis | Host |
|---|---|---|---|---|
| F. graminearum | 31,084 | PH-1 | Wheat/barley | |
| F. verticillioides | 20,956 | 7600 | Maize | |
| F. oxysporum | 34,936 | FOL4287 | Lycopersici race 2 | Lycopersicum |
| 54,003 | MN25 | Lycopersici race 3 | Lycopersicum | |
| 26,381 | CL57 | Radicis-lycopersici | Lycopersicum | |
| 37,622 | HDV247 | Pisi | Pisum | |
| 26,406 | Melonis | Cucurbita | ||
| 54,008 | PHW808 | Conglutinans race 2 | Brassica/Arabidopsis | |
| 54,004 | PHW815 | Raphani | Raphanus/Arabidopsis | |
| 25,433 | Vasinfectum | Gossypium | ||
| 54,002 | Fo47 | Soil (biocontrol agent) | ||
| 32,931 | FOSC 3-a | Human | ||
| 54,006 | II5 | Cubense tropical race 4 | Musa |
aNRRL: Northern Regional Research Laboratory (Agricultural Research Service Culture Collection)
Cryptococcus Genome Sequences
Whole genome sequences and annotation data for C. neoformans serotype A strain H99 and C. gattii serotype B strains R265 and WM276 were obtained from the Broad Institute in January 2012 (http://www.broadinstitute.org, last accessed February 25, 2012). Two genome sequences of C. neoformans var. neoformans serotype D were included. The genome of strain JEC21 was obtained from GenBank (PRJNA13856) as deposited by TIGR. The genome of strain B-3501A was sequenced by the Stanford Genome Technology Center and deposited as PRJNA12386 on GenBank. Illumina whole-genome sequence data for 20 more C. gattii serotype B strains (VGI–III) were obtained from the NCBI Short Read Archive (accessions see table 2) as deposited by Gillece et al. (2011). The strains were obtained from state and local health departments, clinicians, veterinarians, and ongoing environmental studies. The sampling period spans from 2005 to 2010 and samples originated from British Columbia, Canada, and the United States Pacific Northwest (Gillece et al. 2011). The C. gattii isolates were identified by MLST to belong to three of the four known molecular types (VGI–III) with the majority (n = 18) being VGIIa–c (table 2). A genome-wide single nucleotide polymorphism analysis of the isolates confirmed the classification by MLST types and provided a much higher resolution of genotypic differentiation (Gillece et al. 2011).
Table 2.
Cryptococcus Strains Included in the Comparative Genomics Study
| Species | Serotype | MLST Type (VG) | Strain | Geographic Source | Clinical Source | Collection Date | Raw Data | Accessiona | Scaffold N50b |
|---|---|---|---|---|---|---|---|---|---|
| C. neoformans | A (var. grubii) | H99 | Assembled | Broad | |||||
| C. gattii | B | I | B7488 | Oregon | Human | 2009 | 852.9M | SRX105736 | 124,757 |
| WM276 | British Columbia | Human | Assembled | Broad | |||||
| IIa | B7395 | Washington | Dog | 2008 | 752.4M | SRX105724 | 156,865 | ||
| B7467 | Oregon | Porpoise | 2009 | 740.2M | SRX105735 | 151,963 | |||
| B8849 | Oregon | Environmental | 2010 | 1G | SRX057999 | 159,394 | |||
| B8577 | British Columbia | Environmental | 2009 | 793.3M | SRX105743 | 147,949 | |||
| R265 | British Columbia | Human | Assembled | Broad | |||||
| IIb | B7422 | Oregon | Cat | 2009 | 1.8G | SRX105728 | 152,093 | ||
| B7436 | N. California | Alpaca | 2009 | 2.2G | SRX105730 | 148,255 | |||
| B7394 | Washington | Cat | 2008 | 354.2M | SRX105727 | 132,070 | |||
| B7735 | Oregon | Human | 2009 | 789.1M | SRX105737 | 3,278 | |||
| B8554 | Oregon | Dog | 2008 | 1.3G | SRX105740 | 163,409 | |||
| B8828 | Washington | Porpoise | 2010 | 626.7M | SRX105744 | 147,946 | |||
| IIc | B8571 | Washington | Human | 2009 | 880.3M | SRX105741 | 158,987 | ||
| B8843 | Oregon | Human | 2010 | 1.1G | SRX105754 | 157,952 | |||
| B8838 | Washington | Human | 2010 | 1G | SRX105753 | 156,444 | |||
| B7466 | Oregon | Cat | 2008 | 803.6 M | SRX105734 | 159,669 | |||
| B7737 | Oregon | Human | 2009 | 735.8 M | SRX105738 | 162,088 | |||
| B6863 | Oregon | Human | 2005 | 614 M | SRX105723 | 154,100 | |||
| B7390 | Idaho | Human | 2008 | 601.8 M | SRX105726 | 151,536 | |||
| B7432 | Oregon | Human | 2009 | 496.1 M | SRX105729 | 148,648 | |||
| III | B8212 | Oregon | Human | 2007 | 996.6 M | SRX105739 | 3,361 | ||
| C. neoformans | D (var. neoformans) | JEC21 | Assembled | PRJNA13856 | |||||
| D (var. neoformans) | B-3501A | Assembled | PRJNA12386 | ||||||
aNCBI short read archive accessions as deposited by Gillece et al. (2011).
bSee Materials and Methods for details.
The whole-genome sequence data for the 20 strains was generated on an Illumina GAIIx in paired-end mode with a nominal insert length of 450 bp (Gillece et al. 2011). The read length was either 75 or 101 bp. A de novo assembly was produced for each strain by using SOAPdenovo version 1.05 (Li et al. 2010). We explored a range of kmer sizes and determined that a length of 35 was optimal for most isolates (a kmer size of 49 was used for the two strains with lower coverage: B7735 and B8212). The assembly procedure included both scaffolding and gap closing with SOAPdenovo. Scaffold N50 ranged from 124 to 163 kb, except for isolates B7735 and B8212 where the scaffold N50 was 3,278 and 3,361 bp, respectively. The longest scaffolds spanned 425–498 kb, except for B7735 and B8212 where the longest scaffolds were 33,703 and 49,057 bp, respectively.
Phylogenetic Reconstruction within Clades
To reconstruct phylogenetic relationships among all included genomes within each clade, we extracted sequences of the following three standard fungal barcoding genes (James et al. 2006): the elongation factor EF1-alpha, RNA polymerase II largest subunit (RPB1) and second largest subunit (RPB2). We aligned concatenated sequences of all three genes separately for Fusarium and Cryptococcus strains with MAFFT version 6.853 b (Katoh and Toh 2008) with maximum accuracy settings (iterative refinement, maximum 1,000 cycles). In total, the alignment length was 6,011 bp for Cryptococcus and 10,070 bp for Fusarium. We performed phylogenetic reconstruction with maximum-likelihood as implemented in PhyML 3.0 (Guindon and Gascuel 2003) with a general time-reversible model.
Identification of Orthologous Genes
The identification of the most likely orthologs between different species within the Cryptococcus and Fusarium clades was performed with reciprocal BLASTn searches. In the Fusarium clade, orthologous genes were identified by performing a reciprocal best BLASTn search for F. oxysporum f. sp. lycopersici 4287 and F. verticillioides 7600, as well as a reciprocal best BLASTn search for F. oxysporum f. sp. lycopersici 4287 and F. graminearum PH-1. Sets of orthologs were retained if they were identified in both sets of reciprocal best BLASTn searches. Through this procedure, a total of 4,449 sets of most likely orthologs were identified (including 25.1%, 31.4%, and 33.4% of all transcripts in F. oxysporum, F. verticillioides, and F. graminearum, respectively). For Cryptococcus, we identified orthologs by comparing the transcripts predicted by the Broad Institute in two sets comprising either C. neoformans serotype A strain H99 and C. gattii serotype B strain R265 or C. neoformans serotype D strain JEC21 and C. gattii serotype B strain R265. A total of 5,391 transcripts were identified as reciprocal best hit pairs in both sets, including 77.3%, 80.1%, and 86.8% of all C. neoformans serotype A, C. neoformans serotype D and C. gattii transcripts, respectively. The much higher percentage of ortholog sets identified in Cryptococcus compared with Fusarium most likely stems from the longer divergence times among the included Fusarium species compared with the included Cryptococcus species.
We extended the sets of ortholog sequences for genes with variable intron positions among the studied Fusarium genomes by performing reciprocal best BLASTn searches against transcripts of two related fungal species: Nectria haematococca (Coleman et al. 2009) and Trichoderma reesei (Martinez et al. 2008). We obtained ortholog sequences for 66 out of 126 genes from at least one of the two outgroup species. For the studied Cryptococcus genomes, we performed a reciprocal best BLASTn search against transcripts of the wood-decaying jelly fungus Tremella mesenterica (Floudas et al. 2012). Because of the relatively long divergence time between focal Cryptococcus species and T. mesenterica, we identified orthologs for only 3 out of 61 genes with variable intron positions. Phylogenetic relationships of the outgroups compared with Fusarium and Cryptococcus species are shown in supplementary figure S1, Supplementary Material online. Phylogenies for outgroups were reconstructed as described earlier.
Search for Discordant Exon–Exon Boundaries among Genomes
To identify genes having gained or lost introns within the fungal clades, we used one well-annotated focal genome as a reference. For the Cryptococcus clade, we used the nearly finished C. gattii serotype B strain R265 genome and for the Fusarium clade we used F. oxysporum f. sp. lycopersici 4287 as a reference. We searched for discordant exon–exon boundaries among genomes as described in (Torriani et al. 2011). From each reference genome, we artificially created exon sequences from each gene based on the latest available reference genome annotation (downloaded January 2012; http://www.broadinstitute.org, last accessed February 25, 2012). Each exon was mapped to all included genome assemblies within the clade using LASTZ with a 60% sequence identity threshold (Harris 2007). Custom perl scripts were used to discard low scoring hits. Candidate loci for missing introns in mapped genes were identified if neighboring exons mapped at an unusually short distance from each other or if an inserted sequence was detected within a reference exon.
For each candidate locus, a multiple sequence alignment was produced that included mapped gene sequences and approximately 3,000 bp of flanking sequences for all available genomes within the clade, including the reference strain genomic and transcript sequences. We also included identified orthologs and the corresponding transcript from the other reference genomes within the clade (discussed earlier for the identification ortholog triplets). The multiple sequence alignment was performed using MAFFT (Katoh and Toh 2008) with the iterative refinement option enabled (—maxiterate 1,000). We inspected each candidate locus for the following criteria: 1) unambiguous alignment of the coding sequences within the clade, 2) unambiguous alignment of the flanking sequences within species, 3) mapped gene sequences did not show obvious signs of pseudogenization (i.e., 1–2 bp indels), 4) the deletion of the intron sequence was restricted to the exact exon–exon boundaries in the reference genome (i.e., the deletion did not include adjacent exonic sequences), and 5) we checked whether the splicing signals were conserved. For the comparison among orthologs, we required that the position of the intron was conserved within the gene. We discarded the candidate intron position if the intron position was shifted within the gene or if adjacent introns were fused into a single larger intron.
Search for Putative Homologous Intron Sequences
We searched for similarities among intron sequences by using USEARCH 4.2.66 (Edgar 2010) to cluster sequences and employed a length cut-off to exclude intron sequences outside of the range 50–1,000 bp. Introns were required to share at least 80% length identity and at least 80% sequence similarity.
Results
Genome-Wide Identification of Variable Intron Positions in Fusarium Species
The focal genome for the search of intron gains and losses was the tomato pathogen F. oxysporum f.sp. lycopersici 4287. The genome is gene rich with 17,708 gene models carrying a total of 34,137 introns (1.9 introns per gene on average). The genome of the wheat and barley pathogen F. graminearum contained fewer gene models (n = 13,220), but the intron density is very similar with 1.8 introns per gene on average. To compare intron gains and losses in the Fusarium clade, we included the maize pathogen F. verticillioides and 10 additional genomes of F. oxysporum species comprising diverse pathogens of plants and animals (table 1).
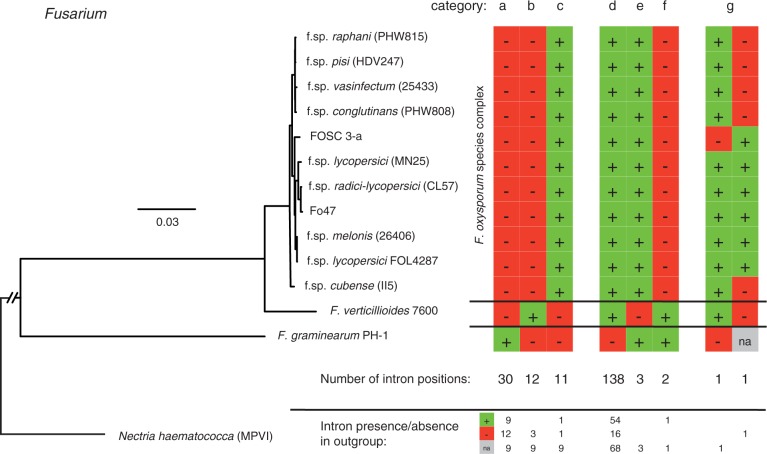
Within the Fusarium clade, we identified a total of 198 intron positions among orthologs where the intron was missing in at least one genome (∼0.5% of all introns in F. oxysporum f.sp. lycopersici 4287). A substantial number of introns were found in only one out of the three included species. The largest number of putatively private introns was found in F. graminearum with 30 introns (fig. 1, category a). The outgroup species N. haematococca contained introns at homologous positions in 9 out of 30 cases, suggesting that these introns were most likely lost in the phylogenetic branch leading to F. verticillioides and F. oxysporum (fig. 1 and supplementary fig. S1, Supplementary Material online). For 12 intron positions, we found no intron at homologous positions in N. haematococca. Hence, these introns may have been gained in F. graminearum. However, independent intron losses in the other species could have produced an identical pattern. We found no homologous intron positions in the second outgroup T. reesei (fig. 1 and supplementary fig. S1, Supplementary Material online). Twelve and 11 introns were found only in F. verticillioides and the FOSC, respectively (fig. 1, categories b and c). At four of these intron positions, the outgroup N. haematococca was lacking an intron and at one position the outgroup was found to have an intron. The largest fraction of the variable intron positions in the Fusarium clade was found to be introns missing only in F. graminearum (fig. 1, category d, 138 positions). Introns missing only in one clade are most likely losses specific to the lineage, as shown by the fact that 54 of these positions were found to contain an intron in N. haematococca. For 16 intron positions, either a parallel loss of introns occurred in N. haematococca and F. graminearum or these introns were gained in the branch leading to the sister species F. verticillioides and F. oxysporum. Low numbers of lineage-specific losses were also found in F. verticillioides and the species complex F. oxysporum, with three and two missing introns, respectively (fig. 1, categories e and f).
Fig. 1.—
Intron evolution in Fusarium species. The phylogenetic tree includes F. graminearum, F. verticillioides and 11 F. oxysporum strains. A strain of N. haematococca (MPVI) was included as an outgroup. F. oxysporum f.sp. lycopersici FOL4287 was the focal genome for our analyses. Vertical columns identify the different categories of intron conservation among strains. Red (“−”) indicates intron absence and green (“+”) indicates intron presence. The total number of intron positions per category is shown below the columns. For Nectria haematococca, a summary of intron presence (“+”) or absence (“−”) is reported for each category. “NA” indicates that no ortholog could be reliable aligned.
Within the FOSC, two intron positions were found to be variable among isolates (fig. 1, category g). One intron was missing in a gene encoding a heat shock 70 kDa protein (FOXG_00795; fig. 2) from the strain FOSC 3-a isolated from human blood. This intron may have been subjected to two independent intron losses, as the same intron was also missing in F. graminearum. The second variable intron was found in a putative amino acid permease gene (FOXG_12112; fig. 2) and only in six monophyletic strains of F. oxysporum infecting mostly tomato. This intron most likely represents a recent intron gain, as it is not fixed in the FOSC and is absent in F. verticillioides. Furthermore, the intron was missing in the outgroup N. haematococca.
Fig. 2.—
Sequence alignments of genes with variable intron positions in F. oxysporum. Sequence alignment of genomic scaffolds and transcript sequences for two genes found with a variable intron position in the FOSC. The schematic drawing shows the relative lengths of exons and introns on the genomic scaffold. The variable intron positions are shown in red for both FOXG_00795 and FOXG_12112. Gene sequences of the variable intron position and neighboring exon sequences are shown as an alignment. Transcript sequences of the orthologs in F. graminearum and F. verticillioides are shown below the genomic sequence (FGSG_00838 and FVEG_00720 for FOXG_00795 and FVEG_10683 for FOXG_12112). The black bar hides an internal section of the intron sequence alignment.
Characterization of Potential Intron Gains in Fusarium Fungi
The most likely intron gains among the Fusarium species are introns found only in a single species (fig. 1, categories a, b, c, and g). Within each category of variable intron positions, we focused only on positions for which the intron state could be ascertained in the outgroup N. haematococca. We distinguished two types of potential intron gains. Either the intron could be gained through insertion of an exogenous sequence or functional splicing evolved in a coding sequence creating a newly spliced intron. Alternatively, in the case of an intron loss rather than a gain, the two different types would represent either a deletion of an intron sequence (likely reverse transcription mediated) or the loss of functional splicing sites creating a novel coding sequence segment. For introns missing in F. verticillioides and F. oxysporum (fig. 1, category a), the majority of introns represent an insertion or deletion event, regardless of the presence or absence of a homologous intron in the outgroup N. haematococca (table 3). The three introns found only in F. verticillioides likely represent an intronization event in two out of three cases (table 3). The two introns found only in F. oxysporum were likely gained through an intronization and an insertion event, respectively. Another intron found in F. oxysporum shares an intron at a homologous position in N. haematococca (table 3). This intron was likely lost independently in F. graminearum and F. verticillioides through loss of functional splicing sites.
Table 3.
Characterization of Intron Gains and Losses in Fusarium
| Categorya | N. haematococca | F. graminearum | F. verticillioides | F. oxysporum | Total (n) | Insertion/Deletionb | Intronization/Exonizationc |
|---|---|---|---|---|---|---|---|
| a | + | + | − | − | 9 | 7 | 2 |
| a | − | + | − | − | 12 | 11 | 1 |
| b | − | − | + | − | 3 | 1 | 2 |
| c | + | − | − | + | 1 | 0 | 1 |
| c | − | − | − | + | 1 | 0 | 1 |
| g | − | N.A. | − | +/− | 1 | 1 | 0 |
aSee figure 1 for more details on the different categories.
bThe intron sequence was inserted or deleted from the gene sequence.
cThe intron sequence either evolved from a coding sequence (“intronization”) or an intron sequence lost functional splicing sites (“exonization”).
Genes with Extensive Changes in Exon–Intron Structure in the Fusarium Clade
Variable intron positions among Fusarium species were found to be nonrandomly distributed among genes. The 198 variable intron positions were identified in 128 distinct genes. Out of these 128 genes, 13 genes showed three variable intron positions. In two genes, we found four variable intron positions and in both cases the variable intron positions were adjacent within the gene. The first gene (FOXG_12247) encodes a peroxisomal copper amine oxidase. Four introns are missing in F. graminearum, suggesting that all four introns were lost in the lineage leading to F. graminearum (fig. 3). One intron was likely lost in F. oxysporum. The second gene (FOXG_13510) was missing four adjacent introns in F. graminearum. In a gene encoding, an ATP-binding cassette transporter (FOXG_02979) all five introns found in F. oxysporum were missing in F. graminearum. The best ortholog in the outgroup species to the studied Fusarium clade, Nectria haematococca (Gene ID: 35 868), was missing the first four of the five introns of F. oxysporum. A particularly intron-rich gene (FOXG_13797, encoding an argininosuccinate synthase) containing 17 introns in F. oxysporum was missing five of these introns in F. graminearum. In the same gene, an intron present in F. graminearum was missing in F. oxysporum (fig. 3). Adjacent changes in intron positions were likely due to intron losses caused by a reverse transcription-related mechanism. Among all variable intron positions, we found 17 instances of a likely parallel loss of two adjacent introns. Furthermore, we found six instances of a likely parallel loss of three adjacent introns and one instance of a likely parallel loss of four adjacent introns (see FOXG_13510, fig. 3).
Fig. 3.—
Extensive changes in the exon–intron structure of Fusarium and Cryptococcus genes. Four genes of F. oxysporum f.sp. lycopersici FOL4287 with the highest number of variable intron positions in the Fusarium clade are shown. Four genes of C. gattii R265 with either three or four variable intron positions in the Cryptococcus clade are shown. Exons (blue) and introns (gray) are drawn to scale according to the genome annotation. Columns above introns indicate a variable intron position among the three main clades of the Fusarium and Cryptococcus species, respectively. Red (“−”) indicates intron absence and green (“+”) indicates intron presence. A rectangle comprising both red and green colors indicates that the intron is not fixed within the clade.
Genome-Wide Identification of Variable Intron Positions in Cryptococcus Species
The C. gattii R265 strain has an intron-rich genome with an average of 5.2 introns per gene (or 32,497 introns among 6,211 gene models). Intron densities in genomes of different Cryptococcus species are similar, as the C. neoformans var. neoformans JEC21 strain genome has an intron density of 5.5 introns per gene (Sharpton et al. 2008). To identify potential intron gain and loss events within and among species of the Cryptococcus species complex, we generated genome assemblies for 18 C. gattii serotype B VGII and 1 of each C. gattii serotype B VGI and B VGIII from Illumina sequence data. The isolates are all from the Pacific Northwest region of Canada and the United States and were isolated from diverse clinical sources (table 2). De novo assemblies efficiently captured the gene space of the C. gattii genome (see Materials and Methods, table 2). We mapped all exon–exon boundaries of the C. gattii R265 genome to genome assemblies of the 20 C. gattii serotype B strains and to the four genome sequences including C. gattii WM276 (a serotype B VGI strain), C. neoformans var. grubii H99 (serotype A), and the two serotype D strains C. neoformans var. neoformans B-3501 and JEC21 (the first sequenced genome of Cryptococcus).
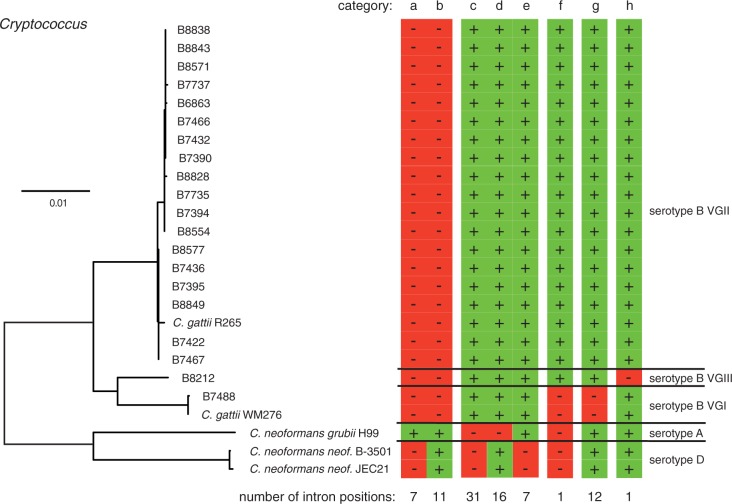
Examination of 88 exon–exon boundaries showed that at least one strain was unambiguously missing the intron sequence among orthologous genes (0.2% of all C. gattii R265 introns). Two of these intron positions showed intron absence in C. neoformans var. grubii H99 and intron presence in all C. gattii serotype B strains; however, the location of the introns in the two C. neoformans var. neoformans strains was not at a homologous position and we therefore excluded these two cases from further analyses. The remaining 86 intron positions were all found in genes with unambiguous sets of orthologs among the three included serotypes. We grouped the intron positions into eight distinct categories based on the pattern of intron presence or absence among the different lineages (fig. 4). The majority of the changes (72 out of 86) were identified among the divergent lineages of Cryptococcus serotypes A, B, and D (fig. 4, categories a–e). At seven homologous intron locations, the intron sequence was present only in C. neoformans var. grubii H99 (fig. 4, category a). In the closest available outgroup species, the jelly fungus T. mesenterica, one out of the seven intron positions showed a reliable ortholog alignment and we found presence of an intron at the homologous position (CNBG_4555). C. neoformans var. grubii H99 is most likely ancestral to the other Cryptococcus serotypes in our analyses (supplementary fig. S1, Supplementary Material online). Hence, the most likely evolutionary reconstruction for this intron position is an intron loss in the lineage leading to serotypes B and D. A total of 16 and 7 intron positions are likely intron losses in C. neoformans var. grubii H99 and C. neoformans var. neoformans, respectively (fig. 4, categories d and e). One gene (CNBG_2928), falling into category d on figure 4, could be scored in T. mesenterica and was found to carry an intron at a homologous position. In 42 cases, we could not resolve whether the introns were gained or lost (fig. 4, categories b and c). The gene CNBG_3052 lacking an intron in serotypes A and D was also lacking an intron in T. mesenterica (fig. 4, category c).
Fig. 4.—
Intron evolution in Cryptococcus neoformans species. The phylogenetic tree includes the major serotypes A, B, and D. The strains of serotype B fall into three major genotypes VGI–III. C. gattii R265 was the focal genome for our analyses. Vertical columns identify the different categories of intron conservation among strains. Red (“−”) indicates intron absence and green (“+”) indicates intron presence. The total number of intron positions per category is shown below the columns.
We identified 14 variable intron positions within the more closely related C. gattii serotype B. One intron position in the gene CNBG_0596 (a predicted vacuolar membrane protein) could either represent a recent intron gain or two independent losses in serotype B VGI and in the lineage leading to serotypes A and D (fig. 4, category f). The same gene lost a second intron in serotype B VGI (fig. 4, category g) and a third intron was lost in C. neoformans var. grubii H99 (fig. 4, category d). In total, 12 introns were recently lost in C. gattii serotype B VGI (fig. 4, category g) and one intron was recently lost in serotype B III (fig. 4, category h).
Genes with Extensive Changes in Exon–Intron Structure in the Cryptococcus Clade
Some genes underwent multiple intron gains or losses, as the 88 variable intron positions were found in 61 distinct genes. Among all variable intron positions, we found four instances of two adjacent intron positions changing concurrently between clades. Six genes were found to have at least three variable intron positions among the different serotypes. CNBG_2832 is missing an intron in serotypes A and D, one intron is missing only in serotype D and one intron is missing in serotype B VGI (fig. 3). CNBG_4556 may have gained an intron in serotype A and lost one intron each in serotype A and B VGI, respectively. CNBG_4964 lost four adjacent introns (out of 11 introns) in serotype A. CNBG_5803 is missing three adjacent intron sequences out of four in total in serotypes A and D (fig. 3).
Locations of Gained and Lost Introns within Genes
Introns are preferentially located closer to the start codon of a gene in F. oxysporum f.sp. lycopersici (fig. 5). Introns that were present only in F. oxysporum showed a tendency to be located closer to the start codon (5′) than the genomic average. However, longer genes (>2,000 bp) did not differ markedly from the genomic average. Similarly, introns missing in F. verticillioides and F. graminearum showed no bias toward the 5′- or 3′-end of the gene.
Fig. 5.—
The relative intron position in genes of the Fusarium and Cryptococcus clades. The gray area represents the distribution (relative position 0–1) of all introns of the focal genomes F. oxysporum f.sp. lycopersici FOL4287 and C. gattii R265. Box plots show the distribution of variable intron positions for different categories of intron conservation (fig. 1 and 2). The thick line inside the box represents the median intron position for each category.
In C. gattii R265, introns are located uniformly throughout the gene and the average insertion point of introns is located toward the center of the gene (fig. 5). Introns that were only found in C. gattii serotype B showed a slight bias toward the stop codon end. Introns that were likely recently lost in C. gattii serotype B VGI showed a stronger bias toward the stop codon end of the gene compared with the genomic average (fig. 5). This bias was found to be strongest for genes longer than 2,000 bp.
Screen for Intron Sequence Characteristics
None of the introns at variable intron positions shared any sequence similarity with other intron sequences according to our search criteria. We did not detect short repeats neighboring splicing sites or inverted repeats in the intron sequence itself.
Discussion
The comparison of genome sequences of members of the same or closely related species provides the opportunity to identify changes in genome architecture over short evolutionary timeframes. Intron gains and losses were thought to be rare among closely related species; however, fungi emerged as highly interesting models with the recent discovery that intron gains and losses may be extensive (Torriani et al. 2011). Our analysis significantly expands on this theme by revealing a large number of variable intron positions within and among species in two very distinct fungal clades, Fusarium and Cryptococcus.
Fusarium species showed a very high number of variable intron positions (n = 198). A study including F. graminearum and three other much more distant ascomycete genomes showed that intron gains and losses were approximately in equilibrium because the split with the other species in the comparison (∼230 Ma) and amounted to approximately 300 positions in total for the lineage leading to Fusarium (Nielsen et al. 2004). The comparably large number of variable intron positions found among Fusarium species in this study may partly be explained by the more extensive set of orthologs that could be screened for intron conservation. By far the largest fraction of polymorphic intron positions differentiated the F. graminearum genome from both the F. verticillioides and F. oxysporum genomes, consistent with the comparably long divergence times between these species. F. graminearum is considered ancestral to the other two species and is more closely related to the distant F. solani [teleomorph: N. haematococca; (Ma et al. 2010)]. We used N. haematococca as the outgroup to identify the sequence of evolutionary events leading either to intron gain or loss. Identifying the most likely intron gains requires balancing the likelihood of multiple independent intron loss events against single intron gain events. Hence, the strongest cases for intron gains are found in categories c and g in figure 1. If N. haematococca was missing an intron at a homologous position, an intron gain event would have to be weighed against a scenario of three independent losses in the lineages N. haematococca, F. graminearum and F. verticillioides. Intron positions found in categories a and b in figure 1, showing absence of a homologous intron in N. haematococca, are difficult to identify as intron gains or losses. Either two independent intron loss events or a single intron gain event could have created the observed pattern among the available lineages. Similarly, for the largest category of variable intron positions, we were also unable to differentiate gains or losses within the framework of our analyses, as they could either represent intron losses in F. graminearum or intron gains prior to the branching of F. verticillioides and F. oxysporum (category d in fig. 1). Ambiguity between intron gains and losses could be resolved using several approaches. First, identifying orthologs in more distant species could be informative about the ancestral state; however, a large proportion of genes are difficult to resolve accurately due to weak orthology and major changes in the gene structure over long divergence times. Second, a probabilistic model of intron evolution could be used instead of parsimony (Nielsen et al. 2004). This approach may, however, be of limited power as the currently available genomes of Fusarium fall into three highly distinct species. The most powerful approach to improve the evolutionary model would be to sequence multiple closely related species and include these in the comparison.
Interestingly, two intron positions were found to be variable within the FOSC. One intron was likely recently gained in a monophyletic group of tomato-infecting strains of F. oxysporum and the second intron likely represents an independent loss both in the F. oxysporum FOSC 3-a strain and the outgroup F. graminearum. F. oxysporum is thought to be asexual with highly differentiated lineages varying in host range and pathogenicity. Unlike the large number of segregating presence–absence polymorphisms at intron positions within populations of the sexual fungus M. graminicola (Torriani et al. 2011), variable intron positions are more likely to be fixed within lineages of F. oxysporum.
Three major serotypes of Cryptococcus were subject to two independent screens for changes in the exon–intron structure (Stajich and Dietrich 2006; Sharpton et al. 2008). Our analyses used C. gattii (serotype B) as the focal species to screen for recent intron gain or loss events among 20 closely related C. gattii isolates. As expected, we detected a substantial number of variable intron positions (n = 72) between the major serotypes A, B, and D. The number of variable intron positions is comparable with Stajich et al. (2006) and higher than reported by Sharpton et al. (2008): 80 and 49 variable intron positions, respectively. The differences may be due to a more sensitive search algorithm and/or the slightly higher number of accepted ortholog triplets. Intron loss dominated the variable intron positions (n = 31) consistent with the earlier studies (Stajich and Dietrich 2006; Sharpton et al. 2008). Interestingly, several cases of recent intron loss were found within serotype B, with losses specific to VGI being the most frequent. A single intron loss was also found in VGIII. Candidates for intron gains were found mostly in C. neoformans var. grubii (n = 7); however, some of these cases might represent parallel losses in the other serotypes as shown by the presence of an intron at one homologous position in the outgroup T. mesenterica. We found a single likely intron gain shared by C. gattii serotype B VGII and VGIII. Combined with two likely intron gains found in C. neoformans var. neoformans identified by Stajich and Dietrich (2006), there is now support for independent intron gains in all major serotypes of Cryptococcus.
The relatively short evolutionary divergence times among the screened genomes in the Fusarium and Cryptococcus clades may have preserved signatures of intron gain and loss mechanisms. We focused on three types of molecular signatures: 1) the sequence similarity of gained or lost introns with other introns, 2) the position of the gained or lost intron within the gene, and 3) extensive changes in the exon–intron structure of particular genes. We did not find any sequence homology between gained or lost introns in C. gattii. Similarly, introns in F. oxysporum did not show any evident sequence similarity. In C. neoformans var. neoformans, a gained intron with close sequence homology to other introns of the same gene was previously identified and was likely due to a repetitive gene structure (Sharpton et al. 2008). The absence of evident signs of recent intron transposition or transposon-related duplication suggests that these phenomena may not be extensive in fungi. The strongest evidence for intron transposition through “reverse splicing” was found in the Mycosphaerella clade, where a large fraction of introns either in transitory stages of gain or loss within the species or recently gained introns showed high sequence homology (Torriani et al. 2011). In multiple-related species of M. graminicola, including the tomato pathogen Cladosporium fulvum, a large proportion of recently gained introns were found to contain recently expanded introner-like elements (van der Burgt et al. 2012). These poorly understood genomic elements share similarity with introner elements previously described in the green algae Micromonas pusilla (Worden et al. 2009). In the genome of M. pusilla, a massive expansion of introns created a large number of nearly identical intron sequences in unrelated genes. These introner elements lacked known features of transposable elements (Worden et al. 2009). The mechanisms driving intron transposition or multiplication remains to be elucidated as the phenomenon occurs in highly diverse organisms such as Tunicates, picoeukaryotes, and Archaea (Worden et al. 2009; Denoeud et al. 2010; Fujishima et al. 2010; Roy and Irimia 2012).
There has been considerable debate about the role of positional biases of introns within genes. The comparison of the average intron position within genes between Fusarium and Cryptococcus corroborates the trend for a broad range of eukaryotic genomes (Jeffares et al. 2006). The intron-poorer F. oxysporum genome shows a 5′-bias in the average intron position, while the intron-rich C. gattii genome shows a relatively even distribution of introns. Analyses of the position of different categories of intron presence–absence patterns among lineages showed no major deviation from the average intron position in the genomes. We found a tendency for introns that were lost in C. gattii VGIII to be located closer to the 3′-end of the gene. A 3′-bias would be expected under the classical model for mRNA-mediated intron loss (Mourier and Jeffares 2003). This model stipulates that a poly-A tail initiated, partial cDNA produced by reverse transcription is re-inserted into the genomic copy through recombination. However, our analyses showed that intron loss was preferentially internal, corroborating the trend identified among distant ascomycete genomes (Nielsen et al. 2004). The most likely mechanism responsible for a bias toward internal intron loss is self-primed reverse transcription (Sharpton et al. 2008). This mechanism requires that the 3′-end of the RNA transcript folds back on itself, creating a hairpin secondary structure. The cDNA generated by reverse transcription would then most likely be initiated internally, instead of being located toward the 3′-end of the transcript. Homologous recombination of the cDNA with the genomic copy of the gene would then most likely result in an internal intron loss (Fedorova and Fedorov 2003). Our data from Cryptococcus and Fusarium indicate that internal intron gain and loss is likely a general trend affecting intron evolution over short evolutionary timeframes.
The most likely cases of intron gains are found in the Fusarium clade, as the outgroup N. haematococca strengthens the parsimony reconstruction. In F. verticillioides, three intron positions were likely gained, as the alternate scenario would require three independent intron losses in the other lineages. We observed both intronization of a coding sequence (Irimia et al. 2008) and insertion of a novel sequence generating these likely new introns. Similarly, the one intron found only in F. oxysporum and absent in all other lineages was likely gained through intronization (fig. 1, category c; table 3). An intron gain was likely caused through insertion of an intron sequence in a subset of F. oxysporum strains, indicating that the intron has not reached fixation within the species. The lack of sequence homology elsewhere in the genome to the newly inserted intron sequences makes it difficult to infer the underlying mechanism in these six cases. The seven introns found only in C. neoformans var. grubii were all created through intronization of a coding sequence or lost in the other clades through the evolution of functional splicing sites. The observation of intronization is consistent with previous findings among Cryptococcus clades (Roy 2009).
In line with these findings, a substantial proportion of recently gained introns in the Mycosphaerella clade were due to intronization (Torriani et al. 2011). The process of intronization is, therefore, a major process driving the evolution of new introns in fungi. Intronization may occur through several scenarios. The emergence of a new intron may be due to neutral processes that fix mutations, which create de novo splicing sites in a coding region. This scenario is likely if the intronization occurred in a region coding for a nonessential domain of the protein. Second, the new intron may be fixed by selection if a part of the coding sequence has negative effects on the function of the encoded protein (e.g., through fixation of a deleterious mutation). In this case, the splicing of the affected sequence may remedy the deleterious effects (Irimia et al. 2008).
In both the Fusarium and Cryptococcus clades, we found genes with substantial changes in their exon–intron structure. One of the four genes with more than three intron gain or loss events within the Fusarium clade had likely lost all the affected introns in a single event in the lineage leading to F. graminearum. The most likely explanation in this case is intron loss through reverse transcription of RNA and genomic integration. All five intron positions in gene FOXG_02979 were found to be variable. We hypothesize that multiple independent losses of adjacent introns generated the currently observed diversity in gene structure. The first two intron positions may represent a rare occurrence of three independent intron losses in the outgroup N. haematococca, F. graminearum, and F. verticillioides. Alternatively, these two introns may have been gained in the lineage leading to F. oxysporum. However, the gain of two unrelated intron sequences at adjacent positions in a gene is highly unlikely. The third and fourth introns were likely independently lost in the outgroup N. haematococca and F. graminearum. The same gene lost the fifth intron in the lineage leading to F. graminearum. In most genes showing multiple intron gains or losses, the affected introns were adjacent, suggesting that the position in the gene played a role in the intron gain or loss. In Cryptococcus, adjacent intron losses were found in multiple genes (see also Stajich and Dietrich 2006; Sharpton et al. 2008). Multiple adjacent intron gains and losses over short evolutionary timeframes seem to be ubiquitous in fungi as similar patterns were also found among closely related species of the Mycosphaerella clade (Torriani et al. 2011). For adjacent intron losses in a gene, the reintegration of cDNA after reverse transcription of spliced RNA provides a convincing mechanism. However, no general model currently exists to explain adjacent spliceosomal intron gains. In mammals, intron losses correlated with increased expression levels as predicted from the standard model of intron loss (Coulombe-Huntington and Majewski 2007). Similarly, intron gains mediated by insertion of exogenous RNA sequences by “reverse splicing” may be favored by higher gene expression levels, increasing the likelihood of “reverse splicing” of the transcript sequences.
Fungi have now emerged as the leading model organisms to study intron gains and losses, shedding light on the processes driving the enormous variation in intron densities among genomes. Unlike in the Mycosphaerella fungal clade, we found no evidence for recent transposition or duplication of intron sequences in these genomes, suggesting that these mechanisms may only be active in restricted phylogenetic groups. The sequencing of larger sets of closely related fungi will largely remove difficulties arising from parsimony reconstruction and provide the much-needed opportunity to further investigate mechanisms of intron gains.
Supplementary Material
Supplementary figure S1 is available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
D.C. and B.A.M. conceived the study. D.C. developed the algorithms, carried out analyses, and drafted the manuscript. D.C. and B.A.M. edited the manuscript. The authors thank Christine Grossen for assistance with data analysis and comments on an earlier version of this manuscript. They thank the reviewers for helpful comments. This work was supported by an ETH Zurich grant [ETH-03 12-1] to D.C. and B.A.M.
Literature Cited
- Agrios GN. Plant pathology. Oxford (UK): Elsevier Academic Press; 2005. [Google Scholar]
- Bartlett KH, Kidd SE, Kronstad JW. The emergence of Cryptococcus gattii in British Columbia and the Pacific Northwest. Curr Infect Dis Rep. 2008;10:58–65. doi: 10.1007/s11908-008-0011-1. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Selfish DNA and the origin of introns. Nature. 1985;315:283–284. doi: 10.1038/315283b0. [DOI] [PubMed] [Google Scholar]
- Chang DC, et al. Multistate outbreak of Fusarium keratitis associated with use of a contact lens solution. JAMA. 2006;296:953–963. doi: 10.1001/jama.296.8.953. [DOI] [PubMed] [Google Scholar]
- Coleman JJ, et al. The genome of Nectria haematococca: contribution of supernumerary chromosomes to gene expansion. PLoS Genet. 2009;5:e1000618. doi: 10.1371/journal.pgen.1000618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins L, Penny D. Complex spliceosomal organization ancestral to extant eukaryotes. Mol Biol Evol. 2005;22:1053–1066. doi: 10.1093/molbev/msi091. [DOI] [PubMed] [Google Scholar]
- Coulombe-Huntington J, Majewski J. Characterization of intron loss events in mammals. Genome Res. 2007;17:23–32. doi: 10.1101/gr.5703406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denoeud F, et al. Plasticity of animal genome architecture unmasked by rapid evolution of a pelagic tunicate. Science. 2010;330:1381–1385. doi: 10.1126/science.1194167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibb NJ. Why do genes have introns? FEBS Lett. 1993;325:135–139. doi: 10.1016/0014-5793(93)81429-4. [DOI] [PubMed] [Google Scholar]
- Duret L, Bucher P. Searching for regulatory elements in human noncoding sequences. Curr Opin Struct Biol. 1997;7:399–406. doi: 10.1016/s0959-440x(97)80058-9. [DOI] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Fedorova L, Fedorov A. Introns in gene evolution. Genetica. 2003;118:123–131. [PubMed] [Google Scholar]
- Floudas D, et al. The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science. 2012;336:1715–1719. doi: 10.1126/science.1221748. [DOI] [PubMed] [Google Scholar]
- Fravel D, Olivain C, Alabouvette C. Fusarium oxysporum and its biocontrol. New Phytol. 2003;157:493–502. doi: 10.1046/j.1469-8137.2003.00700.x. [DOI] [PubMed] [Google Scholar]
- Fujishima K, Sugahara J, Tomita M, Kanai A. Large-scale tRNA intron transposition in the archaeal order Thermoproteales represents a novel mechanism of intron gain. Mol Biol Evol. 2010;27:2233–2243. doi: 10.1093/molbev/msq111. [DOI] [PubMed] [Google Scholar]
- Gillece JD, et al. Whole genome sequence analysis of Cryptococcus gattii from the Pacific Northwest reveals unexpected diversity. PLoS One. 2011;6:e28550. doi: 10.1371/journal.pone.0028550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. System Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Harris RS. University Park (PA): Pennsylvania State University; 2007. Improved pairwise alignment of genomic DNA. [Google Scholar]
- Irimia M, et al. Origin of introns by “intronization” of exonic sequences. Trends Genet. 2008;24:378–381. doi: 10.1016/j.tig.2008.05.007. [DOI] [PubMed] [Google Scholar]
- James TY, et al. Reconstructing the early evolution of fungi using a six-gene phylogeny. Nature. 2006;443:818–822. doi: 10.1038/nature05110. [DOI] [PubMed] [Google Scholar]
- Jeffares DC, Mourier T, Penny D. The biology of intron gain and loss. Trends Genet. 2006;22:16–22. doi: 10.1016/j.tig.2005.10.006. [DOI] [PubMed] [Google Scholar]
- Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinformatics. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- Koonin EV. Intron-dominated genomes of early ancestors of eukaryotes. J Hered. 2009;100:618–623. doi: 10.1093/jhered/esp056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren E, Lev-Maor G, Ast G. The emergence of alternative 3′ and 5′ splice site exons from constitutive exons. PLoS Comput Biol. 2007;3:895–908. doi: 10.1371/journal.pcbi.0030095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2010;20:265–272. doi: 10.1101/gr.097261.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Tucker AE, Sung W, Thomas WK, Lynch M. Extensive, recent intron gains in Daphnia populations. Science. 2009;326:1260–1262. doi: 10.1126/science.1179302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonberg N, Gilbert W. Intron exon structure of the chicken pyruvate-kinase gene. Cell. 1985;40:81–90. doi: 10.1016/0092-8674(85)90311-3. [DOI] [PubMed] [Google Scholar]
- Lynch M. Intron evolution as a population-genetic process. Proc Natl Acad Sci U S A. 2002;99:6118–6123. doi: 10.1073/pnas.092595699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. The origins of eukaryotic gene structure. Mol Biol Evol. 2006;23:450–468. doi: 10.1093/molbev/msj050. [DOI] [PubMed] [Google Scholar]
- Lynch M. The origins of genome architecture. Sunderland (MA): Sinauer Associates; 2007. [Google Scholar]
- Lynch M, Richardson AO. The evolution of spliceosomal introns. Curr Opin Genet Dev. 2002;12:701–710. doi: 10.1016/s0959-437x(02)00360-x. [DOI] [PubMed] [Google Scholar]
- Ma L-J, et al. Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature. 2010;464:367–373. doi: 10.1038/nature08850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez D, et al. Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina) Nat Biotechnol. 2008;26:553–560. doi: 10.1038/nbt1403. [DOI] [PubMed] [Google Scholar]
- Michielse CB, Rep M. Pathogen profile update: Fusarium oxysporum. Mol Plant Pathol. 2009;10:311–324. doi: 10.1111/j.1364-3703.2009.00538.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourier T, Jeffares DC. Eukaryotic intron loss. Science. 2003;300:1393. doi: 10.1126/science.1080559. [DOI] [PubMed] [Google Scholar]
- Nielsen CB, Friedman B, Birren B, Burge CB, Galagan JE. Patterns of intron gain and loss in fungi. PLoS Biol. 2004;2:e422. doi: 10.1371/journal.pbio.0020422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell K, et al. Genetic diversity of human pathogenic members of the Fusarium oxysporum complex inferred from multilocus DNA sequence data and amplified fragment length polymorphism analyses: evidence for the recent dispersion of a geographically widespread clonal lineage and nosocomial origin. J Clin Microbiol. 2004;42:5109–5120. doi: 10.1128/JCM.42.11.5109-5120.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park BJ, et al. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS. 2009;23:525–530. doi: 10.1097/QAD.0b013e328322ffac. [DOI] [PubMed] [Google Scholar]
- Roy S. Intron-rich ancestors. Trends Genet. 2006;22:468–471. doi: 10.1016/j.tig.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Roy SW. Intronization, de-intronization, and intron sliding are rare in Cryptococcus. BMC Evol Biol. 2009;9:192. doi: 10.1186/1471-2148-9-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy SW, Gilbert W. The evolution of spliceosomal introns: patterns, puzzles, and progress. Nat Rev Genet. 2006;7:211–221. doi: 10.1038/nrg1807. [DOI] [PubMed] [Google Scholar]
- Roy SW, Irimia M. Genome evolution: where do new introns come from? Curr Biol. 2012;22:R529–R531. doi: 10.1016/j.cub.2012.05.017. [DOI] [PubMed] [Google Scholar]
- Roy SW, Irimia M. Mystery of intron gain: new data and new models. Trends Genet. 2009;25:67–73. doi: 10.1016/j.tig.2008.11.004. [DOI] [PubMed] [Google Scholar]
- Sharpton TJ, Neafsey DE, Galagan JE, Taylor JW. Mechanisms of intron gain and loss in Cryptococcus. Genome Biol. 2008;9:R24. doi: 10.1186/gb-2008-9-1-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R, Ast G. Intronic sequences flanking alternatively spliced exons are conserved between human and mouse. Genome Res. 2003;13:1631–1637. doi: 10.1101/gr.1208803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stajich JE, Dietrich FS. Evidence of mRNA-mediated intron loss in the human-pathogenic fungus Cryptococcus neoformans. Eukaryot Cell. 2006;5:789–793. doi: 10.1128/EC.5.5.789-793.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stajich JE, Dietrich FS, Roy SW. Comparative genomic analysis of fungal genomes reveals intron-rich ancestors. Genome Biol. 2007;8:R223. doi: 10.1186/gb-2007-8-10-r223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung G-H, Poinar GO, Jr, Spatafora JW. The oldest fossil evidence of animal parasitism by fungi supports a Cretaceous diversification of fungal-arthropod symbioses. Mol Phylogenet Evol. 2008;49:495–502. doi: 10.1016/j.ympev.2008.08.028. [DOI] [PubMed] [Google Scholar]
- Torriani SFF, Stukenbrock EH, Brunner PC, McDonald BA, Croll D. Evidence for extensive recent intron transposition in closely related fungi. Curr Biol. 2011;21:2017–2022. doi: 10.1016/j.cub.2011.10.041. [DOI] [PubMed] [Google Scholar]
- Tseng C-K, Cheng S-C. Both catalytic steps of nuclear pre-mRNA splicing are reversible. Science. 2008;320:1782–1784. doi: 10.1126/science.1158993. [DOI] [PubMed] [Google Scholar]
- van der Burgt A, Severing E, de Wit PJGM, Collémare J. Birth of new spliceosomal introns in fungi by multiplication of introner-like elements. Curr Biol. 2012;22:1260–1265. doi: 10.1016/j.cub.2012.05.011. [DOI] [PubMed] [Google Scholar]
- Worden AZ, et al. Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science. 2009;324:268–272. doi: 10.1126/science.1167222. [DOI] [PubMed] [Google Scholar]
- Xu J, Vilgalys R, Mitchell TG. Multiple gene genealogies reveal recent dispersion and hybridization in the human pathogenic fungus Cryptococcus neoformans. Mol Ecol. 2000;9:1471–1481. doi: 10.1046/j.1365-294x.2000.01021.x. [DOI] [PubMed] [Google Scholar]