Abstract
DNA methylation is an epigenetic modification essential for the regulation of gene expression that has been implicated in many diseases, including cancer. Few studies have investigated the wide range of potential predictors of global DNA methylation, including biomarkers. Here, we investigated associations between DNA methylation and dietary factors, sex-steroid hormones, metabolic, lipid, inflammation, immune and one-carbon biomarkers. Data and baseline biomarker measurements were obtained from 173 overweight/obese postmenopausal women. Global DNA methylation in lymphocyte DNA was measured using the pyrosequencing assay for LINE-1 repeats. We used correlations and linear regression analyses to investigate associations between continuous data and DNA methylation, while t-tests were used for categorical data. Secondary analyses stratified by serum folate levels and multivitamin use were also conducted. There was little variability in LINE-1 methylation (66.3–79.5%). Mean LINE-1 methylation was significantly higher among women with elevated glucose levels. Mean LINE-1 methylation was also higher among women with high CD4+/CD8+ ratio, and lower among women with elevated vitamin B6, but neither reached statistical significance. In analyses stratified by folate status, DNA methylation was negatively associated with sex hormone concentrations (estrone, estradiol, testosterone and sex hormone binding globulin) among women with low serum folate levels (n = 53). Conversely, among women with high serum folate levels (n = 53), DNA methylation was positively associated with several immune markers (CD4/CD8 ratio, NK1656/lymphocytes and IgA). Results from this screening suggest that global DNA methylation is generally stable, with differential associations for sex hormones and immune markers depending on one-carbon status.
Keywords: DNA methylation, epigenetics, epidemiology, folate, glucose, metabolic, immune function, inflammation, post-menopausal, sex hormones, women
Introduction
DNA methylation is a reversible epigenetic process of gene regulation and may also impact genomic stability.1 The mechanism of DNA methylation is particularly important during embryogenesis and carcinogenesis and has been studied mainly in the context of cancer and aging.1-5 Reduced levels of genomic methylation are commonly observed early during carcinogenesis, and this epigenetic dysregulation, jointly with hypermethylation at specific CpG islands, correlates with disease severity and metastatic capabilities in many tumors.6,7 There is also evidence that altered DNA methylation patterns are associated with other pathologies such as autoimmune and cardiovascular diseases.7-9
Studies of DNA methylation distinguish between hypermethylated sequences in CpG islands near promoter sites of specific genes and global hypomethylation at repetitive sequences as separate mechanisms that foster carcinogenesis.10,11 Although global hypomethylation is less well understood than CpG island hypermethylation,12 it has been suggested that a decrease in global DNA methylation may contribute to cancer progression via chromosomal instability and aneuploidy altering chromatin structure.13-15
Several studies have revealed the importance of environmental factors in relation to DNA methylation.16-19 Recent studies have also shown that risk factors across the life course may be associated with adult DNA methylation.17,20 Nevertheless, the impact of many other lifestyle characteristics on DNA methylation has not been well characterized. While some studies have reported associations between age, gender and alcohol and global hypomethylation,16,20-26 others have not.11,26-30 Few studies have investigated associations between global DNA methylation and biomarkers, which may represent the systemic hormonal, immunological or inflammatory milieu. In a recent experimental study, an elevated glucose level was observed to increase DNA methylation by enhancing DNA methyltransferase activity.31 It has been suggested that metabolic control of folate and methyl groups may be under hormonal regulation; however, very little is known about the associations between these hormones and DNA methylation.32 Folate is a critical source of methyl groups for epigenetic changes and plays an important role in global genomic methylation12,21,25,33-39 and DNA methylation levels correlate with folate bioavailability.40
In this exploratory study, we analyzed dietary, metabolic, lipid, inflammation, immune and sex hormone biomarkers to identify factors that may be associated with DNA methylation at long interspersed nucleotide elements (LINE-1). We also investigated the relationship between LINE-1 methylation and biomarkers of interest stratified by serum folate and multivitamin use, in order to account for one-carbon status. While the goal of this study was to perform an overall screen, we also specifically hypothesized that blood glucose and sex hormone concentrations would be positively associated with DNA methylation. In addition, we hypothesized that the associations between dietary, metabolic, lipid, inflammation, immune and sex hormone biomarkers and DNA methylation could depend on one-carbon status; hence, our stratification by serum folate and multivitamin use.
Results
Characteristics of eligible study participants are presented in Table 1. The average age of the participants was 60.6 ± 6.7 y and 86% were Caucasian. All participants were overweight or obese, with a mean body mass index (BMI) of 30.5 ± 3.9 kg/m2, and a mean percentage body fat of 47.4 ± 4.7%.
Table 1. Baseline characteristics of the study participants.
| Characteristic | N | Mean ± SD or % |
|---|---|---|
| Methylation (%) |
173 |
72.5 ± 2.2 |
| Age (yr) |
173 |
60.6 ± 6.7 |
| Weight (kg) |
173 |
81.6 ± 13.0 |
| Alcohol intake (g/day) |
173 |
4.3 ± 0.6 |
| Body mass index (kg/m2) |
173 |
30.5 ± 3.9 |
| Body fat (%) |
173 |
47.4 ± 4.7 |
| Ethnicity: Non-Hispanic white African American Asian/Pacific Islander Hispanic/Latino American Indian Others |
149 7 9 2 2 3 |
86.0% 4.0% 5.0% 1.0% 1.0% 2.0% |
| Ever user of hormonesa |
173 |
42.2% |
| Ever smoked > 100 cigarettesb |
173 |
46.5% |
| Ever user of birth control pills |
172 |
61.6% |
| First-degree relative with cancer |
167 |
59.9% |
| Multivitamin use (≥ 1 time/wk)c | 111 | 51.4% |
a Former users of postmenopausal hormones, as current users were excluded. bFormer smokers, as current users were excluded. cMultivitamin use ≥ 1 time/wk during the past 3 mo. Multivitamin use was only assessed among a subset of study participants who participated in the Immune Exercise (IMEX) study
There were no associations with DNA methylation in analyses conducted using categorical variables (Table 2). Unadjusted t-tests and Pearson’s correlations on log-transformed continuous variables in relation to DNA methylation are presented in Table 3. There were weak negative correlations between DNA methylation and energy intake, total fat intake and sex hormone binding globulin (SHBG) (r = -0.16 to -0.19) and a weak positive correlation with CD4+/CD8+ ratio (r = 0.18) (Table 3). We detected few significant associations with methylation status. DNA methylation was significantly increased among those with higher blood glucose (p = 0.01) and those with a higher CD4+/CD8+ ratio (p = 0.05). A t-test of the median split for DNA methylation with glucose was statistically significant (p = 0.01) with a low mean of 72.0 ± 1.9 (n = 87) and a high mean of 72.9 ± 2.4 (n = 83) for DNA methylation. There was also a significant trend for glucose in quartiles with DNA methylation (p-trend = 0.02) (data not shown). On the other hand, DNA methylation was lower among women with higher dietary vitamin B6 intakes (p = 0.05). There were no significant associations between age, alcohol, BMI, biomarkers of inflammation, sex hormones and methylation status.
Table 2. Associations between categorical variables and DNA methylation at baseline.
| Variable |
No |
Yes |
|
||
|---|---|---|---|---|---|
| N | DNA methylation | N | DNA methylation | P-value | |
| |
|
|
|
|
|
| Ever use of postmenopausal hormonesa |
100 |
72.3 ± 0.2 |
73 |
72.5 ± 0.2 |
0.53 |
| Ever smoke (100+ cigarettes)b |
92 |
72.7 ± 0.2 |
81 |
72.3 ± 0.3 |
0.18 |
| Ever use of birth control pills |
66 |
72.5 ± 0.2 |
106 |
72.4 ± 0.3 |
0.71 |
| First-degree relative with cancer |
67 |
72.4 ± 0.2 |
100 |
72.4 ± 0.2 |
0.99 |
| Multivitamin use (≥ 1 time/wk)c | 54 | 72.4 ± 0.3 | 57 | 72.4 ± 0.3 | 0.91 |
a Former users of postmenopausal hormones, as current users were excluded. bFormer smokers, as current smokers were excluded. cMultivitamin use ≥ 1 time/wk during the past 3 mo, assessed among a subset of study participants who participated in the Immune Exercise (IMEX) study.
Table 3. Associations between age, biomarkers and DNA methylation.
| Markera | N | Mean | Standard Deviation | P value ≥ vs. < medianb |
Corr r (p)c |
|---|---|---|---|---|---|
| Age |
173 |
60.6 |
± 6.7 |
0.77 |
0.07 (0.38) |
|
Insulin/IGF/metabolic hormones |
173 |
|
|
|
|
| Glucose (mg/dl) |
170 |
98.0 |
± 9.1 |
0.01 |
0.12 (0.11) |
| Insulin (µg/ml) |
173 |
20.0 |
± 9.7 |
0.98 |
0.03 (0.72) |
| IGF-1 (ng/ml) |
173 |
107.0 |
± 36.5 |
0.84 |
0.03 (0.74) |
| IGFBP-3 (µg/ml) |
173 |
4.1 |
± 1.1 |
0.64 |
-0.05 (0.53) |
| Ghrelin (pg/ml) |
173 |
613.9 |
± 342.9 |
0.10 |
-0.06 (0.43) |
| Leptin (ng/ml) |
173 |
28.0 |
± 8.4 |
0.99 |
0.02 (0.78) |
|
Lipid metabolism biomarkers |
165 |
|
|
|
|
| Cholesterol (mg/dl) |
165 |
231.6 |
± 39.6 |
0.68 |
0.05 (0.50) |
| HDL (mg/dl) |
165 |
52.1 |
± 12.4 |
0.57 |
0.01 (0.90) |
| LDL (mg/dl) |
165 |
152.2 |
± 38.8 |
0.65 |
0.04 (0.61) |
| VLDL(mg/dl) |
165 |
27.2 |
± 13.1 |
0.57 |
0.01 (0.88) |
| Triglycerides (mg/dl) |
170 |
135.3 |
± 65.2 |
0.53 |
0.02 (0.85) |
|
One-carbon nutrient biomarkers |
106 |
|
|
|
|
| Serum B12 (pg/ml) |
106 |
390.6 |
± 189.5 |
0.85 |
0.03 (0.79) |
| Serum folate (ng/ml) |
106 |
56.2 |
± 34.0 |
0.66 |
0.06 (0.52) |
|
Dietary intake |
168 |
|
|
|
|
| Energy (kcal) |
168 |
1643 |
± 624 |
0.19 |
-0.16 (0.04) |
| Alcohol (gm) |
168 |
4.4 |
± 7.9 |
0.37 |
0.08 (0.64) |
| Total carbohydrates (gm) |
168 |
189.8 |
± 80.3 |
0.64 |
-0.11 (0.14) |
| Daily fruit (med portions) |
168 |
1.8 |
± 1.4 |
0.61 |
-0.10 (0.22) |
| Daily vegetables (med portions) |
168 |
2.1 |
± 1.4 |
0.23 |
-0.08 (0.30) |
| Total fat (gm) BL |
168 |
67.7 |
± 32.8 |
0.07 |
-0.19 (0.01) |
| Folate (µg) BL |
168 |
230.5 |
± 105.6 |
0.29 |
-0.07 (0.35) |
| Vitamin B6 (mg) BL |
168 |
1.6 |
± 0.6 |
0.05 |
-0.10 (0.18) |
| Vitamin B12 (µg) BL |
168 |
5.3 |
± 2.6 |
0.22 |
-0.08 (0.29) |
|
Body composition |
173 |
|
|
|
|
| BMI (kg/m2) |
173 |
30.5 |
± 3.9 |
0.16 |
0.06 (0.45) |
| Percent body fat (%) |
173 |
47.4 |
± 4.7 |
0.33 |
0.03 (0.68) |
|
Immune markers |
115 |
|
|
|
|
| % T cells (% of lymphocytes) |
112 |
70.5 |
± 6.4 |
0.7 |
0.05 (0.57) |
| T Cytotoxic cells (% of lymphocytes) |
113 |
20.5 |
± 8.4 |
0.51 |
-0.16 (0.09) |
| T helper cells (% of lymphocytes) |
114 |
49.3 |
± 8.1 |
0.23 |
0.16 (0.09) |
| CD4+/CD8+ ratio |
113 |
2.9 |
± 1.4 |
0.05 |
0.18 (0.05) |
| NK cells (% lymphocytes) |
115 |
8.6 |
± 4.3 |
0.23 |
0.05 (0.56) |
| B cells (cells/μl) |
115 |
14.3 |
± 4.8 |
0.89 |
-0.09 (0.33) |
| IgA (mg/dl) |
113 |
201.5 |
± 93.4 |
0.1 |
0.13 (0.16) |
| IgG (mg/dl) |
114 |
980.6 |
± 229.2 |
0.8 |
-0.12 (0.20) |
| IgM (mg/dl) |
114 |
117.6 |
± 58.2 |
0.37 |
-0.13 (0.16) |
|
Biomarkers of Inflammation |
115 |
|
|
|
|
| C-Reactive Protein (mg/l) |
114 |
0.4 |
± 0.4 |
0.48 |
-0.02 (0.82) |
| IL-6 (ng/l) |
115 |
3.3 |
± 3.0 |
0.91 |
0.05 (0.60) |
|
Sex Hormone Biomarkers |
173 |
|
|
|
|
| Estradiol (pg/ml) |
173 |
19.3 |
± 8.0 |
0.9 |
-0.05 (0.56) |
| Free Estradiol (pg/ml) |
173 |
0.5 |
± 0.2 |
0.95 |
0.01 (0.92) |
| Bioavailable Estradiol (pg/ml) |
173 |
13.4 |
± 6.0 |
0.32 |
0.01 (0.92) |
| Estrone (pg/ml) |
173 |
46.9 |
± 17.1 |
0.76 |
-0.08 (0.31) |
| Testosterone (pg/ml) |
173 |
234.5 |
± 98.9 |
0.09 |
-0.07 (0.34) |
| Free Testosterone (pg/ml) |
173 |
5.0 |
± 2.1 |
0.73 |
-0.02 (0.82) |
| Bioavailable Testosterone (pg/ml) |
173 |
12.3 |
± 5.2 |
0.83 |
-0.02 (0.82) |
| Androstenedione (ng/ml) |
173 |
597.3 |
± 230.6 |
0.56 |
-0.05 (0.51) |
| DHEA (ng/ml) |
173 |
2.7 |
± 1.4 |
0.27 |
-0.08 (0.31) |
| DHEA-SO4 (ng/ml) |
166 |
71.7 |
± 48.4 |
0.59 |
-0.07 (0.37) |
| Prolactin (ng/ml) |
172 |
8.1 |
± 4.6 |
0.61 |
-0.03 (0.67) |
| Follicle stimulating Hormone (mIu/l) |
173 |
66.4 |
± 23.1 |
0.09 |
-0.09 (0.24) |
| Sex Hormone Binding Globulin (nmol/l) | 173 | 39.5 | ± 20.8 | 0.15 | -0.16 (0.04) |
a All variables are log transformed. bT test compares mean methylation values of groups above and below biomarker median. cCorrelation coefficients presented are from partial correlations adjusted for age.
Stratification
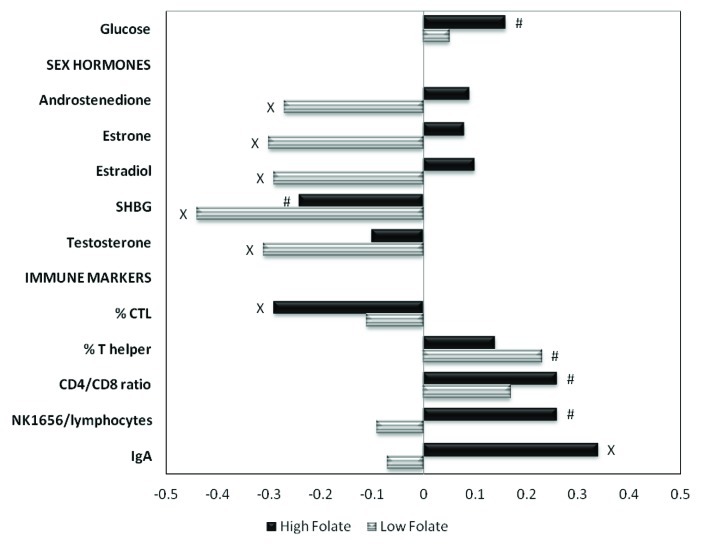
We stratified our biomarker-methylation analyses by serum folate levels and multivitamin use to investigate associations with biomarkers under a high or low one-carbon status. The associations for sex hormones and immune markers differed substantially by serum folate or multivitamin use (Table 4 and Fig. 1). Among women with low serum folate, there were significant inverse correlations between DNA methylation and the following sex hormones: androstenedione (r = -0.27, p = 0.05), estrone (r = -0.30, p = 0.03), estradiol (r = -0.29, p = 0.04), SHBG (r = -0.44, p = 0.001) and testosterone
Table 4. Univariate associations of biomarkers with DNA methylation, stratified by serum folate or multivitamin use.
| Markera |
|
Overalla |
Serum folateb |
Multivitamin use |
||
|---|---|---|---|---|---|---|
| Corr r(p) (n = 106) |
Low Corr r (p) (n = 53) |
High Corr r (p) (n = 53) |
No Corr r (p) (n = 54) |
Yes Corr r (p) (n = 57) |
||
|
Nutritional biomarkers |
|
|
|
|
|
|
| Glucose |
|
0.12 (0.11) |
0.05 (0.74) |
0.16 (0.08) |
0.18 (0.20) |
0.03 (0.82) |
|
Sex hormones |
|
|
|
|
|
|
| Androstenedione |
|
-0.08 (0.38) |
-0.27 (0.05) |
0.09 (0.53) |
-0.22 (0.12) |
0.02 (0.91) |
| Estrone |
|
-0.11 (0.28) |
-0.30 (0.03) |
0.08 (0.55) |
-0.36 (0.01) |
0.08 (0.54) |
| Estradiol |
|
-0.10 (0.31) |
-0.29 (0.04) |
0.10 (0.48) |
-0.34 (0.02) |
0.07 (0.58) |
| SHBG |
|
-0.32 (0.00) |
-0.44 (0.001) |
-0.24 (0.09) |
-0.31 (0.03) |
-0.33 (0.01) |
| Testosterone |
|
-0.21 (0.03) |
-0.31 (0.02) |
-0.10 (0.46) |
-0.24 (0.09) |
-0.17 (0.20) |
|
Immune markers |
|
|
|
|
|
|
| % CTL |
|
-0.21 (0.03) |
-0.11 (0.50)d |
-0.29 (0.04)d |
-0.01 (0.92)e |
-0.34 (0.01)f |
| % T helper |
|
0.18 (0.07) |
0.23 (0.09) |
0.14 (0.32)d |
0.22 (0.13) |
0.16 (0.22)f |
| CD4/CD8 ratio |
|
0.22 (0.03) |
0.17 (0.24)c |
0.26 (0.06)d |
0.10 (0.52) |
0.31 (0.02)f |
| NK1656/lymphocytes |
|
0.11 (0.27) |
-0.09 (0.53) |
0.26 (0.06) |
-0.08 (0.56) |
0.23 (0.08) |
| IgA | 0.17 (0.08) | -0.07 (0.61)c | 0.34 (0.01) | -0.01 (0.93) | 0.38 (0.00) | |
All variables are log transformed. P-values < 0.10 are marked in bold. aRestricted to women with serum folate measurement. bSerum folate: low (15.17 – 49.88 ng/ml), high (50.45 – 258.71 ng/ml). cn = 51; dn = 52; en = 53; fn = 56.
Figure 1. Correlations between markers and DNA methylation, stratified by serum folate concentrations. P values x < 0.05; # ≤ 0.10.
(r = -0.31, p = 0.02). DNA methylation was positively correlated with IgA levels among women with high folate concentrations (r = 0.34, p = 0.01). Among women who did not use multivitamins, there were very similar patterns, with significant negative correlations between DNA methylation and serum estrone, estradiol and SHBG concentrations. Among women who used multivitamins, DNA methylation was negatively correlated with SHBG and % CTL, while being positively correlated with CD4+/CD8+ ratio and IgA levels.
Discussion
This screen of metabolic, immunologic and sex hormone biomarkers associated with global DNA methylation is one of the first comprehensive studies to investigate biologic correlates of DNA methylation. Although there were few associations in the overall analyses (with glucose, vitamin B6 and CD4+/CD8+ ratio), our study revealed many associations between DNA methylation, sex hormones and immune markers when one-carbon status was taken into account.
We observed that individuals with elevated glucose levels had significantly higher mean LINE-1 methylation values. This is similar to what has been recently reported.41 Pearce and colleagues reported that a 10% increase in LINE-1 methylation was associated with an 0.28 mmol/l increase in fasting glucose concentrations.41 Furthermore, global DNA methylation has also been shown to be associated with insulin resistance with a 10% increase in global DNA methylation resulting from a 4.55 unit increase in homeostasis model assessment.42 A potential biological mechanism linking promoter-specific hypermethylation to glucose levels is that age-related increases in methylation may increase susceptibility to hepatic insulin resistance and diabetes.43 This hypothesis is supported by evidence from experimental studies, which indicate that high glucose concentration increases promoter-specific methylation of enzymes such as hepatic glucokinase (GCK) and global DNA hypomethylation in hepatic tissue.31,43
The effect of age on DNA methylation is controversial. Although the mechanism underlying age-related hypomethylation remains unknown, it has been suggested that repeated rounds of DNA replication may result in progressive loss of methylation in repetitive elements since most normal methylation is in repetitive DNA.44 Similar to some previous studies,11,26-30,45-47 we observed no associations between age and DNA methylation. This could be due to the limited age range of the women in our study (55 to 75 y). Bjornsson and colleagues reported time-dependent changes in DNA methylation within the same individual in two longitudinal studies in Iceland and the US.48 Also, other studies have reported age-associated changes in DNA methylation,49,50 but these changes have been rather small. More recently, Zhu and colleagues reported that Alu, but not LINE-1, methylation levels decreased with aging, suggesting that changes in DNA methylation due to aging may not be consistent within the genome.16
Heavy alcohol intake impairs DNA methylation.51 Alcohol may potentially stimulate DNA hypomethylation by competing with folate metabolism and decreasing the synthesis of the methyl donor S-adenosylmethionine (SAM).52 However, similar to our study, most previous studies have not reported any associations between alcohol intake and LINE-1 methylation.16,29,30,37 This may be because in most of these studies alcohol consumption among study participants was low. For example, in our study, mean alcohol intake was 4.3 ± 0.6 g/d, largely because moderate or heavy alcohol drinkers were excluded from participating in the study. It is likely that the impact of alcohol on DNA methylation is more pronounced among populations with higher alcohol intake.
There are indications that the effects of folate status on DNA methylation are complex, depend on cell type and may be gene and site specific.53,54 For instance, Slattery and colleagues reported interactions between folate status, polymorphisms in methylenetetrahydrofolate reductase (MTHFR) gene and DNA methylation, whereby at low folate levels, individuals who were homozygous for the variant TT genotype of the MTHFR gene had DNA hypomethylation.55 Although the results from our study do not provide evidence for any associations between DNA methylation and serum folate and vitamin B12 levels, our data suggest that one-carbon status may modify the effect of sex hormones and immune markers on global DNA methylation. We observed inverse associations between DNA methylation and several sex hormones among women with low serum folate levels and those who did not use multivitamins, but not among women with high folate levels and those who used multivitamins. Hence, it may be important for studies investigating the associations between hormones and DNA methylation to examine for possible interactions.
Few studies have investigated the relationship between global DNA methylation and immune function. Based on our stratified analyses, there are suggestions that one-carbon status modifies the associations between immune markers and global DNA methylation. This is because immune markers were associated with global DNA hypomethylation among women with high serum folate and those who use multivitamin but not among women with low serum folate and those not taking multivitamins. T cell DNA demethylation occurs in lupus and rheumatoid arthritis56 and may be implicated in other immune system disorders. Global age-related hypomethylation occurs in the T-lymphocytes of aging humans,44,57 although data are sparse on how these age-related methylation changes affect the immune system.44 The aging process parallels the deterioration in immune function.14,44 T-cell mediated autoimmune diseases may be included among these age-related diseases that result in changes in the DNA methylation pattern of cells.14,44 These methylation changes are akin to the changes found in malignant cells, with general genome hypomethylation combined with specific gene hypermethylation and gene silencing.14
Gender may be a predictor of LINE-1 methylation, with females having lower methylation levels than males.23,26,30 It has been suggested that the effect of gender on DNA methylation may be mediated through hormonal differences between males and females, wherein estrogen could be responsible for the lower methylation levels in females and testosterone for the higher methylation levels in males.26 However, direct evidence linking both hormones to methylation levels is lacking. Similar to the overall observations in our study, El-Maarri and colleagues reported no associations between estrogen and testosterone at LINE-1, as well as Alu repeat regions.26 Nevertheless, no previous studies have reported on the associations of sex hormones with DNA methylation stratified by serum folate or multivitamin use. In our study, we observed moderate but significant negative associations between sex hormones (estrone, estradiol, androstenedione, testosterone and sex hormone binding globulin) and DNA methylation among women with low serum folate and, with the exception of androstenedione, also among women who did not use multivitamins. Thus, one-carbon status appears to be an important factor in the association between sex hormones and DNA methylation and the associations are only apparent among women who may have lower one-carbon status and thus lower levels of the methyl-donor S-adenosylmethionine.
The strength of our study is that it is one of the first to comprehensively screen metabolic, hormonal and immunological associations with global DNA methylation. The following limitations need to be considered when interpreting the results from our study. Our study was restricted to overweight and obese postmenopausal women; thus, the findings may not be generalizable to other study populations. However, about two-thirds of women in that age group in the US are overweight or obese. Also, the restricted study population may have reduced the variability in methylation values. The LINE-1 pyrosequencing assay used to measure global methylation does not assess site specific methylation. Furthermore, the relationship between the LINE-1 assay and the methyl group acceptance assay used in previous studies is unknown. The level of LINE-1 methylation is significantly correlated with the amount of genome-wide 5-methyl-cytosine when measured by high performance liquid chromatography.58 However, this correlation is not perfect and variables such as folate may affect DNA methylation in genomic regions besides LINE-1. Because this was an exploratory study to generate hypotheses for future studies, we did not adjust for multiple comparisons. Hence, with the large number of variables tested and the moderate sample size, some of the variables may have shown statistical significance by chance. Lastly, due to the cross-sectional nature of the study, the associations between the markers and DNA methylation cannot be interpreted as causal.
In this screening study, we noted a limited number of predictors of global DNA methylation differences among overweight/obese postmenopausal women. However, our study revealed that the associations between several markers, particularly sex hormones and immune markers and global DNA methylation are modified by one-carbon status. This knowledge is essential in epidemiological studies investigating the associations of DNA methylation and health. Overall, our exploratory findings await replication in larger studies.
Materials and Methods
Participants
This study was conducted using data from women (n = 173) participating in the Physical Activity for Total Health Study, an exercise trial among overweight and obese sedentary women (age 55 to 75 y) which has been previously described in detail.59 Participants were eligible if they were not using postmenopausal hormone therapy, sedentary, non-smokers, consumed < 2 alcohol drinks/day, and had no endocrine-related disease or cancer.59 Women were excluded from the study if they had any factors that might affect intervention adherence or retention or factors that might affect study endpoints (e.g., might affect endogenous hormone levels).59 As part of the study, several hormonal, metabolic and immunologic biomarkers were assessed. We used baseline data for investigating cross-sectional associations. Informed consent was obtained from all participants and approved by the Fred Hutchinson Cancer Research Center (FHCRC) Institutional Review Board.
Baseline data collection
Height and weight were measured in duplicate. Total body fat was determined using dual-energy X-ray absorptiometry (Hologic QDR 1500: Hologic Inc.). Information on age, education, income, employment status, marital status, race/ethnicity, use of medications, medical history and smoking history were collected by questionnaire. Measures of dietary and alcohol intake were also evaluated using a 120-item food frequency questionnaire designed and validated at the FHCRC.60 Use of nutritional supplements was ascertained during an in-person interview and by copying nutritional labels.
Methylation
Buffy coats were assayed for global genomic DNA methylation in lymphocytes using LINE-1 (long interspersed nucleotide elements) bisulfite pyrosequencing. Sodium bisulfite modification of lymphocyte DNA was performed as previously described.61 Methylation analysis of LINE-1 promoter (GenBank accession number X58075) was investigated using a pyrosequencing-based methylation analysis reported elsewhere.29
Metabolic measures
Participants provided a 12-h fasting baseline blood sample, which was processed within 1 h of collection. For all assays, laboratory personnel were blinded to the sample identity. Detailed descriptions on assay methods and reproducibility are given in previous publications.62-64 Briefly, fasting glucose was quantified on a clinical chemistry autoanalyzer, measuring the combined catalytic activities of hexokinase and glucose-6-phosphate dehydrogenase.62 Insulin was quantified by a 48-h, polyethylene glycol-accelerated, double-antibody radioimmunoassay.63 Leptin assays were performed using a commercially available radioimmunoassay (Linco Research).63 Ghrelin was measured using a modification of a commercial radioimmunoassay (Phoenix Pharmaceuticals).63 IGF-1 was quantified via a two-site chemiluminescence immunoassay using the Nichols Advantage Specialty System (Nichols Diagnostic Institute).64 IGFBP-3 was quantified via a competitive protein-binding RIA using the IGFBP-3 100T kit (Nichols Diagnostic Institute)64.
Lipid metabolism biomarkers
Total cholesterol (TC), high density lipoproteins (HDL) and triglycerides were measured enzymatically using the Hitachi 917 autoanalyzer at the Northwest Lipid Research Laboratories (University of Washington). All lipoprotein assays were performed using previously described methods.65 We used the Friedewald’s equation method66 to calculate low-density lipoprotein (LDL).
One-carbon nutrient biomarkers
Plasma concentrations of vitamin B12 and total plasma folate were measured in fasting subjects by combined affinity high performance liquid chromatography (HPLC) with electrochemical detection at the Jean Mayer USDA Human Nutrition Aging Center at Tufts University, Boston, MA as described previously.67
Immune function markers
A subset of Physical Activity for Total Health subjects (n = 114) participated in the Immune Exercise (IMEX) study, for which data on markers of immune function were collected. Methods for measuring immune markers including quality control markers are described elsewhere.68,69 Briefly, lymphocytes were isolated from whole blood using a whole-blood lysis technique. A four-color flow cytometer (XL-MCL, Beckman Coulter) was used to enumerate subsets of lymphocytes in blood samples. T lymphocyte proliferation studies were conducted using cryo-preserved PBMC, and were performed by 2 methods: tritiated 3H-thymidine incorporation in response to the mitogen phytohemagglutinin (PHA), and by the cell division tracking method in response to anti-CD3. Immunoglobulin A (IgA), G (IgG), and M (IgM) assays were performed by nephelometry using the Behring Nephelometer II analyzer (Dade-Behring Diagnostics, Deerfield, IL).
Sex hormones
Serum hormone assays were performed at the Reproductive Endocrine Research Laboratory (University of Southern California) using methods described in detail elsewhere.70 Briefly, estradiol, estrone, testosterone, androstenedione, and DHEA were quantified by radioimmunoassay after organic solvent extraction and Celite column partition chromatography.71,72 Chromatographic separation of the steroids was achieved by use of different concentrations of toluene in isooctane and ethyl acetate in isooctane. Sex hormone-binding globulin (SHBG) was quantified via an immunometric assay, and dehydroepiandrosterone sulfate (DHEAS) was quantified via a competitive immunometric assay (Immulite Analyzer, Diagnostic Products Corp.). Free estradiol and testosterone were calculated using methods described and validated elsewhere.73-75
Statistical analysis
The Physical Activity for Total Health study population included 173 women with data on metabolic hormones, lipid metabolism biomarkers, dietary intake, body composition and sex hormones. Missing data for all assays ranged from one to three, except for DHEA-SO4, which was not available for seven women. All biomarkers were log-transformed to reduce departure from normality.
To investigate the associations between biomarkers and DNA methylation, we used Pearson’s correlation (because global DNA methylation was nearly normally distributed), t-tests (grouped by approximate median split) were used for categorical data and linear regression analyses were used for continuous data. Methylation values ranged from 66.30% to 79.45%. We investigated unadjusted and age or
race/ethnicity-adjusted associations between biomarkers for metabolism, lipid levels, one-carbon nutrients, dietary intake, body composition, immune function and sex hormones with DNA methylation. Because the results were similar for the adjusted and unadjusted analyses, we have presented only results for the unadjusted analyses. All analyses were performed using SAS version 9.1 (SAS Institute). A two-sided P value < 0.05 was considered statistically significant.
We investigated the following categorical variables: regular multivitamin use (yes/no), postmenopausal hormone use (ever/never), oral contraceptive use (ever/never), smoking (ever/never) and a first-degree relative with cancer (yes/no). Glucose was analyzed as an indicator variable for quartiles and as a continuous variable.
We measured associations between the following continuous variables and methylation status: metabolic hormones and lipid metabolism biomarkers, one-carbon nutrient biomarkers, dietary intake, body composition, immune function biomarkers and sex hormones.
Stratification
In secondary analyses, we stratified our investigation of the potential biomarker predictors of DNA methylation by serum folate levels (≤ 50 ng/ml vs. > 50 ng/ml), and multivitamin use (no vs. yes) (n = 106, available for this analysis). Stratification by these factors was based on the a priori hypothesis that one-carbon status may influence biomarker-methylation relationships. Serum folate values ranged from 15.2–49.9 ng/ml for the low folate group and 50.5– 258.7 ng/ml for the high folate group (Fig. 1).
Acknowledgments
Supported by grants from the Hartwell Innovation Fund. LP: NCI T32 CA009001 and US. National Institutes of Health, National Cancer Institute R01 CA69334
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Ethical statement
The study was approved by the Fred Hutchinson Cancer Research Center (FHCRC) Institutional Review Board. The study complied with all of the legal requirements.
Footnotes
Previously published online: www.landesbioscience.com/journals/epigenetics/article/21464
References
- 1.Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–70. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 2.Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res. 1998;72:141–96. doi: 10.1016/S0065-230X(08)60702-2. [DOI] [PubMed] [Google Scholar]
- 3.Laird PW, Jaenisch R. DNA methylation and cancer. Hum Mol Genet. 1994;3(Spec No):1487–95. doi: 10.1093/hmg/3.suppl_1.1487. [DOI] [PubMed] [Google Scholar]
- 4.Toyota M, Issa JP. Epigenetic changes in solid and hematopoietic tumors. Semin Oncol. 2005;32:521–30. doi: 10.1053/j.seminoncol.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 5.Choi JY, James SR, Link PA, McCann SE, Hong CC, Davis W, et al. Association between global DNA hypomethylation in leukocytes and risk of breast cancer. Carcinogenesis. 2009;30:1889–97. doi: 10.1093/carcin/bgp143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 7.Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 8.Kim M, Long TI, Arakawa K, Wang R, Yu MC, Laird PW. DNA methylation as a biomarker for cardiovascular disease risk. PLoS One. 2010;5:e9692. doi: 10.1371/journal.pone.0009692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu CC, Ou TT, Wu CC, Li RN, Lin YC, Lin CH, et al. Global DNA methylation, DNMT1, and MBD2 in patients with systemic lupus erythematosus. Lupus. 2011;20:131–6. doi: 10.1177/0961203310381517. [DOI] [PubMed] [Google Scholar]
- 10.Issa JP. CpG island methylator phenotype in cancer. Nat Rev Cancer. 2004;4:988–93. doi: 10.1038/nrc1507. [DOI] [PubMed] [Google Scholar]
- 11.Chalitchagorn K, Shuangshoti S, Hourpai N, Kongruttanachok N, Tangkijvanich P, Thong-ngam D, et al. Distinctive pattern of LINE-1 methylation level in normal tissues and the association with carcinogenesis. Oncogene. 2004;23:8841–6. doi: 10.1038/sj.onc.1208137. [DOI] [PubMed] [Google Scholar]
- 12.Ulrich CM, Grady WM. Linking epidemiology to epigenomics--where are we today? Cancer Prev Res (Phila) 2010;3:1505–8. doi: 10.1158/1940-6207.CAPR-10-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Issa JP, Shen L, Toyota M. CIMP, at last. Gastroenterology. 2005;129:1121–4. doi: 10.1053/j.gastro.2005.07.040. [DOI] [PubMed] [Google Scholar]
- 14.Teitell M, Richardson B. DNA methylation in the immune system. Clin Immunol. 2003;109:2–5. doi: 10.1016/S1521-6616(03)00224-9. [DOI] [PubMed] [Google Scholar]
- 15.Kim YI. Folate and DNA methylation: a mechanistic link between folate deficiency and colorectal cancer? Cancer Epidemiol Biomarkers Prev. 2004;13:511–9. [PubMed] [Google Scholar]
- 16.Zhu ZZ, Hou L, Bollati V, Tarantini L, Marinelli B, Cantone L, et al. Predictors of global methylation levels in blood DNA of healthy subjects: a combined analysis. Int J Epidemiol. 2012;41:126–39. doi: 10.1093/ije/dyq154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borghol N, Suderman M, McArdle W, Racine A, Hallett M, Pembrey M, et al. Associations with early-life socio-economic position in adult DNA methylation. Int J Epidemiol. 2011;41:62–74; . doi: 10.1093/ije/dyr147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tost J. DNA methylation: an introduction to the biology and the disease-associated changes of a promising biomarker. Mol Biotechnol. 2010;44:71–81. doi: 10.1007/s12033-009-9216-2. [DOI] [PubMed] [Google Scholar]
- 19.Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A. 2005;102:10604–9. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Terry MB, Ferris JS, Pilsner R, Flom JD, Tehranifar P, Santella RM, et al. Genomic DNA methylation among women in a multiethnic New York City birth cohort. Cancer Epidemiol Biomarkers Prev. 2008;17:2306–10. doi: 10.1158/1055-9965.EPI-08-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rampersaud GC, Kauwell GP, Hutson AD, Cerda JJ, Bailey LB. Genomic DNA methylation decreases in response to moderate folate depletion in elderly women. Am J Clin Nutr. 2000;72:998–1003. doi: 10.1093/ajcn/72.4.998. [DOI] [PubMed] [Google Scholar]
- 22.Keyes MK, Jang H, Mason JB, Liu Z, Crott JW, Smith DE, et al. Older age and dietary folate are determinants of genomic and p16-specific DNA methylation in mouse colon. J Nutr. 2007;137:1713–7. doi: 10.1093/jn/137.7.1713. [DOI] [PubMed] [Google Scholar]
- 23.El-Maarri O, Becker T, Junen J, Manzoor SS, Diaz-Lacava A, Schwaab R, et al. Gender specific differences in levels of DNA methylation at selected loci from human total blood: a tendency toward higher methylation levels in males. Hum Genet. 2007;122:505–14. doi: 10.1007/s00439-007-0430-3. [DOI] [PubMed] [Google Scholar]
- 24.Choi SW, Stickel F, Baik HW, Kim YI, Seitz HK, Mason JB. Chronic alcohol consumption induces genomic but not p53-specific DNA hypomethylation in rat colon. J Nutr. 1999;129:1945–50. doi: 10.1093/jn/129.11.1945. [DOI] [PubMed] [Google Scholar]
- 25.Schernhammer ES, Giovannucci E, Kawasaki T, Rosner B, Fuchs CS, Ogino S. Dietary folate, alcohol and B vitamins in relation to LINE-1 hypomethylation in colon cancer. Gut. 2010;59:794–9. doi: 10.1136/gut.2009.183707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.El-Maarri O, Walier M, Behne F, van Üüm J, Singer H, Diaz-Lacava A, et al. Methylation at global LINE-1 repeats in human blood are affected by gender but not by age or natural hormone cycles. PLoS One. 2011;6:e16252. doi: 10.1371/journal.pone.0016252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Estécio MR, Gharibyan V, Shen L, Ibrahim AE, Doshi K, He R, et al. LINE-1 hypomethylation in cancer is highly variable and inversely correlated with microsatellite instability. PLoS One. 2007;2:e399. doi: 10.1371/journal.pone.0000399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suter CM, Martin DI, Ward RL. Hypomethylation of L1 retrotransposons in colorectal cancer and adjacent normal tissue. Int J Colorectal Dis. 2004;19:95–101. doi: 10.1007/s00384-003-0539-3. [DOI] [PubMed] [Google Scholar]
- 29.Figueiredo JC, Grau MV, Wallace K, Levine AJ, Shen L, Hamdan R, et al. Global DNA hypomethylation (LINE-1) in the normal colon and lifestyle characteristics and dietary and genetic factors. Cancer Epidemiol Biomarkers Prev. 2009;18:1041–9. doi: 10.1158/1055-9965.EPI-08-0926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang FF, Cardarelli R, Carroll J, Fulda KG, Kaur M, Gonzalez K, et al. Significant differences in global genomic DNA methylation by gender and race/ethnicity in peripheral blood. Epigenetics. 2011;6:623–9. doi: 10.4161/epi.6.5.15335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chiang EP, Wang YC, Chen WW, Tang FY. Effects of insulin and glucose on cellular metabolic fluxes in homocysteine transsulfuration, remethylation, S-adenosylmethionine synthesis, and global deoxyribonucleic acid methylation. J Clin Endocrinol Metab. 2009;94:1017–25. doi: 10.1210/jc.2008-2038. [DOI] [PubMed] [Google Scholar]
- 32.Williams KT, Schalinske KL. New insights into the regulation of methyl group and homocysteine metabolism. J Nutr. 2007;137:311–4. doi: 10.1093/jn/137.2.311. [DOI] [PubMed] [Google Scholar]
- 33.Kim YI. Folate, colorectal carcinogenesis, and DNA methylation: lessons from animal studies. Environ Mol Mutagen. 2004;44:10–25. doi: 10.1002/em.20025. [DOI] [PubMed] [Google Scholar]
- 34.Dolinoy DC, Huang D, Jirtle RL. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc Natl Acad Sci U S A. 2007;104:13056–61. doi: 10.1073/pnas.0703739104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paz MF, Avila S, Fraga MF, Pollan M, Capella G, Peinado MA, et al. Germ-line variants in methyl-group metabolism genes and susceptibility to DNA methylation in normal tissues and human primary tumors. Cancer Res. 2002;62:4519–24. [PubMed] [Google Scholar]
- 36.Curtin K, Slattery ML, Ulrich CM, Bigler J, Levin TR, Wolff RK, et al. Genetic polymorphisms in one-carbon metabolism: associations with CpG island methylator phenotype (CIMP) in colon cancer and the modifying effects of diet. Carcinogenesis. 2007;28:1672–9. doi: 10.1093/carcin/bgm089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pufulete M, Al-Ghnaniem R, Khushal A, Appleby P, Harris N, Gout S, et al. Effect of folic acid supplementation on genomic DNA methylation in patients with colorectal adenoma. Gut. 2005;54:648–53. doi: 10.1136/gut.2004.054718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shelnutt KP, Kauwell GP, Gregory JF, 3rd, Maneval DR, Quinlivan EP, Theriaque DW, et al. Methylenetetrahydrofolate reductase 677C-->T polymorphism affects DNA methylation in response to controlled folate intake in young women. J Nutr Biochem. 2004;15:554–60. doi: 10.1016/j.jnutbio.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 39.Jacob RA, Gretz DM, Taylor PC, James SJ, Pogribny IP, Miller BJ, et al. Moderate folate depletion increases plasma homocysteine and decreases lymphocyte DNA methylation in postmenopausal women. J Nutr. 1998;128:1204–12. doi: 10.1093/jn/128.7.1204. [DOI] [PubMed] [Google Scholar]
- 40.Friso S, Choi SW, Girelli D, Mason JB, Dolnikowski GG, Bagley PJ, et al. A common mutation in the 5,10-methylenetetrahydrofolate reductase gene affects genomic DNA methylation through an interaction with folate status. Proc Natl Acad Sci U S A. 2002;99:5606–11. doi: 10.1073/pnas.062066299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pearce MS, McConnell JC, Potter C, Barrett LM, Parker L, Mathers JC, et al. Global LINE-1 DNA methylation is associated with blood glycaemic and lipid profiles. Int J Epidemiol. 2012;41:210–7. doi: 10.1093/ije/dys020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao J, Goldberg J, Bremner JD, Vaccarino V. Global DNA Methylation Is Associated With Insulin Resistance: A Monozygotic Twin Study. Diabetes. 2011;61:542–6. doi: 10.2337/db11-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang MH, Fei J, Lan MS, Lu ZP, Liu M, Fan WW, et al. Hypermethylation of hepatic Gck promoter in ageing rats contributes to diabetogenic potential. Diabetologia. 2008;51:1525–33. doi: 10.1007/s00125-008-1034-8. [DOI] [PubMed] [Google Scholar]
- 44.Issa JP. Age-related epigenetic changes and the immune system. Clin Immunol. 2003;109:103–8. doi: 10.1016/S1521-6616(03)00203-1. [DOI] [PubMed] [Google Scholar]
- 45.Cho NY, Kim BH, Choi M, Yoo EJ, Moon KC, Cho YM, et al. Hypermethylation of CpG island loci and hypomethylation of LINE-1 and Alu repeats in prostate adenocarcinoma and their relationship to clinicopathological features. J Pathol. 2007;211:269–77. doi: 10.1002/path.2106. [DOI] [PubMed] [Google Scholar]
- 46.Moore LE, Pfeiffer RM, Poscablo C, Real FX, Kogevinas M, Silverman D, et al. Genomic DNA hypomethylation as a biomarker for bladder cancer susceptibility in the Spanish Bladder Cancer Study: a case-control study. Lancet Oncol. 2008;9:359–66. doi: 10.1016/S1470-2045(08)70038-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ogino S, Kawasaki T, Nosho K, Ohnishi M, Suemoto Y, Kirkner GJ, et al. LINE-1 hypomethylation is inversely associated with microsatellite instability and CpG island methylator phenotype in colorectal cancer. Int J Cancer. 2008;122:2767–73. doi: 10.1002/ijc.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bjornsson HT, Sigurdsson MI, Fallin MD, Irizarry RA, Aspelund T, Cui H, et al. Intra-individual change over time in DNA methylation with familial clustering. JAMA. 2008;299:2877–83. doi: 10.1001/jama.299.24.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fuke C, Shimabukuro M, Petronis A, Sugimoto J, Oda T, Miura K, et al. Age related changes in 5-methylcytosine content in human peripheral leukocytes and placentas: an HPLC-based study. Ann Hum Genet. 2004;68:196–204. doi: 10.1046/j.1529-8817.2004.00081.x. [DOI] [PubMed] [Google Scholar]
- 50.Bollati V, Schwartz J, Wright R, Litonjua A, Tarantini L, Suh H, et al. Decline in genomic DNA methylation through aging in a cohort of elderly subjects. Mech Ageing Dev. 2009;130:234–9. doi: 10.1016/j.mad.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Terry MB, Delgado-Cruzata L, Vin-Raviv N, Wu HC, Santella RM. DNA methylation in white blood cells: association with risk factors in epidemiologic studies. Epigenetics. 2011;6:828–37. doi: 10.4161/epi.6.7.16500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hamid A, Wani NA, Kaur J. New perspectives on folate transport in relation to alcoholism-induced folate malabsorption--association with epigenome stability and cancer development. FEBS J. 2009;276:2175–91. doi: 10.1111/j.1742-4658.2009.06959.x. [DOI] [PubMed] [Google Scholar]
- 53.Crider KS, Quinlivan EP, Berry RJ, Hao L, Li Z, Maneval D, et al. Genomic DNA methylation changes in response to folic acid supplementation in a population-based intervention study among women of reproductive age. PLoS One. 2011;6:e28144. doi: 10.1371/journal.pone.0028144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ulrich CM. Nutrigenetics in cancer research--folate metabolism and colorectal cancer. J Nutr. 2005;135:2698–702. doi: 10.1093/jn/135.11.2698. [DOI] [PubMed] [Google Scholar]
- 55.Slattery ML, Potter JD, Samowitz W, Schaffer D, Leppert M. Methylenetetrahydrofolate reductase, diet, and risk of colon cancer. Cancer Epidemiol Biomarkers Prev. 1999;8:513–8. [PubMed] [Google Scholar]
- 56.Richardson B, Scheinbart L, Strahler J, Gross L, Hanash S, Johnson M. Evidence for impaired T cell DNA methylation in systemic lupus erythematosus and rheumatoid arthritis. Arthritis Rheum. 1990;33:1665–73. doi: 10.1002/art.1780331109. [DOI] [PubMed] [Google Scholar]
- 57.Golbus J, Palella TD, Richardson BC. Quantitative changes in T cell DNA methylation occur during differentiation and ageing. Eur J Immunol. 1990;20:1869–72. doi: 10.1002/eji.1830200836. [DOI] [PubMed] [Google Scholar]
- 58.Weisenberger DJ, Campan M, Long TI, Kim M, Woods C, Fiala E, et al. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res. 2005;33:6823–36. doi: 10.1093/nar/gki987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McTiernan A, Ulrich CM, Yancey D, Slate S, Nakamura H, Oestreicher N, et al. The Physical Activity for Total Health (PATH) Study: rationale and design. Med Sci Sports Exerc. 1999;31:1307–12. doi: 10.1097/00005768-199909000-00012. [DOI] [PubMed] [Google Scholar]
- 60.Patterson RE, Kristal AR, Tinker LF, Carter RA, Bolton MP, Agurs-Collins T. Measurement characteristics of the Women’s Health Initiative food frequency questionnaire. Ann Epidemiol. 1999;9:178–87. doi: 10.1016/S1047-2797(98)00055-6. [DOI] [PubMed] [Google Scholar]
- 61.Toyota M, Ho C, Ahuja N, Jair KW, Li Q, Ohe-Toyota M, et al. Identification of differentially methylated sequences in colorectal cancer by methylated CpG island amplification. Cancer Res. 1999;59:2307–12. [PubMed] [Google Scholar]
- 62.Frank LL, Sorensen BE, Yasui Y, Tworoger SS, Schwartz RS, Ulrich CM, et al. Effects of exercise on metabolic risk variables in overweight postmenopausal women: a randomized clinical trial. Obes Res. 2005;13:615–25. doi: 10.1038/oby.2005.66. [DOI] [PubMed] [Google Scholar]
- 63.Foster-Schubert KE, McTiernan A, Frayo RS, Schwartz RS, Rajan KB, Yasui Y, et al. Human plasma ghrelin levels increase during a one-year exercise program. J Clin Endocrinol Metab. 2005;90:820–5. doi: 10.1210/jc.2004-2081. [DOI] [PubMed] [Google Scholar]
- 64.McTiernan A, Tworoger SS, Rajan KB, Yasui Y, Sorenson B, Ulrich CM, et al. Effect of exercise on serum androgens in postmenopausal women: a 12-month randomized clinical trial. Cancer Epidemiol Biomarkers Prev. 2004;13:1099–105. [PubMed] [Google Scholar]
- 65.Mohanka M, Irwin M, Heckbert SR, Yasui Y, Sorensen B, Chubak J, et al. Serum lipoproteins in overweight/obese postmenopausal women: a one-year exercise trial. Med Sci Sports Exerc. 2006;38:231–9. doi: 10.1249/01.mss.0000184584.95000.e4. [DOI] [PubMed] [Google Scholar]
- 66.Friedewald WT, Levy RI, Fredrickson DS. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin Chem. 1972;18:499–502. [PubMed] [Google Scholar]
- 67.Bagley PJ, Selhub J. Analysis of folate form distribution by affinity followed by reversed- phase chromatography with electrical detection. Clin Chem. 2000;46:404–11. [PubMed] [Google Scholar]
- 68.Shade ED, McTiernan A, Wener MH, Wood B, Yasui Y, LaCroix K, et al. Frequent intentional weight loss is associated with lower natural killer cell cytotoxicity constituting possible long-term effects on immune function. J Am Diet Assoc. 2004;104:903–12. doi: 10.1016/j.jada.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 69.Meyers JA, Liu AY, McTiernan A, Wener MH, Wood B, Weigle DS, et al. Serum leptin concentrations and markers of immune function in overweight or obese postmenopausal women. J Endocrinol. 2008;199:51–60. doi: 10.1677/JOE-07-0569. [DOI] [PubMed] [Google Scholar]
- 70.McTiernan A, Tworoger SS, Ulrich CM, Yasui Y, Irwin ML, Rajan KB, et al. Effect of exercise on serum estrogens in postmenopausal women: a 12-month randomized clinical trial. Cancer Res. 2004;64:2923–8. doi: 10.1158/0008-5472.CAN-03-3393. [DOI] [PubMed] [Google Scholar]
- 71.Probst-Hensch NM, Ingles SA, Diep AT, Haile RW, Stanczyk FZ, Kolonel LN, et al. Aromatase and breast cancer susceptibility. Endocr Relat Cancer. 1999;6:165–73. doi: 10.1677/erc.0.0060165. [DOI] [PubMed] [Google Scholar]
- 72.Goebelsmann U, Bernstein GS, Gale GA, Kletzky OA, Nakamura RM, Coulson AH, et al. Serum gonadotropin, testosterone, estradiol and estrone levels prior to and following bilateral vasectomy [Google Scholar]
- 73.Vermeulen A, Verdonck L, Kaufman JM. A critical evaluation of simple methods for the estimation of free testosterone in serum. J Clin Endocrinol Metab. 1999;84:3666–72. doi: 10.1210/jc.84.10.3666. [DOI] [PubMed] [Google Scholar]
- 74.Södergård R, Bäckström T, Shanbhag V, Carstensen H. Calculation of free and bound fractions of testosterone and estradiol-17 beta to human plasma proteins at body temperature. J Steroid Biochem. 1982;16:801–10. doi: 10.1016/0022-4731(82)90038-3. [DOI] [PubMed] [Google Scholar]
- 75.Rinaldi S, Geay A, Déchaud H, Biessy C, Zeleniuch-Jacquotte A, Akhmedkhanov A, et al. Validity of free testosterone and free estradiol determinations in serum samples from postmenopausal women by theoretical calculations. Cancer Epidemiol Biomarkers Prev. 2002;11:1065–71. [PubMed] [Google Scholar]