Abstract
DNA methylation is one of the principal epigenetic signals that participate in cell specific gene expression in vertebrates. DNA methylation plays a quintessential role in the control of gene expression, cellular differentiation and development. It also plays a central role in the preservation of chromatin structure and chromosomal integrity, parental imprinting, X-chromosome inactivation, aging and carcinogenesis. The foremost contributor in the mammalian methylation scheme is DNMT1, a maintenance methyltransferase that faithfully copies the pre-existing methyl marks onto hemimethylated daughter strands during DNA replication to maintain the established methylation patterns across successive cell divisions. The ever-changing cellular physiology and the significant part that DNA methylation plays in genome regulation necessitate rigid management of this enzyme. In mammalian cells, a host of intrinsic and extrinsic mechanisms regulate the expression, activity and stability of DNMT1. Transcriptional regulation, post-transcriptional auto-inhibitory controls and post-translational modifications of the enzyme are responsible for the efficient inheritance of DNA methylation patterns. Also, a large number of intra- and intercellular signaling cascades and numerous interactions with other modulator molecules that affect the catalytic activity of the enzyme at multiple levels function as major checkpoints of the DNMT1 control system. An in-depth understanding of the DNMT1 enzyme, its targeting and function is crucial for comprehending how DNA methylation is coordinated with other critical developmental and physiological processes. This review aims to provide a comprehensive account of the various regulatory mechanisms and interactions of DNMT1 so as to elucidate its function at the molecular level and understand the dynamics of DNA methylation at the cellular level.
Keywords: DNA methylation, DNA methyltransferases, DNMT1, PTMs, epigenetics, intrinsic and extrinsic regulatory mechanisms, non-coding RNA
Introduction
The genetic message is differentially decoded in both time and space as a result of epigenetic constraints on the genome resulting either in gene activation or silencing.1 The machinery behind this cell-specific differential gene expression mainly involves heritable epigenetic modifications and regulations that function without changing the DNA sequence.2,3 The major epigenetic signals include DNA methylation and demethylation, covalent post-translational reversible modifications of the histone proteins, such as acetylation, methylation, phosphorylation and ubiquitination, incorporation of histone variants and gene regulation by non-coding RNAs.4 All of these varied processes nonetheless wield a stabilizing effect to guarantee efficient temporal and spatial gene expression during development. These modifications synchronize the working of various nuclear processes to maintain higher order chromatin architecture. They influence the transcriptional state and regulate the genome for proper cell-cycle progression, stem cell renewal, differentiation, and development. A harmonious and homeostatic balance exists between the various epigenetic manipulations so as to preserve the integrity of the genetic message across several generations yet culminating in functional specialization in a cell and tissue-specific manner.
DNA methylation is a prominent epigenetic programming that facilitates effective cell-specific functional gene expression.4 It is considered to be one of the first steps in epigenetic regulation, acting as a fundamental mechanism in functional organization of the human genome. It is also an attractive and promising target in the development of new drugs for cancer chemotherapy as epigenetic alterations are, in principle, more readily reversible than genetic anomalies.4,5 The complexity in the patterns of methylation, the discrepancy in distribution and abundance of methyl marks in different cell types, makes it necessary to have a comprehensive and precise understanding of the machinery that manages this epigenetic modulator.6 It is thus necessary to study the main perpetuators of DNA methylation—a family of enzymes known as the DNMTs [DNA cytosine-(C5)-methyltransferases] and the intricate web of regulatory mechanisms that target these enzymes to their genomic locations. Of the three prominent DNMTs (DNMT1, DNMT3A and DNMT3B) involved in DNA methylation, DNMT1, the maintenance methyltransferase, occupies an imperial status. It faithfully maintains and propagates the existing methylation marks across successive cell generations, hence becomes essential for the genomic integrity. Various regulatory mechanisms are set into place to monitor the interactions of DNMT1 with other epigenetic manipulators and chromatin remodelers. The enzyme is directed to specific genomic loci in a cell-dependent manner and its enzymatic activity is rigidly controlled to ultimately accomplish its duty as a transcriptional repressor.7 These regulatory partners work in a concerted manner in a multi-tiered organization to supervise the working of DNMT1 so as to maintain a firm control over its functioning. In this review, we focus on the numerous intrinsic and extrinsic factors that coordinate to form an extremely complex and interconnected network for regulating the stability and activity of DNMT1 and also the DNA methylation machinery. Furthermore, we attempt to provide a detailed and up-to-date account of the various check points such as auto-inhibitory controls, allosteric regulators, multiple interactions of DNMT1 with other molecules that participate in DNA methylation, the dynamic interplay between the various inter-reliant post-translational modifications of DNMT1, post-transcriptional and post-translational targeting of DNMT1 by RNAs so as to have a better understanding of this epigenetic reprogrammer. The detailed investigation of these expression control systems is vital to understand the role of DNA methylation and that of DNMT1 in regulating gene expression during development and in disease. A comprehensive knowledge regarding the functional intricacy and enzymatic activity of DNMT1 as well as its potential restraining points will help in designing novel mechanistic based drugs targeting DNMT1 for effective cancer therapy in the future.
DNA Methylation—The Epigenetic Writer
Methylated cytosine in the genome was first discovered in calf thymus DNA by Hotchkiss, in 1948.8 In prokaryotes, DNA methylation takes place on both cytosine and adenine bases and constitutes a branch of the host restriction system to distinguish self and non-self DNA. However in multicellular eukaryotes this modification is limited to cytosine bases only and acts mainly as a repressive tag resulting in silent chromatin state and transcriptional repression.9,10 Methylation of DNA is a post synthetic process catalyzed by a family of dedicated enzymes known as DNMTs. DNMT1, DNMT3A and DNMT3B methylate the cytosine residue in the presence of cofactor SAM (S-Adenosyl methionine), which donates the –CH3 group and is converted to SAH (S-Adenosyl homocysteine).2,11 The genomic methylation blueprint is created during two developmental periods—in germ cells and pre-implantation embryos—generating cells with broad developmental potential.12,13 This system of establishing de novo methylation pattern where new methyl marks are added to previously unmethylated cytosines is mainly chaperoned by the de novo methyltransferases DNMT3A and DNMT3B. Maintenance methylation by DNMT1 then ensures that hemimethylated daughter strands in somatic differentiated cells get methylated to faithfully propagate the proper DNA methylation patterns across successive cell generations.13 DNA methylation obstructs transcriptional activity via two general schemes. First, methylated cytosine bases can hinder the interaction of transcriptional factors and RNA polymerase II with their cognate DNA recognition sequences. Second, methyl-CpG-binding proteins, such as MBD (Methyl-CpG-Binding Domain) proteins, associate with methylated DNA and result in heterochromatin formation by recruiting HDACs (Histone Deacetylases).10,14 DNA methylation is intricately involved in gene regulation and silencing in eukaryotic cells, making significant contributions to the cell phenotype. It plays an important role in regulation of parental imprinting and stabilization of X-chromosome inactivation as well as maintenance of the genome integrity through protection against endogenous retroviruses and selfish genetic elements like transposons. It is also implicated in the development of the immune system, in brain function and behavior as well as in cellular reprogramming and induction of stem cell differentiation. Alternatively, abnormal DNA methylation patterns are associated with several human diseases, including psychiatric diseases and diseases of the immune system.9 Most essentially, a paradoxical alteration in the established DNA methylation patterns involving both gene-specific hypermethylation and genome-wide hypomethylation is a characteristic hallmark of cancer cells, implicating DNA methylation as a major culprit in tumorigenesis.15
Structure and Mechanism of DNMT1
DNMT1 is the principal custodian of genomic integrity in higher eukaryotes as it participates in preserving and faithfully propagating the existing methylation patterns in normal cells. DNMT1 was the first mammalian DNA methyltransferase enzyme to be cloned and biochemically characterized.4 It demonstrates optimal methyltransferase activity on hemimethylated DNA rather than unmethylated DNA and localizes at the DNA replication foci during S phase, properties that make it suitable for maintenance methylation. DNMT1 is a multi-domain large enzyme, comprising ~1,620 amino acids in humans (see Fig. 1). Its N- and C-terminals are joined by a series of KG (Lysine-Glycine) repeats. The N-terminal part of DNMT1 enzyme is composed of several functional domains such as a charge-rich domain, a DMAP1 (DNA methyltransferase associated protein 1) interaction domain, a PBD domain [proliferating cell nuclear antigen (PCNA) binding domain], at least three independent functional NLS (nuclear localization signal) sequences, a RFTS [replication foci targeting sequence; also called TS (targeting sequence)] domain, a zinc domain, also called as CXXC domain, and a PBHD domain (polybromo homology domain), containing two adjacent BAH domains [BAH1 and BAH2 (bromo-adjacent homology 1 and 2)].16,17 The DMAP1 domain at the very N-terminus (residues 12–105) is involved in the interaction of DNMT1 with the transcriptional repressor DMAP1 and helps in the stability and binding of DNMT1 to DNA at CpG sites. The PBD domain mediates the interaction of DNMT1 with PCNA and targets DNMT1 to the replication foci. The RFTS domain targets the DNMT1 to replication foci and also mediates dimerization of DNMT1. The CXXC domain contains eight conserved cysteine residues clustered in two CXXCXXC repeats and is necessary for binding to unmethylated CpGs and influencing the catalytic activity of the enzyme. The PBHD domain has been proposed to act as a protein–protein interaction module specialized in gene silencing.4 Thus, the N-terminal regulatory domain contains part of a sequence that binds the methylation target site (DMAP1, PBD and RFTS domains), an allosteric site that binds nucleic acids (CXXC domain) and sequence motifs that support interaction with other proteins (PBHD domains), thus making DNMT1 ideally suited for its role as the maintenance methyltransferase.
Figure 1. The schematic diagram of the different structural domains of DNMT1 and their functions. The N-terminal domain contains (1) a charge-rich DMAP domain which helps in interaction with the transcriptional repressor DMAP1 as well as controls the stability and binding of DNMT1 to DNA at CpG sites, (2) a PBD domain (Proliferating Cell Nuclear Antigen (PCNA) Binding Domain), which mediates the interaction of DNMT1 with PCNA and targets DNMT1 to the replication foci, (3) at least three independent functional NLS (Nuclear Localization Signal) sequences, (4) a RFTS [Replication Foci Targeting Sequence,also called TS (Targeting Sequence)] domain that targets the DNMT1 to replication foci and also mediates dimerization of DNMT1, (5) a zinc domain, also called as CXXC domain which is necessary for the catalytic activity of the enzyme and (6) a PBHD domain (Polybromo Homology Domain) containing two adjacent BAH sequence [BAH1 and BAH2 (Bromo-adjacent homology 1 and 2)]. The PBHD domain has been proposed to act as a protein–protein interaction module specialized in gene silencing. The C-terminal catalytic domain of DNMT1 contains ten characteristic sequence motifs (i.e., conserved motifs I–X), and the spacing sequences between VIII and IX motif is referred to as TRD. The coordination between N-terminal and C-terminal domains is essential for efficient catalytic activity as well as for its interactions with other protein regulators.
The C-terminal domain of DNMT1 contains ten characteristic sequence motifs (i.e., conserved motifs I–X) with high similarity to prokaryotic DNA (cytosine-5) methyltransferases. Several lines of evidence further indicate that about six of the conserved motifs, that is motifs I, IV, VI, VIII, IX and X, might be highly conserved in mammalian DNA methyltransferases. The C-terminal catalytic region of DNMT1 could be oriented and folded into two domains—the large and small domains, separated by a huge cleft. The large domain encompasses the most conserved motifs, including motifs I–VIII and the most C-terminal part of motif X, which participates in SAM cofactor binding, substrate (cytosine) targeting and other essential catalysis events. The small domain comprises an extremely long variable region between conserved motif VIII and IX, known as the TRD (Target Recognition Domain), conserved motif IX and partial N-terminal region of conserved motif X. The catalytic domain provides a station for binding of the target DNA in the active site and various other regulatory molecules in the allosteric sites, thus presenting avenues for regulation of the enzyme at multiple levels.4,18 The catalytic process of DNA methylation involves a nucleophilic attack of the enzyme on the C6 of the target cytosine. The attack is performed by the thiol group of the cysteine residue in a PCQ motif conserved in the active site of cytosine-C5-MTases (motif IV). The formation of the covalent bond activates the C5 atom toward electrophilic attack and leads to the addition of the methyl group from SAM to C5. Abstraction of the proton from 5C followed by β-elimination allows reformation of the 5C = 6C double bond and release of the enzyme and substrate DNA with a methylated cytosine. The glutamic acid of the amino acid motif ENV (motif VI) is important to stabilize the DNA-protein complex. The methyl group of SAM is bound to a sulphonium atom, which thermodynamically destabilizes the molecule and makes the relatively inert methyl thiol of the methionine moiety very reactive toward nucleophilic attack by the activated C atom (carbanion) of cytosine.8,18,19
Regulation of DNMT1: A Mechanistic Overview
Owing to the immense role of DNMT1 in the maintenance of methylation patterns and subsequent control of gene expression, it becomes vital to know the various mechanisms that control the expression, stability and activity of this enzyme.20 A number of intrinsic and extrinsic factors participate in this regulation; a detailed study will provide a clear insight into these factors and their effect on the activity of DNMT1.
Intrinsic factors regulating DNMT1 activity and stability
During the course of evolution the cell has developed a great repertoire of intrinsic regulatory mechanisms that control the activity of DNMT1. These factors include transcriptional regulation via cellular signaling networks, post-transcriptional auto-inhibitory controls that inactivate the enzyme and allosteric modulation of its activity. All of these factors contribute toward efficient functioning of DNMT1 and thus are of considerable importance in regulating DNA methylation.
Transcriptional regulation of DNMT1 activity by cellular signaling networks
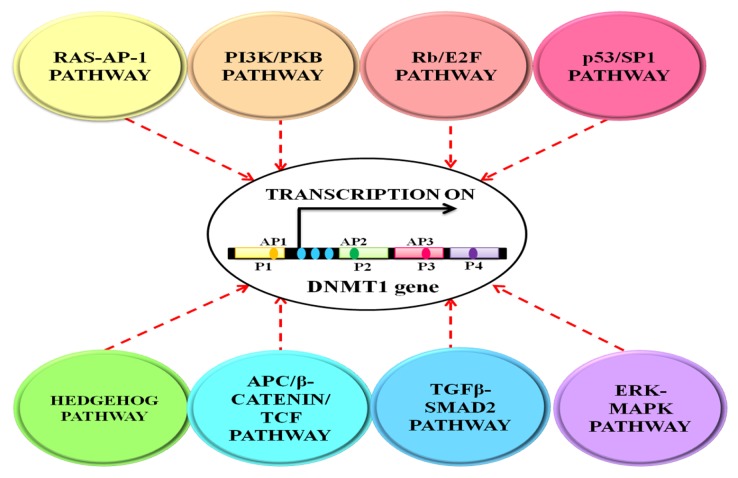
A complex three-dimensional interplay between the epigenomic modulators, the ever-changing environmental cues and the numerous developmentally inclined signaling cascades determine the fate of cellular differentiation.21 Each cell communicates with its environment, receives and sends appropriate signals to the epigenome, which is then translated into stable heritable propagation of gene activity. The expression and activity of DNMT1 is regulated by a number of signal transduction pathways, the more prominent ones among them being RAS- AP-1, PI3/PKB, Rb/E2F and p53/SP122 (see Fig. 2). These signaling systems induce higher expression of the enzyme in a programmed manner in distinct biological settings. Therefore malfunctioning, especially upregulation of these pathways as seen in tumorigenic conditions results in constitutive activation of DNMT1 enzyme leading to aberrant regional hypermethylation of the genome in cancer.
Figure 2. The figure depicts transcriptional regulation of DNMT1 expression by different cellular signaling pathways. The expression and activity of DNMT1 is synchronized by a number of signal transduction pathways that include cancer prone cellular signaling pathways such as RAS and PI3K/PKB as well as developmentally inclined networks such as HH and TGFβ-SMAD pathways. These signaling pathways control the expression of DNMT1 and, consequently, the functioning of the DNA methylation machinery to maintain a homeostatic pattern of gene expression in accordance to the cellular needs. P1, P2, P3 and P4 represents the transcription initiation sites and AP1, AP2, AP3 represents the three AP (activator protein) sites. Blue dots downstream of P1 site represent the three c-Jun-dependent enhancers.
DNMT1 usually shows constitutive yet low level of expression in a normal or resting cell. This basal state of expression of the enzyme is altered by different cellular signaling pathways.23 RAS-AP-1 pathway is one of the important nodal cellular regulatory programs that play a principal role in DNMT1 expression vis-a-vis transcriptional activation and mRNA stabilization in the S-phase of cell cycle.24 The DNMT1 gene contains one major and three minor transcription initiation sites (P1–P4) regulated by independent enhancers and promoter sequences. P1, like other housekeeping genes, is housed within a CG-rich area; P2–P4 are found in CG-poor areas. Three c-Jun-dependent enhancers can be found downstream to P1 and upstream to P2–P4, which explains the receptiveness of DNMT1 gene toward RAS oncogenic signaling pathway.25 Also, DNMT1 promoter region bears a number of AP-1 (activator protein 1) sites (one each in P1, P2 and P3), which prompt its activity via the RAS-Jun signaling pathway.23,25,26 Overexpression of endogenous RAS protein induces a cascade of protein kinases, which in turn activates and phosphorylates AP-1 proteins such as c-Fos/c-Jun. This results in transcriptional stimulation of DNMT1 promoter and an increase in DNA methylation activity.23,24,26,27 A fascinating hypothesis that explains maintenance of steady-state of methylation in normal conditions or during challenges such as global hypomethylation is that DNMT1 gene is regulated by negative feedback methylation.28 The AP-1 regulatory element present in the CG rich P1 promoter sequence is normally heavily methylated, preventing access of AP-1 proteins (c-Fos/c-Jun) to this region and resulting in low basal transcriptional activity as mentioned earlier. As demethylation occurs, interaction of AP-1 with the regulatory element induces DNA methyltransferase activity and re-methylation of the regulatory region resulting in a return to basal level expression. Thus, methylation seems to function as a universal mechanism to regulate gene expression when there is CG sequence (for example, DNMT1 gene itself is regulated by DNA methylation).28
The PI3K/PKB (phosphatidylinositol 3-kinase/protein kinase B) pathway, mainly responsible for cell growth, viability, and metabolism, is also intimately associated with regulation of DNMT1. It is perhaps the only pathway till date that affects DNMT1 activity by modulating the stability of the enzyme. PI3K/PKB pathway elevates the cellular DNMT1 protein level by inhibiting the ubiquitin-proteasome mediated degradation of DNMT1 via Gsk3β (glycogen synthase kinase 3β).29 Rb/E2F pathway is another well characterized cell-cycle monitoring pathway that participates in cellular proliferation, differentiation, survival/apoptosis and senescence.30 In principle, this pathway must therefore play a supervisory role in the transcriptional regulation of DNMT1 gene that is intricately associated with the cell cycle control. During the G1 phase, unphosphorylated Rb (Retinoblastoma) protein binds to E2F, recruits HDAC to this complex and prevents turning on of the cell cycle proteins such as cyclin E, PCNA, pol α, E2F-1 and DNMT1 to maintain basal level expression of DNMT1.22,30,31 As the cell passes into the S phase, increased DNA synthesis initiates a need for maintaining the methyl marks on hemimethylated daughter strands. Consequently, CDK (cyclin dependent kinases) mediated phosphorylation of Rb proteins disrupts the binding of Rb with E2F, releasing the transcription factor and resulting in functional-level expression of DNMT1.31 Progression into G2 phase results in dephosphorylation of the Rb protein and subsequent inactivation of the DNMT1 gene.22,30 p53 (also known as protein 53 or tumor protein 53) mediated regulation of DNMT1 is another route of exercising control over DNMT1 mRNA expression.32 In the absence of genotoxic stress, p53 localizes to the nucleus and binds to the consensus sites in the DNMT1 promoter, thus preventing expression of DNMT1 gene. However, when DNA damage occurs, activation of p53 signaling pathway eliminates the inhibitory effect of p53 on DNMT1 mainly via post-translational modifications, or interactions with other transcriptional factors, and leads to an upregulation in DNMT1 expression.32 As mentioned above, one such transcriptional co-regulator of p53 affecting DNMT1 expression is SP1 (specificity protein 1). There are three SP1 putative binding sites on the DNMT1 promoter, one of them (+7 to +20) being proximal to the p53 putative binding site (+30 to +56).33 The stoichiometric ratio between p53 and SP1 determines the state of expression of the DNMT1 promoter. In low levels, SP1 transcription factor interacts with p53 and assists repression of the DNMT1 gene. However, higher expression of SP1 results in MDM2 (murine double minute 2)-mediated ubiquitination and proteasomal degradation of p53, ensuing in overexpression of DNMT1.22,33
The observation that DNMT1 activity is affected by both proto-oncogenic (RAS-Jun) and tumor suppressor (p53/SP1, Rb/E2F) signaling pathways creates an interesting paradox. Regulation of an enzyme by opposing signaling pathways may provide some hitherto unknown insights into the documented co-existence of global hypomethylation and gene-specific hypermethylation as well as increased DNMT1 expression during tumorigenesis. It also supports the fact that DNMT1 plays an important role in malignant transformation triggered by either inhibition of tumor suppressors or activation of proto-oncogenes.25 In addition to the above described pathways, ERK-MAPK (extracellular signal-regulated kinase/mitogen-activated protein kinase) pathway,34 TGFβ-SMAD2 pathway,35 APC/β-catenin/TCF pathway3 and HH (Hedgehog) pathway36 also play an important part in regulating DNMT1 expression. These signaling pathways modulate the expression of DNMT1 and, consequently, the functionality of the DNA methylation machinery to maintain a homeostatic pattern of gene expression in accordance to the cellular needs.35 The essential participation of cancer prone cellular signaling pathways, such as RAS and PI3K/PKB, as well as developmentally inclined networks, such as HH and TGFβ-SMAD pathways, in the regulation of DNA methylation presents an opportunity for therapeutic interventions for effective cancer treatment.
Auto-inhibitory regulation of DNMT1 activity for faithful maintenance methylation
One of the primary obligations of a maintenance methyltransferase is the conservation of the existing array of methyl tags in the genome to ensure that the hemimethylated regions after replication retain their designated methylation patterns and at the same time, the methylation free genomic locations remain as such.37 For this purpose, an auto-inhibitory regulatory scheme is set into place which thwarts any unforeseen and unprogrammed de novo methylation by DNMT1. This arrangement acts as a failsafe against uncontrolled methylation by DNMT1 to protect the genome from being transcriptionally silenced at several gene loci which are crucial for growth and differentiation.
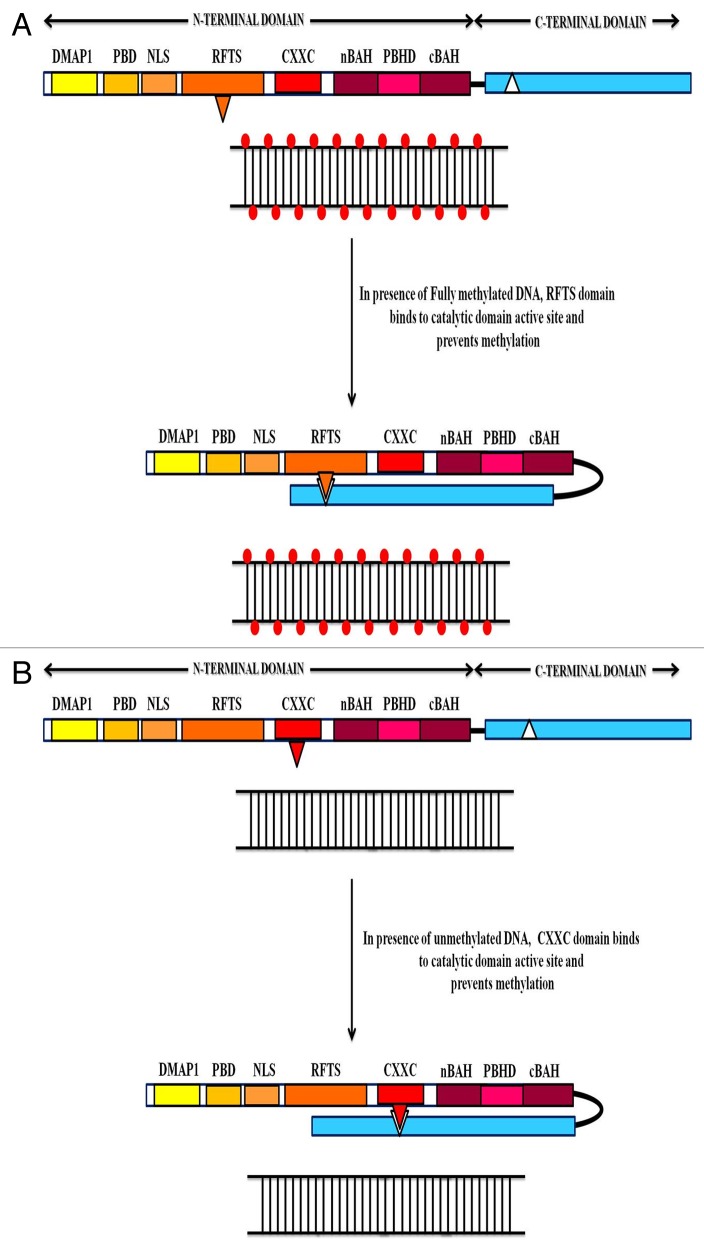
The N-terminal regulatory part of DNMT1 contains many independent compartments and interspersing flexible linkers with functional interactions between adjacent sub-domains, as well with the C-terminal catalytic lobe.38,39 The RFTS domain is positioned deep inside the catalytic domain in the place where the hemimethylated DNA is expected to fit.40 Complementary electrostatic potential and hydrogen bonds between the surfaces of the RFTS domain (net negative charge) and the catalytic core (net positive charge) facilitates this anchoring.37,41 Additionally, amino acids (F631, F634 and F635) present in the linker stretching between RFTS and CXXC domain hydrophobically interact with those situated in vicinity of the PCQ loop in the catalytic core (Y1243 and F1246) and constrict the entrance of the DNA-binding pocket. This strategy stabilizes the RFTS domain in the substrate-DNA binding pocket, effectively masks the catalytic core and prevents the enzyme from needlessly methylating the genome de novo40,41(see Fig. 3). The CXXC domain, a zinc-finger like motif in the N-terminus of DNMT1 functions as yet another auto-inhibitory overseer of the enzyme activity.42 This regulatory domain of DNMT1 binds specifically to unmethylated CpG sites as the CpG dinucleotides emerge from the replication complex.38,42-44 Binding of the CXXC domain acts as a sensor for the methylation status of the substrate, excludes the substrate from the catalytic center, thus averting any erroneous methylation in previously unmethylated regions (see Fig. 3). More recently, studies on crystal structure of DNMT1 and biochemical analyses have uncovered an acidic auto-inhibitory linker between the CXXC and the succeeding BAH1 domains.43,44 The interaction between CXXC domain and the unmethylated substrate activates this linker which then occupies the catalytic pocket in between the substrate DNA and the active site of the enzyme thus interfering in its function.
Figure 3. The figure represents the various auto-inhibitory mechanisms that prevent any unforeseen and un-programmed de novo methylation by DNMT1.This arrangement acts as a failsafe against uncontrolled methylation by DNMT1 to protect the genome from being transcriptionally silenced at several gene loci that are crucial for growth and differentiation. (A) When DNMT1 bind with fully methylated DNA, the RFTS domain in the N-terminal region is inserted deeply into the active site in the catalytic region; thus, inhibiting the enzyme from needlessly methylating the genome de novo. (B) When DNMT1 binds to an unmethylated site, the CXXC domain acts as a sensor for the methylation status of the substrate excludes the substrate from the catalytic center, thus avoiding any erroneous methylation in previously unmethylated regions.
These auto-inhibitory mechanisms for all intents and purposes serve the purpose of maintenance methylation through two distinct yet mutually inter-reliant means. DNMT1 is tethered to the replication fork and as unmethylated DNA emerge, the auto-inhibitory mechanisms are set into motion in order to prevent unauthorized de novo methylation of the DNA. However, as hemimethylated sites are released from the replication complex, the TRD region in the C-terminal domain in association with UHRF1 (ubiquitin-like with PHD and ring finger domains 1) recognizes and marks them as prime targets for methylation, thus perpetuating the methylation patterns faithfully across successive generations.44 These observations point toward a multi-tiered system of post-transcriptional auto-regulation that modulates the activity of DNMT1. Sequence-specific interaction of the substrate DNA with the active center of the enzyme results in numerous structural alterations and methylation-specific conformational changes.38-41 Nonetheless, several important issues need to be addressed such as (1) whether these auto-inhibitory mechanisms are mutually complementary and co-operative? (2) What other domains in the N-terminal region play a role in auto-inhibitory regulation? And (3) which of these mechanisms is circumvented during deregulation of the DNMT1 activity in tumorigenic condition? A clear understanding of these complexities may go a long way in interpreting the dynamics of DNA methylation and regulation of DNMT1. Co-crystal structures of the full-length DNMT1 and hemimethylated DNA may shed some light on the yet unknown roles of the N-terminus in enzyme activity and regulation.
Allosteric regulation: A binary switch controlling the activity of DNMT1
Allosteric regulation of DNMT1 activity acts mainly as a binary switch to guarantee the faithful imitation of methylation patterns on to hemimethylated DNA as well as preservation of methylation-free regions of the DNA.45 DNMT1 actively methylates and enriches the already existing, highly methylated CpG regions but becomes inactivated on poorly methylated or unmethylated sites, thus generating a bimodal methylation pattern in the human genome.4 This monitoring system employs a double sieve mechanism to control the activity of DNMT1. Allosteric inhibition by excess of unmethylated DNA substrate and allosteric activation by fully methylated DNA work independently and processively to moderate the enzymatic reaction.39
DNMT1 has multiple DNA-binding sites, two in the N-terminal domain and one in the C-terminal domain.38,46 The first DNA-binding site resides within the NLS containing domain comprising 261–356 amino acid residues, particularly the amino acids Lys 284, Lys 285, His 286 and Arg 287.46 This site is mainly responsible for mediating allosteric activation of the enzyme by fully methylated substrates. The second DNA-binding site is positioned within the amino acid segment 580–697 in the Zn-finger domain and is the allosteric site that leads to inhibition by unmethylated DNA.39,46 These two sites combine to form the allosteric activation domain, which interacts with different regulatory molecules and controls the catalysis. The C-terminal domain of DNMT1 encloses the third DNA binding region and forms the active site for catalysis.46 During the process of methylation, the occupancy of the allosteric site(s) in the N-terminal part of the enzyme determines the enzyme’s commitment to catalysis at the active site in the catalytic domain.5,47,48 The methylation reaction catalyzed by DNMT1 can take place on varied DNA substrates, mainly unmethylated and pre-methylated dsDNA (which includes hemimethylated dsDNA, proximally methylated dsDNA, fully methylated dsDNA and methylated ssDNA), where each type of substrate can be combined with one of four different allosteric regulators: fully methylated dsDNA, hemimethylated dsDNA, unmethylated dsDNA, methylated ssDNA.5 Depending on the nature of the substrates and the ability of the allosteric modulator, the kinetic behavior of the enzyme varies greatly, resulting either in stimulation and high processivity or full/partial inhibition of methylation (see Table 1).5,49
Table 1. The different allosteric modulators and substrates for methylation that affect the kinetic behavior of the enzyme DNMT1 resulting either in stimulation and high processivity or full/partial inhibition of methylation.
| Allosteric regulator | Substrate for methylation | State of the enzyme | References |
|---|---|---|---|
| Unmethylated dsDNA |
Unmethylated DNA |
Partial Inhibition |
5, 51 |
| Pre-methylated DNA |
Full Inhibition |
5, 51 |
|
| Fully Methylated dsDNA |
Unmethylated DNA |
Full Activation |
5, 51 |
| Pre-methylated DNA |
Full Activation |
5, 51 |
|
| Hemimethylated dsDNA |
Unmethylated DNA |
Full Activation |
5, 51 |
| Pre-methylated DNA |
Full Activation |
5, 51 |
|
| Proximally methylated dsDNA |
Unmethylated DNA |
Full Activation |
5, 51 |
| Pre-methylated DNA |
Full Activation |
5, 51 |
|
| Methylated ssDNA |
Unmethylated DNA |
Full Inhibition |
5, 51 |
| Pre-methylated DNA |
Full Inhibition |
5, 51 |
|
| Poly (ADP-ribose) | Unmethylated DNA |
Full Inhibition |
5, 90 |
| Pre-methylated DNA | Full Inhibition | 5, 90 |
Pre-methylated DNA means either hemimethylated dsDNA or proximally methylated dsDNA, or fully methylated dsDNA, or methylated ssDNA.
Allosteric inhibition of DNMT1 enzyme averts any wanted methylation and ensures that the methylation-free regions remain as such. As the CpG dinucleotides emerge from the replication complex, the allosteric site in the CXXC domain (510–789 residues) of the enzyme reads the methylation status of the substrate DNA and accordingly sets the course of the catalysis. When unmethylated substrate lodges in the N-terminal allosteric site, the acidic linker connecting CXXC and BAH1 domain occupies the active site of the enzyme in the catalytic pocket preventing DNA binding39,43,44 and inactivating the enzyme. However, when pre-methylated DNA occupies the regulatory site, the CXXC domain (which specifically recognizes unmethylated CpG sites) does not bind to the substrate.38,42-44 The subsequent slow conformational changes exposes the catalytically active site for better access of the co-factor SAM and the substrate DNA resulting in a slow relief from the allosteric inhibition and start of the catalysis.39,46,47,50,51 The activation of DNMT1 by methylated cytosines warrants that methylation is stimulated wherever the complementary strand is methylated. Thus, pre-methylated parental strand bound at the N-terminal allosteric activation pocket during DNA replication acts as an allosteric activator to direct methylation on the unmethylated daughter strand in the carboxy terminal domain.46,50 The functional significance of this phenomenon can be appreciated during somatic cell division when DNMT1, an enzyme with low turnover numbers, is allosterically manipulated for rapid DNA methylation by hemimethylated DNA.46 DNMT1 selectivity for pre-methylated substrates is a consequence of allosteric inhibition by unmethylated substrates, rather than allosteric activation by pre-methylated substrates.51
DNMT1 performs methylation in two different modes depending upon the substrate DNA: on hemimethylated DNA, the enzyme works quickly, processively and accurately, whereas on unmethylated DNA, its activity slows down and is dependent upon stimulation by allosteric activation by nearby methylated sites.52 Thus, allosteric regulation ensures that DNA methylation performs like an all or none switch to create a biphasic distribution of methylation levels49 and to increase the efficiency of DNA methylation, switching gene expression on or off.52 A complete understanding of the allosteric regulation of DNMT1 requires clearing up of some questions such as, (1) how many different allosteric sites besides the ones discussed above exist on DNMT1? (2) Is there functional cooperativity between the different allosteric sites? And, (3) which of these allosteric regulatory sites is dysfunctional during tumorigenesis?
Extrinsic factors regulating DNMT1 activity and stability
The cell survives not only by efficient internal physiological homeostasis but also as a result of its interaction with the outer environment. DNMT1 participates in numerous interactions with other regulatory molecules and is in turn affected by regulation through post-translational modifications and post-transcriptional control by RNA. Thus, there seems to exist a mutually symbiotic relationship between DNMT1 and the extrinsic milieu of biomolecules that supervise its activity and maintain steady methylation levels in the cell.
Multiple interactions of DNMT1: An intricate web of extrinsic regulators
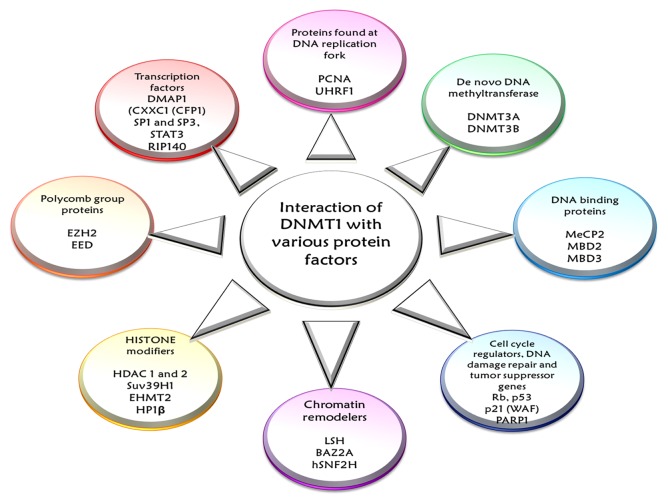
DNMT1, the main architect in the epigenomic landscaping, participates in a number of chromatin modifying processes, such as in cell cycle controlling programs, damage repair schemes and in many RNA directed mechanisms.13,40 Studies in the recent years provide evidence about multiple interactions of DNMT1 with a host of regulatory proteins and a myriad nuclear re-programmers. DNMT1 works in concert with proteins found at DNA replication forks, proteins participating in chromatin re-organization, DNA binding proteins, proteins associated with cell cycle regulation or response to DNA damage and tumor suppressors and, finally, a number of transcription factors and regulators involved in DNA methylation inheritance (see Fig. 4).5,40,53 In essence, this multitude of interactions of DNMT1 can (1) stimulate or inhibit DNMT1 activity, (2) guide DNMT1 to methylation sites, (3) facilitate dissociation from target-DNA sites, (4) increase or decrease the efficiency of the DNA methylation machinery as a whole and (5) maintain the pre-existing, site-specific patterns of DNA methylation.39,54
Figure 4. The figure shows the various interactions of DNMT1. DNMT1 participates in a number of collaborations with a host of regulatory proteins and nuclear re-programmers that simultaneously affect DNMT1 activity as well as its functional exchange. DNMT1 works in concert with proteins found at DNA replication forks—PCNA, proteins participating in chromatin re-organization—DNA methyltransferase DNMT3A and DNMT3B, HDAC1, HDAC2, DNA binding proteins—MeCp2, MBD2, MBD3 and other chromatin binding proteins—UHRF1 and polycomb proteins, proteins associated with cell cycle regulation or response to DNA damage and tumor suppressors—p21 (WAF), Rb protein, p53 protein, PARP1—and, finally, a number of transcription factors and regulators involved in DNA methylation inheritance.
DNMT1 carries out post-replicative maintenance of genomic methylation patterns in mammalian cells by directly binding to PCNA, which enhances the methylation efficiency by about 2-fold.40,53,55 However, this association is not obligatory and helps to accommodate the diverse kinetics of DNA replication and methylation while contributing to faithful propagation of epigenetic information.40,56 DNMT1 binds with de novo methyltransferases DNMT3A and DNMT3B, demonstrating an intricate networking between the DNMTs for efficient methylation of target DNA.40,57 This interaction might answer some of the key questions about the alleged classification of these enzymes, as the maintenance methyltransferase (DNMT1) and as de novo methyltransferases (DNMT3A and DNMT3B).39 DNMT1 also interacts with chromatin modifiers HDAC1 and HDAC2 (Histone Deacetylase 1 and 2) and DNA binding proteins, MeCP2, MBD2 and MBD3 (Methyl-CpG-Binding Domain proteins). DNMT1 interacts with HDAC1 and HDAC2 to bring about transcriptional repression 39, 58. DNMT1 interacts with MeCP2 via its TRD and induces chromatin compaction, whereas binding of MBD2 and MBD3 to methylated DNA results in recruitment of HDAC1 and HDAC2 and subsequent interaction with DNMT1. The association of MBD proteins and HDACs may enrich DNMT1 in highly methylated regions to increase local methylation efficiency and lead to heterochromatin formation.39,40 DNMT1 also allies with major eukaryotic histone methyltransferases Suv39H1 (suppressor of variegation 3–9 homolog 1) and EHMT2 (euchromatic histone-lysine N-methyltransferase 2; also known as G9a), which are essential for H3K9 methylation, and HP1β (meterochromatin binding protein 1), for the establishment and maintenance of heterochromatin structure.16,40,59,60 In addition, DNMT1 interacts with PcG (polycomb group) proteins such as EZH2 (enhancer of zeste homolog 2) that recruits DNMT1 to target genes and thereby mediates promoter methylation.40,61 DNMT1 also works together with chromatin remodeling ATPases like LSH (lymphoid specific helicases) and BAZ2A (bromodomain adjacent to zinc finger domain 2A; also known as TIP5) and chromatin remodelers like hSNF2H (also known as SMARCA5 (SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily A, member 5).39,40,62 These interactions enhance the binding affinity of DNMT1 to mononucleosomes and help to better access the substrate sites in heterochromatic regions.39,40 Besides these indirect alliances for transcriptional silencing, DNMT1 exhibits direct association with transcription regulators and factors such as DMAP1 (DNA methyltransferase 1 associated protein 1), CXXC1 (CFP1) (CXXC finger protein 1), SP1 and SP3, STAT3 (signal transducer and activator of transcription 3), RIP140 (also called NRIP1; nuclear receptor interacting protein 1). These liaisons help in regulation of cell signaling and induction of cell transformation.40,54 Recently, studies have shown that, DNMT1 interacts with zinc finger factor SNAIL1, a potent transcriptional repressor of the E-cadherin gene and induces active gene expression of E-cadherin. This observation points toward the fact that DNMT1 may be directly involved in epithelial-mesenchymal transitions, an activity entirely different from it assigned biological function.63 Additional DNMT1 interactions have been reported with various tumor suppressor genes including WT1 (Wilms tumor 1), Rb and p53.40 The Rb tumor suppressor gene product associates with DNMT1 and the transcription factor E2F1, resulting in transcriptional repression of E2F1- responsive promoters,64 whereas the interaction with p53 stimulates DNMT1 activity leading to hypermethylation (see Fig. 4).40
The miscellany of proteins identified to interact with DNMT1 provides insight into the complexity of the epigenetic network and highlights the central role of DNMT1 in these regulatory pathways.40 A common result of many of these interactions is transcriptional repression, which places DNMT1 at the epicenter of a complex network relating two epigenetic pathways, DNA methylation and histone modification for the establishment and maintenance of transcriptionally inactive chromatin.53 The sheer number of all reported DNMT1 interactions makes it improbable to occur all at one time; thus, it becomes necessary to determine the precise order of these interactions and their functional consequences.53 The interaction of DNMT1 with an impressive array of extrinsic regulators may provide an explanation for the specific patterns of DNA methylation seen in different cells in an individual organism. It may also present functional points for targeting DNMT1 activity in order to design novel mechanism-based inhibitors of the enzyme. Moreover, this collaborative and interactive ability of DNMT1 with different nuclear factors and a variety of regulator proteins may also enable the enzyme to perform certain biological functions in the cell nucleus that are independent of its enzymatic activity.63
Post-translational modifications: A dynamic scheme regulating DNMT1 activity and stability
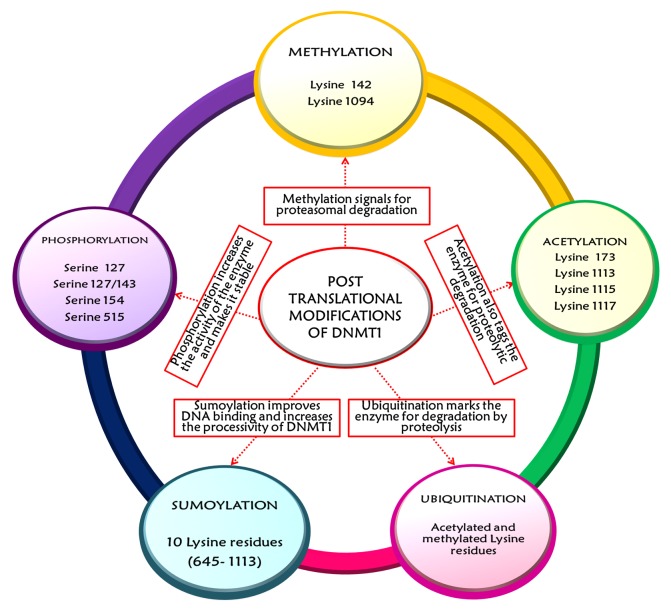
Covalent reversible post-translational modifications, such as methylation, acetylation, phosphorylation, sumoylation and ubiquitination, augment the functional potential of proteins. PTMs (post-translational modifications) of catalytically active DNMT1 influence to a great extent its activity, stability and interaction with other proteins.22 The first studied PTM affecting DNMT1 activity was phosphorylation of DNMT1 at Ser 515 residue present in the RFTS domain.40,65 This modification facilitates the interaction between the N-terminal domain of DNMT1 and its catalytic C-terminal domain to increase the activity of the enzyme.66 DNMT1 is also phosphorylated at residues Ser 127/143 and Ser 127 by AKT1 (v-akt murine thymoma viral oncogene homolog 1) and PKC (protein kinase C), respectively, contributing toward its stability. Phosphorylation in these sites lowers the interaction of DNMT1 with PCNA and UHRF1 at the replication fork in early and mid S-phase. This leads to inactivation of the enzyme to prevent erroneous methylation of DNA in late S- and early G2-phase when no hemimethylated targets are available.13 A recent study by Lavoie and St-Pierre has shown that CDKs such as CDK1, CDK2 and CDK5 can phosphorylate human DNMT1 at Ser154 residue. The Ser154 phosphorylation affects domain-domain interactions between N-and C-termini and consequently controls DNMT1 activity and protein stability. This post-translational manipulation of the enzyme is of paramount significance as deregulation of CDKs can cause aberrant phosphorylation of DNMT1 and lead to DNA hypermethylation, as seen during tumorigenic progression.67 Lysine methylation of DNMT1 is another PTM of considerable importance for regulation of stability and activity of the enzyme. Methylated DNMT1 is less stable and this biochemical tag appears to be a signal for proteasomal degradation of the protein through ubiquitination.22,68,69 The prominent lysine residues of the enzyme that are methylated include Lys 142 and Lys 1094. Lys 142 of DNMT1 is an in vivo methylation target of SET7 (SET domain containing lysine methyltransferase 7, also known as SET7/9 and KMT7). In turn, LSD1 (lysine specific demethylase, also called KDM1A) removes these methyl marks and antagonizes SET7 activity.13 Similarly, methylation of Lys 1094 residue of DNMT1 by SET7/9 helps in destabilization of the enzyme. The corresponding demethylation by KDM1A increases the stability of the enzyme and maintains the DNA methylation patterns.40,70,71 A delicate balance between these two epigenetic modulators, SET7 and LSD1, affects the methylation status of the enzyme and subsequently regulates gene expression (see Fig. 5).68
Figure 5. The figure depicts the various post-translational modifications that affect the stability and activity of DNMT1. The major PTMs include methylation, acetylation, ubiquitination, sumoylation and phosphorylation. All of these post-translational regulatory operations are interlinked to maintain a rigid control over the stability, abundance and activity of DNMT1 in a cell-cycle dependent manner.
The stability and functional availability of DNMT1 protein is also regulated by sequentially coordinated acetylation and acetylation-driven ubiquitination with respect to the different stages of cell cycle40 (Fig. 5). The acetyltransferase Tip60 [also called KAT5, K-(lysine) acetyltransferase 5] acetylates DNMT1 at Lysine residues 173, 1113, 1115 and 111713,72 and sets it for proteasomal degradation. UHRF1, an E3 ubiquitin ligase, plays a dual role in the maintenance of DNMT1 function. First, it binds to hemimethylated DNA and guides DNMT1 to these sites72-75 and, second, it ubiquitinates DNMT1 and triggers its proteolytic destruction. However, during G1/S-phase transition when DNMT1 is required for methylation of newly synthesized DNA strands, the ubiquitin ligase activity of UHRF1 is curtailed by HDAC1 and HAUSP (herpes virus-associated ubiquitin-specific protease, also known as USP7; ubiquitin specific peptidase 7) by deacetylation and deubiquitination, respectively.40,72,74,76 USP7, a deubiquitinase, increases the stability of the protein by protecting the enzyme from proteolysis and also stimulates both the maintenance and the de novo DNA methylation activity of DNMT1 by a factor of two.74,76 Thus, the destroyers (Tip60 and UHRF1) and the protectors (HAUSP and HDAC1) form a multi-protein complex along with DNMT1 at the replication fork in association with PCNA and carry out their function in a series of post-translational events.77 Finally, the most recently recognized PTM affecting DNMT1 activity and stability is sumoylation, which is very much similar to ubiquitination in terms of biochemistry but differs in terms of downstream functions.22 This modification is necessary for protein–protein interactions, enzyme activity, protein stability, DNA binding and sub-cellular localization of proteins.13,22,78 Sumoylation of endogenous DNMT1 by Ubc9, an E2 conjugating enzyme also called UBE2I (ubiquitin-conjugating enzyme 2I), on several lysine residues (645–1113) in the BAH domains enhances the catalytic action of DNMT1 on genomic DNA78 (see Fig. 5). The increase in DNMT1 activity can be assumed to be a result of conformation changes induced by sumoylation that improves the DNMT1-DNA binding and stimulates methylation by making the enzyme more processive.22,78
Interestingly, all of these post-translational regulatory operations are interlinked to maintain a stiff control over the stability, abundance and activity of DNMT1 in a cell-cycle dependent manner. Examples of such dynamic cross talking include the “methylation and phosphorylation” switch65 and the “acetylation and ubiquitination” module.72,77 These mutually exclusive epigenetic switches orchestrate the stability of DNMT1 and have profound impact upon the global genome methylation patterns.13 Phosphorylation of Ser 143 residue by AKT1 kinase during early and mid-S phase stabilizes DNMT1, and prevents Lys 142 methylation by SET7 and subsequent proteolytic degradation during late S- and G2 phase.40,65 Similarly, Tip60 mediated sequential acetylation of lysine residues leads to proteasomal degradation of the enzyme by UHRF1 arbitrated ubiquitination.72 Thus, a delicate equilibrium between the different post-translational manipulations helps maintain a systematic mode of gene expression during normal development. However, several questions need to be answered, such as, (1) which PTMs, other than acetylation, methylation, phosphorylation and ubiquitination, influence the activity of DNMT1? (2) Lysine and, in a few cases, serine seem to be the preferred substrate for a majority of PTMs, such as methylation, acetylation, ubiquitination and sumoylation. Which other amino acid residue is subjected to PTMs? (3) Besides affecting the stability and degradation of the enzyme, in what other ways do the PTMs regulate DNMT1 activity? (4) Which of these PTMs is mostly interrupted during malignant initiation and progression? Thus, a comprehensive directory of all PTMs and their mode of regulation of DNMT1 activity in various cellular contexts and phenotypes will go a long way in understanding the DNA methylation machinery more clearly.
RNA-mediated post-transcriptional and post-translational targeting of DNMT1
RNA directed regulation of DNMT1 expression and activity is another important control point of the DNA methylation machinery. Post-transcriptional gene silencing by miRNAs (microRNAs) and post-translational targeting of DNMT1 by different non-coding regulatory RNA molecules could be the missing pieces of the puzzle that could explain formation of cell-specific differential methylation patterns.5 miRNAs, the most acknowledged class of ncRNA (non-coding RNAs) are small, typically 21–25 nucleotides long. They regulate gene expression by targeting protein-coding mRNAs and inducing their translational repression, de-adenylation or degradation.13 miRNAs either increase or decrease the transcriptional activity of a gene by preventing the target mRNA from producing a protein.13,79 Several families of miRNA have been shown to affect DNMT1 expression, either directly by binding to the 3′-UTR (3′-untranslated region) or the coding sequence of DNMT1 mRNA and decreasing the enzyme levels or indirectly by interacting with transcription factors that control the DNMT1 expression.22 DNMT1 expression is directly affected miR-148a, which targets the protein-coding region of its transcript. Dysregulation, especially overexpression of miR-148, leads to a decrease in DNMT1 protein levels, resulting in DNA hypomethylation and an increase in expression of critical methylation-sensitive genes, such as CD70 in CD4+ T cells and RASSF1A in cholangiocarcinoma.13,22,80,81 On the other hand, the majority of miRNA seems to target the 3′-UTR of DNMT1, for example, miR-152 directly binds to the 3′-UTR of DNMT1. Its downregulation in HBV (Hepatitis B Virus)-related HCC (Hepatocellular Carcinoma) causes global DNA hypermethylation and increases the methylation levels of two tumor suppressor genes, GSTP1 (Glutathione S-Transferase pi 1) and CDH1 (E-cadherin 1).13,82 Similarly, miR-185 is also predicted to bind to the 3′-UTR of DNMT1. Lowered levels of miR-185 expression may result in the abnormal expression of DNMT1 and aberrant DNA methylation, contributing to the development of human glioma.13,83 miR-126 also inhibits DNMT1 translation via an interaction with its 3′-UTR and overexpression of miR-126 in CD4+ T cells induces demethylation and consequent upregulation of genes encoding CD11a and CD70, thereby causing T and B cell hyperactivity.13,84 Some miRNAs, however, indirectly affect the expression and activity of DNMT1, for example, miR-29b in AML (Acute Myeloid Leukemia) cells downregulates SP1, a known transcriptional activator of DNMT1, thereby interfering with the SP1-dependent transcription of DNMT1.13,85 The miR-290 family of miRNAs regulates Rb proteins, especially RbL2 (retinoblastoma-like 2) and therefore affects the Rb-E2F pathway mediated expression of DNMT1.22,86
Post-translational targeting of DNMT1 by non-coding regulatory RNA molecules was probably the first reported interaction of DNMT1 with other molecules.39,87 DNMT1 exhibits physical or functional interaction with RNA polymerase II39 and, in cells, with several RNA binding proteins such as MBD proteins and AUF1 [AU-rich element/ poly (U)-binding/ degradation factor 1].39,88 The interaction between MBD proteins and DNMT1 facilitates the easy access of the enzyme to methylated DNA sites and orchestrates efficient gene expression.89 DNMT1 is essential for devotedly copying the epigenetic message during each cell division; hence, it becomes crucial that its expression is tightly coordinated with the state of cell division. AUF1 interacts with an AU-rich conserved element in the 3′-UTR of DNMT1 and negatively affect its expression by proteolytically destroying the DNMT1 mRNA. AUF1 protein levels are inversely regulated by the cell cycle specific proteasomal degradation, resulting in destabilization of DNMT1 mRNA. The cell specific targeting of DNMT1 mRNA to the exosome and its subsequent degradation by AUF1 is perhaps the first example of a cell cycle dependent regulation of an mRNA involving the exosome machinery.88 Thus, RNA seems to play a very crucial role in guiding DNMT1 to specific genomic loci and, consequently, in establishing distinct DNA methylation patterns. However, the functional significance of RNA directed DNA methylation in the context of development and disease still remains in a shroud of darkness. It is possible that several other categories of both small and long ncRNAs associated with the regulation of DNMT1 will be unearthed in the near future which will provide avenues for designing of rational and novel RNA based modulators of DNMT1.
Conclusions
In this review we attempt to provide a detailed account of the various interrelated and dynamically functioning regulatory mechanisms that systematize the working of DNMT1. With increasingly acknowledged role of DNA methylation in genome maintenance as well as in cancer, it becomes necessary to understand the structural, molecular and biochemical specificities of the DNMT1 enzyme and elucidate its function more minutely. Perceptions regarding DNMT1 function and design of its inhibitors and activators will strongly rest upon building a correlation between the available structural and functional data and various signaling pathways that direct the enzyme toward aberrant DNA methylation. It is also essential to understand the fundamental control systems that coordinate the effectiveness of DNMT1 so that futuristic implications, such as use of DNMT1 as an epigenetic drug target, can be realized. A precise and comprehensive analysis of the numerous intrinsic and extrinsic factors might help in unraveling many unanswered mysteries. What are the epigenetic triggers that set into motion the extremely fine-tuned and elaborately executed DNA methylation machinery? Is there a redundancy in the control of DNMT1 regulation and activity? Are different regulatory mechanisms working in tandem or differentially in different cellular contexts? Which one of the supervisory networks is majorly impaired during malignant transformation? Which checkpoints can be considered as the most suitable ones for designing of novel and rational mechanistic based drugs? Questions such as these can only be explained with in-depth analyses of the DNA methylation phenomenon and study of the factors that influence the regulation of DNMT1.
Glossary
Abbreviations:
- 3′-UTR
3′-untranslated region
- AKT1
v-akt murine thymoma viral oncogene homolog 1
- AML
acute myeloid leukemia
- AP-1
activator protein 1
- APC
adenomatous polyposis coli
- AUF1
AU-rich element/poly (U)-binding/degradation factor 1
- BAH
bromo-adjacent homology
- BAH1 and BAH2
bromo-adjacent homology 1 and 2
- BAZ2A
bromodomain adjacent to zinc finger domain
- CDH1
E-cadherin 1
- CDK
cyclin-dependent kinases
- CXXC1/CFP1
CXXC finger protein 1
- DMAP1
DNA methyltransferase 1 associated protein 1
- DNMTs
DNA Methyltransferases
- EHMT2
euchromatic histone-lysine N-methyltransferase 2
- ERK-MAPK
extracellular signal-regulated kinase/mitogen-activated protein kinase
- EZH2
enhancer of zeste homolog 2
- Gsk3β
glycogen synthase kinase 3β
- GSTP1
glutathione S-transferase pi 1
- HAUSP
herpes virus-associated ubiquitin-specific protease
- HBV
hepatitis B virus
- HCC
hepatocellular carcinoma
- HDAC
histone deacetylase
- HH
hedgehog
- HP1β
heterochromatin binding protein 1 β
- hSNF2H/SMARCA5
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily A, member 5
- KAT5
K-(lysine) acetyltransferase 5
- KG
lysine-glycine
- LSD1/KDM1A
lysine specific demethylase 1/lysine (K)-specific demethylase 1A
- LSH
lymphoid specific helicases
- MDM2
murine double minute 2
- MeCp2
MBD2, MBD3, methyl-CpG-binding domain proteins
- miRNAs
microRNAs
- ncRNA
non-coding RNAs
- NLS
nuclear localization signal
- PARP1
poly (ADP-ribose) polymerase 1
- PBD
proliferating cell nuclear antigen (PCNA) binding domain
- PBHD
polybromo homology domain
- PcG
polycomb group
- PCNA
proliferating cell nuclear antigen
- PI3K/PKB
phosphatidylinositol 3-kinase/protein kinase B
- PKC
protein kinase C
- PTM
post-translational modifications
- RAS
rat sarcoma
- Rb
retinoblastoma
- RbL2
retinoblastoma-like 2
- RFTS/TS
replication foci targeting sequence/targeting sequence
- RIP140/ NRIP1
nuclear receptor interacting protein 1
- SAH
S-Adenosyl homocystein
- SAM
S-adenosyl methionine
- SET7/SET7/9/KMT7
SET domain containing (lysine methyltransferase) 7
- SP1 and SP3
specificity protein 1 and 3
- STAT3
signal transducer and activator of transcription 3
- Suv39H1
suppressor of variegation 3-9 homolog 1
- TCF
T-cell Factor
- TGFβ
transforming growth factor β
- TRD
target recognition domain
- Ubc9
ubiquitin-conjugating enzyme
- UHRF1
ubiquitin-like with PHD and ring finger domains 1
- USP7
ubiquitin specific peptidase 7
- WT1
Wilms Tumor 1
Footnotes
Previously published online: www.landesbioscience.com/journals/epigenetics/article/21568
References
- 1.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–54. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 2.Patra SK, Deb M, Patra A. Molecular marks for epigenetic identification of developmental and cancer stem cells. Clin Epigenetics. 2011;2:27–53. doi: 10.1007/s13148-010-0016-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell PM, Szyf M. Human DNA methyltransferase gene DNMT1 is regulated by the APC pathway. Carcinogenesis. 2003;24:17–24. doi: 10.1093/carcin/24.1.17. [DOI] [PubMed] [Google Scholar]
- 4.Jurkowska RZ, Jurkowski TP, Jeltsch A. Structure and function of mammalian DNA methyltransferases. Chembiochem. 2011;12:206–22. doi: 10.1002/cbic.201000195. [DOI] [PubMed] [Google Scholar]
- 5.Svedruzić ZM. Mammalian cytosine DNA methyltransferase Dnmt1: enzymatic mechanism, novel mechanism-based inhibitors, and RNA-directed DNA methylation. Curr Med Chem. 2008;15:92–106. doi: 10.2174/092986708783330700. [DOI] [PubMed] [Google Scholar]
- 6.Jeltsch A. Reading and writing DNA methylation. Nat Struct Mol Biol. 2008;15:1003–4. doi: 10.1038/nsmb1008-1003. [DOI] [PubMed] [Google Scholar]
- 7.Robertson KD. DNA methylation and chromatin - unraveling the tangled web. Oncogene. 2002;21:5361–79. doi: 10.1038/sj.onc.1205609. [DOI] [PubMed] [Google Scholar]
- 8.Hotchkiss RD. The quantitative separation of purines, pyrimidines, and nucleosides by paper chromatography. J Biol Chem. 1948;175:315–32. [PubMed] [Google Scholar]
- 9.Jeltsch A. Beyond Watson and Crick: DNA methylation and molecular enzymology of DNA methyltransferases. Chembiochem. 2002;3:274–93. doi: 10.1002/1439-7633(20020402)3:4<274::AID-CBIC274>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 10.Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 11.Lan J, Hua S, He X, Zhang Y. DNA methyltransferases and methyl-binding proteins of mammals. Acta Biochim Biophys Sin (Shanghai) 2010;42:243–52. doi: 10.1093/abbs/gmq015. [DOI] [PubMed] [Google Scholar]
- 12.Jones PA, Liang G. Rethinking how DNA methylation patterns are maintained. Nat Rev Genet. 2009;10:805–11. doi: 10.1038/nrg2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Denis H, Ndlovu MN, Fuks F. Regulation of mammalian DNA methyltransferases: a route to new mechanisms. EMBO Rep. 2011;12:647–56. doi: 10.1038/embor.2011.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buryanov YI, Shevchuk TV. DNA methyltransferases and structural-functional specificity of eukaryotic DNA modification. Biochemistry (Mosc) 2005;70:730–42. doi: 10.1007/s10541-005-0178-0. [DOI] [PubMed] [Google Scholar]
- 15.Luczak MW, Jagodziński PP. The role of DNA methylation in cancer development. Folia Histochem Cytobiol. 2006;44:143–54. [PubMed] [Google Scholar]
- 16.Cheng X, Blumenthal RM. Coordinated chromatin control: structural and functional linkage of DNA and histone methylation. Biochemistry. 2010;49:2999–3008. doi: 10.1021/bi100213t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dhe-Paganon S, Syeda F, Park L. DNA methyl transferase 1: regulatory mechanisms and implications in health and disease. Int J Biochem Mol Biol. 2011;2:58–66. [PMC free article] [PubMed] [Google Scholar]
- 18.Xu F, Mao C, Ding Y, Rui C, Wu L, Shi A, et al. Molecular and enzymatic profiles of mammalian DNA methyltransferases: structures and targets for drugs. Curr Med Chem. 2010;17:4052–71. doi: 10.2174/092986710793205372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patra SK, Bettuzzi S. Epigenetic DNA-(cytosine-5-carbon) modifications: 5-aza-2′-deoxycytidine and DNA-demethylation. Biochemistry (Mosc) 2009;74:613–9. doi: 10.1134/S0006297909060042. [DOI] [PubMed] [Google Scholar]
- 20.Turek-Plewa J, Jagodziński PP. The role of mammalian DNA methyltransferases in the regulation of gene expression. Cell Mol Biol Lett. 2005;10:631–47. [PubMed] [Google Scholar]
- 21.Mohammad HP, Baylin SB. Linking cell signaling and the epigenetic machinery. Nat Biotechnol. 2010;28:1033–8. doi: 10.1038/nbt1010-1033. [DOI] [PubMed] [Google Scholar]
- 22.Kinney SR, Pradhan S. Regulation of expression and activity of DNA (cytosine-5) methyltransferases in mammalian cells. Prog Mol Biol Transl Sci. 2011;101:311–33. doi: 10.1016/B978-0-12-387685-0.00009-3. [DOI] [PubMed] [Google Scholar]
- 23.Rouleau J, MacLeod AR, Szyf M. Regulation of the DNA methyltransferase by the Ras-AP-1 signaling pathway. J Biol Chem. 1995;270:1595–601. doi: 10.1074/jbc.270.4.1595. [DOI] [PubMed] [Google Scholar]
- 24.Patra SK. Ras regulation of DNA-methylation and cancer. Exp Cell Res. 2008;314:1193–201. doi: 10.1016/j.yexcr.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 25.Bigey P, Ramchandani S, Theberge J, Araujo FD, Szyf M. Transcriptional regulation of the human DNA Methyltransferase (dnmt1) gene. Gene. 2000;242:407–18. doi: 10.1016/S0378-1119(99)00501-6. [DOI] [PubMed] [Google Scholar]
- 26.MacLeod AR, Rouleau J, Szyf M. Regulation of DNA methylation by the Ras signaling pathway. J Biol Chem. 1995;270:11327–37. doi: 10.1074/jbc.270.19.11327. [DOI] [PubMed] [Google Scholar]
- 27.Deng C, Yang J, Scott J, Hanash S, Richardson BC. Role of the ras-MAPK signaling pathway in the DNA methyltransferase response to DNA hypomethylation. Biol Chem. 1998;379:1113–20. doi: 10.1515/bchm.1998.379.8-9.1113. [DOI] [PubMed] [Google Scholar]
- 28.Slack A, Cervoni N, Pinard M, Szyf M. Feedback regulation of DNA methyltransferase gene expression by methylation. Eur J Biochem. 1999;264:191–9. doi: 10.1046/j.1432-1327.1999.00603.x. [DOI] [PubMed] [Google Scholar]
- 29.Sun L, Zhao H, Xu Z, Liu Q, Liang Y, Wang L, et al. Phosphatidylinositol 3-kinase/protein kinase B pathway stabilizes DNA methyltransferase I protein and maintains DNA methylation. Cell Signal. 2007;19:2255–63. doi: 10.1016/j.cellsig.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 30.McCabe MT, Davis JN, Day ML. Regulation of DNA methyltransferase 1 by the pRb/E2F1 pathway. Cancer Res. 2005;65:3624–32. doi: 10.1158/0008-5472.CAN-04-2158. [DOI] [PubMed] [Google Scholar]
- 31.Kimura H, Nakamura T, Ogawa T, Tanaka S, Shiota K. Transcription of mouse DNA methyltransferase 1 (Dnmt1) is regulated by both E2F-Rb-HDAC-dependent and -independent pathways. Nucleic Acids Res. 2003;31:3101–13. doi: 10.1093/nar/gkg406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peterson EJ, Bögler O, Taylor SM. p53-mediated repression of DNA methyltransferase 1 expression by specific DNA binding. Cancer Res. 2003;63:6579–82. [PubMed] [Google Scholar]
- 33.Lin RK, Wu CY, Chang JW, Juan LJ, Hsu HS, Chen CY, et al. Dysregulation of p53/Sp1 control leads to DNA methyltransferase-1 overexpression in lung cancer. Cancer Res. 2010;70:5807–17. doi: 10.1158/0008-5472.CAN-09-4161. [DOI] [PubMed] [Google Scholar]
- 34.Lu R, Wang X, Chen ZF, Sun DF, Tian XQ, Fang JY. Inhibition of the extracellular signal-regulated kinase/mitogen-activated protein kinase pathway decreases DNA methylation in colon cancer cells. J Biol Chem. 2007;282:12249–59. doi: 10.1074/jbc.M608525200. [DOI] [PubMed] [Google Scholar]
- 35.Papageorgis P, Lambert AW, Ozturk S, Gao F, Pan H, Manne U, et al. Smad signaling is required to maintain epigenetic silencing during breast cancer progression. Cancer Res. 2010;70:968–78. doi: 10.1158/0008-5472.CAN-09-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He S, Wang F, Yang L, Guo C, Wan R, Ke A, et al. Expression of DNMT1 and DNMT3a are regulated by GLI1 in human pancreatic cancer. PLoS One. 2011;6:e27684. doi: 10.1371/journal.pone.0027684. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Frauer C, Leonhardt H. Twists and turns of DNA methylation. Proc Natl Acad Sci U S A. 2011;108:8919–20. doi: 10.1073/pnas.1105804108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fatemi M, Hermann A, Pradhan S, Jeltsch A. The activity of the murine DNA methyltransferase Dnmt1 is controlled by interaction of the catalytic domain with the N-terminal part of the enzyme leading to an allosteric activation of the enzyme after binding to methylated DNA. J Mol Biol. 2001;309:1189–99. doi: 10.1006/jmbi.2001.4709. [DOI] [PubMed] [Google Scholar]
- 39.Svedružić ZM. Dnmt1 structure and function. Prog Mol Biol Transl Sci. 2011;101:221–54. doi: 10.1016/B978-0-12-387685-0.00006-8. [DOI] [PubMed] [Google Scholar]
- 40.Qin W, Leonhardt H, Pichler G. Regulation of DNA methyltransferase 1 by interactions and modifications. Nucleus. 2011;2:392–402. doi: 10.4161/nucl.2.5.17928. a. [DOI] [PubMed] [Google Scholar]
- 41.Takeshita K, Suetake I, Yamashita E, Suga M, Narita H, Nakagawa A, et al. Structural insight into maintenance methylation by mouse DNA methyltransferase 1 (Dnmt1) Proc Natl Acad Sci U S A. 2011;108:9055–9. doi: 10.1073/pnas.1019629108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pradhan M, Estève PO, Chin HG, Samaranayke M, Kim GD, Pradhan S. CXXC domain of human DNMT1 is essential for enzymatic activity. Biochemistry. 2008;47:10000–9. doi: 10.1021/bi8011725. [DOI] [PubMed] [Google Scholar]
- 43.Song J, Rechkoblit O, Bestor TH, Patel DJ. Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation. Science. 2011;331:1036–40. doi: 10.1126/science.1195380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Song J, Teplova M, Ishibe-Murakami S, Patel DJ. Structure-based mechanistic insights into DNMT1-mediated maintenance DNA methylation. Science. 2012;335:709–12. doi: 10.1126/science.1214453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reed MC, Lieb A, Nijhout HF. The biological significance of substrate inhibition: a mechanism with diverse functions. Bioessays. 2010;32:422–9. doi: 10.1002/bies.200900167. [DOI] [PubMed] [Google Scholar]
- 46.Pradhan S, Estève PO. Allosteric activator domain of maintenance human DNA (cytosine-5) methyltransferase and its role in methylation spreading. Biochemistry. 2003;42:5321–32. doi: 10.1021/bi034160+. [DOI] [PubMed] [Google Scholar]
- 47.Bacolla A, Pradhan S, Larson JE, Roberts RJ, Wells RD. Recombinant human DNA (cytosine-5) methyltransferase. III. Allosteric control, reaction order, and influence of plasmid topology and triplet repeat length on methylation of the fragile X CGG.CCG sequence. J Biol Chem. 2001;276:18605–13. doi: 10.1074/jbc.M100404200. [DOI] [PubMed] [Google Scholar]
- 48.Svedruzić ZM, Reich NO. Mechanism of allosteric regulation of Dnmt1’s processivity. Biochemistry. 2005;44:14977–88. doi: 10.1021/bi050988f. b. [DOI] [PubMed] [Google Scholar]
- 49.Jeltsch A. On the enzymatic properties of Dnmt1: specificity, processivity, mechanism of linear diffusion and allosteric regulation of the enzyme. Epigenetics. 2006;1:63–6. doi: 10.4161/epi.1.2.2767. [DOI] [PubMed] [Google Scholar]
- 50.Bacolla A, Pradhan S, Roberts RJ, Wells RD. Recombinant human DNA (cytosine-5) methyltransferase. II. Steady-state kinetics reveal allosteric activation by methylated dna. J Biol Chem. 1999;274:33011–9. doi: 10.1074/jbc.274.46.33011. [DOI] [PubMed] [Google Scholar]
- 51.Svedruzić ZM, Reich NO. DNA cytosine C5 methyltransferase Dnmt1: catalysis-dependent release of allosteric inhibition. Biochemistry. 2005;44:9472–85. doi: 10.1021/bi050295z. a. [DOI] [PubMed] [Google Scholar]
- 52.Hermann A, Goyal R, Jeltsch A. The Dnmt1 DNA-(cytosine-C5)-methyltransferase methylates DNA processively with high preference for hemimethylated target sites. J Biol Chem. 2004;279:48350–9. doi: 10.1074/jbc.M403427200. b. [DOI] [PubMed] [Google Scholar]
- 53.Spada F, Rothbauer U, Zolghadr K, Schermelleh L, Leonhardt H. Regulation of DNA methyltransferase 1. Adv Enzyme Regul. 2006;46:224–34. doi: 10.1016/j.advenzreg.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 54.Hervouet E, Vallette FM, Cartron PF. Dnmt1/Transcription factor interactions: an alternative mechanism of DNA methylation inheritance. Genes Cancer. 2010;1:434–43. doi: 10.1177/1947601910373794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spada F, Haemmer A, Kuch D, Rothbauer U, Schermelleh L, Kremmer E, et al. DNMT1 but not its interaction with the replication machinery is required for maintenance of DNA methylation in human cells. J Cell Biol. 2007;176:565–71. doi: 10.1083/jcb.200610062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schermelleh L, Haemmer A, Spada F, Rösing N, Meilinger D, Rothbauer U, et al. Dynamics of Dnmt1 interaction with the replication machinery and its role in postreplicative maintenance of DNA methylation. Nucleic Acids Res. 2007;35:4301–12. doi: 10.1093/nar/gkm432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim GD, Ni J, Kelesoglu N, Roberts RJ, Pradhan S. Co-operation and communication between the human maintenance and de novo DNA (cytosine-5) methyltransferases. EMBO J. 2002;21:4183–95. doi: 10.1093/emboj/cdf401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Robertson KD, Ait-Si-Ali S, Yokochi T, Wade PA, Jones PL, Wolffe AP. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat Genet. 2000;25:338–42. doi: 10.1038/77124. [DOI] [PubMed] [Google Scholar]
- 59.Fuks F, Hurd PJ, Deplus R, Kouzarides T. The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res. 2003;31:2305–12. doi: 10.1093/nar/gkg332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Estève PO, Chin HG, Smallwood A, Feehery GR, Gangisetty O, Karpf AR, et al. Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication. Genes Dev. 2006;20:3089–103. doi: 10.1101/gad.1463706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Viré E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–4. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 62.Myant K, Stancheva I. LSH cooperates with DNA methyltransferases to repress transcription. Mol Cell Biol. 2008;28:215–26. doi: 10.1128/MCB.01073-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Espada J, Peinado H, Lopez-Serra L, Setién F, Lopez-Serra P, Portela A, et al. Regulation of SNAIL1 and E-cadherin function by DNMT1 in a DNA methylation-independent context. Nucleic Acids Res. 2011;39:9194–205. doi: 10.1093/nar/gkr658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pradhan S, Kim GD. The retinoblastoma gene product interacts with maintenance human DNA (cytosine-5) methyltransferase and modulates its activity. EMBO J. 2002;21:779–88. doi: 10.1093/emboj/21.4.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Estève PO, Chang Y, Samaranayake M, Upadhyay AK, Horton JR, Feehery GR, et al. A methylation and phosphorylation switch between an adjacent lysine and serine determines human DNMT1 stability. Nat Struct Mol Biol. 2011;18:42–8. doi: 10.1038/nsmb.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Goyal R, Rathert P, Laser H, Gowher H, Jeltsch A. Phosphorylation of serine-515 activates the Mammalian maintenance methyltransferase Dnmt1. Epigenetics. 2007;2:155–60. doi: 10.4161/epi.2.3.4768. [DOI] [PubMed] [Google Scholar]
- 67.Lavoie G, St-Pierre Y. Phosphorylation of human DNMT1: implication of cyclin-dependent kinases. Biochem Biophys Res Commun. 2011;409:187–92. doi: 10.1016/j.bbrc.2011.04.115. [DOI] [PubMed] [Google Scholar]
- 68.Estève PO, Chin HG, Benner J, Feehery GR, Samaranayake M, Horwitz GA, et al. Regulation of DNMT1 stability through SET7-mediated lysine methylation in mammalian cells. Proc Natl Acad Sci U S A. 2009;106:5076–81. doi: 10.1073/pnas.0810362106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pradhan S, Chin HG, Estève PO, Jacobsen SE. SET7/9 mediated methylation of non-histone proteins in mammalian cells. Epigenetics. 2009;4:383–7. doi: 10.4161/epi.4.6.9450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hotz HR, Peters AHFM. Protein demethylation required for DNA methylation. Nat Genet. 2009;41:10–1. doi: 10.1038/ng0109-10. [DOI] [PubMed] [Google Scholar]
- 71.Wang J, Hevi S, Kurash JK, Lei H, Gay F, Bajko J, et al. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat Genet. 2009;41:125–9. doi: 10.1038/ng.268. [DOI] [PubMed] [Google Scholar]
- 72.Du Z, Song J, Wang Y, Zhao Y, Guda K, Yang S, et al. DNMT1 stability is regulated by proteins coordinating deubiquitination and acetylation-driven ubiquitination. Sci Signal. 2010;3:ra80. doi: 10.1126/scisignal.2001462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bostick M, Kim JK, Estève PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–4. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 74.Felle M, Joppien S, Németh A, Diermeier S, Thalhammer V, Dobner T, et al. The USP7/Dnmt1 complex stimulates the DNA methylation activity of Dnmt1 and regulates the stability of UHRF1. Nucleic Acids Res. 2011;39:8355–65. doi: 10.1093/nar/gkr528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hong Q, Shao ZM. Ubiquitination/deubiquitination and acetylation/deacetylation: making DNMT1 stability more coordinated. Acta Pharmacol Sin. 2011;32:139–40. doi: 10.1038/aps.2011.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Qin W, Leonhardt H, Spada F. Usp7 and Uhrf1 control ubiquitination and stability of the maintenance DNA methyltransferase Dnmt1. J Cell Biochem. 2011;112:439–44. doi: 10.1002/jcb.22998. b. [DOI] [PubMed] [Google Scholar]
- 77.Bronner C. Control of DNMT1 abundance in epigenetic inheritance by acetylation, ubiquitylation, and the histone code. Sci Signal. 2011;4:pe3. doi: 10.1126/scisignal.2001764. [DOI] [PubMed] [Google Scholar]
- 78.Lee B, Muller MT. SUMOylation enhances DNA methyltransferase 1 activity. Biochem J. 2009;421:449–61. doi: 10.1042/BJ20090142. [DOI] [PubMed] [Google Scholar]
- 79.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–14. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 80.Braconi C, Huang N, Patel T. MicroRNA-dependent regulation of DNA methyltransferase-1 and tumor suppressor gene expression by interleukin-6 in human malignant cholangiocytes. Hepatology. 2010;51:881–90. doi: 10.1002/hep.23381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pan W, Zhu S, Yuan M, Cui H, Wang L, Luo X, et al. MicroRNA-21 and microRNA-148a contribute to DNA hypomethylation in lupus CD4+ T cells by directly and indirectly targeting DNA methyltransferase 1. J Immunol. 2010;184:6773–81. doi: 10.4049/jimmunol.0904060. [DOI] [PubMed] [Google Scholar]
- 82.Huang J, Wang Y, Guo Y, Sun S. Down-regulated microRNA-152 induces aberrant DNA methylation in hepatitis B virus-related hepatocellular carcinoma by targeting DNA methyltransferase 1. Hepatology. 2010;52:60–70. doi: 10.1002/hep.23660. [DOI] [PubMed] [Google Scholar]
- 83.Zhang Z, Tang H, Wang Z, Zhang B, Liu W, Lu H, et al. MiR-185 targets the DNA methyltransferases 1 and regulates global DNA methylation in human glioma. Mol Cancer. 2011;10:124. doi: 10.1186/1476-4598-10-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhao S, Wang Y, Liang Y, Zhao M, Long H, Ding S, et al. MicroRNA-126 regulates DNA methylation in CD4+ T cells and contributes to systemic lupus erythematosus by targeting DNA methyltransferase 1. Arthritis Rheum. 2011;63:1376–86. doi: 10.1002/art.30196. [DOI] [PubMed] [Google Scholar]
- 85.Garzon R, Liu S, Fabbri M, Liu Z, Heaphy CEA, Callegari E, et al. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood. 2009;113:6411–8. doi: 10.1182/blood-2008-07-170589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Benetti R, Gonzalo S, Jaco I, Muñoz P, Gonzalez S, Schoeftner S, et al. A mammalian microRNA cluster controls DNA methylation and telomere recombination via Rbl2-dependent regulation of DNA methyltransferases. Nat Struct Mol Biol. 2008;15:268–79. doi: 10.1038/nsmb.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bolden A, Ward C, Siedlecki JA, Weissbach A. DNA methylation. Inhibition of de novo and maintenance methylation in vitro by RNA and synthetic polynucleotides. J Biol Chem. 1984;259:12437–43. [PubMed] [Google Scholar]
- 88.Torrisani J, Unterberger A, Tendulkar SR, Shikimi K, Szyf M. AUF1 cell cycle variations define genomic DNA methylation by regulation of DNMT1 mRNA stability. Mol Cell Biol. 2007;27:395–410. doi: 10.1128/MCB.01236-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jeffery L, Nakielny S. Components of the DNA methylation system of chromatin control are RNA-binding proteins. J Biol Chem. 2004;279:49479–87. doi: 10.1074/jbc.M409070200. [DOI] [PubMed] [Google Scholar]
- 90.Reale A, Matteis GD, Galleazzi G, Zampieri M, Caiafa P. Modulation of DNMT1 activity by ADP-ribose polymers. Oncogene. 2005;24:13–9. doi: 10.1038/sj.onc.1208005. [DOI] [PubMed] [Google Scholar]