Abstract
Rationale and Objectives
Quantitative measurement provides essential information about disease progression and treatment response in patients with Glioblastoma multiforme (GBM). The goal of this paper is to present and validate a software pipeline for semi-automatic GBM segmentation, called AFINITI (Assisted Follow-up in NeuroImaging of Therapeutic Intervention), using clinical data from GBM patients.
Materials and Methods
Our software adopts the current state-of-the-art tumor segmentation algorithms and combines them into one clinically usable pipeline. Both the advantages of the traditional voxel-based and the deformable shape-based segmentation are embedded into the software pipeline. The former provides an automatic tumor segmentation scheme based on T1- and T2-weighted MR brain data, and the latter refines the segmentation results with minimal manual input.
Results
Twenty six clinical MR brain images of GBM patients were processed and compared with manual results. The results can be visualized using the embedded graphic user interface (GUI).
Conclusion
Validation results using clinical GBM data showed high correlation between the AFINITI results and manual annotation. Compared to the voxel-wise segmentation, AFINITI yielded more accurate results in segmenting the enhanced GBM from multimodality MRI data. The proposed pipeline could be used as additional information to interpret MR brain images in neuroradiology.
Keywords: Glioblastoma multiforme, segmentation, clinical validation
1. Introduction
Despite the best available standard therapies, including surgery, radiation, and chemotherapy, the survival in patients diagnosed with Glioblastoma multiforme (GBM) remains dismal at 14 months (1). Newer therapeutic strategies aiming at targeting specific molecules are being developed and tested in clinical trials (2). Temozolamide chemoradiation has significantly prolonged survival but produces pseudoprogression that is difficult or impossible to distinguish from recurrence in 30-50% of patients (1, 3). In addition, antiangiogenic therapies have been used in combination with conventional chemotherapy in patients with recurrent GBM, demonstrating radiographic response rates of 35%–50% (4-6). These agents improve significantly patient quality of life but alter the pattern of recurrence by a potent effect on tumor permeability, suppressing enhancement within a solid tumor with a resulting increase in the frequency of infiltrative recurrence (7).
These therapy-induced alterations in the natural history and imaging appearance of treated GBM have made imaging follow-up by conventional MRI difficult, which motivates widespread ongoing research to discover additional imaging biomarkers and has led to a revision in response criteria. Although the most commonly used imaging criteria for evaluating treatment response are still based on measurement of enhancing tumor (the Macdonald Criteria) (8), the increase in infiltrative recurrence and the difficulty in distinguishing recurrence from progression has led to proposal of a new criteria for tumor response that includes abnormality on T2-weighted or fluid-attenuated inversion recovery (FLAIR) images as additional markers for progression (the RANO criteria) (9). The RANO criteria also recommends the use of volumetric measurements of enhancing tumor because reliance on cross product diameters is problematic and highly operator dependent in cases of irregularly shaped tumor, multifocal tumor, or tumor with cystic or necrotic components. Recently, volumetric measures were found comparable (10) or superior (11-12) to linear diameter measures as indicators of tumor evaluation.
Volumetric methods have the advantage of more reproducibly and precisely measuring the size of tumor and are being increasingly used in clinical settings. For example, volumetric measurement of both the enhancing and non-enhancing tumor have been correlated to progression free survival (PFS) and overall survival (OS) (7, 13). To date, the major barrier to widespread adoption of these methods in clinical neuro-oncology has been that manual and assisted manual segmentation methods are quite time consuming for the operator. Due to presence of heterogeneous signal intensity in necrotic or cystic tumor and at the margin of infiltrative tumors, it is difficult to segment a tumor by hand, and the development of automatic or semi-automatic software tools that can provide efficient volumetric measurement and assist longitudinal shape analysis for follow-up studies could provide significant benefit.
Tumor segmentation algorithms are classified into voxel-based or deformable shape model-based methods. Fuzzy clustering methods (voxel-based) are among the more popular approaches (14-18) and classify each voxel into either one of the normal brain tissues (gray matter, white matter, and CSF) or tumor tissues. The algorithm developed by Philips et al. (18) can differentiate clinically vital boundaries of tumor and edema from hemorrhage in multimodal MRI. The performance of multimodal intensity-based clustering can be limited by overlapping of intensity between tumor and normal tissues (19-20). To account for this, additional features such as multi-dimensional intensity vectors have been designed for the clustering. Clark et al. (21) has integrated knowledge-based techniques and multimodality clustering to segment GBM tumors. Fletcher-Heath et al. (17) presented the first tumor segmentation for non-enhancing MR data, including T1, T2, and proton density weighted images, to track tumor size over time. Prastawa et al. (22-23) designed a knowledge-based tumor segmentation algorithm that learns voxel-intensity distributions from normal brain and detects outlying tumor voxels. Kaus et al. developed a spatially varying statistical classification algorithm using a template to moderate the segmentation obtained by statistical classification (24-25).
A second class of algorithms use deformable shape models to segment tumor from normal brain. These methods are derived from the traditional Snake model (26) that uses surfaces to match tumor boundaries. The concept of these techniques is the use of energy function and various shape models: the external energy derived from the matching degree between the shape and the image features is used to distinguish tumor from normal tissues, and the internal energy is used to constrain the tumor shape. In order to adjust for the change of topology, implicit models such as level sets (27-32) can be used. Intensity distributions within and outside tumor region have been used for level set segmentation (33-35).
Voxel-based segmentation algorithms can better adapt the segmented tumor shape to local image, and deformable model-based segmentation schemes are more robust but generally need proper initialization. To take the advantages of both algorithms, we propose an AFINITI pipeline for segmenting MR images by combining them. In the first stage, the voxel-based segmentation using the FSL FAST (36-37) is performed automatically for initial tumor segmentation from T1-weighted images. The T2-weighted images are also automatically segmented and combined with the T1 segmentation results. Then, a level-set-based segmentation is used to refine the segmentation results with minimal manual input by embedding the major functions of ITK-SNAP (38). These tools are integrated into one pipeline with a single GUI. We validate the AFINITI pipeline by applying the software to 26 clinical GBM cases by comparing the results with those obtained using manual segmentation.
2. Methods
a. Patients and Data
The protocol was approved by the institutional review boards for retrospective retrieval and analysis of patient clinical and imaging data. Serial MRI scans from 26 consecutive patients with diagnosis of GBM at Brigham Womens Hospital between 2004 and 2009 who had interpretable high resolution MR scans were retrieved.
All MR scans were performed on 1.5T or 3T MRI scanners, and the imaging protocol contains at least an axial 3D SPGR T1-weighted series covering the whole brain acquired at a 5-10 minute delay after the intravenous administration of 0.1-0.2 mmol/kg gadopentetate dimeglumine contrast agent, and an axial 2D T2-weighted MR sequences. The slice thickness in all cases was between 1.0 and 1.5mm for 3D SPGR sequences and 6mm for the 2D FSE T2 weighted sequences. The typical 1.5T 3D SPGR parameters were set as: TR=25ms, TE=6ms, FOV=200mm×240mm, Matrix=224×224.
Manual segmentation was performed under the supervision of two practicing faculty neuroradiologists (GY & RH) by a research associate trained in neuro-anatomy and neuroimaging (KS), and the final segmentations were reviewed and corrected jointly by GY and RH to minimize the bias among ratters. Segmentation was performed using ITK-Snap by manually tracing the boundary between the areas of abnormal enhancement and normal tissue on the 3D SPGR images, excluding non-enhancing, presumably necrotic or cystic portions of tumor but including areas of heterogeneous enhancement felt to represent tumor.
b. The AFINITI Software Pipeline
The goal of AFINITI is to seamlessly implement the state-of-the-art neuroimaging tools into one package to facilitate clinical quantification of GBM. The GUI for implementing and visualizing the image processing modules was developed based on the ITK-SNAP framework. Additional modules were added to assist users selecting input images, specifying and adjusting segmentation parameters, as well as examining, editing and saving results. To make the software user-friendly it takes DICOM series inputs for processing in Section 2c. DICOM series with overlapping ROIs were written with modified DICOM tags as the outputs so that the results can be presented on different viewing workstations and distinguished from the original images. The AFINITI software pipeline was written using c++ and MS DOS batch scripting languages. The major tools embedded in the software pipeline included preprocessing, the FSL FAST, and the modified level-set method in the ITK-SNAP package.
c. Brain Tumor Segmentation Pipeline
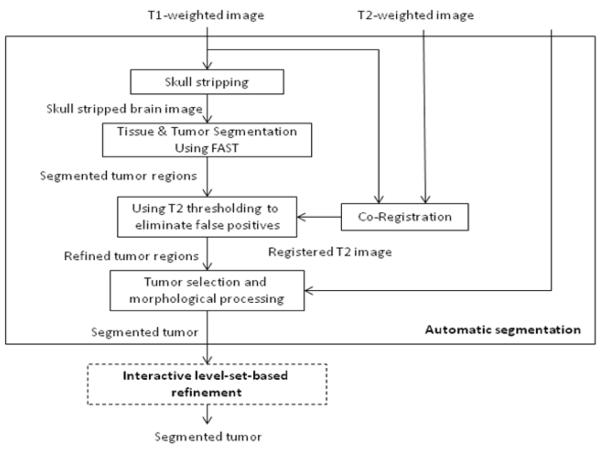
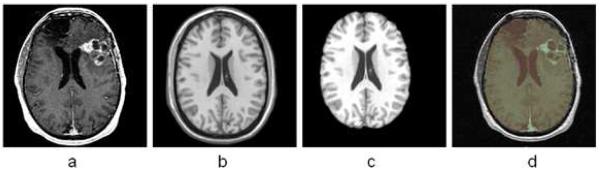
Fig. 1 shows the framework of AFINITI. The preprocessing step mainly strips skulls from T1-weighted images and co-registers T1-weighted images to T2-weighted images. A deformable brain image registration was used to automatically align the skull-stripped template image onto each individual image to remove the skulls (Fig. 2). The FSL FLIRT was embedded to automatically calculate the transformation (degree of freedom was set to 6) between T2- and T1-weighted images for each patient.
Fig. 1.
Brain tumor segmentation pipeline.
Fig. 2.
Registration-based skull stripping method. a. Original image to be processed; b. the template image; c. the segmented brain region of the template image; d. overlapping the brain region onto the original image.
Automatic segmentation step
In the software pipeline, we integrated the FAST tool (39) for initial brain tumor segmentation. We classified the tissues into not only the normal tissue types (WM, GM, and CSF) but also active tumor tissues (14-18). Fig. 3 shows an example of segmentation using FAST. It can be seen that the segmentation results can be noisy with many small vascular spots in normal tissues that were classified as tumor tissue.
Fig. 3.
Some examples of tumor segmentation results after FAST. (a) and (c) Input images; (b) and (d) FAST segmentation results.
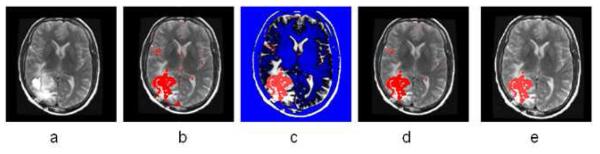
To reduce the false positive spots, we developed a T2 mapping method to remove them. As shown in Fig. 4.b, the T2-weighted image (Fig. 4a) was first registered onto the T1-weighted image with initial segmentation of FAST overlapped. A low-bound intensity thresholding was then applied to the registered T2-weighted image and only high intensity regions covering all the segmented tumor regions but not the false-positive regions were kept. Combining the T1 and T2 segmentation results and performing morphological shape corrections, we obtained the final automatic tumor segmentation. Then, the tumor can be automatically selected by using morphological operations. Therefore regions with small volumes or flat shapes were deleted by applying 3D open operation. If the tumor seed points are provided, a region grow operation will be performed to further remove false positive regions. Fig. 4.e gives the final automatic segmentation of the tumor.
Fig. 4.
Illustration of the combined T1 and T2 segmentation. (a) T2 image registered onto T1 image; (b) overlaying the segmentation result from T1 image onto T2 image; (c) thresholding T2 image (blue shows the ROI filtered out); by adjusting the threshold we can eliminate the enhanced big vessels close to the tumor; (d) after applying T2 thresholding, the majority of false positive spots were removed; (e) other isolated spots are removed using morphological operations.
Interactive processing step
After automatic segmentation, level-set-based segmentation can be further performed to interactively refine the results by visualization and manually correction (40). These tasks were accomplished by embedding ITK-SNAP into our software pipeline. ITK-SNAP is a software application used to segment structures in 3D medical images. It provides semi-automatic segmentation using level set methods, as well as manual delineation and image navigation.
Finally, after the segmented tumor result is accepted, the tumor volume, tumor center point location, and the overlapping of the segmented tumor region on the original images were automatically generated in the original DICOM format using a new series number. According to the current neuroradiology workflow, the new data series can be uploaded to PACS server so radiologists can interpret the films of GBM patients by referring to the segmentation results.
3. Results
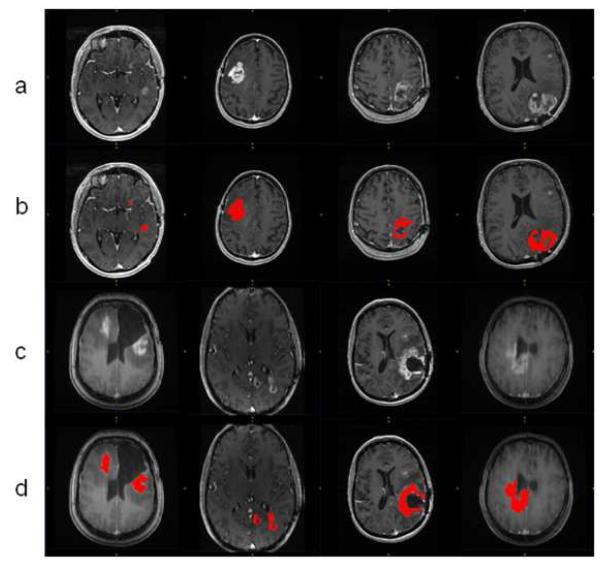
All the GBM tumor patient cases were processed using the proposed AFINITI software pipeline. Of all 26 cases, 24 were visually inspected and found satisfactory by GY, and 2 cases were further refined using the second step, namely level-set-based refinement. The average time for the two cases using level-set-based refinement was approximately 4 minutes. Fig. 5 shows eight examples of the segmentation results. The first and the third rows show the original T1-weighted images and the other rows give corresponding segmentation results. Satisfactory results were obtained from clinical cases both before and after tumor resection.
Fig.5.
Representative segmentation results. Row (a) and (c) are original images; row (b) and (d) are the corresponding segmentation results.
Quantitative measurements from manual segmentation and the results of AFINITI software were compared (Table 1). Denoting the results of AFINITI as set A and those of manual segmentation as M, the first and the second columns list the volume of the tumor segmented (unit in ml) using AFINITI and manual methods, respectively. The third and the fourth columns show the volumes of the intersection and union of the two results, i.e. ∣A⋂M∣ and ∣AUM∣, respectively. The fifth column gives the ratio of the intersection vesus AFINITI results, i.e. ∣A⋂M∣/∣A∣, and the sixth column gives the ratio of the intersection vesus the union, i.e. ∣A⋂M∣/∣AUM∣. Ideally, the results in the last two columns should be close to 100%. However, the Jaccard index is relatively small and the intersections match AFINITI results better, indicating that the manual segmentation results in larger regions for the datasets. To further investigate the performance, we correlated the AFINITI results with manual results (Figure 6). The Pearson correlation coefficient for all the 26 cases is 0.96.
Table 1.
Detailed AFINITI segmentation results (unit in ml)
| AFINITI results |
Manual results |
Intersection | Union | Intersection/AFINITI | Intersection /Union (Jaccard index) |
|---|---|---|---|---|---|
| 6.57 | 7.26 | 6.16 | 7.67 | 93.7% | 80.2% |
| 20.63 | 27.60 | 16.47 | 31.76 | 79.8% | 51.9% |
| 28.95 | 41.32 | 24.38 | 45.88 | 84.2% | 53.1% |
| 2.52 | 4.27 | 2.49 | 4.30 | 98.6% | 57.8% |
| 53.91 | 81.80 | 51.81 | 83.91 | 96.1% | 61.8% |
| 22.84 | 28.03 | 22.72 | 28.14 | 99.5% | 80.7% |
| 10.35 | 19.65 | 10.26 | 19.74 | 99.1% | 52.0% |
| 21.54 | 20.79 | 17.00 | 25.33 | 78.9% | 67.1% |
| 4.63 | 13.96 | 4.30 | 14.29 | 92.8% | 30.1% |
| 6.49 | 21.09 | 6.45 | 21.13 | 99.4% | 30.2% |
| 1.22 | 2.27 | 1.18 | 2.32 | 96.5% | 51.1% |
| 3.73 | 6.62 | 3.71 | 6.64 | 99.5% | 55.9% |
| 40.71 | 58.44 | 40.67 | 58.49 | 99.9% | 69.5% |
| 8.19 | 17.13 | 7.71 | 17.61 | 94.2% | 43.8% |
| 35.61 | 45.60 | 34.22 | 46.99 | 96.1% | 72.8% |
| 15.24 | 19.99 | 14.79 | 20.44 | 97.1% | 72.4% |
| 12.49 | 14.74 | 12.08 | 15.15 | 96.8% | 79.8% |
| 7.59 | 20.48 | 7.54 | 20.53 | 99.3% | 36.7% |
| 1.36 | 2.34 | 1.31 | 2.39 | 96.6% | 54.9% |
| 10.14 | 16.04 | 8.10 | 18.08 | 79.9% | 44.8% |
| 17.98 | 43.75 | 15.90 | 45.83 | 88.4% | 34.7% |
| 64.66 | 90.07 | 64.16 | 90.56 | 99.2% | 70.8% |
| 1.42 | 3.13 | 1.40 | 3.15 | 98.5% | 44.5% |
| 39.76 | 51.85 | 35.93 | 55.68 | 90.4% | 64.5% |
| 9.24 | 25.32 | 6.86 | 27.70 | 74.3% | 24.8% |
| 14.90 | 44.10 | 13.91 | 45.09 | 93.3% | 30.8% |
Fig. 6.
Correlation of the segmentation results.
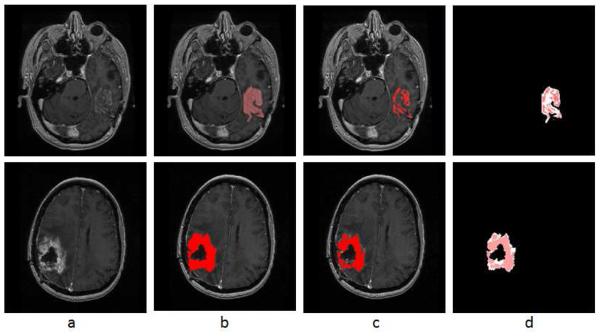
It can be seen from Table 1 that there are a few cases where the automatic results yielded lower volume measures than the manual results. Take the last two rows of Table 1 as example, Fig. 7 shows the original images, the manual and AFINITI segmentation results, and the overlapping images of these two cases. The tumor sizes measured from AFINITI were generally smaller than those obtained from manual segmentation, suggesting that human raters tend to over-segment tumor. During manual selection of the tumor regions, the bright enhanced regions were marked while the small dark regions inside and close to the tumor boundary were not selected. Therefore, although AFINITI results generated a more detailed boundary of the tumor (Fig. 7.c), the volumes were slightly smaller. Considering the Pearson correlation coefficient between AFINITI and manual results, they are highly correlated, suggesting comparable performance for the proposed automatic tumor segmentation pipeline.
Fig. 7.
Difference between manual and semi-automatic segmentations: a. origal image; b. manual segmentaion; c. semi-automatic segmentation; d. difference between manual segmentation (background white shape) and semi-auto segmentation (highlighted red shape).
We also compared AFINITI segmentation against voxel-based segmentation (FSL FAST). Because the segmentation results of the voxel-based segmentation lacked spatial continuity (see Fig. 3.d for example), they were further processed with tumor selection and morphological operation for the comparison. No manual interaction was involved in any of the 26 cases for the FAST algorithm. Table 2 lists the detailed segmentation results. The student T-Test of the Jaccard indexes of AFINITI and voxel-based results (p-value = 0.0071) showed the advantage of AFINITI.
Table 2.
Detailed voxel-based segmentation results (unit in ml)
| FAST results |
Manual results |
Intersection | Union | Intersection/FAST | Intersection/Union (Jaccard index) |
|---|---|---|---|---|---|
| 6.60 | 7.26 | 6.16 | 7.69 | 93.4% | 80.1% |
| 61.03 | 27.60 | 17.04 | 71.58 | 27.9% | 23.8% |
| 80.03 | 41.32 | 27.39 | 93.96 | 34.2% | 29.2% |
| 2.52 | 4.27 | 2.49 | 4.30 | 98.6% | 57.8% |
| 73.75 | 81.80 | 54.34 | 101.21 | 73.7% | 53.7% |
| 23.16 | 28.03 | 23.03 | 28.16 | 99.4% | 81.8% |
| 10.91 | 19.65 | 10.72 | 19.83 | 98.3% | 54.1% |
| 27.81 | 20.79 | 17.20 | 31.39 | 61.9% | 54.8% |
| 10.19 | 13.96 | 5.70 | 18.45 | 55.9% | 30.9% |
| 7.01 | 21.09 | 6.54 | 21.57 | 93.2% | 30.3% |
| 1.26 | 2.27 | 1.21 | 2.32 | 96.1% | 52.0% |
| 3.73 | 6.62 | 3.71 | 6.64 | 99.5% | 55.9% |
| 40.93 | 58.44 | 40.86 | 58.51 | 99.8% | 69.8% |
| 10.75 | 17.13 | 8.19 | 19.70 | 76.2% | 41.6% |
| 37.40 | 45.60 | 35.71 | 47.29 | 95.5% | 75.5% |
| 17.59 | 19.99 | 15.50 | 22.08 | 88.1% | 70.2% |
| 12.55 | 14.74 | 12.14 | 15.15 | 96.7% | 80.1% |
| 7.59 | 20.48 | 7.54 | 20.53 | 99.3% | 36.7% |
| 1.38 | 2.34 | 1.33 | 2.39 | 96.7% | 55.8% |
| 15.32 | 16.04 | 9.03 | 22.34 | 58.9% | 40.4% |
| 18.34 | 43.75 | 16.11 | 45.99 | 87.8% | 35.0% |
| 65.10 | 90.07 | 64.51 | 90.65 | 99.1% | 71.2% |
| 1.48 | 3.13 | 1.46 | 3.15 | 98.4% | 46.2% |
| 85.36 | 51.85 | 40.13 | 97.08 | 47.0% | 41.3% |
| 0.01 | 25.32 | 0.00 | 25.33 | 0.0% | 0.0% |
| 9.48 | 44.10 | 9.42 | 44.15 | 99.4% | 21.3% |
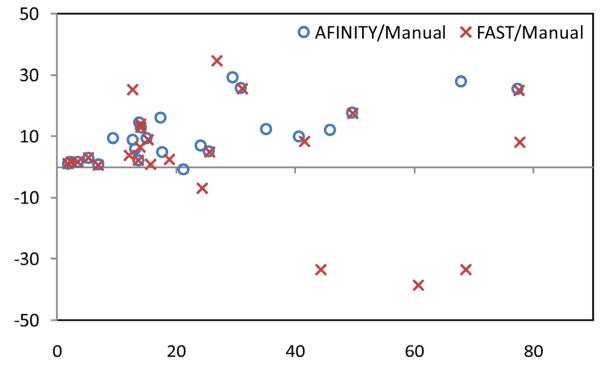
To further evaluate the agreement and systematic differences between segmentation methods and manual segmentation, the Bland-Altman plots of the volume measures between AFINITI and manual results, and between FAST and manual results, were shown in Fig. 8. The horizontal axis is the average of the volume measure, and the vertical axis indicates the difference between manual and automatic measures (manual – automatic). It can be seen that FAST generated relatively large errors as compared to AFINITI. Meanwhile, as stated in the above discussions, AFINITI tends to yield less volume because of the detailed segmentation of enhanced tumor regions (see Fig. 7 for details).
Fig. 8.
Bland-Altman plots of the volume measures between AFINITI and manual results, and between FAST and manual results, respectively.
Finally, it is worth noting that the automatic process of AFINITI took approximately 20 minutes for each dataset using a workstation with 1.86G Hz Intel Core 2 CPU and 2 GB RAM, and the average interactive refinement process took approximately 4 minutes of operator time. In contrast, the time required for manual segmentation of the dataset varied considerably, depending on the attributes of the tumor, ranging from 30 minutes to 90 minutes. In the proposed clinical workflow, the scanned data would be first automatically routed to the AFINITI workstation for data processing prior to study interpretation, and the segmented AFINITI output would be transferred as a new additional series of images onto the PACS server along with the quantitative measurement of the tumor. The interpreting radiologists would examine the overlays to confirm the accuracy of automatic segmentation and incorporate the volumetric output into the clinical report as part of the standard workflow.
4. Discussion
We present a software pipeline, an efficient toolkit for clinical GBM quantification, to segment MR brain images of high grade brain tumor by combining the voxel-based and deformable model (level set)-based algorithms. Evaluation of the software by comparison of volumetric output with manual segmentation of the same clinical dataset showed a significant linear correlation and high degree of overlapping. Although the process time used for manual operation was not rigorously evaluated, use of the system reduced clinical operator time required for segmentation and quantitation from approximately 30-90 minutes per case to less than 4 minutes in the roughly 8% of cases that required correction and to less than one minute in the majority of cases where the AFINITI output did not require correction. It is not clear how the average processing time using software based on the AFINITI method would compare with currently available commercial software for assisted manual segmentation. Clearly, recently released commercial tools would be expected to substantially decrease operator time compared with the open-source tools used in this study, but it seems unlikely that even these tools could achieve a lower average operator time for segmentation and quantitation that the AFINITI method provides. The software package was evaluated using the typical clinical MRI data, although ideally the robustness of the algorithm can be tested with different protocols, due to the availability of manual results we did not compare the accuracy for different groups of protocols. This could be a future work by using clinical studies from multiple scanners.
Nevertheless, the fraction of cases that require operator correction are clearly an area that requires further development, possibly with incorporation of additional data types. In addition, the processing time of 20 minutes is significant, although since this is proposed to occur offline prior to expert interpretation, it is not a critical problem for current diagnostic imaging workflow. This processing speed and the user-friendliness of the graphical interface need to be further improved for full clinical translation of the methods presented as will production of FDA approved software tools. Both of these improvements and full translation will require product development resources that are beyond the scope of our lab, so we are releasing the AFINITI software pipeline free of charge on our website http://www.cbi-tmhs.org/AFINITI/ in the hope that other groups will be able to extend our work and that its availability will motivate the development of fully translatable clinical tools.
An additional significant weakness is that 2D input data was used for the T2-weighted image analysis, which has larger between slice thickness (5~6mm). 3D T2-weighted whole brain image data was not available routinely at the time that the initial datasets were acquired for this study, but because of rapid progress in 3.0T MRI, they are now routinely available for clinical brain imaging. Adaptation of the software to perform segmentation on high resolution 3D T2-weighted image datasets is important because inclusion of such datasets would allow automated assessment of infiltrative progression as required for use of the recently proposed RANO criteria for brain tumor follow-up, and would allow automated computation of recently proposed image metrics such at rNTR that may be more predictive of patient outcome (7).
The automatic tumor delineation pipeline could be applied in clinics by first automatically routing the scanned data to the AFINITI workstation for automatic data processing prior to study interpretation. After this, the segmented AFINITI output will be superimposed on the original MRI images, and new images are generated with new series numbers under the same patient ID. The data are then transferred onto the PACS server along with the quantitative measurement of the tumor. The interpreting radiologists would examine the overlay images together with the original sequences for diagnosis and for incorporating the volumetric measures into the clinical report as part of the standard workflow.
Currently, after segmentation, quantitative measures of tumor can be obtained, relieving the tedious work load for manual segmentation. The quantitative measures can be used in a follow-up study that precisely gives the temporal changes for tumor evaluation. In the long run, we foresee that computational image analysis could provide more detailed measures after segmentation, such as heterogeneity measures, transition from enhanced tumor to neucrosis, and transition from enhanced tumor to edema. Generally, in contrast enhanced MR images, GBM consists of enhancing tumor, necrosis, and non-enhancing tumor, including edema. In multimodal MR, enhanced tumor can be clearly seen from T1 post-contrast images but non-enhanced tumor often overlaps with edema by visual assessment. Quantitative segmentation is thus important to segment GBM in detail and to help neuroradiologists determine the tumor margin and determine the aggressiveness of GBM.
5. Conclusion
We presented a software pipeline for GBM segmentation and demonstrated its applicability to clinical data. The software adopts the current state-of-the-art tumor segmentation algorithms and combines the advantages of the traditional voxel-based and deformable shape-based segmentation methods. This provides automatic tumor segmentation based on both T1- and T2-weighted MR brain data, with graphical and numerical output that can be visualized and interactively refined using the embedded GUI based on the ITK-SNAP framework. Finally, the software was incorporated into a conventional PACS-based MRI interpretation workflow. Validation of results using clinical GBM data showed high correlation between the AFINITI results and manual annotation and suggested significant reduction in operator time for performing volumetric quantitation of GBM. The AFINITI software pipeline is freely available from our public website.
Acknowledgements
The research is supported by Ting Tsung and Wei Fong Chao Foundation, and NIH G08 LM008937.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–96. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 2.Van Meir EG, Hadjipanayis CG, Norden AD, Shu HK, Wen PY, Olson JJ. Exciting new advances in neuro-oncology: the avenue to a cure for malignant glioma. CA Cancer J Clin. 60(3):166–93. doi: 10.3322/caac.20069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Topkan E, Topuk S, Oymak E, Parlak C, Pehlivan B. Pseudoprogression in Patients With Glioblastoma Multiforme After Concurrent Radiotherapy and Temozolomide. Am J Clin Oncol. 2012 doi: 10.1097/COC.0b013e318210f54a. in press. [DOI] [PubMed] [Google Scholar]
- 4.Pope WB, Lai A, Nghiemphu P, Mischel P, Cloughesy TF. MRI in patients with high-grade gliomas treated with bevacizumab and chemotherapy. Neurology. 2006;66(8):1258–60. doi: 10.1212/01.wnl.0000208958.29600.87. [DOI] [PubMed] [Google Scholar]
- 5.Vredenburgh JJ, Desjardins A, Herndon JE, 2nd, et al. Phase II trial of bevacizumab and irinotecan in recurrent malignant glioma. Clin Cancer Res. 2007;13(4):1253–9. doi: 10.1158/1078-0432.CCR-06-2309. [DOI] [PubMed] [Google Scholar]
- 6.Vredenburgh JJ, Desjardins A, Herndon JE, 2nd, et al. Bevacizumab plus irinotecan in recurrent glioblastoma multiforme. J Clin Oncol. 2007;25(30):4722–9. doi: 10.1200/JCO.2007.12.2440. [DOI] [PubMed] [Google Scholar]
- 7.Norden AD, Young GS, Setayesh K, et al. Bevacizumab for recurrent malignant gliomas: efficacy, toxicity, and patterns of recurrence. Neurology. 2008;70(10):779–87. doi: 10.1212/01.wnl.0000304121.57857.38. [DOI] [PubMed] [Google Scholar]
- 8.Macdonald DR, Cascino TL, Schold SC, Jr., Cairncross JG. Response criteria for phase II studies of supratentorial malignant glioma. J Clin Oncol. 1990;8(7):1277–80. doi: 10.1200/JCO.1990.8.7.1277. [DOI] [PubMed] [Google Scholar]
- 9.Wen PY, Macdonald DR, Reardon DA, et al. Updated response assessment criteria for high-grade gliomas: response assessment in neuro-oncology working group. J Clin Oncol. 28(11):1963–72. doi: 10.1200/JCO.2009.26.3541. [DOI] [PubMed] [Google Scholar]
- 10.Galanis E, Buckner JC, Maurer MJ, et al. Validation of neuroradiologic response assessment in gliomas: measurement by RECIST, two-dimensional, computer-assisted tumor area, and computer-assisted tumor volume methods. Neuro Oncol. 2006;8(2):156–65. doi: 10.1215/15228517-2005-005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sorensen AG, Patel S, Harmath C, et al. Comparison of diameter and perimeter methods for tumor volume calculation. J Clin Oncol. 2001;19(2):551–7. doi: 10.1200/JCO.2001.19.2.551. [DOI] [PubMed] [Google Scholar]
- 12.Gladwish A, Koh E, Hoisak J, et al. Evaluation of early imaging response criteria in glioblastoma multiform. Radiation Oncology. 2011;6:1–7. doi: 10.1186/1748-717X-6-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellingson BM, Cloughesy TF, Lai A, Nghiemphu PL, Mischel PS, Pope WB. Quantitative volumetric analysis of conventional MRI response in recurrent glioblastoma treated with bevacizumab. Neuro Oncol. 13(4):401–9. doi: 10.1093/neuonc/noq206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corso JJ, E Sharon, S Dube, El-Saden S, Sinha U, Yuille A. Efficient multilevel brain tumor segmentation with integrated bayesian model classification. IEEE transactions on medical imaging. 2008;27(5):629–40. doi: 10.1109/TMI.2007.912817. [DOI] [PubMed] [Google Scholar]
- 15.Vaidyanathan M, Clarke LP, Hall LO, et al. Monitoring brain tumor response to therapy using MRI segmentation. Magnetic resonance imaging. 1997;15(3):323–34. doi: 10.1016/s0730-725x(96)00386-4. [DOI] [PubMed] [Google Scholar]
- 16.Vaidyanathan M, Clarke LP, Velthuizen RP, et al. Comparison of supervised MRI segmentation methods for tumor volume determination during therapy. Magnetic resonance imaging. 1995;13(5):719–28. doi: 10.1016/0730-725x(95)00012-6. [DOI] [PubMed] [Google Scholar]
- 17.Fletcher-Heath LM, Hall LO, Goldgof DB, Murtagh FR. Automatic segmentation of non-enhancing brain tumors in magnetic resonance images. Artificial intelligence in medicine. 2001;21(1-3):43–63. doi: 10.1016/s0933-3657(00)00073-7. [DOI] [PubMed] [Google Scholar]
- 18.Phillips WE, 2nd, Velthuizen RP, Phuphanich S, Hall LO, Clarke LP, Silbiger ML. Application of fuzzy c-means segmentation technique for tissue differentiation in MR images of a hemorrhagic glioblastoma multiforme. Magnetic resonance imaging. 1995;13(2):277–90. doi: 10.1016/0730-725x(94)00093-i. [DOI] [PubMed] [Google Scholar]
- 19.Moonis G, Liu J, Udupa JK, Hackney DB. Estimation of tumor volume with fuzzy-connectedness segmentation of MR images. AJNR Am J Neuroradiol. 2002;23(3):356–63. [PMC free article] [PubMed] [Google Scholar]
- 20.Emblem KE, Nedregaard B, Hald JK, Nome T, Due-Tonnessen P, Bjornerud A. Automatic glioma characterization from dynamic susceptibility contrast imaging: brain tumor segmentation using knowledge-based fuzzy clustering. J Magn Reson Imaging. 2009;30(1):1–10. doi: 10.1002/jmri.21815. [DOI] [PubMed] [Google Scholar]
- 21.Clark MC, Hall LO, Goldgof DB, Velthuizen R, Murtagh FR, Silbiger MS. Automatic tumor segmentation using knowledge-based techniques. IEEE transactions on medical imaging. 1998;17(2):187–201. doi: 10.1109/42.700731. [DOI] [PubMed] [Google Scholar]
- 22.Prastawa M, Bullitt E, Ho S, Gerig G. A brain tumor segmentation framework based on outlier detection. Medical image analysis. 2004;8(3):275–83. doi: 10.1016/j.media.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 23.Prastawa M, Bullitt E, Moon N, Van Leemput K, Gerig G. Automatic brain tumor segmentation by subject specific modification of atlas priors. Academic radiology. 2003;10(12):1341–8. doi: 10.1016/s1076-6332(03)00506-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaus MR, Warfield SK, Nabavi A, Black PM, Jolesz FA, Kikinis R. Automated segmentation of MR images of brain tumors. Radiology. 2001;218(2):586–91. doi: 10.1148/radiology.218.2.r01fe44586. [DOI] [PubMed] [Google Scholar]
- 25.Warfield SK, Kaus M, Jolesz FA, Kikinis R. Adaptive, template moderated, spatially varying statistical classification. Medical image analysis. 2000;4(1):43–55. doi: 10.1016/s1361-8415(00)00003-7. [DOI] [PubMed] [Google Scholar]
- 26.Kass M, Witkin A, Terzopoulos D. Snakes: Active Contour Models. International Journal of Computer Vision. 1988:321–31. [Google Scholar]
- 27.Rivest-Henault D, Cheriet M. Unsupervised MRI segmentation of brain tissues using a local linear model and level set. Magnetic resonance imaging. 2011;29(2):243–59. doi: 10.1016/j.mri.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 28.Wang L, Chen Y, Pan X, Hong X, Xia D. Level set segmentation of brain magnetic resonance images based on local Gaussian distribution fitting energy. Journal of neuroscience methods. 188(2):316–25. doi: 10.1016/j.jneumeth.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 29.Chen Y, Zhang J, Macione J. An improved level set method for brain MR images segmentation and bias correction. Comput Med Imaging Graph. 2009;33(7):510–9. doi: 10.1016/j.compmedimag.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 30.Hu S, Collins DL. Joint level-set shape modeling and appearance modeling for brain structure segmentation. NeuroImage. 2007;36(3):672–83. doi: 10.1016/j.neuroimage.2006.12.048. [DOI] [PubMed] [Google Scholar]
- 31.Cheng L, Yang J, Fan X, Zhu Y. A generalized level set formulation of the Mumford-Shah functional for brain MR image segmentation. Inf Process Med Imaging. 2005;19:418–30. doi: 10.1007/11505730_35. [DOI] [PubMed] [Google Scholar]
- 32.Vese LA, Chan TF. A multiphase level set framework for image segmentation using the Mumford and Shah model. International Journal of Computer Vision. 2002;50(3):271–93. [Google Scholar]
- 33.Cheng LS, Fan X, Yang J, Zhu Y. A generalized level set formulation of the Mumford-Shah functional with shape prior for medical image segmentation. Computer Vision for Biomedical Image Applications, Proceedings. 2005;3765:61–71. doi: 10.1007/11505730_35. [DOI] [PubMed] [Google Scholar]
- 34.Dydenko I, Jamal F, Bernard O, D’hooge J, Magnin IE, Friboulet D. A level set framework with a shape and motion prior for segmentation and region tracking in echocardiography. Medical image analysis. 2006;10(2):162–77. doi: 10.1016/j.media.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 35.Rousson M, Cremers D. Efficient kernel density estimation of shape and intensity priors for level set segmentation. Med Image Comput Comput Assist Interv. 2005;8(Pt 2):757–64. doi: 10.1007/11566489_93. [DOI] [PubMed] [Google Scholar]
- 36.Smith SM, Jenkinson M, Woolrich MW, et al. Advances in functional and structural MR image analysis and implementation as FSL. NeuroImage. 2004;23(Suppl 1):S208–19. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- 37.Woolrich MW, Jbabdi S, Patenaude B, et al. Bayesian analysis of neuroimaging data in FSL. NeuroImage. 2009;45(1 Suppl):S173–86. doi: 10.1016/j.neuroimage.2008.10.055. [DOI] [PubMed] [Google Scholar]
- 38.Yushkevich PA, Piven J, Hazlett HC, et al. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. NeuroImage. 2006;31(3):1116–28. doi: 10.1016/j.neuroimage.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE transactions on medical imaging. 2001;20(1):45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]
- 40.Xue X, Xue Z, Cao F, et al. PICE: Prior Information Constrained Evolution for 3-D and 4-D Brain Tumor Segmentation. IEEE International Symposium on Biomedical Imaging. 2010:840–43. [Google Scholar]