Abstract
Admixture is of great relevance to the clinical application of pharmacogenetics and personalized medicine. Preliminary findings in Puerto Ricans further substantiate the argument for admixture as a critical covariate in a customized DNA-guided warfarin dosing algorithm. To this purpose, a genome-wide approach that incorporates admixture as an independent predictor of dose variability in DNA-guided algorithms has been postulated. Admixture is expected to be able to reveal some relevant associations in the genetic epidemiology of Hispanics and will be indispensable to assure that pharmacogenomic research can be pursued in such mixed populations. Consequently, the clinical utility of knowing an individual’s genotype before initiating drug treatment in Puerto Ricans, and Hispanics in general, will finally be untangled by developing a “Genetic Prescription Model” that takes admixture into consideration. This approach will help lead physicians and patients to their desired treatment goal, resulting in more effective healthcare in admixed people.
Keywords: Admixture, Personalized medicine, Puerto ricans, Pharmacogenetics, Warfarin
Introduction
A major advance in healthcare would be a transition from the current empirical approaches in drug therapy to a genetically predictive framework for determining the individual patient’s response to medicines. The richer mixture of genetic variants in admixed populations, including that observed in Puerto Ricans, is likely to contribute substantially to wider variability in response to drug therapies within these populations, a component that will be missed by traditional studies in more homogeneous populations. This addressable oversight is of great concern, since it will tend to exacerbate the healthcare disparity already experienced by Hispanic populations in the USA. Accordingly, an understanding of how human genetic diversity and admixture in Hispanics is structured is not only of anthropological importance, but also of medical relevance.
At the writing of this commentary, PubMed database lists only 20 entries, including letter to editors and reviews, for a query combining the terms “pharmacogenetics/genomics and admixture”. Lerman [1] argued that explanations for the lack of consistency in the results of genetic association studies for smoking cessation treatments include biases in ethnic admixture [1]. Corvol and co-workers of the GALA and SAGE projects found an interleukin 6 (IL6) and interleukin 6 receptor (IL6R) pharmacogenetic gene-gene interaction for the bronchodilator responsiveness to albuterol that was significantly modified by the Native American ancestry of individuals [2]. To control for possible marginal effects of ancestry on drug response, authors included genetic ancestry as an independent variable in the regression model used to test association between individual’s IL6 and IL6R SNPs and bronchodilator effect. Interestingly, the mean bronchodilator response for the pharmacogenetic interaction increased with increasing amounts of Native American ancestry; whereas, the drug response among asthmatic patients for the same pharmacogenetic interaction decreased with increasing amounts of European ancestry [2]. Differences observed among Mexican, Puerto Ricans and African-Americans may be explained by the different proportions of Native American and European ancestries in these three ethnically diverse populations. Polymorphisms of IL6 and IL6R occurring together resulted in lower drug response in African-Americans given their almost null Native American ancestry, but resulted in higher drug response in Latinos (better in Mexican than Puerto Ricans, which is likely a consequence of a relatively higher contribution of Native American ancestry to Mexicans and, conversely, a relatively higher European proportions in Puerto Ricans).
Suarez-Kurtz and co-workers [3–5] have recently shown that the distribution of polymorphisms in various genes of pharmacological relevance, such as ABCB1, CYP3A5, GNB3, and GSTM1, among the population of Rio de Janeiro in Brazil, is best fit by a continuous function of the proportion of individual African ancestry, irrespective of self-reported race/color categories. It is reasonable to anticipate that the continuum in genetic ancestry within admixed populations like Hispanics will be reflected in the frequency distribution of pharmacogenetic polymorphisms. Perini et al. [6] observed that the required weekly dose of warfarin to achieve the target INR varied over tenfold within each race group in Brazilian patients [6].
Bryc et al. [7] found evidence of a significant sex bias in admixture proportions of Hispanics that is consistent with disproportionate contribution of European male and Amerindian or African female ancestry to present populations [7]. These authors also suggested that future genome-wide association studies in Hispanics will require correction for local genomic ancestry at a sub-continental scale [7].
Taken altogether, these still limited numbers of reports are suggesting that populations with varying levels of admixture can be particularly useful for pharmacogenetics/genomics and genetic-epidemiologic studies. For example, the self-reported degree of white admixture was found strongly correlated with protection from type 2 diabetes mellitus in the Pima Indian population [8]; differences in relative European ancestry proportions correlate with higher susceptibility in Puerto Ricans to asthma as compared with Mexicans [9]; and the increased risk of cardiovascular disease and hypertension in Puerto Ricans from Boston with greater African ancestry [10].
Backgrounds
Recently, the pharmacogenetic analysis of a significant physiogenomic (PG) data from Puerto Ricans has been accomplished to infer admixture structure and trichotomous ancestry in such a population by using an array of 384 SNPs (Single Nucleotide Polymorphisms) in 222 cardio-metabolic and neuro-endocrine candidate genes coding for key pharmaceutical targets. The genotypes and allele frequencies are publically available as an online supplement to the publication of this study [11].
The Puerto Rican population represents different admixtures of 3 major ethno-geographic groups (i.e., Amerindians, Europeans and West-Africans). Each subject in the study population was a ‘genetic mosaic’, with contributions from each of these three clusters, but in widely different proportions. Results also indicate that the admixture in the Puerto Rican population exists in the form of a continuous gradient with varying levels of admixture [11]. Such admixture results in a rich repertoire of combinatorial genotypes for key pharmacological, biochemical and physiological pathways, rendering Puerto Ricans a better resource to develop DNA-guided systems for clinical management of cardio-metabolic diseases (e.g., diabetes), urgent medical needs not only in Hispanic Puerto Ricans but also in the population at large. Accordingly, within the broad personalization paradigm, we have an enormous opportunity to turn our genetic variability into a clinical asset [12].
Figure 1 depicts the structure triangle derived from a Bayesian clustering analysis of a sample (n=100) representative of the Puerto Rican population, using the PG-array and the STRUCTURE v2.2 software package [13–14] in order to describe different levels of substructure in the data. This program takes genotype data from individuals without considering their origin and infers populations to which the individuals are assigned in such a way that linkage disequilibrium and Hardy-Weinberg disequilibrium are minimized within each inferred population. Each analyzed individual can belong to one or several inferred populations according to its “coefficients of ancestry”. This analysis is to determine how many ancestral groups contribute to the study population. However, it is important to mention that the assignment of the individuals in different inferred populations cannot be easily interpreted in terms of population history because STRUCTURE does not give any indication of the relevance of these units nor of their differentiation with regard to the other inferred populations.
Figure 1.
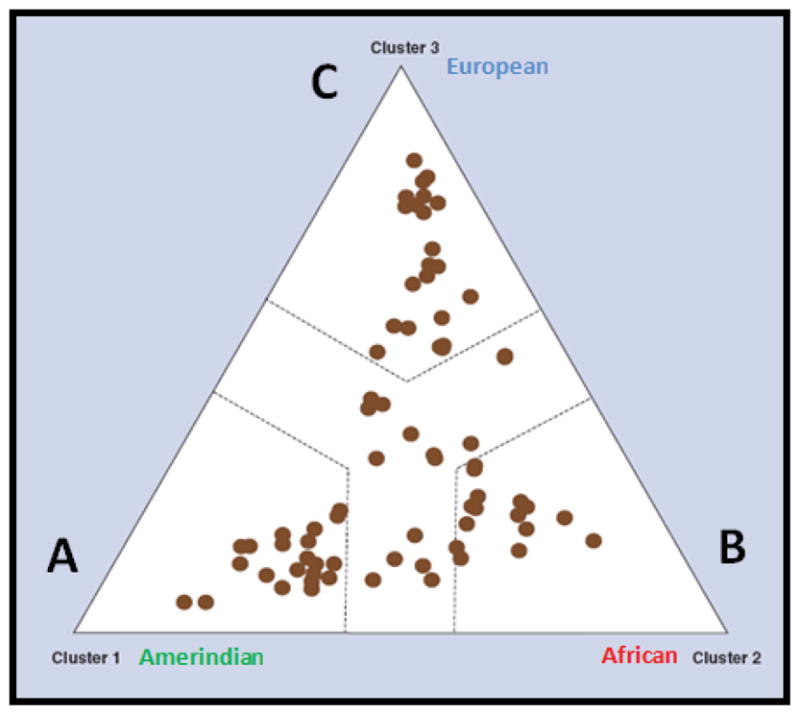
Population stratification of the Puerto Rican sample as depicted by the Structure triangle plot The three main, historical ancestral groups (i.e., Amerindians, Africans and Europeans) that contributed to the genetic background of the Puerto Rican population are represented. Adapted from Pharmacogenomics 2009; 10(4): 565–577 [8], with permission of Future Medicine Ltd.
Each individual was represented by a dot in the Figure 1. We performed a series of 3 Markov chain Monte Carlo simulation runs, ranging from K=1 to 5 with 80,000 iterations, as optimization algorithm for choosing a cluster size. Log-likelihood for each run was calculated as an a posteriori probability. Overall, results pointed toward a good fit for three clusters. Assuming K=3, proximity value (between 0 and 1) corresponding to each of 3 vertices of the triangle were calculated to each dot in the triangle. Closer proximity to any vertex resulted in proportionally higher value. We then assigned each individual to one of the three clusters by determining which of the three proximity values was greatest. A categorical criterion of proximity was added requiring that in order for an individual to be assigned to a specific cluster, its higher proximity value must be greater than the two others by at least 0.2. Thus excluding individuals not clearly positioned towards any vertex but rather located near the middle of the triangle. We added these dashed lines to separate the regions out each other. The degree of admixture in the study sample was evident as no one plotted dot was clearly placed on any of the three vertices of the structure triangle but rather positioned toward the borderline of the categorical criterion of proximity or the middle of the triangle. Some individuals overlapped in their position in the triangle.
In contrast to the results observed from a reference population of Kentucky (results not showed), most individuals in the Puerto Rican sample were found to be highly admixed between the three inferred populations (i.e., A thru C, Figure 1). 30 Caucasians from the Kentucky population showed a practically null admixture of the inferred populations A and C, whereas 10 African-Americans from Kentucky exhibited a more moderate admixture from the inferred populations A and B. It is worth noting that the assignments of individuals to inferred populations were not stable when the number of inferred populations is changed. That is, some individuals drastically changed their coefficients of ancestry between inferred populations when the number of inferred populations was increased from two to three. This is because we used a model in which the allele frequencies in the inferred populations at each locus are correlated with each other. The choice of this model is based on the assumption that all human populations originated from a single ancestral source population quite recently.
The majority of genetic analyses concerning population structure have assigned individuals into a small number of clusters, corresponding to four or five major racial/ethnic categories, but such classification is not completely discontinuous given the continuous intermixing among groups. In particular, genetic admixture has been well documented in different populations and, today, it is well accepted that admixture represent genetic gradations (geographic clines) [15]. In the Americas, the proportions of admixture will vary according to geographical region with geographic gradients from north to southeast. Although all individuals show mixed ancestry composed of mainly three inferred population (tri-hybrid model), Mexican individuals are closer to Asians and, therefore, present higher amount of Native American ancestry, whereas Brazil are at the other extreme of the gradient, with Puerto Rican individuals in an intermediate position.
The first step in developing the genetic rules for personalized medicine is clinical research to “train the algorithms” supporting medical decision making [12]. Admixture introduces distinct levels of population sub-structure or stratum, with marked variations in individual ancestry among the members of a particular population or ethnic group, depending on the dynamics of the process. Accordingly, any extrapolations of clinically relevant pharmacogenetic data from non-admixed to admixed groups will be plagued with uncertainty as exemplified by Suarez-Kurtz [16]. The predictive power of previously published DNA-guided warfarin dosing algorithms for admixed populations like Puerto Ricans is inadequate. We have conducted an exploratory study to test association between combinatorial CYP2C9 and VKORC1 genotypes, INR measurements and warfarin dosing in 167 warfarin-treated Puerto Rican patients recruited from San Juan, Puerto Rico and Hartford, CT, and gauge the impact of these polymorphisms on warfarin dose using the published algorithm by the International Warfarin Pharmacogenetics Consortium (IWPC) [17]. The algorithm performed poorly when applied to our Puerto Rican patient cohort, with significant scatter, low correlation coefficient (r2=0.36), high mean bias (25%), and high absolute difference (AD) values of 7.9 mg/week, respectively.
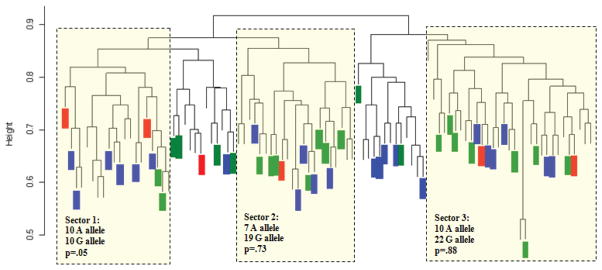
Recently, Villagra et al. [18] provided valuable evidence on the importance of controlling for admixture in pharmacogenetic studies of Puerto Rican Hispanics and stress the argument for incorporating admixture-matching in order to probe variations in warfarin response across different genotypes (i.e., stratification of the Puerto Rican population) [18]. Figure 2 depicts the increased prevalence of VKORC1-1639G>A allele in sector 1 of the dendrogram versus the rest of the population, which was consistent with previous assignment of sector 1 as primarily of Amerindian descent. The higher proportions of individuals carrying the VKORC1 haplotype A in sector 1 are requiring reduced warfarin doses in comparison to individuals assigned to the other two sectors, according to their respective ancestral coefficients. In this study we observed that inter-individual variations in ancestral contributions of Puerto Ricans will have significant implications for the way each individual responds to warfarin therapy [18]. Therefore, admixture may explain deviations from published algorithms. The results of admixture analysis can thus be utilized to parameterize population structure appropriately and account for it as a covariate in the corresponding association studies.
Figure 2.
Individual VKORC1(1639 G→A) genotypes, overlaid on the genetic distance dendrogram for a Puerto Rican population (where sectors 1, 2 and 3 correspond to Amerindian, Caucasian, and West African heritage, respectively). Green color represents G/G genotype; whereas, blue and red colors are for the G/A and A/A genotypes, respectively. P-values were calculated by a chi-squared test comparing observed allele frequencies with expected frequencies given the overall allelic ratios. The VKORC1 SNP 1639 G→A is in high linkage disequilibrium with haplotype A, which has been associated with a significant decrease in the warfarin dose per allele. Reprinted from Clin Chimica Acta 2010; 411:1306–11 [16], with permission of Elsevier Ltd.
It is also important to recognize that some minority groups in the U.S. (e.g., Hispanics) might be highly underrepresented in typical clinical pharmacogenetic trials, such as those conducted to derive DNA-guided dosing algorithms for warfarin, relative to the real impact of these groups on the current U.S. population. Thus, most of the previously published models might not be optimal for validating a globally derived algorithm. Because of the heterogeneity and extensive admixture of the Puerto Rican population, extrapolation on a global scale of data derived from well-defined ethnic groups (i.e., Caucasians) is clearly not applicable to the majority of Puerto Ricans.
Future Work
Available data suggest that allele frequency of polymorphisms in genes of pharmacological relevance may differ significantly among populations. However, even larger intra-ethnic variability is also evident, particularly in admixed groups [16]. As pharmacogenomic-guided multi-gene models are developed to predict drug response, the range of possible allelic combinations in the Puerto Rican population is certain to exceed that in populations without such degree of admixture. When treating individual Puerto Ricans, the focus must be shifted from population to the inherent genetic individuality that result from unique mosaics of variable combinatorial genotypes, a notion particularly important for admixed populations, in which stratification and substructure created by inter-ethnic gene flow further increases the fluidity of ethnic labels.
Due to a current lack of sufficient data in this admixed population, we believe that the clinical utility of knowing an individual’s genotype before initiating warfarin treatment in Puerto Ricans is still an open question. On the other hand, known or cryptic population stratification created by admixture can have a strong confounding effect on genetic association analysis of clinical drug responses. It further substantiates the argument for including admixture as a critical covariate in future customized warfarin dosing algorithm. That is, rather than allocate the subject to a single stratum in the analysis, it is desirable to construct a new covariate for each stratum, giving the corresponding ancestral proportion derived from physiogenomic-driven STRUCTURE-based clustering analysis and then include these covariates as adjustment variables in multistep regression models to develop a DNA-guided algorithm for warfarin dosing.
Another potential use of admixture analysis in the context of pharmacogenomic studies is to gather valuable information on groups that are usually either excluded or underrepresented in clinical drug trials (i.e., Hispanics). Moreover, gene flow between two or more genetically distinct populations result in admixture linkage disequilibrium (ALD) stretches among all loci (linked and unlinked) that have different allele frequencies in the founding populations, in a longer proportion than isolated populations, which enable fewer markers to be used in genetic association studies [19].
Admixture analysis can also be used to reflect relevant environmental and socioeconomic differences in the Puerto Rican population across the island. Although usually classified as a single ethnic group by researchers, Hispanics are heterogeneous from cultural, socioeconomic, and genetic perspectives. We envisage that the admixture analysis will illustrate the unique opportunity Puerto Ricans offer for studying the interaction between racial, genetic, socioeconomic and environmental contributions to drug response. Strikingly, Choudhry et al. [20] using 44 AIMs found that among Puerto Ricans, ancestry was associated with socioeconomic status. That is, Puerto Ricans reporting “upper” socioeconomic status had 9.1% lower African ancestry and 9.2% higher European ancestry than individuals reporting “moderate” and “middle” socioeconomic status [20]. In addition, correlations may be apparent with socioeconomic history, for example between African ancestry and location of sugar mills, plantations and slave ports. Using this admixture analysis, the history of migration and mixture in the Island of Puerto Rico can be reconstructed. Thus, future research among Puerto Rican and other Hispanic admixed populations should complement clinical genetic data with measures of ancestry, environmental and socioeconomic status.
A long-term goal is to extend the analysis of admixture in Puerto Ricans to genome wide (GW) arrays encompassing polymorphisms in all genes with 1.2 million SNP genotyping for fine resolution mapping of admixture. Admixture studies at this resolution afford delineation of candidate genes for pharmacogenetics not only for cardiovascular disease and diabetes, but also for other medical conditions of higher prevalence or incidence in Hispanics. The extremely high resolution of GW population analysis is able to resolve regional gradients in genetic dissimilarity. In a study of Europeans, it was possible to predict from GW genotype data the geographical point of origin of individuals to within a few hundred kilometers [21]. We will perform a similar analysis to examine whether there is a correlation between genetic and geographic distances, identify possible genetic gradients that will be useful in association studies and determine geographic stratification in the Island of Puerto Rico.
Conclusions
The population of the Americas carries a genomic legacy resulting from the European colonization and African slavery. In those parts of the Americas where admixture took hold, the populations manifest the combined anthropological heritage in their genomes, lifestyle, diet, and even socioeconomic status. Hispanics in the USA stand to gain the most from the revolution in personalized medicine, given longstanding disparities in healthcare afflicting them. At the same time, admixture analysis could become indispensable for the globalization of pharmacogenetic research beyond the Americas, to Africa, Asia and Europe, the continents whose populations contributed to the admixture in the first place.
Rather than ignoring admixture, pharmacogeneticists should consider it as starting point for better understanding the underlying basis of the observed wider variability in drug responses among patients of mixed populations. Such understanding provides the opportunity to develop strategies for leapfrogging the healthcare standards of these populations.
Acknowledgments
We acknowledge the support of National Institute of Health, National Center for Research Resources RCMI Award G12RR-03051; the Puerto Rican Newborn Screening Program and Genomas internal research and development funds.
References
- 1.Lerman C, Niaura R. Applying genetic approaches to the treatment of nicotine dependence. Oncogene. 2002;21:7412–7420. doi: 10.1038/sj.onc.1205801. [DOI] [PubMed] [Google Scholar]
- 2.Corvol H, De Giacomo A, Eng C, Seibold M, Ziv E, et al. Genetic ancestry modifies pharmacogenetic gene-gene interaction for asthma. Pharmacogenet Genomics. 2009;19:489–496. doi: 10.1097/FPC.0b013e32832c440e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suarez-Kurtz G, Vargens DD, Struchiner CJ, Bastos-Rodrigues L, Pena SD. Self-reported skin color, genomic ancestry and the distribution of GST polymorphisms. Pharmacogenet Genomics. 2007;17:765–771. doi: 10.1097/FPC.0b013e3281c10e52. [DOI] [PubMed] [Google Scholar]
- 4.Suarez-Kurtz G, Perini JA, Bastos-Rodrigues L, Pena SD, Struchiner C. Impact of population admixture on the distribution of the CYP3A5*3 polymorphism. Pharmacogenomics. 2007;8:1299–1306. doi: 10.2217/14622416.8.10.1299. [DOI] [PubMed] [Google Scholar]
- 5.Estrela RC, Ribeiro FS, Carvalho RS, Gregório SP, Dias-Neto E, et al. Distribution of ABCB1 polymorphisms among Brazilians: impact of population admixture. Pharmacogenomics. 2008;9:267–276. doi: 10.2217/14622416.9.3.267. [DOI] [PubMed] [Google Scholar]
- 6.Perini JA, Struchiner CJ, Silva-Assuncao E, Santana IS, Rangel F, et al. Pharmacogenetics of warfarin: development of a dosing algorithm for Brazilian patients. Clin Pharmacol Ther. 2008;84:722–728. doi: 10.1038/clpt.2008.166. [DOI] [PubMed] [Google Scholar]
- 7.Bryc K, Velez C, Karafet T, Moreno-Estrada A, Reynolds A, et al. Colloquium paper: genome-wide patterns of population structure and admixture among Hispanic/Latino populations. Proc Natl Acad Sci U S A. 2010;107:8954–8961. doi: 10.1073/pnas.0914618107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Williams RC, Long JC, Hanson RL, Sievers ML, Knowler WC. Individual estimates of European genetic admixture associated with lower body-mass index, plasma glucose, and prevalence of type 2 diabetes in Pima Indians. Am J Hum Genet. 2000;66:527–538. doi: 10.1086/302773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salari K, Choudhry S, Tang H, Naqvi M, Lind D, et al. Genetic admixture and asthma-related phenotypes in Mexican American and Puerto Rican asthmatics. Genet Epidemiol. 2005;29:76–86. doi: 10.1002/gepi.20079. [DOI] [PubMed] [Google Scholar]
- 10.Lai CQ, Tucker KL, Choudhry S, Parnell LD, Mattei J, et al. Population admixture associated with disease prevalence in the Boston Puerto Rican health study. Hum Genet. 2009;125:199–209. doi: 10.1007/s00439-008-0612-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruaño G, Duconge J, Windemuth A, Cadilla CL, Kocherla M, et al. Physiogenomic Analysis of the Puerto Rican Population. Pharmacogenomics. 2009;10:565–577. doi: 10.2217/pgs.09.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruaño G. Bringing DNA-Guided Medicine to the Hispanic Population. P R Health Sci J. 2009;28:266–267. [PubMed] [Google Scholar]
- 13.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.The University of Chicago Department of Biological Sciences, Pritchard Lab. http://pritch.bsd.uchicago.edu.
- 15.Serre D, Pääbo S. Evidence for Gradients of Human Genetic Diversity Within and Among Continents. Genome Res. 2004;14:1679–1685. doi: 10.1101/gr.2529604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suarez-Kurtz G. Pharmacogenomics in admixed populations. Trends Pharmacol Sci. 2005;26:196–201. doi: 10.1016/j.tips.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 17.Klein TE, Altman RB, Eriksson N, Gage BF, Kimmel SE, et al. Estimation of the warfarin dose with clinical and pharmacogenetic data. N Engl J Med. 2009;360:753–764. doi: 10.1056/NEJMoa0809329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Villagra D, Duconge J, Windemuth A, Cadilla CL, Kocherla M, et al. CYP2C9 and VKORC1 genotypes in Puerto Ricans: A case for admixture-matching in clinical pharmacogenetic studies. Clin Chim Acta. 2010;411:1306–1311. doi: 10.1016/j.cca.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pfaff CL, Parra EJ, Bonilla C, Hiester K, McKeigue PM, et al. Population structure in admixed populations: effect of admixture dynamics on the pattern of linkage disequilibrium. Am J Hum Genet. 2001;68:198–207. doi: 10.1086/316935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choudhry S, Coyle NE, Tang H, Salari K, Lind D, et al. Population stratification confounds genetic association studies among Latinos. Hum Genet. 2006;118:652–664. doi: 10.1007/s00439-005-0071-3. [DOI] [PubMed] [Google Scholar]
- 21.Novembre J, Johnson T, Bryc K, Kutalik Z, Boyko AR, et al. Genes mirror geography within Europe. Nature. 2008;456:98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]