Abstract
In this issue Orans et al. (2011) and Tsutakawa et al. (2011) report exciting insights into the molecular principles governing diverse endo- and exonucleolytic cleavage specificities of members of the RAD2/FEN superfamily of nucleases, which have critical roles in DNA replication and maintenance.
The nucleases that cleave the phosphodiester backbones of DNA and RNA have many cellular functions, including RNA processing and maturation, RNA interference, apoptosis, cellular pathogen defense, and genome replication and maintenance. To carry out these functions, nucleases with a wide array of protein folds have evolved diverse mechanisms for recognizing and cleaving nucleic acids of specific sequence (e.g. restriction endonucleases), length (e.g. Dicer cleavage of RNA) or structure (eg., Mre11 nuclease processing of DNA double strand breaks) (Yang)Williams et al, 2008). The structure-specific nucleases include a conserved superfamily of endo- and exonucleases whose eukaryotic members comprise FEN1, EXO1, GEN1, and XPG (Figure 1A). These nucleases have critical functions, as revealed by the fact that mutations in the genes encoding them result in cellular stress and genome instability, and for FEN1 and XPG, increased cancer susceptibility (DePamphilis, 2006; Zheng et al.). In order to avoid cell death and/or genome instability, these nucleases cooperate with other proteins in multistep transactions in order to cleave their substrates precisely and only when needed. This raises the question of how they find their cognate substrates, how their substrate cleavage specificities are determined and how they are regulated. Although important insights on these issues have come from earlier structural studies (for instance see (Chapados et al., 2004; Hosfield et al., 1998), the underlying mechanisms of FEN-family substrate binding and specificity have remained puzzling because structures of the enzymes bound to their cognate substrates have been lacking. Now, elegant structure-function studies of human FEN1 (Tsutakawa et al., 2011) and EXO1 (Orans et al., 2011) reveal a common mechanism of DNA binding and cleavage, and provide new insights into how substrate specificities are determined and hints as to how the activities of these nucleases may be regulated.
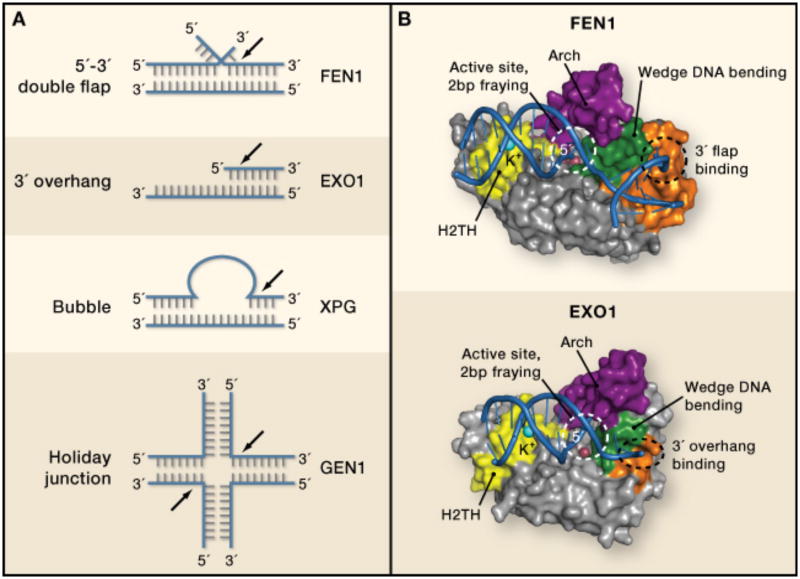
Figure 1. Molecular basis for FEN family nuclease specificities.
(A) Structures of preferred substrates for FEN family nucleases. As members of the same superfamily,FEN1, EXO1, GEN1 and XPG are homologous, and they all cleave DNA on the 5′ end of a substrate(black arrows). However, the structures on the 5′ and 3′ sides of the cleavage sites vary drastically, suchthat substrates can contain 5′ flaps, 3′ flaps, bubbles, gaps, and Holliday junctions. GEN1 (gapendonuclease 1) participates in resolving Holliday junctions generated during recombination, and XPG(Xeroderma pigmentosum complementation group G protein) cleaves bubble substrates arising duringnucleotide excision repair of DNA lesions.
(B) Structural comparison of FEN1/DNA and EXO1 DNA complexes. Purple: Arch region, also calledthe microdomain in EXO1 or helical gateway in FEN1. Yellow: H2TH DNA binding motif. Green:helical wedge. Orange: specificity binding pocket for double stranded DNA bearing either a 3′ flap(FEN1) or a 3′ overhang (EXO1).
The two enzymes have distinct biological functions. FEN1 (flap endonuclease 1) has primary responsibility for removing the estimated 50 million short DNA flaps created during replication of mammalian nuclear genomes. This already huge workload is further increased by the need to remove flaps produced during base excision repair of DNA lesions. EXO1 (exonuclease 1) has several important but different functions, including exonucleolytic digestion of DNA during repair of potentially lethal double strand DNA breaks and during repair of replication errors. Despite these differences, the two nucleases use a surprisingly common strategy to bind and cleave DNA. Duplex DNA is bound via a H2TH (helix-two turn-helix) potassium-binding fold (yellow in Figure 1B) on one side of the active site and by additional interactions (orange) on the other side. Using helical wedges (green) to dramatically splay apart the DNA duplex, both enzymes physically segregate the 5′ and 3′ flanking DNA structures. These interactions result in a marked 90-100° DNA bend proximal to the cleavage site, and DNA 5′ end fraying of two nucleotides. This fraying provides access to the scissile phosphodiester bond, which is cleaved by a common two metal ion catalytic mechanism (red balls, Figure 1B).
In addition to these common features, differences in substrate specificity are partly rationalized by surface pockets unique to each enzyme that interrogate the surrounding nucleic acid environment (orange surfaces, Figure 1B). For EXO1, which prefers 3′ overhangs and gapped structures, the 3′ overhand is bound via sequence non- specific contacts to a positively charged groove adjacent to the active site. By comparison, FEN1's defining specificity for processing Okazaki fragments bearing equilibrating 5′ and 3′ flaps is dictated by a 3′ flap-binding pocket that cradles the unpaired 3′ end and is formed by added helical and loop elements that are not found in Exo1.
Both EXO1 and FEN1 harbor 5′ flap endonuclease activity, and in both structures the 5′ end is directed into an active site covered by a helical “arch” structure (purple). Determinants for binding and substrate engagement on the 5′ end, in particular 5′ flap binding for FEN1 cleavage, has been a matter of debate. Two mechanisms of flap engagement have been proposed. One involves tracking, in which the enzymes slide down single stranded DNA (ssDNA) to engage the ssDNA/double stranded DNA (dsDNA) junction at a flap. The other involves threading, where the 5′ flap is pushed through a hole in the protein. Based on structural observations indicating that the arch helicies can unfold, and in support of a threading mechanism, Tainer and colleagues (Tsutakawa et al., 2011) suggest the arch collapses with a disorder-to-order transition upon engagement of the 5′ end during a binding and threading event. Orans et al (Orans et al., 2011) suggest a third alternative involving conformational clamping by the arch helicies. The clamping model is attractive for circumstances in which free DNA 5′ ends may not be present, such as in bubbles and Holiday junctions. Threading the arch in FEN1, and the presence of its 3′ flap-binding pocket (Figure 1B), might reflect specific mechanistic elaborations adopted by FEN1 to convert equilibrating 5′ flaps to ligatable nicks during DNA lagging strand replication (Jin et al., 2001). Deciphering flap-binding mechanism(s) undoubtedly will involve capturing molecular snapshots of FEN-family members bound to additional DNA substrates, including intact, un-cleaved flaps.
Excision repair of DNA lesions requires multiple enzymatic steps, with the product of one enzyme serving as the substrate for the next. Because repair “intermediates” can potentially be more lethal or mutagenic than the original lesion, a prevailing hypothesis (Parikh et al., 1999; Wilson and Kunkel, 2000) is that coordinated “hand-offs” occur among repair proteins to avoid release of toxic intermediates. Nucleases can destroy DNA and so must be tightly regulated. How do cells guard against inappropriate catalysis? A great case-in point is EXO1, whose involvement i(Tumbale et al.)n excising replication errors requires interaction with the mismatch recognition complex MutSα (Genschel et al., 2002). Beese and colleagues (Orans et al., 2011) propose that the C-terminus of EXO1 is an auto-inhibitory domain that prevents EXO1 from excising new, correctly replicated DNA. Inhibition is relieved when MutSα detects a mismatch and travels along the DNA until it encounters EXO1 bound at a nick. When the two proteins interact, EXO1 activation may occur via a conformational change in the 2-helix “microdomain” (also known as the arch) in EXO1 that positions the scissile bonds over the metal center. Thus EXO1 is activated for mismatch repair only when a mismatch is detected (see Figure 6 in (Orans et al., 2011). Similarly, FEN1 binds many other proteins and is post-translationally modified by phosphorylation, acetylation, and methylation (Zheng et al.), potentially providing many opportunities to regulate FEN1 activities. Deciphering the molecular basis of these controls will be important for understanding the roles for FEN1 activity in multiple pathways, including telomere maintenance, apoptosis, replication fork rescue, and long patch base excision repair (Zheng et al.).
As with all seminal advances, these studies establish a testable platform for future studies to address a number of important questions. Do FENs thread the arch? Precisely how do XPG and GEN1 bind substrate? Do these mechanistic concepts extend to the FEN-related RNA metabolizing enzymes (XRN1 and XRN2) (Yang, 2011)? How do protein partnerships and covalent modifications regulate nuclease activity? In a broader sense, in our ever-expanding “-omics” world, these elegant studies are the end products of two technical triumphs, and underscore the time-tested value of focused, “low-throughput”, integrated structure-function approaches for understanding key cellular functions.
References
- Chapados BR, Hosfield DJ, Han S, Qiu J, Yelent B, Shen B, Tainer JA. Structural basis for FEN-1 substrate specificity and PCNA-mediated activation in DNA replication and repair. Cell. 2004;116:39–50. doi: 10.1016/s0092-8674(03)01036-5. [DOI] [PubMed] [Google Scholar]
- DePamphilis M. DNA Replication and Human Disease. Cold Spring Harbor Monograph. 2006;46 [Google Scholar]
- Genschel J, Bazemore LR, Modrich P. Human exonuclease I is required for 5′ and 3′ mismatch repair. J Biol Chem. 2002;277:13302–13311. doi: 10.1074/jbc.M111854200. [DOI] [PubMed] [Google Scholar]
- Hosfield DJ, Mol CD, Shen B, Tainer JA. Structure of the DNA repair and replication endonuclease and exonuclease FEN-1: coupling DNA and PCNA binding to FEN-1 activity. Cell. 1998;95:135–146. doi: 10.1016/s0092-8674(00)81789-4. [DOI] [PubMed] [Google Scholar]
- Jin YH, Obert R, Burgers PM, Kunkel TA, Resnick MA, Gordenin DA. The 3′-->5′ exonuclease of DNA polymerase delta can substitute for the 5′ flap endonuclease Rad27/Fen1 in processing Okazaki fragments and preventing genome instability. Proc Natl Acad Sci U S A. 2001;98:5122–5127. doi: 10.1073/pnas.091095198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orans J, McSweeney E, Iyer R, Hast M, Hellinga H, Modrich P, Beese L. Structures of human exonuclease I DNA complexes suggest a unified mechanism for nuclease family. Cell. 2011 doi: 10.1016/j.cell.2011.03.005. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh SS, Mol CD, Hosfield DJ, Tainer JA. Envisioning the molecular choreography of DNA base excision repair. Curr Opin Struct Biol. 1999;9:37–47. doi: 10.1016/s0959-440x(99)80006-2. [DOI] [PubMed] [Google Scholar]
- Tsutakawa S, Classen S, Chapados B, Arvai A, Finger LD, Guenther G, Tomlinson C, Thompson P, Sarkar A, Shen B, et al. Human flap endonuclease structures, DNA double base flipping and a unified understanding of the FEN1 superfamily. Cell. 2011 doi: 10.1016/j.cell.2011.03.004. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson SH, Kunkel TA. Passing the baton in base excision repair. Nat Struct Biol. 2000;7:176–178. doi: 10.1038/73260. [DOI] [PubMed] [Google Scholar]
- Yang W. Nucleases: diversity of structure function and mechanism. Q Rev Biophys. 44:1–93. doi: 10.1017/S0033583510000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Jia J, Finger LD, Guo Z, Zer C, Shen B. Functional regulation of FEN1 nuclease and its link to cancer. Nucleic Acids Res. 39:781–794. doi: 10.1093/nar/gkq884. [DOI] [PMC free article] [PubMed] [Google Scholar]