Summary
Retinoic acid inducible gene-I (RIG-I) is a key intracellular immune receptor for pathogenic RNAs, particularly from RNA viruses. Here we report the crystal structure of human RIG-I bound to a 5′tri-phosphorylated RNA hairpin and ADP nucleotide at 2.8Å resolution. The RNA ligand contains all structural features that are essential for optimal recognition by RIG-I, as it mimics the panhandle-like signatures within the genome of negative stranded RNA viruses. RIG-I adopts an intermediate, semi-closed conformation in this product state of ATP hydrolysis. The structure of this complex allows us to visualize the first steps in RIG-I recognition and activation upon viral infection.
Keywords: ATP hydrolysis, RNA virus, innate immunity, RNA helicase, X-ray crystallography
Introduction
Pathogen recognition receptors (PRRs) are signalling proteins that continually survey cells for the presence of pathogen associated molecular patterns (PAMPs). Retinoic acid inducible gene I (RIG-I) is a major cellular PRR that senses viral RNA PAMPs in the cytoplasm of infected cells (Kato et al., 2011; Yoneyama et al., 2004). RIG-I recognizes a broad spectrum of viruses, including the negative-stranded vesicular stomatitis virus, influenza, and rabies viruses; and also positive-stranded viruses such as dengue and hepatitis C virus (Kawai and Akira, 2007; Ramos and Gale, 2011). Defective viral replication by Sendai virus and influenza virus generates short sub-genomic RNAs that may be a principal ligand for RIG-I during viral infection (Baum and Garcia-Sastre, 2011; Baum et al., 2011). At the molecular level, RIG-I preferentially recognizes double stranded RNAs that contain a tri-phosphate moiety at the 5′end, exemplified by the panhandle-like RNAs of negative strand viruses such as influenza (Hornung et al., 2006; Pichlmair et al., 2006; Schlee et al., 2009). Recent biochemical and structural studies have shown that the C terminal domain (CTD) of RIG-I recognizes duplex termini, interacting specifically with terminal 5′-tri-phosphate moieties (Cui et al., 2008; Lu et al., 2010; Wang et al., 2010). The central SF2 helicase domain (HEL) binds internally to the dsRNA backbone (Jiang et al., 2011; Kowalinski et al., 2011; Luo et al., 2011). A novel pincer domain connects the CTD and the HEL domains and provides mechanical support for coordinated RNA recognition by the two domains (Luo et al., 2011). The N terminal tandem caspase activation and recruitment domains (CARDs) are responsible for downstream signaling, leading to the expression of anti-viral interferon-stimulated genes(Jiang and Chen, 2011; Ramos and Gale, 2011).
The current model of RIG-I activation suggests that the binding of RNA by the HEL and CTD generates a nano-mechanical force that releases an inhibitory conformation imposed by the CARD domains, a process that also requires ATPase activity through an unknown mechanism(Kowalinski et al., 2011; Luo et al., 2011). Identifying the molecular determinants for RNA recognition and understanding how RIG-I distinguishes viral RNA from cellular RNA represent important unanswered questions in the field of innate immunity. Here we report the crystal structure of RIG-I in complex with a 5′tri-phosphorylated double stranded RNA and adenosine nucleotide, thereby providing the first biologically-relevant snapshot of viral PAMP recognition by RIG-I. We show that binding of different ATP analogs induces specific conformational changes within the protein, verifying the structural observations and supporting a tightly-regulated, multi-step activation mechanism of RIG-I.
Results and Discussion
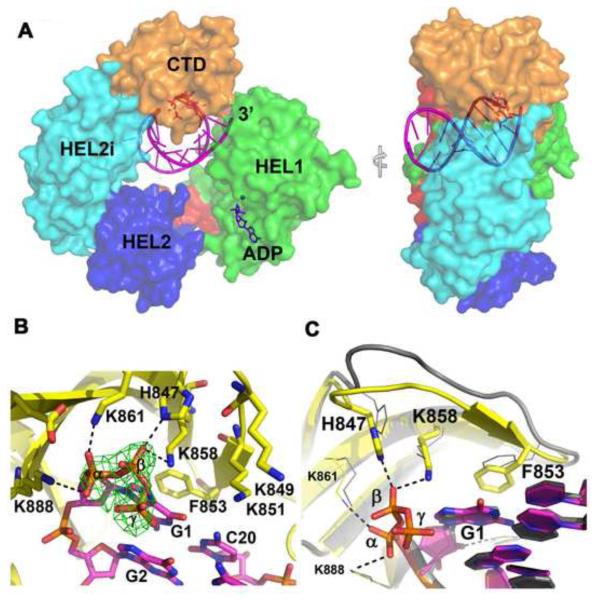
To unravel the molecular details of viral PAMP recognition by RIG-I, we designed a hairpin RNA (hereafter named as 5′ppp8L which contains a 5′ tri-phosphate moiety and a stem of 8 base pairs that is terminated by a UUCG tetra loop) that mimics the panhandle-like genome of negative-stranded RNA viruses. We co-crystallized 5′ppp8L with a human RIG-I construct that lacks the CARD domains (RIG-I (ΔCARDs:1-238), (Fig. 1)). All atoms of the RNA hairpin are observed and unambiguously built into the 2.8Å density map (Fig. 1 Tab. 1, and Fig. S1).
Figure 1. Ternary complex of RIG-I (ΔCARDs 1-238): 5′ppp8L: ADP-Mg2+.
(A) Structure of the 5′ tri-phosphorylated hairpin RNA (5′ppp8L, in purple with 5′ GTP in red) bound at the center of the RIG-I (ΔCARDs). Bound ADP-Mg2+ is in purple. (B) The 5′tri-phosphate binding site at CTD. Fo-Fc omit map is in green and contoured at 3.5 σ. (C) Superposition of RIG-I with 5′ tri-phosphorylated hairpin RNA and RIG-I with 5′ hydroxyl dsRNA in grey (PDB: 2ykg). See also Fig. S1.
Table 1.
Crystallographic statistics
| Data collection | |
| Structure | RIG-I (ΔCARDs 1-238): 5’ppp8L: ADP-Mg2+ |
| Space group | P212121 |
| Cell dimensions (Å) | 47.7, 76.2, 221.2 |
| Resolution (Å) | 47.7 - 2.8 (2.95 - 2.8)a |
| R merge (%) | 13.2 (61.4) |
| I / σ | 12.7 (3.7) |
| Completeness (%) | 98.5 (98.7) |
| Redundancy | 3.8 (3.9) |
| Refinement | |
| Resolution (Å) | 24.9 - 2.8 |
| R work / R free (%) | 21.8 / 28.6 |
| No. atoms | 5,542 |
| Macromolecules | 5,411 |
| Ligands | 61 |
| Water | 70 |
| B-factors (Å2) | 54.2 |
| Macromolecules | 54.4 |
| Solvent | 35.2 |
| Ramachandran analysis | |
| Favored | 93 |
| Additionally allowed | 6.2 |
| Not favored | 0.8 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.008 |
| Bond angles (°) | 1.15 |
Highest resolution shell is shown in parenthesis.
The overall structure of the complex (RIG-I (ΔCARDs: 1-238): 5′ppp8L: ADP-Mg2+) is similar to the RIG-I:dsRNA10 structure reported previously (RMSD = 0.38Å for 559 superimposed Ca atoms)(Luo et al., 2011). However, in the new structure, the CTD encapsulates the 5′ tri-phosphate moiety at the duplex terminus. Functional groups along the RNA duplex interact with the HEL1 and HEL2i domains as observed previously. Importantly, one can now observe the position of bound nucleotide, revealing that ADP interacts exclusively with conserved ATPase motifs localized in HEL1 (Fig. 1A). HEL2 is not involved in RNA binding or ADP binding (Fig. 1). The protein conformation observed in this structure is likely to be biologically relevant because we observe that 5′ppp8L RNA readily stimulates efficient ATP hydrolysis by RIG-I (Fig. S3 and Tab. S1).
The RNA tri-phosphate is specifically recognized by the RIG-I CTD, which forms a network of electrostatic and hydrophobic interactions (Fig. 1B and 1C). Specifically, the α-phosphate interacts with K861 and K888 and the β-phosphate interacts with H847 and K858. Intriguingly, the γ phosphate (for which there is strong electron density) does not form any direct contacts with the protein in this structure, suggesting that it is not a major recognition determinant. If the tri-phosphate moiety were to adopt a more extended configuration in an alternative conformational state, the γ phosphate would be likely to establish interactions with the K849 and K851 residues, as hypothesized in structural studies of the isolated CTD in complex with tri-phosphorylated RNA (Fig. S1B) (Lu et al., 2010; Wang et al., 2010). The structure of the intact complex (RIG-I (ΔCARDs: 1-238): 5′ppp8L: ADP-Mg2+) indicates that the α and β phosphates at the 5′-RNA terminus are particularly critical for RIG-I recognition. This may be due to the fact that RNA γ phosphates in the cell are often hydrolyzed by host and viral RNA tri-phosphatases (Decroly et al., 2012), perhaps necessitating that RIG-I evolve primary binding to a 5′-diphosphate. Interactions involving the β phosphate appear to be particularly important, as they have global consequences for the structure of the complex. Specifically, contacts with H847 and K858 rigidify the intervening loop and deliver it to the blunt end of the tri-phosphorylated RNA, enabling aromatic loop residue F853 to stack on the first base pair of the duplex and form energetically favorable π-π interactions (Fig. 1C). Mutations that disrupt this interdigitated network of contacts weaken tri-phosphorylated RNA binding by RIG-I (Fig. S4)(Wang et al., 2010). Together, they help RIG-I to select the correct pathogenic RNA from the vast pool of capped cellular RNAs.
Backbone atoms of the RNA duplex form an extensive set of interactions with the HEL2i domain, providing further insights into the mechanism of duplex recognition by RLR proteins. The shape-selective RNA interface explains why RIG-I is capable of binding to double-stranded RNAs from diverse viruses (Fig.1A and Fig. S2). Significantly, the UUCG tetraloop at the hairpin terminus is absorbed into an RNA binding tunnel and does not establish any base specific contacts with RIG-I. The structure demonstrates that a variety of RNA motifs, including mismatches and ordered loops, would be readily accommodated at the “far end” of the RIG-I RNA binding tunnel (i.e. the end opposite 5′ppp binding). This is likely to be particularly important for RIG-I detection of negative-sense viral genomic RNAs, including influenza, rabies, para-influenza, and respiratory syncytial virus, which also form short terminal duplexes capped by loops (Fig. S2).
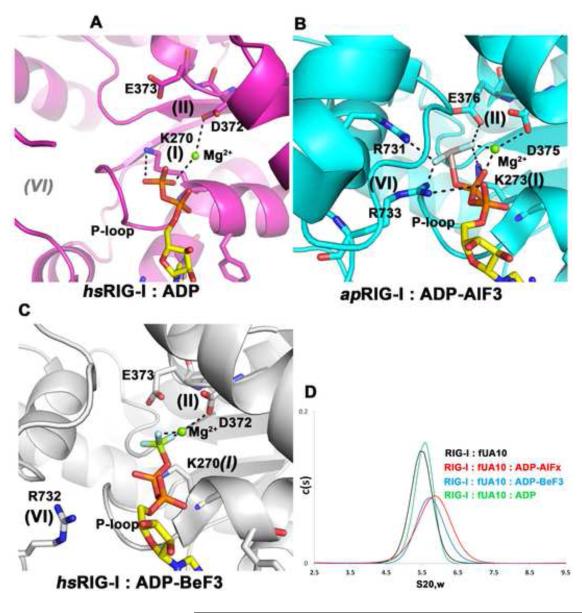
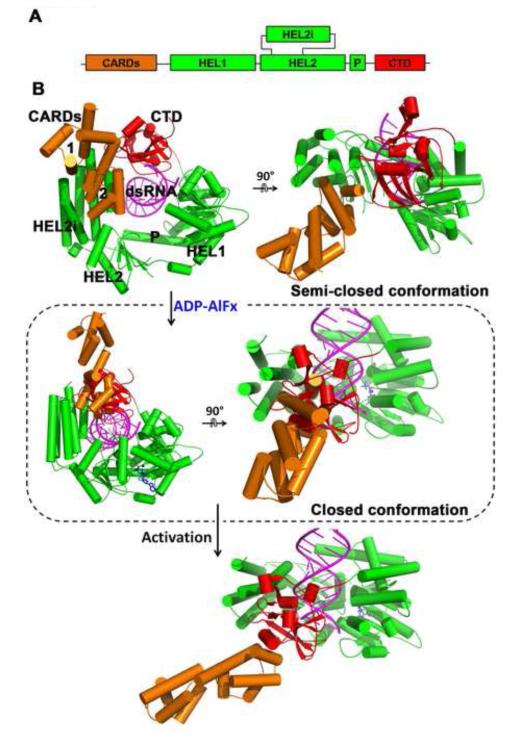
In addition to RNA recognition, the structure of the complex (which contains ADP-Mg2+) provides new insights into RIG-I recognition of bound nucleotide. The phosphates of ADP interact with K270 and T271 (motif I) and with D372 (motif II) through a bridging Mg2+ (Fig. 2A). The adenine nucleobase is recognized by Q247 (Q motif) and stacks between R244 and F241. A comparison of available RIG-I:nucleotide structures reveals that RIG-I (and perhaps related RLRs and DEAD-box proteins) has a distinctive strategy for binding and activating nucleotide ligands. Similar to DEAD box proteins, the helicase domain of RIG-I is in an open conformation in the absence of RNA substrate (Kowalinski et al., 2011; Luo et al., 2011; Pyle, 2008). In the presence of RNA and the ATP analog ADP-AlF3, the helicase domain adopts the closed conformation, bringing motifs I and VI into proximity 14. In complex with ADP-Mg2+, as observed here, RIG-I adopts an intermediate, semi-closed state that lacks contacts with motif VI from HEL2 (Fig. 2B). Interestingly, a similar semi-closed conformation was reported in the structure of RIG-I with RNA and ADP-BeF3 (Fig. 2C)(Jiang et al., 2011), which may represent a transient state prior to a completely closed ATP-bound state. Taken together, these structures show that a bona-fide closed conformation of the helicase core is only captured in the presence of both dsRNA and ADP-AlF3 and in the absence of CTD14, indicating that RIG-I conformation is exceptionally sensitive to ATP binding, hydrolysis and product release. Importantly, the process of ATP hydrolysis moves the CTD and HEL2i in opposite directions (Fig. 3 and Mov. S1), which likely allows the CARDs to be released from HEL2i (Kowalinski et al., 2011; Luo et al., 2011). This provides a striking example of the conversion of chemical energy into mechanical force and activation of a signaling relay.
Figure 2. ATP binding and hydrolysis by RIG-I.
(A) Interactions between human RIG-I and ADP-Mg2+. (B) Duck RIG-I with ADP-AlF3-Mg2+ (PDB: 4A36). (C) Human RIG-I with ADP-BeF3-Mg2+ (PDB: 3TMI). (D) Hydrodynamic analysis using sedimentation velocity. Shown are the calculated distribution c(s) versus s20,w of RIG-I:fUA10, RIG-I:fUA10:ADP-AlFx (red), RIG-I:fUA10 : ADP-BeF3 (blue), RIG-I:fUA10 : ADP (green). The peak values for the c(S) distributions are 5.49S, 5.87S, 5.71S, and 5.60S, which correspond to frictional coefficients of 1.58, 1.47, 1.51, and 1.54 respectively. See also Fig. S3 and Tab.S1.
Figure 3. Sequential activation of RIG-I by RNA and ATP.
(A) Schematic representation of RIG-I protein. (B) ADP-AlFx binding induced conformational changes of RIG-I. Conformational changes upon ADP-AlFx binding is modeled based on the following crystal structures: human RIG-I:dsRNA binary complex (PDB: 2ykg), duck RIG-I apo enzyme (PDB: 4a2w), and duck RIG-I:dsRNA:ADP-AlFx ternary complex (PDB:4a36)(Kowalinski et al., 2011; Luo et al., 2011). The binding of ADP-AlFx (blue) causes the helicase domain to close and moves the CTD (red) and HEL2i (green) in opposite directions. This directional movement probably allows the CARDs (orange) to be released from HEL2i which otherwise would clash with CTD (circled box). As a result, the structure is likely to reorganize, reorienting the relative positions of the CARDs and HEL2i. This structural arrangement may allow the CARDs to gain access to polyubiquitins, making it available for MAVS activation (Jiang et al., 2012; Zeng et al., 2010). See also Mov. S1.
To examine these nucleotide-dependent conformational changes in solution, we performed a hydrodynamic analysis of the RIG-I-RNA complex using sedimentation velocity analytical ultracentrifugation (SV-AUC). We observe a large shift in the sedimentation coefficient upon ADP-AlFx binding to the complex (6.9% change in peak S value, Fig. 2D). By contrast, binding of ADP-BeF3 or ADP increases the peak S value only 4% and 2% relative to the nucleotide-free state, respectively (Fig. 2D). An increase in S value indicates compaction of the hydrodynamic radius of the complex, and this correlates well with available structural data(Jiang et al., 2011; Kowalinski et al., 2011; Luo et al., 2011), as the greatest structural compaction is observed in the presence of ADP-AlF3 (Fig 2D, data shown for the full length RIG-I). We suggest that ADP-AlFx mimics the transition-state of ATP hydrolysis, while ADP-BeF3 likely mimics the initial ATP binding to the RecA-like HEL1 domain. ADP is obviously the product bound state during the ATP hydrolysis cycle of RIG-I. Importantly, we do not observe functional interactions between RIG-I protein molecules in the presence or absence of RNA. RIG-I and its coupling cycle are therefore likely to be different from the homologous MDA5, which cooperatively binds RNA (Berke and Modis, 2012; Peisley et al., 2012).
In conclusion, it is now possible to visualize the conformational response of RIG-I to binding of its two ligands, tri-phosphorylated duplex RNA and nucleotide, and to envision the resultant influence on antiviral signaling. While intriguing in their dynamic implications, these snapshots also provide vital information for the rational design of therapeutics that modulates RIG-I mediated immune responses.
Experimental Procedures
Cloning, expression and purification
The full length RIG-I and N-terminal CARDs (1-238) deletion constructs, hereafter named RIG-I (ΔCARDs 1-238), was cloned into the pET-SUMO vector (Invitrogen). Transformed Rosetta II (DE3) E. coli cells (Novagen) were grown at 37 °C in LB medium supplemented with 40 μg ml−1 kanamycin and 34 μg ml−1 chloramphenicol to an OD600nm of 0.6 to 0.8. Protein expression was induced at 18°C by adding isopropyl-β-D-thiogalactopyranoside (IPTG) to a final concentration of 0.5 mM. After 20 hours growth, cells were harvested by centrifugation at 8,000 × g for 10 min at 4°C and stored at −20°C. Cells resuspended in buffer A (25 mM Hepes, pH 8.0, 0.5 M NaCl, 10 mM imidazole, 10% glycerol, 5 mM β-ME) were lysed by passing three times through a MicroFluidizer at 15,000 psi and the lysate was clarified by centrifugation at 15,000 × g for 60 minutes at 4°C. The supernatant was purified by batch binding with Qiagen Ni-NTA beads. The beads were collected in Biorad polyprep columns and the SUMO-tagged proteins were eluted with buffer B (25 mM Hepes, pH 8.0, 0.3 M NaCl, 10 % glycerol, 5 mM β-ME, 200 mM imidazole). The fraction containing His6-Sumo-RIG-I was then digested with ulp protease (Invitrogen), 4°C overnight. The cleavage mixture was loaded onto a HisTrap HP column to remove the His6-Sumo protein and ulp protease from the mixture. The recombinant protein was then further purified by using a HiTrap Heparin HP column (GE Healthcare) by running buffer C with an additional 1 M NaCl gradient. Concentrated proteins were subjected to a final gel-filtration purification step through a HiPrep 16/60 Superdex 200 column (Amersham Bioscience) in buffer D (25 mM Hepes, pH 7.4, 150 mM NaCl, 2 mM MgCl2, 5 % glycerol, 5 mM β-ME). Fractions containing monomeric RIG-I were pooled, concentrated and stored at −80°C. Recombinant protein RIG-I (ΔCARDs:1-238) was expressed and purified using the same method. The concentrations of the proteins were determined by measuring the absorbance at 280 nm by using extinction coefficients of 95300 M−1 cm−1 for full length RIG-I and 60040 M−1 cm−1 for RIG-I (ΔCARDs:1-238).
RNA preparation
The 5′ tri-phosphorylated RNA hairpin (here after named 5′ppp8L) was produced by in vitro transcription using a synthetic dsDNA template (Top strand: 5′- GTAATACGACTCACTATA GG CGCGGC ttcg GCCGCG CC -3′) and purified by gel extraction (20% PAGE with 8M urea).
Crystallization and data collection
To grow the crystals of the ternary complex of RIG-I (ΔCARDs:1-238) : 5′ppp8L : ADP-Mg2+, RIG-I (ΔCARDs:1-238) at 2.5 mg ml−1 was preassembled with 5′ppp8L at 50 uM and with 2.5 mM ADP, 2.5 mM MgCl2, 2.5 mM BeCl2, 12.5 mM NaF on ice for 1 hour. The complex solution was then mixed with equal volumes of precipitating solution (0.1 M Bicine, pH 9.0, 26-28 % polyethylene glycol 6,000) and then grown at 13 °C. Crystals also grew into needle clusters within three days and were harvested within two weeks. Crystals were soaked in a cryoprotecting solution containing 0.1 M Bicine, pH 9.0, 30 % polyethylene glycol 6,000 briefly before being flash frozen with liquid nitrogen. Diffraction intensities were recorded at NE-CAT beamline ID-24 at the Advanced Photon Source (Argonne National Laboratory, Argonne, Illinois). Integration, scaling and merging of the intensities were carried out by using the programs XDS (Kabsch, 2010) and SCALA (Evans, 2006).
Structure determination and refinement
The structures were determined through molecular replacement with the program Phaser (McCoy, 2007) by using the structure of RIG-I (ΔCARDs:1-229) : 5′OH-GC10 (PDB: 2ykg) as search model. Refinement cycles were carried out by using Phenix Refine (Adams et al., 2010) and REFMAC5 (Murshudov et al., 1997) with the TLS (translation, liberation, screw-rotation displacement) refinement option with four TLS groups (HEL1: aa 239-455, HEL2-HEL2i: aa 456-795, CTD: aa 796-922, and dsRNA). Refinement cycles were interspersed with model rebuilding by using Coot (Emsley and Cowtan, 2004). The quality of the structures was analyzed by using MolProbity(Davis et al., 2007). A summary of the data collection and structure refinement statistics is given in Tab.1. Figures were prepared by using the program Pymol (DeLano, 2002).
Sedimentation velocity studies
Samples were prepared by mixing 3 μM 5′-Dylight 547-U10:A10 duplex RNA with 7.5 μM of full length RIG-I protein in a buffer containing 25 mM HEPES, 150 mM NaCl, 0.5% glycerol, 5 mM β-ME, 2.5 mM MgCl2, pH 7.4, in addition to the respective ATP analogs (ADP-AlFx: 2.5 mM ADP, 2.5 mM MgCl2, 2.5 mM AlCl3, 12.5 mM NaF; ADP-BeF3: 2.5 mM ADP, 2.5 mM MgCl2, 2.5 mM BeCl3, 12.5 mM NaF; ADP: 2.5 mM ADP, 2.5 mM MgCl2). The samples were then incubated on ice for 1 hour. SV experiments were performed at 20°C in a Beckman Optima XL-I analytical ultracentrifuge. A four-position AN 60 Ti rotor, together with Epon 12-mm double-sector centerpieces, was used at 40,000 rpm. Radial absorption scans were measured at 547 nm with a radial increment of 0.003 cm. Data analyses were performed in Sedfit 8.0 (http://www.analyticalultracentrifugation.com)(Schuck et al., 2002). Sedimentation coefficients at the experimental temperature, buffer density, and viscosity were corrected to standard conditions (s20, w) using the program SEDNTERP (http://jphilo.mailway.com).
Supplementary Material
Highlights.
Crystallographic structure of RIG-I with 5′tri-phosphorylated RNA duplex and ADP.
Solution biophysical analysis of ATP-dependent RIG-I conformational change.
The structural basis for ATP-stimulated RIG-I activation.
ACKNOWLEDGEMENTS
We thank members of Pyle Lab for their generous help and insightful discussions. We thank Dr Steve Ding for providing 5′-Dylight 547-U10 RNA. We thank scientists from APS NECAT 24-ID for the beam line access and technical support. This research was funded by Howard Hughes Medical Institute. DL is a postdoctoral associate and AMP is an investigator with the Howard Hughes Medical Institute.
Footnotes
Accession code The atomic coordinates and structure factors of the ternary complex of RIG-I (ΔCARDs:1-238) : 5′ppp8L : ADP-Mg2+ have been deposited with the RCSB Protein Data Bank under the accession codes 4ay2.
Competing financial interests The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum A, Garcia-Sastre A. Differential recognition of viral RNA by RIG-I. Virulence. 2011;2:166–169. doi: 10.4161/viru.2.2.15481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum A, Sachidanandam R, Garcia-Sastre A. Preference of RIG-I for short viral RNA molecules in infected cells revealed by next-generation sequencing. Proc Natl Acad Sci U S A. 2011;107:16303–16308. doi: 10.1073/pnas.1005077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berke IC, Modis Y. MDA5 cooperatively forms dimers and ATP-sensitive filaments upon binding double-stranded RNA. Embo J. 2012 doi: 10.1038/emboj.2012.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui S, Eisenacher K, Kirchhofer A, Brzozka K, Lammens A, Lammens K, Fujita T, Conzelmann KK, Krug A, Hopfner KP. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol Cell. 2008;29:169–179. doi: 10.1016/j.molcel.2007.10.032. [DOI] [PubMed] [Google Scholar]
- Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decroly E, Ferron F, Lescar J, Canard B. Conventional and unconventional mechanisms for capping viral mRNA. Nat Rev Microbiol. 2012;10:51–65. doi: 10.1038/nrmicro2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL User’s Manual: DeLano Scientific. Palo Alto, CA, USA: 2002. [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. ActaCrystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- Jiang F, Ramanathan A, Miller MT, Tang GQ, Gale M, Jr., Patel SS, Marcotrigiano J. Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature. 2011;479:423–427. doi: 10.1038/nature10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang QX, Chen ZJ. Structural insights into the activation of RIG-I, a nanosensor for viral RNAs. EMBO Rep. 2011;13:7–8. doi: 10.1038/embor.2011.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Kinch LN, Brautigam CA, Chen X, Du F, Grishin NV, Chen ZJ. Ubiquitin-Induced Oligomerization of the RNA Sensors RIG-I and MDA5 Activates Antiviral Innate Immune Response. Immunity. 2012 doi: 10.1016/j.immuni.2012.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Takahasi K, Fujita T. RIG-I-like receptors: cytoplasmic sensors for non-self RNA. Immunol Rev. 2011;243:91–98. doi: 10.1111/j.1600-065X.2011.01052.x. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S. Antiviral signaling through pattern recognition receptors. J Biochem. 2007;141:137–145. doi: 10.1093/jb/mvm032. [DOI] [PubMed] [Google Scholar]
- Kowalinski E, Lunardi T, McCarthy AA, Louber J, Brunel J, Grigorov B, Gerlier D, Cusack S. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147:423–435. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- Lu C, Xu H, Ranjith-Kumar CT, Brooks MT, Hou TY, Hu F, Herr AB, Strong RK, Kao CC, Li P. The structural basis of 5′ triphosphate double-stranded RNA recognition by RIG-I C-terminal domain. Structure. 2010;18:1032–1043. doi: 10.1016/j.str.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D, Ding SC, Vela A, Kohlway A, Lindenbach BD, Pyle AM. Structural insights into RNA recognition by RIG-I. Cell. 2011;147:409–422. doi: 10.1016/j.cell.2011.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Peisley A, Lin C, Wu B, Orme-Johnson M, Liu M, Walz T, Hur S. Cooperative assembly and dynamic disassembly of MDA5 filaments for viral dsRNA recognition. Proc Natl Acad Sci U S A. 2012;108:21010–21015. doi: 10.1073/pnas.1113651108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- Pyle AM. Translocation and Unwinding Mechanisms of RNA and DNA Helicases. Annual Review of Biophysics. 2008;37:317–336. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- Ramos HJ, Gale M., Jr. RIG-I Like Receptors and Their Signaling Crosstalk in the Regulation of Antiviral Immunity. Curr Opin Virol. 2011;1:167–176. doi: 10.1016/j.coviro.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlee M, Roth A, Hornung V, Hagmann CA, Wimmenauer V, Barchet W, Coch C, Janke M, Mihailovic A, Wardle G, et al. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009;31:25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck P, Perugini MA, Gonzales NR, Howlett GJ, Schubert D. Size-distribution analysis of proteins by analytical ultracentrifugation: strategies and application to model systems. Biophys J. 2002;82:1096–1111. doi: 10.1016/S0006-3495(02)75469-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Ludwig J, Schuberth C, Goldeck M, Schlee M, Li H, Juranek S, Sheng G, Micura R, Tuschl T, et al. Structural and functional insights into 5′-ppp RNA pattern recognition by the innate immune receptor RIG-I. Nat Struct Mol Biol. 2010;17:781–787. doi: 10.1038/nsmb.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- Zeng W, Sun L, Jiang X, Chen X, Hou F, Adhikari A, Xu M, Chen ZJ. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 2010;141:315–330. doi: 10.1016/j.cell.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.