Abstract
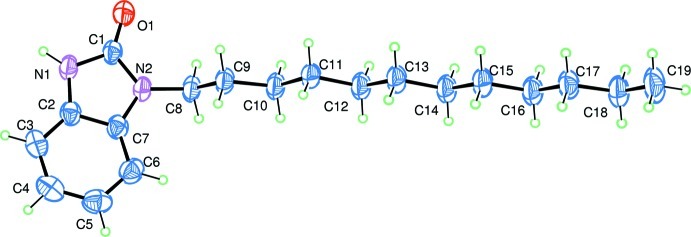
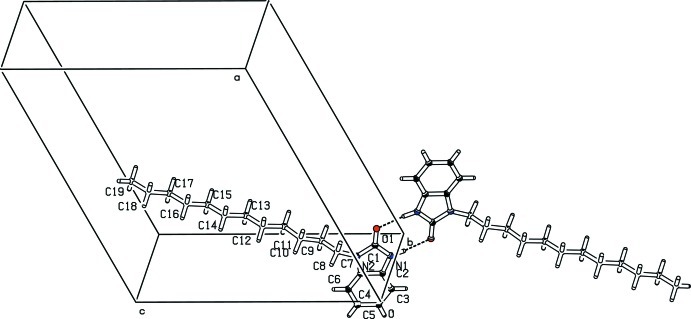
In the title compound, C19H30N2O, the fused ring system is essentially planar, the maximum deviation from the mean plane being 0.013 (2) Å for the N atom bearing the dodecyl chain. The 1-dodecyl group is almost perpendicular to the 1H-benzo[d]imidazol-2(3H)-one plane as indicated by the dihedral angle of 82.9 (2)°between planes through the fused ring system and the first three C atoms of the chain. The C—C—C—C torsion angles (about ±179°) of the dodecyl group indicate an antiperiplanar conformation. In the crystal, inversion dimers are formed by pairs of N—H⋯O hydrogen bonds.
Related literature
For pharmacological and biochemical properties of benzimidazoles and their derivatives, see: Al Muhaimeed (1997 ▶); Scott et al. (2002 ▶); Nakano et al. (2000 ▶); Zhu et al. (2000 ▶); Zarrinmayeh et al. (1998 ▶). For compounds with closely related structures, see: Ouzidan et al. (2011 ▶); Kandri Rodi et al. (2011 ▶); Belaziz et al. (2012 ▶).
Experimental
Crystal data
C19H30N2O
M r = 302.45
Monoclinic,

a = 38.3223 (14) Å
b = 4.8318 (2) Å
c = 21.9831 (8) Å
β = 117.843 (2)°
V = 3599.3 (2) Å3
Z = 8
Mo Kα radiation
μ = 0.07 mm−1
T = 296 K
0.47 × 0.31 × 0.14 mm
Data collection
Bruker X8 APEX Diffractometer
29002 measured reflections
4637 independent reflections
3179 reflections with I > 2σ(I)
R int = 0.028
Refinement
R[F 2 > 2σ(F 2)] = 0.045
wR(F 2) = 0.141
S = 1.01
4637 reflections
199 parameters
H-atom parameters constrained
Δρmax = 0.21 e Å−3
Δρmin = −0.21 e Å−3
Data collection: APEX2 (Bruker, 2005 ▶); cell refinement: SAINT (Bruker, 2005 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 for Windows (Farrugia, 1997 ▶); software used to prepare material for publication: PLATON (Spek, 2009 ▶) and publCIF (Westrip, 2010 ▶).
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S1600536812041189/im2402sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812041189/im2402Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536812041189/im2402Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1⋯O1i | 0.86 | 1.97 | 2.815 (1) | 168 |
Symmetry code: (i) 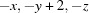 .
.
Acknowledgments
The authors thank the Unit of Support for Technical and Scientific Research (UATRS, CNRST) for the X-ray measurements.
supplementary crystallographic information
Comment
Benzimidazoles and their derivatives exhibit a number of important pharmacological properties, such as antihistaminic (Al Muhaimeed, 1997) anti-ulcerative (Scott et al., 2002) and antiallergic (Nakano et al., 2000). In addition, benzimidazole derivatives are effective against the human cytomegalovirus (HCMV) (Zhu et al., 2000) and are also efficient selective neuropeptide Y Y1 receptor antagonists (Zarrinmayeh et al., 1998).
In a previous study, we reacted benzimidazol-2-one with octyl bromide in the presence of a catalytic quantity of tetra-n-butylammonium bromide under mild conditions to form 1-octyl-1H-benzo[d]imidazol-2(3H)-one (Belaziz et al., 2012). The study has been extended to the synthesis of a new benzimidazol-2-one derivative by action of dodecyl bromide with 1H-benzo[d]imidazol-2(3H)-one to form the title compound (Scheme 1).
The molecular structure of 1-dodecyl-1H-benzo[d]imidazol-2(3H)-one is built up from fused six-and five-membered rings linked to a C12H25 chain as shown in Fig. 1. The fused-ring system is essentially planar, with a maximum deviation of -0.013 (2) Å for N2. The dodecyl group is almost perpendicular to the 1H-benzo[d]imidazol-2(3H)-one plane as indicated by the dihedral angle between planes (C8 C9 C10) and (N1 N2 C1 to C7) of 82.9 (2)° and by the torsion angle (C7 N2 C8 C9) = -84.3 (2)°. In the crystal structure, inversion dimers are formed by N—H···O hydrogen bonds. in the way to form dimers (Fig. 2).
The structure of the title compound is almost identical to that observed for the following molecules: 1-nonyl-1H-benzimidazol-2(3H)-one, 1-octyl-1H-benzimidazol-2(3H)-one and 5-chloro-1-nonyl-1H-benzimidazol-2(3H)-one (Ouzidan et al., 2011, Kandri Rodi et al. 2011). Nevertheless, the different lengths of the chains leads to different unit cells with different crystal symmetry.
Experimental
To 1H-benzo[d]imidazol-2(3H)-one (0.2 g, 1.49 mmol), potassium carbonate (0.41 g, 2.98 mmol) and tetra-n-butylammonium bromide (0.05 g, 0.15 mmol) in DMF (15 ml) was added dodecyl bromide (0.30 ml, 1.78 mmol). Stirring was continued at room temperature for 6 h. The salt was removed by filtration and the filtrate concentrated under reduced pressure. The residue was separated by chromatography on a column of silica gel with ethyl acetate/hexane (1/2) as eluent (yield: 65%). The compound was recrystallized from hexan/acetate to give colourless crystals.
Refinement
H atoms were located in a difference map and treated as riding with N—H = 0.86 Å, C—H = 0.93 Å (aromatic), C—H = 0.97 Å (methylene) and C—H = 0.96 Å (methyl) with Uiso(H) = 1.2 Ueq (N—H, aromatic, methylene) and Uiso(H) = 1.5 Ueq(methyl).
Figures
Fig. 1.
Molecular structure of the title compound with displacement ellipsoids drawn at the 50% probability level. H atoms are represented as small circles.
Fig. 2.
Inversion dimer with molecules linked by N—H···O hydrogen bonds.
Crystal data
| C19H30N2O | F(000) = 1328 |
| Mr = 302.45 | Dx = 1.116 Mg m−3 |
| Monoclinic, C2/c | Melting point: 346.5 K |
| Hall symbol: -C 2yc | Mo Kα radiation, λ = 0.71073 Å |
| a = 38.3223 (14) Å | Cell parameters from 4637 reflections |
| b = 4.8318 (2) Å | θ = 2.4–28.7° |
| c = 21.9831 (8) Å | µ = 0.07 mm−1 |
| β = 117.843 (2)° | T = 296 K |
| V = 3599.3 (2) Å3 | Needle, colourless |
| Z = 8 | 0.47 × 0.31 × 0.14 mm |
Data collection
| Bruker X8 APEX Diffractometer | 3179 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.028 |
| Graphite monochromator | θmax = 28.7°, θmin = 2.4° |
| φ and ω scans | h = −51→51 |
| 29002 measured reflections | k = −6→6 |
| 4637 independent reflections | l = −29→28 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.045 | Hydrogen site location: difference Fourier map |
| wR(F2) = 0.141 | H-atom parameters constrained |
| S = 1.01 | w = 1/[σ2(Fo2) + (0.0721P)2 + 1.030P] where P = (Fo2 + 2Fc2)/3 |
| 4637 reflections | (Δ/σ)max < 0.001 |
| 199 parameters | Δρmax = 0.21 e Å−3 |
| 0 restraints | Δρmin = −0.21 e Å−3 |
Special details
| Geometry. All s.u.'s (except the s.u. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell s.u.'s are taken into account individually in the estimation of s.u.'s in distances, angles and torsion angles; correlations between s.u.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell s.u.'s is used for estimating s.u.'s involving l.s. planes. |
| Refinement. Refinement of F2 against all reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on all data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| N2 | 0.03233 (3) | 0.5579 (2) | 0.13178 (5) | 0.0394 (2) | |
| N1 | −0.01601 (3) | 0.7314 (2) | 0.03782 (5) | 0.0398 (2) | |
| H1 | −0.0289 | 0.8297 | 0.0014 | 0.048* | |
| O1 | 0.04750 (2) | 0.91076 (19) | 0.07646 (4) | 0.0472 (2) | |
| C11 | 0.13888 (4) | 0.6345 (3) | 0.37748 (6) | 0.0440 (3) | |
| H11A | 0.1187 | 0.6113 | 0.3920 | 0.053* | |
| H11B | 0.1428 | 0.8315 | 0.3747 | 0.053* | |
| C7 | −0.00182 (3) | 0.4187 (2) | 0.12105 (6) | 0.0386 (3) | |
| C9 | 0.08398 (3) | 0.6105 (3) | 0.25353 (6) | 0.0431 (3) | |
| H9A | 0.0644 | 0.5596 | 0.2679 | 0.052* | |
| H9B | 0.0844 | 0.8108 | 0.2510 | 0.052* | |
| C15 | 0.24328 (4) | 0.5988 (3) | 0.62951 (6) | 0.0514 (3) | |
| H15A | 0.2457 | 0.7986 | 0.6294 | 0.062* | |
| H15B | 0.2233 | 0.5564 | 0.6435 | 0.062* | |
| C12 | 0.17715 (4) | 0.5077 (3) | 0.43133 (6) | 0.0484 (3) | |
| H12A | 0.1737 | 0.3089 | 0.4317 | 0.058* | |
| H12B | 0.1977 | 0.5418 | 0.4182 | 0.058* | |
| C1 | 0.02366 (3) | 0.7522 (2) | 0.08097 (5) | 0.0371 (3) | |
| C13 | 0.19082 (4) | 0.6180 (3) | 0.50358 (6) | 0.0498 (3) | |
| H13A | 0.1705 | 0.5816 | 0.5171 | 0.060* | |
| H13B | 0.1940 | 0.8170 | 0.5032 | 0.060* | |
| C8 | 0.07235 (3) | 0.4891 (3) | 0.18297 (6) | 0.0424 (3) | |
| H8A | 0.0906 | 0.5557 | 0.1671 | 0.051* | |
| H8B | 0.0749 | 0.2894 | 0.1870 | 0.051* | |
| C2 | −0.03246 (3) | 0.5297 (2) | 0.06114 (6) | 0.0383 (3) | |
| C10 | 0.12435 (4) | 0.5075 (3) | 0.30657 (6) | 0.0463 (3) | |
| H10A | 0.1232 | 0.3081 | 0.3104 | 0.056* | |
| H10B | 0.1433 | 0.5476 | 0.2902 | 0.056* | |
| C17 | 0.29647 (4) | 0.5852 (3) | 0.75462 (6) | 0.0513 (3) | |
| H17A | 0.2983 | 0.7854 | 0.7541 | 0.062* | |
| H17B | 0.2769 | 0.5392 | 0.7693 | 0.062* | |
| C14 | 0.22938 (4) | 0.4922 (3) | 0.55680 (6) | 0.0517 (3) | |
| H14A | 0.2497 | 0.5298 | 0.5433 | 0.062* | |
| H14B | 0.2262 | 0.2930 | 0.5567 | 0.062* | |
| C16 | 0.28233 (4) | 0.4785 (3) | 0.68205 (6) | 0.0522 (3) | |
| H16A | 0.2798 | 0.2788 | 0.6823 | 0.063* | |
| H16B | 0.3022 | 0.5199 | 0.6678 | 0.063* | |
| C6 | −0.00851 (4) | 0.2120 (3) | 0.15747 (7) | 0.0493 (3) | |
| H6 | 0.0118 | 0.1406 | 0.1977 | 0.059* | |
| C3 | −0.07051 (4) | 0.4301 (3) | 0.03570 (7) | 0.0490 (3) | |
| H3 | −0.0910 | 0.5015 | −0.0044 | 0.059* | |
| C5 | −0.04689 (4) | 0.1145 (3) | 0.13174 (7) | 0.0553 (4) | |
| H5 | −0.0524 | −0.0248 | 0.1552 | 0.066* | |
| C18 | 0.33590 (4) | 0.4716 (3) | 0.80666 (7) | 0.0616 (4) | |
| H18A | 0.3555 | 0.5176 | 0.7921 | 0.074* | |
| H18B | 0.3341 | 0.2714 | 0.8072 | 0.074* | |
| C19 | 0.34981 (5) | 0.5790 (4) | 0.87888 (7) | 0.0774 (5) | |
| H19A | 0.3750 | 0.4990 | 0.9090 | 0.116* | |
| H19B | 0.3310 | 0.5294 | 0.8944 | 0.116* | |
| H19C | 0.3522 | 0.7768 | 0.8791 | 0.116* | |
| C4 | −0.07710 (4) | 0.2207 (3) | 0.07186 (8) | 0.0554 (4) | |
| H4 | −0.1024 | 0.1494 | 0.0556 | 0.066* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| N2 | 0.0357 (5) | 0.0440 (5) | 0.0272 (4) | 0.0014 (4) | 0.0053 (4) | 0.0030 (4) |
| N1 | 0.0355 (5) | 0.0458 (6) | 0.0282 (4) | 0.0032 (4) | 0.0065 (4) | 0.0038 (4) |
| O1 | 0.0404 (4) | 0.0546 (5) | 0.0378 (4) | −0.0026 (4) | 0.0110 (4) | 0.0066 (4) |
| C11 | 0.0434 (6) | 0.0463 (7) | 0.0299 (5) | −0.0011 (5) | 0.0067 (5) | 0.0014 (5) |
| C7 | 0.0416 (6) | 0.0397 (6) | 0.0304 (5) | 0.0009 (5) | 0.0134 (5) | −0.0038 (4) |
| C9 | 0.0410 (6) | 0.0450 (7) | 0.0305 (6) | 0.0040 (5) | 0.0061 (5) | 0.0011 (5) |
| C15 | 0.0462 (7) | 0.0619 (8) | 0.0314 (6) | −0.0014 (6) | 0.0057 (5) | −0.0013 (5) |
| C12 | 0.0457 (7) | 0.0524 (7) | 0.0306 (6) | 0.0017 (6) | 0.0040 (5) | −0.0013 (5) |
| C1 | 0.0373 (6) | 0.0405 (6) | 0.0267 (5) | 0.0026 (5) | 0.0092 (4) | −0.0012 (4) |
| C13 | 0.0460 (7) | 0.0582 (8) | 0.0308 (6) | −0.0006 (6) | 0.0058 (5) | −0.0010 (5) |
| C8 | 0.0366 (6) | 0.0470 (7) | 0.0302 (6) | 0.0069 (5) | 0.0044 (5) | 0.0026 (5) |
| C2 | 0.0396 (6) | 0.0404 (6) | 0.0309 (5) | 0.0014 (5) | 0.0131 (5) | −0.0049 (5) |
| C10 | 0.0432 (6) | 0.0482 (7) | 0.0300 (6) | 0.0046 (5) | 0.0025 (5) | 0.0010 (5) |
| C17 | 0.0441 (7) | 0.0647 (9) | 0.0326 (6) | −0.0039 (6) | 0.0075 (5) | −0.0003 (6) |
| C14 | 0.0478 (7) | 0.0579 (8) | 0.0314 (6) | −0.0002 (6) | 0.0034 (5) | −0.0018 (5) |
| C16 | 0.0473 (7) | 0.0597 (8) | 0.0330 (6) | −0.0001 (6) | 0.0048 (5) | −0.0016 (6) |
| C6 | 0.0591 (8) | 0.0471 (7) | 0.0393 (6) | 0.0016 (6) | 0.0210 (6) | 0.0022 (5) |
| C3 | 0.0392 (6) | 0.0564 (8) | 0.0443 (7) | −0.0002 (5) | 0.0135 (5) | −0.0078 (6) |
| C5 | 0.0690 (9) | 0.0500 (8) | 0.0572 (8) | −0.0089 (7) | 0.0382 (7) | −0.0030 (6) |
| C18 | 0.0494 (8) | 0.0736 (10) | 0.0394 (7) | 0.0005 (7) | 0.0021 (6) | 0.0011 (7) |
| C19 | 0.0585 (9) | 0.1111 (14) | 0.0367 (7) | −0.0096 (9) | 0.0006 (7) | 0.0009 (8) |
| C4 | 0.0502 (8) | 0.0569 (8) | 0.0632 (9) | −0.0117 (6) | 0.0300 (7) | −0.0127 (7) |
Geometric parameters (Å, º)
| N2—C1 | 1.3759 (15) | C8—H8A | 0.9700 |
| N2—C7 | 1.3899 (15) | C8—H8B | 0.9700 |
| N2—C8 | 1.4550 (14) | C2—C3 | 1.3816 (17) |
| N1—C1 | 1.3688 (14) | C10—H10A | 0.9700 |
| N1—C2 | 1.3827 (15) | C10—H10B | 0.9700 |
| N1—H1 | 0.8600 | C17—C18 | 1.5095 (18) |
| O1—C1 | 1.2314 (14) | C17—C16 | 1.5156 (18) |
| C11—C10 | 1.5184 (16) | C17—H17A | 0.9700 |
| C11—C12 | 1.5187 (16) | C17—H17B | 0.9700 |
| C11—H11A | 0.9700 | C14—H14A | 0.9700 |
| C11—H11B | 0.9700 | C14—H14B | 0.9700 |
| C7—C6 | 1.3771 (18) | C16—H16A | 0.9700 |
| C7—C2 | 1.3986 (16) | C16—H16B | 0.9700 |
| C9—C8 | 1.5177 (16) | C6—C5 | 1.3885 (19) |
| C9—C10 | 1.5218 (16) | C6—H6 | 0.9300 |
| C9—H9A | 0.9700 | C3—C4 | 1.381 (2) |
| C9—H9B | 0.9700 | C3—H3 | 0.9300 |
| C15—C16 | 1.5157 (18) | C5—C4 | 1.383 (2) |
| C15—C14 | 1.5195 (18) | C5—H5 | 0.9300 |
| C15—H15A | 0.9700 | C18—C19 | 1.511 (2) |
| C15—H15B | 0.9700 | C18—H18A | 0.9700 |
| C12—C13 | 1.5180 (17) | C18—H18B | 0.9700 |
| C12—H12A | 0.9700 | C19—H19A | 0.9600 |
| C12—H12B | 0.9700 | C19—H19B | 0.9600 |
| C13—C14 | 1.5186 (17) | C19—H19C | 0.9600 |
| C13—H13A | 0.9700 | C4—H4 | 0.9300 |
| C13—H13B | 0.9700 | ||
| C1—N2—C7 | 109.94 (9) | N1—C2—C7 | 106.97 (10) |
| C1—N2—C8 | 123.45 (10) | C11—C10—C9 | 114.11 (11) |
| C7—N2—C8 | 126.15 (10) | C11—C10—H10A | 108.7 |
| C1—N1—C2 | 110.23 (9) | C9—C10—H10A | 108.7 |
| C1—N1—H1 | 124.9 | C11—C10—H10B | 108.7 |
| C2—N1—H1 | 124.9 | C9—C10—H10B | 108.7 |
| C10—C11—C12 | 113.25 (11) | H10A—C10—H10B | 107.6 |
| C10—C11—H11A | 108.9 | C18—C17—C16 | 114.41 (12) |
| C12—C11—H11A | 108.9 | C18—C17—H17A | 108.7 |
| C10—C11—H11B | 108.9 | C16—C17—H17A | 108.7 |
| C12—C11—H11B | 108.9 | C18—C17—H17B | 108.7 |
| H11A—C11—H11B | 107.7 | C16—C17—H17B | 108.7 |
| C6—C7—N2 | 131.94 (11) | H17A—C17—H17B | 107.6 |
| C6—C7—C2 | 121.55 (11) | C13—C14—C15 | 114.31 (12) |
| N2—C7—C2 | 106.51 (10) | C13—C14—H14A | 108.7 |
| C8—C9—C10 | 111.39 (10) | C15—C14—H14A | 108.7 |
| C8—C9—H9A | 109.3 | C13—C14—H14B | 108.7 |
| C10—C9—H9A | 109.3 | C15—C14—H14B | 108.7 |
| C8—C9—H9B | 109.3 | H14A—C14—H14B | 107.6 |
| C10—C9—H9B | 109.3 | C17—C16—C15 | 114.33 (12) |
| H9A—C9—H9B | 108.0 | C17—C16—H16A | 108.7 |
| C16—C15—C14 | 114.07 (12) | C15—C16—H16A | 108.7 |
| C16—C15—H15A | 108.7 | C17—C16—H16B | 108.7 |
| C14—C15—H15A | 108.7 | C15—C16—H16B | 108.7 |
| C16—C15—H15B | 108.7 | H16A—C16—H16B | 107.6 |
| C14—C15—H15B | 108.7 | C7—C6—C5 | 117.30 (12) |
| H15A—C15—H15B | 107.6 | C7—C6—H6 | 121.3 |
| C13—C12—C11 | 114.18 (11) | C5—C6—H6 | 121.3 |
| C13—C12—H12A | 108.7 | C4—C3—C2 | 117.68 (12) |
| C11—C12—H12A | 108.7 | C4—C3—H3 | 121.2 |
| C13—C12—H12B | 108.7 | C2—C3—H3 | 121.2 |
| C11—C12—H12B | 108.7 | C4—C5—C6 | 121.22 (13) |
| H12A—C12—H12B | 107.6 | C4—C5—H5 | 119.4 |
| O1—C1—N1 | 127.98 (10) | C6—C5—H5 | 119.4 |
| O1—C1—N2 | 125.67 (10) | C17—C18—C19 | 114.10 (14) |
| N1—C1—N2 | 106.35 (10) | C17—C18—H18A | 108.7 |
| C12—C13—C14 | 113.68 (12) | C19—C18—H18A | 108.7 |
| C12—C13—H13A | 108.8 | C17—C18—H18B | 108.7 |
| C14—C13—H13A | 108.8 | C19—C18—H18B | 108.7 |
| C12—C13—H13B | 108.8 | H18A—C18—H18B | 107.6 |
| C14—C13—H13B | 108.8 | C18—C19—H19A | 109.5 |
| H13A—C13—H13B | 107.7 | C18—C19—H19B | 109.5 |
| N2—C8—C9 | 113.73 (10) | H19A—C19—H19B | 109.5 |
| N2—C8—H8A | 108.8 | C18—C19—H19C | 109.5 |
| C9—C8—H8A | 108.8 | H19A—C19—H19C | 109.5 |
| N2—C8—H8B | 108.8 | H19B—C19—H19C | 109.5 |
| C9—C8—H8B | 108.8 | C3—C4—C5 | 121.52 (13) |
| H8A—C8—H8B | 107.7 | C3—C4—H4 | 119.2 |
| C3—C2—N1 | 132.32 (11) | C5—C4—H4 | 119.2 |
| C3—C2—C7 | 120.71 (12) | ||
| C1—N2—C7—C6 | −179.18 (12) | N2—C7—C2—C3 | 178.75 (10) |
| C8—N2—C7—C6 | 8.5 (2) | C6—C7—C2—N1 | 179.65 (10) |
| C1—N2—C7—C2 | 0.61 (13) | N2—C7—C2—N1 | −0.17 (12) |
| C8—N2—C7—C2 | −171.76 (10) | C12—C11—C10—C9 | 174.05 (11) |
| C10—C11—C12—C13 | −176.14 (11) | C8—C9—C10—C11 | 176.53 (11) |
| C2—N1—C1—O1 | −179.33 (11) | C12—C13—C14—C15 | −179.52 (12) |
| C2—N1—C1—N2 | 0.70 (12) | C16—C15—C14—C13 | −178.39 (12) |
| C7—N2—C1—O1 | 179.22 (11) | C18—C17—C16—C15 | −178.41 (13) |
| C8—N2—C1—O1 | −8.16 (18) | C14—C15—C16—C17 | 179.67 (12) |
| C7—N2—C1—N1 | −0.81 (12) | N2—C7—C6—C5 | −179.26 (12) |
| C8—N2—C1—N1 | 171.81 (10) | C2—C7—C6—C5 | 0.99 (18) |
| C11—C12—C13—C14 | −179.29 (11) | N1—C2—C3—C4 | 179.30 (12) |
| C1—N2—C8—C9 | 104.30 (13) | C7—C2—C3—C4 | 0.70 (18) |
| C7—N2—C8—C9 | −84.31 (15) | C7—C6—C5—C4 | 0.1 (2) |
| C10—C9—C8—N2 | 174.01 (10) | C16—C17—C18—C19 | 179.99 (13) |
| C1—N1—C2—C3 | −179.08 (12) | C2—C3—C4—C5 | 0.4 (2) |
| C1—N1—C2—C7 | −0.33 (13) | C6—C5—C4—C3 | −0.9 (2) |
| C6—C7—C2—C3 | −1.43 (18) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1···O1i | 0.86 | 1.97 | 2.815 (1) | 168 |
Symmetry code: (i) −x, −y+2, −z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: IM2402).
References
- Al Muhaimeed, H. (1997). J. Int. Med. Res. 25, 175–181. [DOI] [PubMed]
- Belaziz, D., Kandri Rodi, Y., Essassi, E. M. & El Ammari, L. (2012). Acta Cryst. E68, o1276. [DOI] [PMC free article] [PubMed]
- Bruker (2005). APEX2 and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Farrugia, L. J. (1997). J. Appl. Cryst. 30, 565.
- Kandri Rodi, Y., Ouazzani Chahdi, F., Essassi, E. M., Luis, S. V., Bolte, M. & El Ammari, L. (2011). Acta Cryst. E67, o3340–o3341. [DOI] [PMC free article] [PubMed]
- Nakano, H., Inoue, T., Kawasaki, N., Miyataka, H., Matsumoto, H., Taguchi, T., Inagaki, N., Nagai, H. & Satoh, T. (2000). Bioorg. Med. Chem. 8, 373–380. [DOI] [PubMed]
- Ouzidan, Y., Kandri Rodi, Y., Butcher, R. J., Essassi, E. M. & El Ammari, L. (2011). Acta Cryst. E67, o283. [DOI] [PMC free article] [PubMed]
- Scott, L. J., Dunn, C. J., Mallarkey, G. & Sharpe, M. (2002). Drugs, 62, 1503–1538. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
- Zarrinmayeh, H., Nunes, A. M., Ornstein, P. L., Zimmerman, D. M., Arnold, M. B., Schober, D. A., Gackenheimer, S. L., Bruns, R. F., Hipskind, P. A., Britton, T. C., Cantrell, B. E. & Gehlert, D. R. (1998). J. Med. Chem. 41, 2709–2719. [DOI] [PubMed]
- Zhu, Z., Lippa, B., Drach, J. C. & Townsend, L. B. (2000). J. Med. Chem. 43, 2430–2437. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S1600536812041189/im2402sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812041189/im2402Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536812041189/im2402Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report