Abstract
Endometrial cancer is the 6th most commonly diagnosed cancer among women worldwide, causing ~74,000 deaths annually 1. Serous endometrial cancers are a clinically aggressive subtype with a poorly defined genetic etiology 2-4. We used whole exome sequencing (WES) to comprehensively search for somatic mutations within ~22,000 protein-encoding genes among 13 primary serous endometrial tumors. We subsequently resequenced 18 genes that were mutated in more than one tumor, and/or were genes that formed an enriched functional grouping, from 40 additional serous tumors. We identified high frequencies of somatic mutations in CHD4 (17%), EP300 (8%), ARID1A (6%), TSPYL2 (6%), FBXW7 (29%), SPOP (8%), MAP3K4 (6%) and ABCC9 (6%). Overall, 36.5% of serous tumors had mutated a chromatin-remodeling gene and 35% had mutated a ubiquitin ligase complex gene, implicating the frequent mutational disruption of these processes in the molecular pathogenesis of one of the deadliest forms of endometrial cancer.
We performed targeted exon capture and next generation sequencing on DNA from 13 primary serous endometrial tumors with high neoplastic cellularity (Supplementary Table 1) and matched normal DNA. The mean depth of coverage for aligned reads was 102.6× and on average 89.5% of targeted bases had sufficient coverage and quality for variant calling (Supplementary Table 2). Using stringent filtering criteria, which included an empirically determined threshold that accounted for read quality and depth and provided an optimal balance between positive predictive value (86.1%) and sensitivity (97.3%) (Materials and Methods), we identified 1,522 exonic somatic mutations (1,183 nonsynonymous: 339 synonymous) and 22 splice junction mutations within the protein-encoding genes of the 13 tumors (Supplementary Figure 1). One tumor had an apparent hypermutable phenotype with a greater number of mutations and a different mutation signature than the other 12 tumors (Supplementary Figure 2; Supplementary Table 3 and Supplementary Table 4) and was excluded from subsequent analyses.
Among the remaining 12 tumors we identified 516 exonic mutations (380 nonsynonymous and 136 synonymous) and 11 splice junction mutations (Supplementary Table 5). We could orthogonally assess 510 of the 527 exonic/splice junction mutations by Sanger sequencing; 86.1% (439 of 510) of mutations confirmed as somatic (Supplementary Table 5 and Supplementary Table 6). The validated somatic mutations included 321 nonsynonymous mutations (279 missense, 86.9%; 19 nonsense, 5.9%; 17 frameshift, 5.3%; 6 in-frame insertions/deletions, 1.9%) and 9 splice junction mutations among 304 protein-encoding genes, and 109 synonymous mutations (Supplementary Table 5). There was a mean of 27.5 validated nonsynonymous and splice junction mutations per tumor (range 5-55) (Supplementary Table 6). The predicted functional impact of the validated missense mutations was assessed in silico; 34.4% of 241 missense mutations that could be assessed by both the SIFT and Mutation Assessor algorithms are predicted to impact protein function (Supplementary Table 5).
To prioritize our search for novel driver mutations in serous endometrial cancer, we focused on the nine genes that had validated nonsynonymous somatic mutations in more than one tumor (Supplementary Table 7). We resequenced these genes in a prevalence screen of 40 additional serous endometrial tumors. Three of the nine genes (TP53, PIK3CA, and PPP2R1A) have established roles in the pathogenesis of serous endometrial cancer 5-8. Among the 52 serous tumors in our study, TP53, PIK3CA, and PPP2R1A were mutated, respectively, in 71%, 31%, and 25% of tumors, as reported here and in a previous study from our group 6 (Supplementary Table 8 and Supplementary Table 9). The other six genes (CHD4, SPOP, FBXW7, ABCC9, CYP4X1, MAP3K4) have no previously reported role in serous endometrial cancer. The combined discovery and prevalence screens revealed high frequency somatic mutations in CHD4 (17%), FBXW7 (29%), SPOP (8%), MAP3K4 (6%), ABCC9 (6%), and CYP4X1 (4%) (Table 1, Figure 1, Supplementary Table 9, Supplementary Figure 3, and Supplementary Figure 4). The mutation rates for CHD4, FBXW7, and SPOP were significantly higher than the background mutation rate (q ≤0.0353) (Supplementary Table 10).
Table 1.
Somatic mutations identified in the discovery and prevalence screens of CHD4, FBXW7, SPOP, MAP3K4, ABCC9, and CYP4X1.
| Gene Name (RefSeq ID; UCSC Transcript Accession ID^) |
Tumor | Histology | Nucleotide Change | Amino Acid Change | Mutation type |
Mutation Assessor Predicted Effect on Function |
|---|---|---|---|---|---|---|
|
CHD4 (NM_001273; uc001qpo.2) |
T3 (OM1323) |
Serous | c.2450G>A | p.Arg817Gln | Missense | Medium |
| T3 (OM1323) |
Serous | c.4709G>A | p.Ser1570Asn | Missense | Low | |
| T10 (OM2009) |
Serous | c.4013G>A | p.Arg1338Lys | Missense | Medium | |
| T29 | Serous | c.3314G>A | p.Arg1105Gln | Missense | High | |
| T50 | Serous | c.3452A>C | p.His1151Arg | Missense | High | |
| T78 | Serous | c.2734T>C | p.Leu912Val | Missense | High | |
| T165 | Serous | c.4001G>T | p.Gly1334Val | Missense | Medium | |
| T33 | Serous | c.3485G>A | p.Arg1162Gln | Missense | High | |
| T154 | Serous | c.1672G>T | p.Val558Phe | Missense | Medium | |
| T56 | Serous | c.3460_3462delATT | p.del1152Asn | In-frame deletion | - | |
| T59 | Clear cell | c.2629C>T | p.Arg877Trp | Missense | High | |
| T97 | Endometrioid | c.1715G>A | p.Arg572Gln | Missense | Medium | |
| T115 | Endometrioid | c.4946C>T | p.Ala1649Val | Missense | Low | |
| T120 | Endometrioid | c.2620_2622delGAA | p.del874Glu | In-frame deletion | - | |
| T126 | Endometrioid | c.5704G>A | p.Glu1902Lys | Missense | Low | |
| T133 | Endometrioid | c.3484C>T | p.Arg1162Trp | Missense | High | |
| T173 | Mixed | c.3644T>C | p.Leu1215Pro | Missense | High | |
| T179 | Mixed | c.218delA | p.Lys72Argfs*128 | Frameshift | - | |
|
FBXW7 (NM_018315; uc003imr.2) |
T3 (OM1323) |
Serous | c.193G>T | p.Glu65* | Nonsense | - |
| T113 | Clear cell | c.207_208insA | p.Asn69Lysfs*26 | Frameshift | - | |
| (NM_033632; uc003ims.2) |
T3 (OM1323) | Serous | c.288A>C | p.Glu96Asp | Missense | Neutral |
| T43 | Serous | c.1590_1608delTGAAACCTGTCTAC ACACG |
p.Glu531Cysfs*19 | Frameshift | - | |
| T50 | Serous | c.1436G>A | p.Arg479Gln | Missense | Medium ¶ | |
| T53 | Serous | c.1393C>T | p.Arg465Cys | Missense | Medium ¶ | |
| T74 | Serous | c.1394G>A | p.Arg465His | Missense | High ¶ | |
| T78 | Serous | c.1394G>A | p.Arg465His | Missense | High ¶ | |
| T79 | Serous | c.504G>T | p.Met168Ile | Missense | Low | |
| T79 | Serous | c.41G>A | p.Arg14Gln | Missense | Neutral | |
| T79 | Serous | c.1436G>A | p.Arg479Gln | Missense | Medium ¶ | |
| T114 | Serous | c.1513C>T | p.Arg505Cys | Missense | Medium ¶ | |
| T162 | Serous | c.1436G>A | p.Arg479Gln | Missense | Medium ¶ | |
| T166 | Serous | c.1634A>T | p.Tyr545Cys | Missense | Medium | |
| T167 | Serous | c.2065C>T | p.Arg689Trp | Missense | Medium | |
| T29 | Serous | c.1387A>G | p.Thr463Ala | Missense | Medium | |
| T80 | Serous | c.1385C>T | p.Ser462Phe | Missense | Medium | |
| T80 | Serous | c.1322G>C | p.Arg441Pro | Missense | Medium | |
| T47 | Serous | c.1436G>A | p.Arg479Gln | Missense | Medium ¶ | |
| T69 | Serous | c.2065C>T | p.Arg689Trp | Missense | Medium | |
| T32 | Clear cell | c.1199_1201delACA | p.del401Asn | In-frame deletion |
- | |
| T77 | Clear cell | c.1513C>T | p.Arg505Cys | Missense | Medium ¶ | |
| T84 | Endometrioid | c.1268G>T | p.Gly423Val | Missense | Medium | |
| T85 | Endometrioid | c.1436G>A | p.Arg479Gln | Missense | Medium ¶ | |
| T88 | Endometrioid | c.994G>T | p.Glu332* | Nonsense | - | |
| T88 | Endometrioid | c.1452G>T | p.Arg484Ser | Missense | High | |
| T88 | Endometrioid | c.1629A>C | p.Arg543Ser | Missense | Low | |
| T94 | Endometrioid | c.1393C>T | p.Arg465Cys | Missense | Medium ¶ | |
| T97 | Endometrioid | c.1436G>A | p.Arg479Gln | Missense | Medium ¶ | |
| T115 | Endometrioid | c.1508C>T | p.Ala503Val | Missense | Medium | |
| T119 | Endometrioid | c.1513C>T | p.Arg505Cys | Missense | Medium ¶ | |
| T178 | Mixed | c.1508C>T | p.Ala503Val | Missense | Medium | |
| T179 | Mixed | c.791A>G | p.Gln264Arg | Missense | Medium | |
| T179 | Mixed | c.1972C>T | p.Arg658* | Nonsense | - | |
|
SPOP (NM_001007229; uc002ipf.2) |
T25 | Serous | c.240C>G | p.Ser80Arg | Missense | Medium |
| T56 | Serous | c.240C>G | p.Ser80Arg | Missense | Medium | |
| T80 | Serous | c.280C>G | p.Pro94Ala | Missense | Low | |
| T154 | Serous | c.362G>A | p.Arg121Gln | Missense | Low | |
| T110 | Clear cell | c.351G>A | p.Met117Ile | Missense | Low | |
| T153 | Clear cell | c.139G>A | p.Glu47Lys | Missense | Medium | |
|
MAP3K4 (NM_006724; uc003qtn.2) |
T79 | Serous | c.1837G>A | p.Glu613Lys | Missense | Low |
| T79 | Serous | c.3928G>T | p.Glu1310* | Nonsense | - | |
| T80 | Serous | c.4165G>A | p.Glu1389Lys | Missense | High | |
| T65 | Serous | c.4077delC | p.Cys1359Alafs*8 | Frameshift | - | |
|
ABCC9 (NM_020297; uc001rfh.2) |
T3 (OM1323) |
Serous | c.2666G>A | p.Ser889Asn | Missense | Low |
| T3 (OM1323) | Serous | c.2868G>T | p.Glu956Asp | Missense | Low | |
| T70 | Serous | c.2185G>T | p.Val729Phe | Missense | Medium | |
| T75 | Serous | c.3381G>A | p.Met1127Ile | Missense | Low | |
|
CYP4X1 (NM_178033; uc001cqr.1) |
T75 | Serous | c.1036G>C | p.Val346Leu | Missense | Neutral |
| T56 | Serous | c.211G>A | p.Glu71Lys | Missense | Neutral |
Known loss-of-function mutations in FBXW7
()Not evaluated by Mutation Assessor
UCSC transcript IDs are based on the hg18 assembly of the human genome sequence
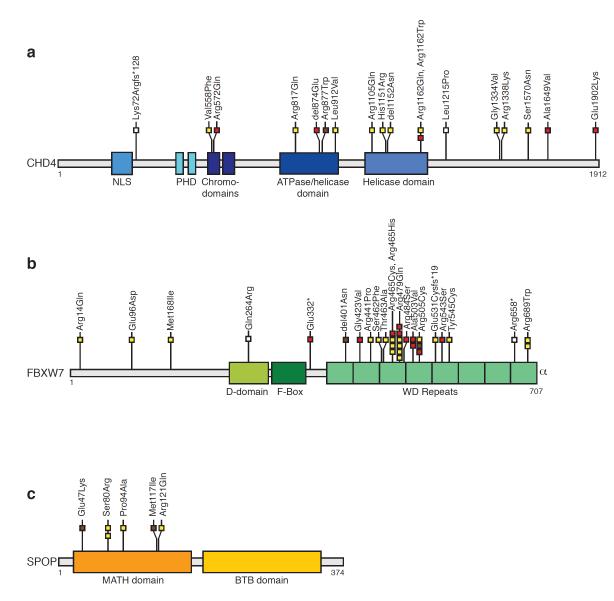
Figure 1. Somatic mutations in CHD4, FBXW7, and SPOP cluster within important functional domains of the encoded proteins.
Schematic representation of the CHD4, FBXW7, and SPOP proteins showing the positions of individual somatic mutations identified among primary endometrial tumors relative to important functional domains. Mutations in serous (yellow boxes), clear cell (brown boxes), endometrioid (red boxes) and mixed histology (white boxes) endometrial tumors are indicated. (a) 50% of all CHD4 mutations localize within the ATPase/helicase and helicase domains (top). (b) The majority of FBXW7 mutations in endometrial cancer cluster within the WD repeats. The FBXW7-Glu65* and -Lys70fs*27 mutations (not displayed) are within an alternate isoform. (c) All SPOP mutations in endometrial cancer localized to the MATH domain. BTB domain, Broad-complex, Tramtrack and Bric-a-brac domain; D-domain, Dimerization domain; MATH, Meprin and TRAF Homology; NLS, nuclear localization signal; PHD, Plant Homeo Domain-type zinc fingers; WD repeats, tryptophan-aspartic acid repeats.
In addition to serous tumors, the major histological subtypes of endometrial cancer are clear cell and endometrioid, with overall 5-year relative survival rates of 45%, 65% and 91%, respectively 9. For comparison across subtypes, we resequenced CHD4, FBXW7, and SPOP from 23 clear cell, 67 endometrioid, and 18 mixed histology endometrial tumors. Collectively, these three genes were somatically mutated in 40% of serous, 26% of clear cell, 15% of endometrioid, and 17% of mixed histology endometrial cancers (Supplementary Table 9). There was no significant association between mutations in CHD4, FBXW7 or SPOP and microsatellite instability or MSH6 mutations (Supplementary Table 11, Supplementary Table 12, Supplementary Figure 5).
CHD4 (Chromodomain-helicase-DNA-binding protein 4) is a catalytic subunit of the NuRD complex, which regulates transcriptional repression, chromatin assembly, and the DNA damage response 10-19. We confirmed endogenous CHD4 expression in endometrial cancer cells (Supplementary Figure 6). CHD4 was highly mutated in serous tumors (17%), and was also mutated in clear cell (4%), endometrioid (7%), and mixed histology (11%) tumors (Supplementary Table 9). Eighty-percent of CHD4 missense mutations, including an arginine-1162 (CHD4Arg1162) hotspot, are predicted to impact protein function (Table 1, and Figure 1). Most CHD4 mutations were missense mutations; 83% of all CHD4 mutations affected residues that are highly conserved throughout evolution or across closely related family members (Supplementary Figure 7 and Supplementary Figure 8).
Half of all CHD4 mutations localized to the ATPase/helicase domains (Figure 1a); two-thirds (6 of 9) of those mutations localized to conserved residues that undergo germline or de novo pathogenic mutations in SMARCAL1, SMARCA4, or SMARCA2 causing Schimke immune-osseous dysplasia, Coffin-Siris syndrome, and Nicolaides-Baraitser syndrome (Supplementary Figure 9) 20-22. This observation leads us to speculate that somatic mutations in the ATPase/helicase domain of CHD4 may be driver mutations in endometrial cancer.
Other frequently mutated genes in our study were FBXW7 and SPOP. The FBXW7 (F-box and WD repeat domain containing 7) tumor suppressor is a component of the FBXW7-SKP1-CUL1 ubiquitin ligase complex, which mediates ubiquitination and proteosomal degradation of phosphoprotein substrates including CYCLIN-E, NOTCH, JUN, and C-MYC 23. Previous reports of FBXW7 mutations in endometrial cancer either did not include serous and clear cell tumors or did not report the histology of mutated cases 24-27. We identified FBXW7 mutations in 29% of serous, 13% of clear cell, 10% of endometrioid, and 11% of mixed histology endometrial cancers (Supplementary Table 9). The mutation frequency was significantly higher among serous tumors than high-grade endometrioid tumors (29% versus 0%, P=0.0146 Fisher’s 2-tailed Exact test of significance). Most FBXW7 mutations localized to the substrate binding WD-repeats (Figure 1b), consistent with the mutation spectrum in other cancers 28. Forty-six percent of FBXW7 missense mutations in our study are known loss-of-function mutations; another 39% of missense mutations are predicted to affect function 29 (Table 1). Our findings may be clinically relevant since loss of FBXW7 function correlates with resistance to antitubulin chemotherapeutics 30 and sensitivity to an HDAC inhibitor 31.
SPOP (Speckle-type POZ Protein) was somatically mutated in serous (8%) and clear cell tumors (9%) but was not mutated in endometrioid or mixed histology tumors (Supplementary Table 9). The mutation frequency was statistically significantly higher in serous than in endometrioid tumors (8% versus 0%, P=0.0341, Fisher’s 2-tailed Exact test of significance). SPOP forms part of a multi-subunit CULLIN3 (CUL3)-dependent ubiquitin ligase complex, and has recently been shown to be mutated at high frequency in prostate cancer 32. All SPOP mutations in endometrial cancer, including a recurrent SPOPSer80Arg mutant, localized to highly evolutionarily conserved residues within the MATH domain that acts as the substrate recognition domain to bind proteins targeted for ubiquitin-mediated degradation (Figure 1c). This localization is strikingly similar to the localization of loss-of-function mutations to the substrate recognition domain of FBXW7 (Figure 1b). Thus, we predict that SPOP mutations in endometrial cancer are likely to be loss-of-function mutants with impaired substrate binding. Intriguingly, the SRC-3/AIB1 oncoprotein, an SPOP substrate 33, is overexpressed in endometrial cancer independent of gene amplification 34.
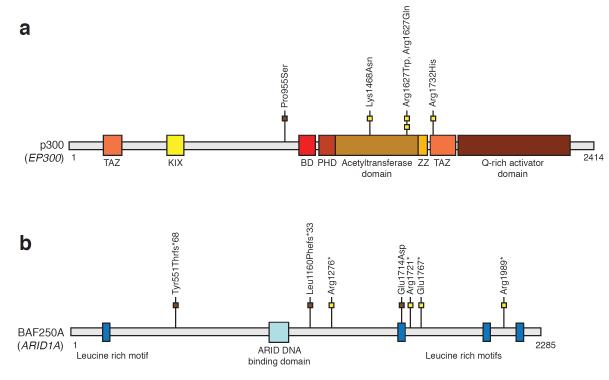
To identify additional candidate driver genes for serous endometrial cancer, we evaluated the functional relationships of the 304 protein-encoding genes that had orthogonally validated mutations (nonsynonymous or splice junction) in the discovery screen (Supplementary Table 13 and Supplementary Table 14) 35,36. One of the enriched functional groupings was chromatin modification, which was formed by CHD4 and ten other genes (EP300, ARID1A, TSPYL2, KDM4B, TRIM16, HDAC7, CTCF, YEATS4, TRRAP, and BAZ1B) (Supplementary Table 13). Although this enriched grouping did not achieve statistical significance following multiple testing correction, we focused on it because it contained CHD4, one of the most highly mutated genes identified in our study, and because chromatin-remodeling genes are a frequent target of somatic mutations in other types of cancer 37-50. We therefore resequenced the ten additional chromatin-remodeling genes from 40 additional serous and 23 clear cell endometrial tumors. In the combined discovery and prevalence screens, the 11 chromatin-remodeling genes were somatically mutated in 36.5% of serous tumors and 22% of clear cell tumors (Table 1, Table 2, and Figure 2). Two of the mutated genes, EP300 (E1A binding protein p300) and ARID1A (AT rich interactive domain 1A, SWI-like), are consensus cancer genes. EP300 and ARID1A were mutated, respectively, in 8% and 6% of serous tumors and in 4% and 13% of clear cell tumors (Table 2). Most EP300 mutations localized within the histone acetyltransferase domain of p300, a global transcriptional co-activator (Figure 3a). p300Arg1627 formed a mutation hotspot in EC; the p300Arg1627Trp mutation has also been described in lymphoma 51. ARID1A encodes the BAF250A tumor suppressor, a subunit of the SWI/SNF-A chromatin-remodeling complex 43,49,52-54. Most ARID1A mutations we uncovered are predicted to truncate BAF250A (Figure 3b), consistent with the mutation spectrum in other tumors 43,49,55. Our finding of ARID1A mutations in 6% of serous endometrial cancers is consistent with a recent study by McConechy et al., which documented ARID1A mutations in 11% of serous endometrial tumors 56. To our knowledge, this is the first report of ARID1A mutations in clear cell endometrial cancer, substantiating previous reports of loss of BAF250A expression in this histotype 57.
Table 2.
Somatic mutations identified in the discovery and prevalence screens of ten chromatin-remodeling genes
| Gene Name (RefSeq ID; UCSC Transcript Accession ID^) |
Mutation frequency in serous tumors |
Mutation frequency in clear cell tumors |
Tumor | Histology | Nucleotide Change | Amino Acid Change |
Mutation type |
Mutation Assessor Predicted Effect on Function |
|---|---|---|---|---|---|---|---|---|
|
EP300 (NM_001429; uc003azi.3) |
8% (4/52) | 4% (1/23) | T56 | Serous | c.5195G>A | p.Arg1732His | Missense | Medium |
| T3 (OM1323) |
Serous | c.4879C>T | p.Arg1627Trp | Missense | Medium | |||
| T74 | Serous | c.4880G>A | p.Arg1627Gln | Missense | Medium | |||
| T79 | Serous | c.4404A>C | p.Lys1468Asn | Missense | Medium | |||
| T113 | Clear cell | c.2863C>T | p.Pro955Ser | Missense | Neutral | |||
|
ARID1A (NM_006015; uc001bmv.1) |
6% (3/52) | 13% (3/23) | T69 | Serous | c.5161C>T | p.Arg1721* | Nonsense | - |
| T74 | Serous | c.3826C>T | p.Arg1276* | Nonsense | - | |||
| T79 | Serous | c.5299G>T | p.Glu1767* | Nonsense | - | |||
| T79 | Serous | c.5965C>T | p.Arg1989* | Nonsense | - | |||
| T61 | Clear cell | c.1645delC | p.Tyr551Thrfs*68 | Frameshift | - | |||
| T82 | Clear cell | c.3478_3479insT | p.Leu1160Phefs*33 | Frameshift | - | |||
| T113 | Clear cell | c.5142G>T | p.Glu1714Asp | Missense | Low | |||
|
TSPYL2 (NM_022117; uc004drx.1) |
6% (3/52) | 0% (0/23) | T70 | Serous | c.752A>C | p.His251Pro | Missense | - |
| (NM_022117; uc004drw.2) |
T3 (OM1323) |
Serous | c.1636G>T | p.Gly546Cys | Missense | Neutral | ||
| T30 | Serous | c.1952G>C | p.Gly651Ala | Missense | Neutral | |||
|
KDM4B (NM_015015; uc002mbq.3) |
4% (2/52) | 0% (0/23) | T154 | Serous | c.2138C>T | p.Pro713Leu | Missense | Neutral |
| T3 (OM1323) |
Serous | c.964G>A | p.Val322Met | Missense | Low | |||
| T3 (OM1323) |
Serous | c.1855C>T | p.Pro619Ser | Missense | Low | |||
|
TRIM16 (NM_006470; uc002gox.2) |
2% (1/52) | 0% (0/23) | T80 | Serous | c.1319delC | p.Tyr440Metfs*3 | Frameshift | - |
|
CTCF (NM_006565; uc002etl.2) |
2% (1/52) | 0% (0/23) | T70 | Serous | c.1094A>C | p.Lys365Thr | Missense | Neutral |
|
HDAC7 (NM_015401; uc001rqi.2) |
2% (1/52) | 0% (0/23) | T75 | Serous | c.1024G>A | p.Glu342Lys | Missense | Low |
|
TRRAP (NM_003496; uc003.upp.1) |
2% (1/52) | 0% (0/23) | T47 | Serous | c.5035G>A | p.Ala1679Thr | Missense | Low |
|
YEATS4 (NM_006530; uc001sux.1) |
2% (1/52) | 0% (0/23) | T27 | Serous | c.617delA | p.Thr206Leufs*1 | Frameshift | - |
|
BAZ1B (NM_032408; uc003tyc.1) |
4% (2/52) | 4% (1/23) | T65 | Serous | c.136A>C | p.Ser46Arg | Missense | Low |
| T3 (OM1323) |
Serous | c.3448G>A | p.Glu1150Lys | Missense | Neutral | |||
| T77 | Clear cell | c.3167G>A | p.Arg1056His | Missense | Low |
()Not evaluated by Mutation Assessor
UCSC transcript IDs are based on the hg18 assembly of the human genome sequence
Figure 2. Oncoprint showing the distribution of nonsynonymous somatic mutations in ubiquitin ligase complex genes and chromatin-remodeling genes among 26 serous and 8 clear cell endometrial tumors, and in TP53, PPP2R1A, and PIK3CA among 26 serous tumors.
Individual tumors are indicated by blue bars. Nonsynonymous somatic mutations are indicated by yellow bars. Only tumors that had somatically mutated the ubiquitin ligase complex genes and chromatin-remodeling genes are shown. Collectively, the two ubiquitin ligase complex genes that regulate ubiquitin (Ub) mediated proteolysis were mutated in 35% (18 of 52) of serous endometrial tumors and 22% (5 of 23) of clear cell endometrial tumors; the 11 chromatin-remodeling genes were mutated in 36.5% (19 of 52) of serous endometrial tumors and 22% (5 of 23) of clear cell endometrial tumors. (*) The PIK3CA mutation pattern was previously reported in Rudd et al 6.
Figure 3. Somatic mutations in the consensus cancer genes EP300, and ARID1A, relative to the functional domains of the encoded proteins.
Schematic representation of the (a) p300 and (b) BAF250A proteins, showing the positions of individual somatic mutations identified among primary endometrial tumors. Mutations in serous (yellow boxes) and clear cell (brown boxes) endometrial tumors are distinguished. Known functional domains of each protein are indicated. ARID, AT-rich Interaction Domain; BD, Bromodomain; KIX, KIX domain; PHD, Plant Homeo Domain Finger; TAZ, zinc finger TAZ-type; ZZ, zinc finger ZZ type.
Our study provides novel insights into the somatic mutations present in serous endometrial cancer exomes. However, it is important to acknowledge that our discovery screen is underpowered to detect all somatically mutated genes that drive serous tumors. For example, PIK3R1, which we previously found to be somatically mutated in 8% of serous endometrial tumors 58, was not somatically mutated among the tumors that formed our discovery screen. We estimate that for genes that are mutated in 8% of all serous endometrial cancers, a discovery screen of 12 tumors has 25% power to detect two mutated tumors and 63% power to detect one mutated tumor (Supplementary Table 15); for genes that are mutated in 20% of all serous endometrial cancers, our discovery screen has an estimated 72.5% power to detect two mutated tumors and 93% power to detect one mutated tumor. Massively parallel sequencing of additional cases will undoubtedly yield deeper insights into the mutational landscape of serous endometrial cancer.
Herein we report the first exome sequence analysis of serous endometrial cancers, which are clinically aggressive tumors that have been poorly characterized genomically. Our findings implicate the disruption of chromatin-remodeling and ubiquitin ligase complex genes in 50% of serous endometrial tumors and 35% of clear cell endometrial tumors (Figure 2). The high frequency and specific distribution of mutations in CHD4, FBXW7, and SPOP strongly suggests these are likely to be driver events in serous endometrial cancer.
Methods
Clinical material
Anonymized, snap-frozen primary tumor tissues, corresponding hematoxylin and eosin-stained tumor sections, matched normal tissues (uninvolved reproductive tissue or whole blood), and clinicopathological information were obtained from the Cooperative Human Tissue Network, which is funded by the National Cancer Institute. The NIH Office of Human Subjects Research determined that this research was not “human subjects research” per the Common Rule (45 CFR 46), and therefore that no IRB review was required for whole exome sequencing of these samples. A small number of samples in the prevalence screen were obtained from the Biosample Repository at Fox Chase Cancer Center, or from Oncomatrix, Inc (San Marcos, CA). Tumor specimens were collected at surgical resection, before treatment. Histological classifications were based upon the entire specimen at time of diagnosis. Tumors consisted of 53 serous cases, 23 clear cell cases, 67 endometrioid cases and 18 cases of mixed histology. A pathologist reviewed hematoxylin and eosin-stained sections of banked tumor tissues, to verify that they were representative of the original histological classification, and to delineate regions of high tumor cell content (>70%) for macrodissection.
DNA extraction and identity testing
Genomic DNA was isolated from macrodissected tumor tissues and normal tissues using the PUREGENE kit (Qiagen), followed by phenol-chloroform purification. Tumor-normal pairs were typed using the Coriell Identity Mapping kit (Coriell). Genotyping fragments were resolved on an ABI-3730xl DNA analyzer (Applied Biosystems) and scored using GeneMapper software.
Exome capture, library construction, and next generation sequencing
Exomes of tumor-normal pairs were captured using the Agilent SureSelect Human All Exon Kit (3 pairs) or the Agilent SureSelect Human All Exon 50 Mb Target Enrichment kit (10 pairs) according to the manufacturer’s instructions. DNAs captured using the SureSelect Human All Exon Kit were run on an Illumina GAIIx platform with version 4 chemistry and version 2 flowcells; DNAs captured using the SureSelect Human All Exon 50 Mb Target Enrichment kit were run on an Illumina GAIIx platform with version 5 chemistry and version 4 flowcells, according to the manufacturer’s instructions to generate 75- or 100-base paired end reads.
Read mapping and genotype calling
Next generation sequence reads were initially mapped to the human reference sequence NCBI build 36 (hg18) using the Illumina ELAND alignment algorithm. When at least one read in a pair mapped to a unique location in the genome, that read and its pair were subjected to a more accurate gapped alignment to the 100kb region surrounding the location with cross_match (URL: http://www.phrap.org/phredphrapconsed.html). Alignments were stored in BAM format, and fed as input to bam2mpg (URL: http://research.nhgri.nih.gov/software/bam2mpg/), to call genotypes at all covered positions using a probabilistic Bayesian algorithm referred to as the most-probable-genotype (MPG) algorithm 59. Highly reliable genotypes have an MPG score ≥10 59. For tumor samples, bam2mpg was run with the --score_variant option, in order to calculate a “most probable variant” (MPV) score, which assesses the posterior probability of the existence of any variant at a position, and therefore is more sensitive than the MPG score at positions for which there is uncertainty about whether a variant is heterozygous or homozygous non-reference. Additional information on the MPG and MPV scores is provided in the Supplementary Note.
Filtering of variant calls
We used a number of steps to filter nucleotide variants identified in the whole exome screen. Germline variants called in paired tumor-normal samples were excluded from further analysis. Variants that were present within dbSNP build 132, but which were not annotated as pathogenic or probable pathogenic variants within dbSNP, were also excluded. We compared the remaining variants in each tumor exome to the variants in all 13 normal exomes sequenced in this study; variants called in both a tumor exome and a normal exome were excluded. Variants representing probable mapping ambiguities were also excluded. All remaining variants were considered to be potential somatic mutations and were annotated using the VarSifter software package 60 into bins representing mutations in exons, introns, splice junctions, UTRs, and non-coding RNAs.
Establishment of filtering criteria based on read score and read depth
After filtering the exome data to exclude germline variants and probable mapping ambiguities, 798 somatic variants were called in the exons and splice junctions of 12 tumors (Supplementary Figure 1 and Supplementary Table 16). We were able to orthogonally assess 730 of the 798 variants by Sanger sequencing; PCR products could not be generated for the remaining 68 variants. Of the 730 variants tested by Sanger sequencing, 451 variants orthogonally validated as somatic (present in tumor and absent from matched normal DNA) yielding a positive predictive value of 61.8% (451 of 730). The remaining variants were either not detected within the tumor or were germline (present in both tumor and matched normal DNA).
Because a positive predictive value of 61.8% was unacceptably low, we sought to empirically establish filtering criteria, based on sequence quality (MPV/MPG score) and read depth (coverage), to achieve an optimal balance between accuracy and sensitivity of mutation detection. We observed (Supplementary Figure 10) that the majority of mutations that did not orthogonally validate as somatic by Sanger sequencing had (i) < 5 reads in the tumor or normal samples; (ii) an MPG score < 10 in the normal sample and/or an MPV score <10 in the tumor sample, (iii) an MPG:COV ratio of <0.5 in the normal sample.
We therefore retrospectively imposed filtering criteria of (i) at least 5 reads in the tumor and normal samples and (ii) an MPG score of ≥ 10 in the normal sample and an MPV score of ≥ 10 in the tumor sample; and (iii) an MPG:COV ratio ≥0.5 in the normal sample, to the 798 somatic variants called by exome sequencing; 527 variants were retained after filtering (Supplementary Table 5). Filtering on score and read depth attained a positive predictive value of 86.1% (510 of 527 retained variants could be assessed by Sanger sequencing; 439 of 510 (86.1%) assessed variants orthogonally validated as somatic mutations) and a sensitivity of 97.3% (439 of 451 orthogonally validated mutations observed before applying the coverage-score filter were retained after filtering).
Filtering criteria of (i) at least 5 reads in the tumor sample and normal sample and (ii) an MPV score of ≥ 10 in the tumor sample and an MPG score of ≥ 10 in the normal sample; (iii) an MPG:COV ratio ≥ 0.5 in the normal sample, was also applied to the 1,042 somatic variants called in T155 by exome sequencing (Supplementary Table 17); 1,017 variants were retained after filtering (Supplementary Table 4).
Predicting the functional significance of missense mutations
Nonsynonymous missense mutations called by whole exome sequencing were evaluated in silico using the SIFT (http://sift.jcvi.org/) and Mutation Assessor (http://mutationassessor.org/) algorithms to predict their impact on protein function. A SIFT prediction of “deleterious” and a Mutation Assessor predictions of “medium impact” or “high impact” were considered to predict an impact on protein function.
Power of study design
Assuming N tumor samples are sequenced in a discovery screen, and a fraction X of all tumor samples have gene G mutated, the probability that the N samples sequenced contain 0 samples with the gene mutation is (1-X)N. Therefore, the probability of observing gene G mutated at least once in the discovery screen is 1 - (1-X)N, which is 93% for 12 discovery samples, assuming X = 0.20, or 20%. Likewise, the probability of observing 0 or 1 samples with a mutation in gene G is (1-X)N + N(1-X)(N-1)X, giving the probability of seeing the mutation twice or more as 1 - (1-X)N – N(1-X)(N-1)X, which is 73% for 12 tumors, assuming X=0.20.
PCR amplification and Sanger sequencing
Genomic DNA (5ng) was amplified using M13-tailed primers (Supplementary Table 18) in a 10μl polymerase chain reaction (PCR) containing 1X AmpliTaq Gold PCR buffer (Applied Biosystems), 1.5mM MgCl2, 75mM dNTP, 400nM sense primer, 400nM antisense primer, and 0.5 units of AmpliTaq Gold DNA polymerase. PCR amplification was performed on a GeneAmp® PCR System 9700 (Applied Biosystems). PCR amplicons were purified using exonuclease I (Epicentre Biotechnologies) and shrimp alkaline phosphatase (USB Corporation) and bidirectionally sequenced using the Big Dye Terminator v.3.1 kit (Applied Biosystems) and M13 primers. Cycle sequencing products were run on an ABI-3730xl DNA analyzer (Applied Biosystems). Tumor and reference sequences were aligned, and compared using in-house software to determine the genotype of variant nucleotide positions. Non-pathogenic variants present in dbSNP were excluded from further analysis. True somatic mutations were confirmed by reamplification and sequencing of matched tumor-normal DNAs and analyzed using Sequencher software (Gene Codes Corporation).
Determination of mutation rates and statistical analyses
The somatic mutation rate was determined by dividing the total number of exonic mutations present within a tumor (after filtering on quality score and coverage) by the number of the exonic bases that had adequate quality and coverage in both the tumor sample (MPV score ≥ 10 and at least 5 reads) and paired normal sample (MPG score ≥10, MPG:COV ratio ≥0.5, and at least 5 reads). A Grubb’s test was used to calculate an approximate p-value for each tumor to identify outliers. A uniform background mutation rate equal to the rate observed in the discovery phase was assumed, and a Poisson distribution function was used to calculate p-values for the observed number of mutations in each gene. False discovery rates were calculated using the Benjamini Hochberg method 61, correcting for 21,441 genes tested. This method is a simplified version of the CaMP (cancer mutation prevalence) scoring method 62 including subsequently suggested corrections 63.
Functional enrichment analyses in silico
The 304 somatically mutated protein-encoding genes identified and orthogonally validated in the discovery screen were analyzed for enriched functional groupings using the Database for Annotation Visualization and Integrated Discovery (DAVID) 35,36, and Ingenuity Systems Pathway Analysis (IPA) in silico tools (www.ingenuity.com). The Bonferroni, Benjamini, and FDR values, computed within DAVID, were assessed for significance.
Determination of defective mismatch repair
Tumor-normal DNA pairs were screened for the presence of microsatellite instability (MSI) using the Promega Microsatellite Instability Analysis System v1.2 (Promega) according to the manufacturer’s instructions. All coding exons of MSH6 were PCR amplified and Sanger sequenced.
Cell lines
Endometrial cancer cell lines (RL-95-2, HEC1A, HEC1B, KLE, ANC3A) were obtained from the American Type Culture Collection, or the NCI Developmental Therapeutics Program cell line repository. ARK1 and ARK2 serous endometrial cancer cell lines were kindly provided by Dr. Alessandro Santin (Yale School of Medicine).
Immunoblotting
Cells were lysed in RIPA buffer (Thermo Scientific) containing 1mM Naorthovanadate, 10mM NaF, and 1X protease inhibitor cocktail (Roche). Lysates were centrifuged followed by denaturing at 95°C in 2X SDS sample buffer (Sigma) prior to SDS-PAGE and transfer to PVDF membranes (Bio-Rad). Primary and HRP-conjugated secondary antibodies were: CHD4 (Cell Signaling), β-Actin (Sigma), goat anti-mouse HRP (Cell Signaling), and goat anti-rabbit HRP (Cell Signaling). Immunoreactive proteins were visualized with enhanced chemiluminescence (Pierce).
Supplementary Material
Acknowledgments
We thank our colleagues for critical reading of the manuscript; R. Travis Moreland and Niraj Trivedi of the NHGRI Bioinformatics and Scientific Programming Core for, respectively, performing in silico PCR and advice on statistics; and Dr. Jamie Teer for sharing expertise on VarSifter. Dr. Alessandro Santin kindly provided the ARK1 and ARK2 cell lines. Funded in part by the Intramural Program of the National Human Genome Research Institute, National Institutes of Health (DWB and JCM); NIH grant R01CA112021 (DCS); the Avon Foundation (DCS); NIH grant R01CA140323 (AKG); the Ovarian Cancer Fund (AKG); PH is supported by grants from the NIH grant (CA016519) and by the Canadian Institutes for Health Research (MOP-38096).
Footnotes
Author Contributions
D.W.B. designed and directed the study, and wrote the manuscript. A.K.G. contributed clinical specimens. M.J.M. and D.C.S. conducted pathological review of clinical specimens. M.L.R. prepared DNA samples, performed identity testing and microsatellite instability analysis. NISC performed library construction and whole exome sequencing. NISC and N.F.H. performed variant calling. M.L.G. and A.O.H. curated and orthogonally validated exome sequencing data. M.L.G., A.O.H. and D.W.B. interpreted the exome data and established filtering criteria. M.L.G., A.O.H., M.L.R., J.C.P., B.M.E., S.Z. and D.W.B. designed, performed, analyzed, and interpreted the mutation prevalence screens. A.O.H. and M.L.G. analyzed MSH6. M.E.U. and M.L.G. generated sequence conservation alignments. M.E.U. performed cell culture and immunoblotting. N.F.H. M.L.G. and J.C.M. performed statistical analyses. N.F.H. performed the power calculation. D.W.B., M.E.U., M.L.G., M.L.R., A.O.H., N.F.H., J.C.M., A.K.G., P.H. and N.O’N. edited and commented on the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Ferlay J, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Sherman ME. Theories of endometrial carcinogenesis: a multidisciplinary approach. Mod Pathol. 2000;13:295–308. doi: 10.1038/modpathol.3880051. [DOI] [PubMed] [Google Scholar]
- 3.Hamilton CA, et al. Uterine papillary serous and clear cell carcinomas predict for poorer survival compared to grade 3 endometrioid corpus cancers. Br J Cancer. 2006;94:642–6. doi: 10.1038/sj.bjc.6603012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hendrickson M, Ross J, Eifel P, Martinez A, Kempson R. Uterine papillary serous carcinoma: a highly malignant form of endometrial adenocarcinoma. Am J Surg Pathol. 1982;6:93–108. doi: 10.1097/00000478-198203000-00002. [DOI] [PubMed] [Google Scholar]
- 5.McConechy MK, et al. Subtype-specific mutation of PPP2R1A in endometrial and ovarian carcinomas. J Pathol. 2011;223:567–73. doi: 10.1002/path.2848. [DOI] [PubMed] [Google Scholar]
- 6.Rudd ML, et al. A unique spectrum of somatic PIK3CA (p110α) mutations within primary endometrial carcinomas. Clin Cancer Res. 2011;17:1331–40. doi: 10.1158/1078-0432.CCR-10-0540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shih Ie M, et al. Somatic mutations of PPP2R1A in ovarian and uterine carcinomas. Am J Pathol. 2011;178:1442–7. doi: 10.1016/j.ajpath.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayes MP, Douglas W, Ellenson LH. Molecular alterations of EGFR and PIK3CA in uterine serous carcinoma. Gynecol Oncol. 2009;113:370–3. doi: 10.1016/j.ygyno.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ries LAG, et al. Patient and tumor characteristics. National Cancer Institute, SEER Program; Bethesda, MD: 2007. SEER Survival Monograph: Cancer survival among adults: U.S. SEER Program, 1988-2001. NIH Pub. No. 07-6215. 2007. [Google Scholar]
- 10.Zhang Y, LeRoy G, Seelig HP, Lane WS, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998;95:279–89. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- 11.Li J, Lin Q, Wang W, Wade P, Wong J. Specific targeting and constitutive association of histone deacetylase complexes during transcriptional repression. Genes Dev. 2002;16:687–92. doi: 10.1101/gad.962502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tong JK, Hassig CA, Schnitzler GR, Kingston RE, Schreiber SL. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature. 1998;395:917–21. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- 13.Xue Y, et al. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–61. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- 14.Hall JA, Georgel PT. CHD proteins: a diverse family with strong ties. BCB. 2007;85:463–76. doi: 10.1139/O07-063. [DOI] [PubMed] [Google Scholar]
- 15.Lai AY, Wade PA. Cancer biology and NuRD: a multifaceted chromatin remodelling complex. Nat Rev Cancer. 2011;11:588–96. doi: 10.1038/nrc3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Polo SE, Kaidi A, Baskcomb L, Galanty Y, Jackson SP. Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. EMBO J. 2010;29:3130–9. doi: 10.1038/emboj.2010.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smeenk G, et al. The NuRD chromatin-remodeling complex regulates signaling and repair of DNA damage. J Cell Biol. 2010;190:741–9. doi: 10.1083/jcb.201001048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chou DM, et al. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc Natl Acad Sci USA. 2010;107:18475–80. doi: 10.1073/pnas.1012946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larsen DH, et al. The chromatin-remodeling factor CHD4 coordinates signaling and repair after DNA damage. J Cell Biol. 2010;190:731–40. doi: 10.1083/jcb.200912135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boerkoel CF, et al. Mutant chromatin remodeling protein SMARCAL1 causes Schimke immuno-osseous dysplasia. Nat Genet. 2002;30:215–20. doi: 10.1038/ng821. [DOI] [PubMed] [Google Scholar]
- 21.Tsurusaki Y, et al. Mutations affecting components of the SWI/SNF complex cause Coffin-Siris syndrome. Nat Genet. 2012;44:376–8. doi: 10.1038/ng.2219. [DOI] [PubMed] [Google Scholar]
- 22.Van Houdt JK, et al. Heterozygous missense mutations in SMARCA2 cause Nicolaides-Baraitser syndrome. Nat Genet. 2012;44:445–9. doi: 10.1038/ng.1105. [DOI] [PubMed] [Google Scholar]
- 23.Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer. 2008;8:83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- 24.Cassia R, et al. Cyclin E gene (CCNE) amplification and hCDC4 mutations in endometrial carcinoma. J Pathol. 2003;201:589–95. doi: 10.1002/path.1474. [DOI] [PubMed] [Google Scholar]
- 25.Dutt A, et al. Drug-sensitive FGFR2 mutations in endometrial carcinoma. Proc Natl Acad Sci U S A. 2008;105:8713–7. doi: 10.1073/pnas.0803379105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spruck CH, et al. hCDC4 gene mutations in endometrial cancer. Cancer Res. 2002;62:4535–9. [PubMed] [Google Scholar]
- 27.Suehiro Y, et al. Aneuploidy predicts outcome in patients with endometrial carcinoma and is related to lack of CDH13 hypermethylation. Clin Cancer Res. 2008;14:3354–61. doi: 10.1158/1078-0432.CCR-07-4609. [DOI] [PubMed] [Google Scholar]
- 28.Forbes SA, et al. The Catalogue of Somatic Mutations in Cancer (COSMIC) Curr Protoc Hum Genet. 2008 doi: 10.1002/0471142905.hg1011s57. Chapter 10, Unit 10 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39:e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wertz IE, et al. Sensitivity to antitubulin chemotherapeutics is regulated by MCL1 and FBW7. Nature. 2011;471:110–4. doi: 10.1038/nature09779. [DOI] [PubMed] [Google Scholar]
- 31.Garnett MJ, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–5. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barbieri CE, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44:685–9. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li C, et al. Tumor-suppressor role for the SPOP ubiquitin ligase in signal-dependent proteolysis of the oncogenic co-activator SRC-3/AIB1. Oncogene. 2011;30:4350–6. doi: 10.1038/onc.2011.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Glaeser M, Floetotto T, Hanstein B, Beckmann MW, Niederacher D. Gene amplification and expression of the steroid receptor coactivator SRC3 (AIB1) in sporadic breast and endometrial carcinomas. Horm Metab Res. 2001;33:121–6. doi: 10.1055/s-2001-14938. [DOI] [PubMed] [Google Scholar]
- 35.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 36.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dalgliesh GL, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463:360–3. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fujimoto A, et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet. 2012;44:760–4. doi: 10.1038/ng.2291. [DOI] [PubMed] [Google Scholar]
- 39.Grasso CS, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–43. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gui Y, et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat Genet. 2011;43:875–8. doi: 10.1038/ng.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guichard C, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet. 2012;44:694–8. doi: 10.1038/ng.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiao Y, et al. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science. 2011;331:1199–203. doi: 10.1126/science.1200609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jones S, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330:228–31. doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ong CK, et al. Exome sequencing of liver fluke-associated cholangiocarcinoma. Nat Genet. 2012;44:690–3. doi: 10.1038/ng.2273. [DOI] [PubMed] [Google Scholar]
- 45.Parsons DW, et al. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011;331:435–9. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwartzentruber J, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–31. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 47.Shain AH, et al. Convergent structural alterations define SWItch/Sucrose NonFermentable (SWI/SNF) chromatin remodeler as a central tumor suppressive complex in pancreatic cancer. Proc Natl Acad Sci USA. 2012;109:E252–9. doi: 10.1073/pnas.1114817109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Varela I, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469:539–42. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wiegand KC, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med. 2010;363:1532–43. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang K, et al. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nat Genet. 2011;43:1219–23. doi: 10.1038/ng.982. [DOI] [PubMed] [Google Scholar]
- 51.Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–9. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 52.Huang J, Zhao YL, Li Y, Fletcher JA, Xiao S. Genomic and functional evidence for an ARID1A tumor suppressor role. Gene Chromosome Canc. 2007;46:745–50. doi: 10.1002/gcc.20459. [DOI] [PubMed] [Google Scholar]
- 53.Guan B, Wang TL, Shih IM. ARID1A, a factor that promotes formation of SWI/SNF-mediated chromatin remodeling, is a tumor suppressor in gynecologic cancers. Cancer Res. 2011;71:6718–27. doi: 10.1158/0008-5472.CAN-11-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011;21:396–420. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guan B, et al. Mutation and loss of expression of ARID1A in uterine low-grade endometrioid carcinoma. Am J Surg Pathol. 2011;35:625–32. doi: 10.1097/PAS.0b013e318212782a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McConechy MK, et al. Use of mutation profiles to refine the classification of endometrial carcinomas. J Pathol. 2012 doi: 10.1002/path.4056. doi: 10.1002/path.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wiegand KC, et al. Loss of BAF250a (ARID1A) is frequent in high-grade endometrial carcinomas. J Pathol. 2011;224:328–33. doi: 10.1002/path.2911. [DOI] [PubMed] [Google Scholar]
- 58.Urick ME, et al. PIK3R1 (p85-alpha/p85{alpha}) is somatically mutated at high frequency in primary endometrial cancer. Can Res. 2011;71:4061–7. doi: 10.1158/0008-5472.CAN-11-0549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Teer JK, et al. Systematic comparison of three genomic enrichment methods for massively parallel DNA sequencing. Genome Res. 2010;20:1420–31. doi: 10.1101/gr.106716.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Teer JK, Green ED, Mullikin JC, Biesecker LG. VarSifter: visualizing and analyzing exome-scale sequence variation data on a desktop computer. Bioinformatics. 2012;28:599–600. doi: 10.1093/bioinformatics/btr711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc. 1995;57:289–300. [Google Scholar]
- 62.Sjoblom T, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–74. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 63.Rubin AF, Green P. Comment on “The consensus coding sequences of human breast and colorectal cancers”. Science. 2007;317:1500. doi: 10.1126/science.1138956. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.